Abstract
RNA interference is widely distributed in eukaryotes and has a variety of functions, including antiviral defence and gene regulation1,2. All RNA interference pathways use small single-stranded RNA (ssRNA) molecules that guide proteins of the Argonaute (Ago) family to complementary ssRNA targets: RNA-guided RNA interference1,2. The role of prokaryotic Ago variants has remained elusive, although bioinformatics analysis has suggested their involvement in host defence3. Here we demonstrate that Ago of the bacterium Thermus thermophilus (TtAgo) acts as a barrier for the uptake and propagation of foreign DNA. In vivo, TtAgo is loaded with 5′-phosphorylated DNA guides, 13–25 nucleotides in length, that are mostly plasmid derived and have a strong bias for a 5′-end deoxycytidine. These small interfering DNAs guide TtAgo to cleave complementary DNA strands. Hence, despite structural homology to its eukaryotic counterparts, TtAgo functions in host defence by DNA-guided DNA interference.
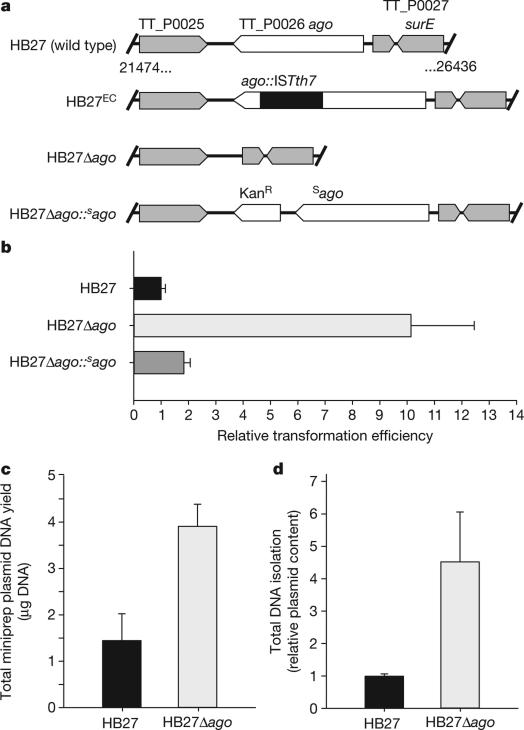
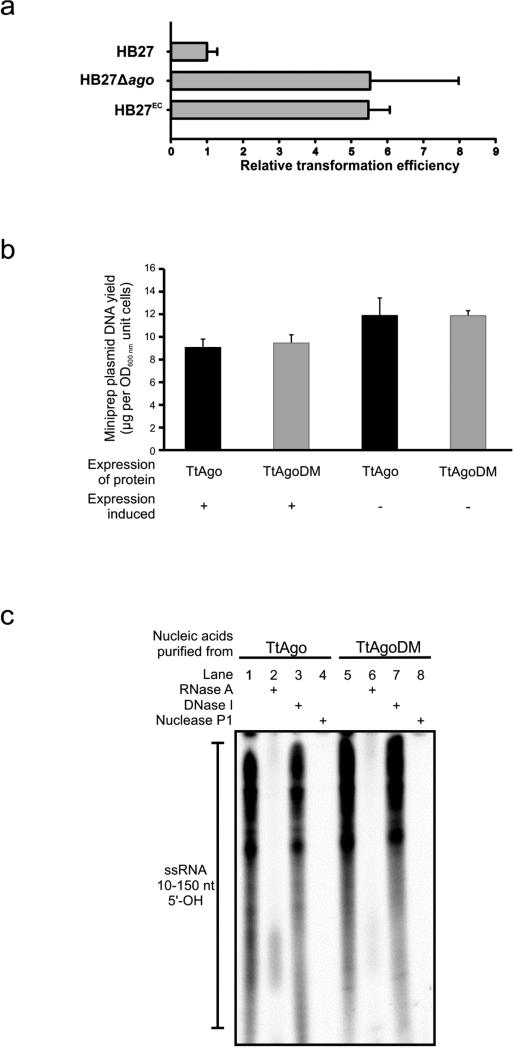
To elucidate the physiological role of Ago in prokaryotes, we studied Ago from T. thermophilus. Comparison of the ago genes of the type strain HB27 (refs 4, 5) and a derivative with enhanced competence (HB27EC; Fig. 1a and Extended Data Fig. 1a), revealed that an insertion sequence (ISTth7)6 disrupts ago in HB27EC. In line with a role of TtAgo in reducing competence, a generated Δago mutant (HB27Δago; Fig. 1a) has a natural transformation efficiency that is a factor of ten higher than the wild-type HB27 (P < 0.02, Fig. 1b). Complementation of the knockout strain with ago (HB27Δago::sago (HB27Δago complemented with a strep(II)-tag-ago gene fusion insert); Fig. 1a, b) almost completely restores the wild-type phenotype. Moreover, isolation of plasmid and total DNA from the wild-type and the ago knockout strains revealed lower plasmid yields from the wild-type strain, indicating that TtAgo reduces the intracellular plasmid concentration (P < 0.02, Fig. 1c; P < 0.02, Fig. 1d).
Figure 1. TtAgo interferes with plasmid DNA.
a, Overview of ago gene loci of T. thermophilus strains: HB27 (wild type), HB27EC (spontaneous derivative with enhanced competence), HB27Δago (knockout), and HB27Δago::sago (HB27Δago complemented with a strep(II)-tag-ago gene fusion insert). KanR, kanamycin resistance marker. b, Transformation efficiency of T. thermophilus strains on transformation with the plasmid pMHPnqosGFP (Extended Data Table 5). Error bars indicate standard deviations of biological duplicates. c, Yield of pMHPnqosGFP plasmid mini preparation (miniprep) of HB27 and HB27Δago. Error bars indicate standard deviations of biological triplicates. d, Plasmid content of total DNA purified from HB27Δago relative to that from HB27, as quantified by Genetools (Syngene) after resolving the DNA on a 0.8% agarose gel. Error bars indicate standard deviations of biological triplicates.
We performed transcriptome analysis of HB27 and HB27Δago to determine whether TtAgo-mediated interference proceeds directly by targeting plasmid DNA, or indirectly by regulating gene expression. Although the comparison revealed pleiotropic changes in gene expression (Extended Data Fig. 2), we did not observe substantial differential expression of genes involved in plasmid uptake or host defence (Extended Data Table 1). Hence, RNA sequencing (RNA-seq) analysis suggests that TtAgo does not influence plasmid uptake and plasmid copy number at the level of transcriptional control.
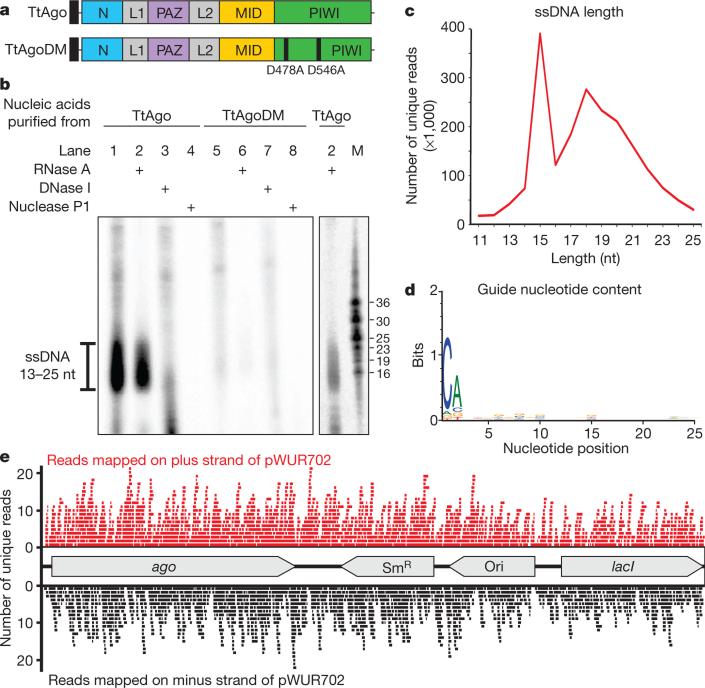
We therefore studied whether TtAgo interacts with plasmid DNA. In agreement with the RNA-seq analysis (Extended Data Fig. 2), affinity-purified TtAgo expressed from the chromosome of HB27Δago::sago could be detected by protein mass spectrometry (Extended Data Table 2). Unfortunately, molecular analysis of TtAgo expressed in T. thermophilus was hampered by the low TtAgo yield, and attempts to overexpress TtAgo in T. thermophilus from a plasmid were unsuccessful. By contrast, expression of Strep(II)-tagged TtAgo (Fig. 2a) in Escherichia coli was successful when performed at 20 °C. Under these conditions, TtAgo has no effect on plasmid content (Extended Data Fig. 1b). Analysis of co-purified nucleic acids revealed that TtAgo-associated RNA (10–150 nucleotides) is preferentially 32P-labelled in a polynucleotide kinase (PNK) forward reaction, indicating the presence of 5′ hydroxyl groups (Extended Data Fig. 1c). By contrast, co-purified DNA has a more defined length (13–25 nucleotides), and is preferentially labelled in a PNK exchange reaction, indicating phosphorylated 5′ ends (Fig. 2b). A 5′ phosphate group is a general feature of Ago guides7–11.
Figure 2. TtAgo guides are 5′-phosphorylated DNA molecules.
a, Schematic representation of TtAgo and TtAgoDM proteins used for all experiments (N, PAZ, MID, and PIWI are structural domains, L1 and L2 are linkers8). The amino-terminal Strep(II)-tag is indicated as a black square. b, Co-purified nucleic acids from TtAgo and TtAgoDM are labelled with [γ-32P]ATP after phosphate exchange by PNK from bacteriophage T4, and treated with enzymes as indicated. M, custom ssDNA marker; nt, nucleotides. c, Length distribution of unique ssDNA sequences co-purified with TtAgo. d, Nucleotide composition of unique ssDNA sequences co-purified with TtAgo. e, Unique reads of TtAgo co-purified ssDNA molecules mapped on the TtAgo expression vector pWUR702.
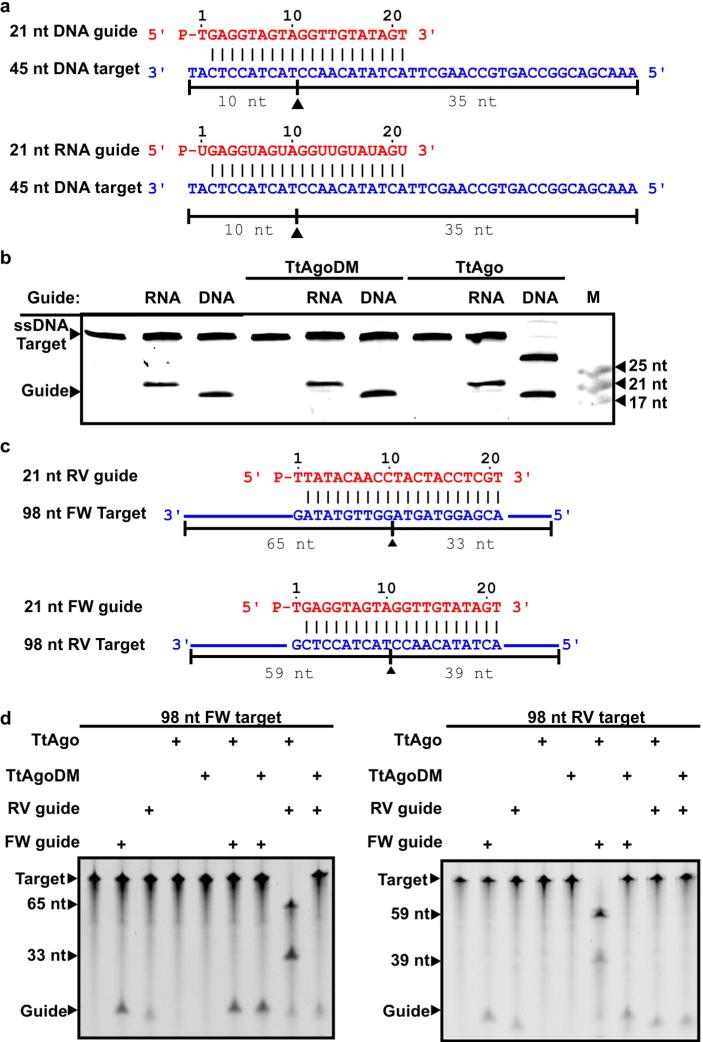
Whereas eukaryotic Ago proteins exclusively use ssRNA guides, some prokaryotic Ago proteins have a higher affinity for single-stranded DNA (ssDNA) guides9,10. Moreover, the characteristics of the small DNAs that associate with TtAgo in vivo are in agreement with previously described in vitro guide requirements8,12,13. TtAgo catalyses cleavage of ssDNA targets in vitro when supplied with complementary 5′-phosphorylated 21-nucleotide ssDNA guides, but not when supplied with analogous ssRNA guides8,12,13 (Extended Data Fig. 3). During isolation of an active site double mutant, TtAgoDM (TtAgo(D478A,D546A); Fig. 2a), only RNAs co-purify (10–150 nucleotides; Extended Data Fig. 1c). This suggests that active site residues are involved in processing and/or binding of the ssDNA molecules.
Cloning and sequencing of TtAgo-bound DNA molecules resulted in 70.6 million sequences, of which 65% can be mapped on the TtAgo expression plasmid pWUR702, 3% on the plasmid pRARE, and 32% on the chromosome of E. coli K12 (Extended Data Table 3). Remarkably, when normalized for the DNA content in each cell, TtAgo predominantly co-purifies with guides complementary to pWUR702 and pRARE (approximately 54 and 8.8 times more frequently, respectively), rather than with guides complementary to the E. coli K12 chromosome (Extended Data Table 3).
More detailed analysis of unique guide sequences revealed two populations of DNA guides: one 15-nucleotides long, and the other ranging from 13 to 25 nucleotides in length (Fig. 2c). No obvious bias towards specific regions of the plasmids or the chromosome was detected: the guides target coding and non-coding regions on both strands independent of GC content (Fig. 2e). Some guides map on one of the plasmids as well as on the chromosome of E. coli (for example, on lacI and proL). The fact that these guides do not seem to be under-represented compared with other plasmid-targeting guides indicates that there is no selection against chromosome-targeting guides, but rather that the differential guide loading (Extended Data Table 3) is a result of preferential acquisition of guides from plasmids.
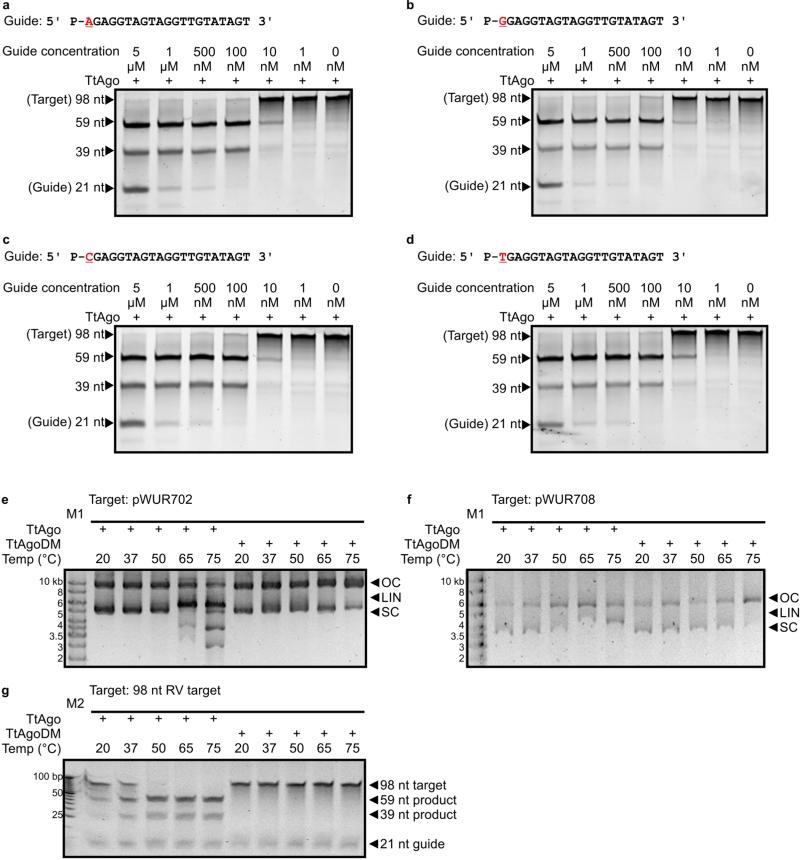
Interestingly, 89% of the DNA guides have a deoxycytidine (dC) at the first position at the 5′ end and 72% have a deoxyadenosine (dA) at the second position (Fig. 2d). Despite this bias, identical TtAgo cleavage activities are observed with DNA guides containing a 5′ dC, dT, dA or dG (Extended Data Fig. 4a–d). The 5′ dC preference may result from specific guide processing, or from preferential 5′ nucleoside selection by TtAgo. A bias for specific 5′ nucleosides also occurs in certain eukaryotic Ago proteins14,15.
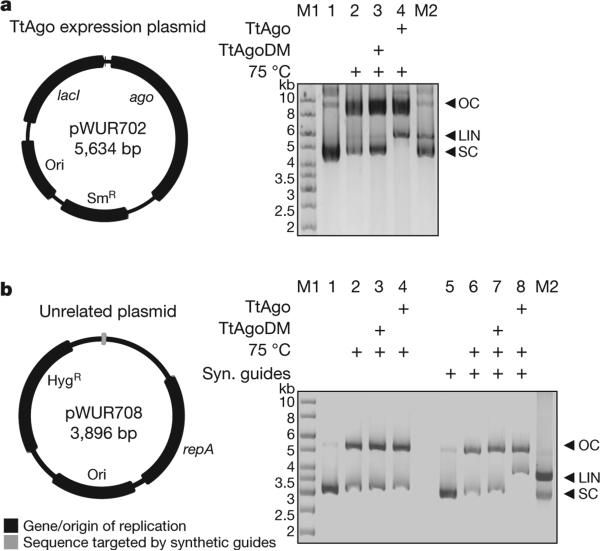
We performed activity assays to investigate whether the in vivo plasmid-derived ssDNAs are functional guides that enable TtAgo to cleave double-stranded DNA (dsDNA) targets (expression plasmid pWUR702). Purified TtAgo linearizes or nicks pWUR702, resulting in linear or open circular plasmid DNA, respectively (Fig. 3a, lane 4), whereas TtAgoDM does not show this activity (Fig. 3a, lane 3). The cleavage activity of TtAgo is strongly temperature dependent: whereas ssDNA is cleaved at temperatures ≥20 °C, plasmid DNA is only cleaved at temperatures ≥65 °C (Extended Data Fig. 4e, f). This agrees with the observation that during TtAgo expression in E. coli at 20 °C, plasmid concentrations are not decreased (Extended Data Fig. 1b). Purified TtAgo is unable to cleave plasmids that have no sequence similarity to pWUR702 or pRARE (for example, pWUR708; Fig. 3b, lane 4). However, when supplied with two synthetic 5′-phosphorylated ssDNA guides that target both strands of the plasmid at the same locus (Fig. 4b), TtAgo was able to linearize or nick pWUR708 (Fig. 3b, lane 8). These findings, together with the guide sequence data, indicate that the in vivo acquired DNA molecules guide TtAgo to cleave dsDNA targets. We propose to refer to these guides of TtAgo as small interfering DNAs (siDNAs).
Figure 3. TtAgo cleaves plasmids complementary to its guides.
a, b, Untreated target plasmid (lane 1, 5), plasmid incubated at 75 °C in the absence of proteins (lane 2, 6), or in the presence of TtAgoDM (lane 3, 7) or TtAgo (lane 4, 8) purified from E. coli, resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); M2, linearized and untreated target plasmid; OC, open circular; SC, supercoiled plasmid. a, TtAgo expression vector pWUR702. b, Target plasmid pWUR708, which shares no sequence identity with expression vector pWUR702 or pRARE. Additionally, synthetic (Syn.) ssDNA guides were added to the reactions with pWUR708 (lane 5–8).
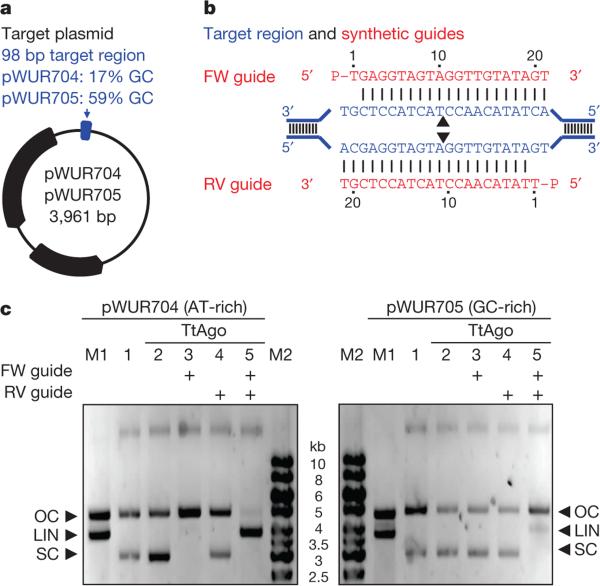
Figure 4. TtAgo cleaves plasmids by nicking two strands.
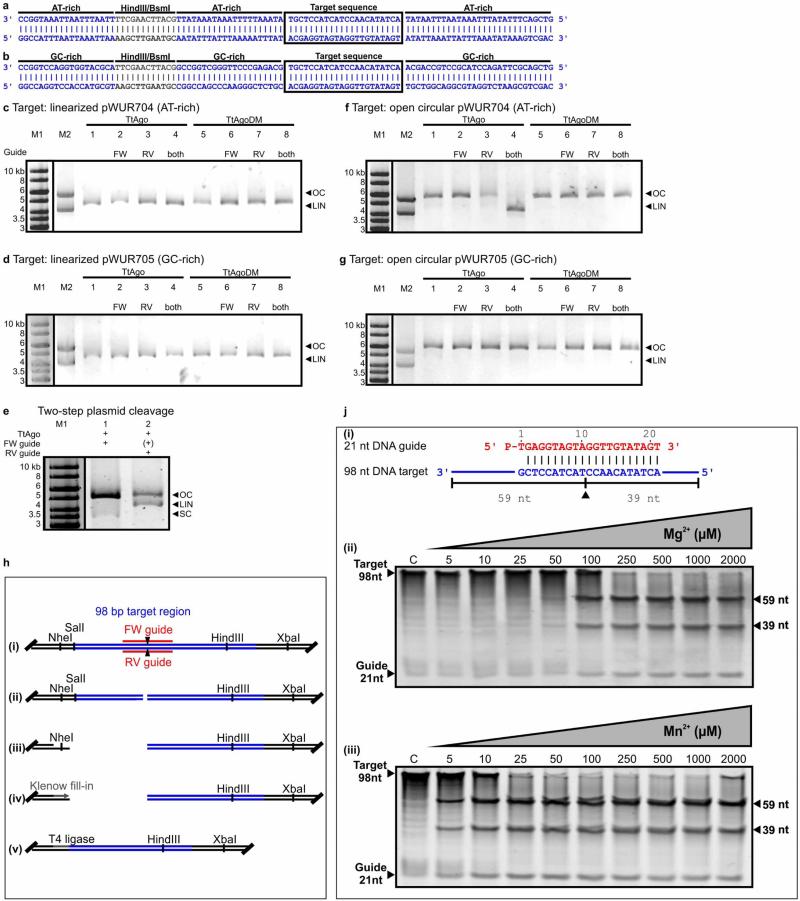
a, Plasmids pWUR704 and pWUR705 contain a 98 bp target region with a GC content of 17% or 59%, respectively, as indicated in blue (for details, see Extended Data Fig. 5a, b). b, Part of the pWUR704 and pWUR705 target site (indicated in blue) and complementary ssDNA guides used in this experiment (indicated in red). Black triangles indicate predicted cleavage sites. c, 0.8% agarose gels loaded with pWUR704 and pWUR705 plasmids that were incubated without proteins (lane 1), or with TtAgo (lane 2), TtAgo–forward (FW) guide complex (lane 3), TtAgo–reverse (RV) guide complex (lane 4), or TtAgo–FW and TtAgo–RV guide complexes. LIN, linear; M1, open circular and linear pWUR704 or pWUR705; M2, 1 kb Generuler marker (Fermentas); OC, open circular; SC, supercoiled plasmid.
To gain insight into the molecular mechanism of dsDNA cleavage by TtAgo, we performed additional in vitro plasmid cleavage assays using purified TtAgo loaded with synthetic siDNAs. Negatively supercoiled plasmids (isolated from E. coli) were used, because at least 95% of all plasmids isolated from T. thermophilus have a negatively supercoiled topology16,17. Negative supercoiling facilitates melting of the DNA duplex, especially at elevated temperatures18–20. Target plasmids pWUR704 and pWUR705 are identical except for the flanking regions of the target site (AT-rich or GC-rich; Fig. 4a). Both plasmids share no sequence similarity with TtAgo expression plasmid pWUR702, and they are not cleaved by TtAgo unless complementary siDNAs are added (Fig. 4c). When supplied with a single 21-nucleotide siDNA, TtAgo nicks the negatively supercoiled plasmid (Fig. 4c, lanes 3, 4), and when supplied with a mixture of two 21-nucleotide siDNAs that target both DNA strands at the same locus, TtAgo linearizes the plasmid (Fig. 4b, c, lane 5). Both nicking and dsDNA cleavage are more efficient when the target sequence is flanked by AT-rich regions (Fig. 4a, c and Extended Data Fig. 5a, b). Interestingly, the same TtAgo–siDNA complexes are not able to cleave linearized plasmids (Extended Data Fig. 5c, d). This suggests that cleavage of dsDNA by TtAgo depends on the negatively supercoiled topology of the target DNA.
Subsequent analysis revealed that the TtAgo–siDNA complex is able to linearize a relaxed, nicked plasmid if its target site is directly opposite the first nick (Extended Data Fig. 5e). If the nicked site is located further away (33 bp) from the target site, linearization of the nicked plasmid occurs only if the target region is AT-rich (Extended Data Fig. 5f, g). Thus, although the negatively supercoiled topology of the plasmid is lost after the primary nick, the nick facilitates local melting of the dsDNA (especially in AT-rich DNA), which allows TtAgo–siDNA complexes to nick the second strand, resulting in a dsDNA break. Like eukaryotic Ago proteins21, the TtAgo–siDNA complex cleaves a phosphate ester bond between the target nucleotides that base pair with guide nucleotides 10 and 11 (ref. 22). Sequence analysis of a cleaved dsDNA target (Extended Data Fig. 5h) demonstrated that dsDNA breaks also result from nicking both strands at the canonical Ago cleavage site.
While this manuscript was under revision, a characterization of a prokaryotic Ago protein from Rhodobacter sphaeroides (RsAgo) was published23. Despite similarities in the overall domain architecture of TtAgo and RsAgo, there are major functional differences between these proteins. RsAgo acquires mRNA-derived RNA guides with a 5′ uridine (U), whereas TtAgo acquires DNA guides with a 5′ dC. In both proteins, guides complementary to plasmids are over-represented. However, RsAgo lacks a functional catalytic site and functions by target-binding alone. TtAgo, on the other hand, harbours a functional catalytic site allowing cleavage of both single- and double-stranded targets.
On the basis of our findings, we propose a model for DNA interference by TtAgo. On the entry of plasmid DNA into the cell, TtAgo acquires siDNA guides (13–25 nucleotides in length) from the invader. Although the mechanism of guide acquisition by TtAgo is unknown, the requirement of an intact catalytic site suggests involvement of the nuclease itself. TtAgo is loaded with siDNAs that are preferentially derived from plasmids; as such, single guides may allow for neutralization of multi-copy invaders. Combining our in vivo and in vitro data, we speculate that TtAgo uses siDNA guides to specifically cleave ssDNA targets, such as DNA taken up by the natural competence system5 or replication intermediates. The siDNA–TtAgo complex also targets negatively supercoiled dsDNA, which results in plasmid nicking. Especially in the case of plasmid DNA, single-strand breaks will result in loss of the supercoiled topology and, as such, in decreased transcription levels24. Furthermore, if the nick site is located in an AT-rich region, TtAgo loaded with an siDNA that targets the opposite strand may generate a dsDNA break, potentially leading to degradation of the plasmid by other nucleases. The observation that invading DNA elements generally have a lower GC content than their hosts25 may explain self/non-self discrimination by TtAgo. Whereas the eukaryotic Ago protein is a key component of sophisticated multi-enzyme systems for RNA-guided RNA interference, we reveal the biochemical activity and functional importance of an evolutionarily related enzyme in prokaryotes that protects its host against mobile genetic elements through DNA-guided DNA interference.
METHODS
Strains
For in vivo experiments, T. thermophilus HB27 (ATCC BAA-163, DSM 7039 and NBRC 101085) was used, which is referred to in this manuscript as HB27 or wild type. Furthermore, HB27EC, and two genomic variants of the HB27 strain, HB27Δago (knockout strain) and HB27Δago::sago (knockout strain complemented with strep(II)-tag-ago fusion and kanamycin resistance marker insert), were used (Fig. 1 and Extended Data Table 4a).
Genomic mutants
HB27 genomic DNA including megaplasmid pTT27 was purified using the FastDNA SPIN Kit for Soil (MP Biomedicals). The genomic regions directly upstream (1 kb) and downstream (2.4 kb) of the ago gene (TT_P0026) were PCR amplified from T. thermophilus HB27 genomic DNA. These genomic regions contained pTT27 base-pair positions 26047–25061 (upstream sequence) and 22996–20583 (downstream sequence). The amplified DNA was cloned into the pUC18 vector (Extended Data Table 5), and the insert was transferred to pK18 (ref. 26) to generate pWUR701 (Extended Data Table 5). Strain HB27 was grown to an OD600nm of 0.4 in TTH medium (0.8% (w/v) bacto-tryptone, 0.4% (w/v) yeast extract, 51.3 mM NaCl, pH to 7.5 with NaOH, dissolved in mineral water (Evian)). 0.5 ml of the culture was transferred to a new tube and naturally transformed by the addition of 1 μg plasmid pWUR701. The culture was incubated overnight in a shaker incubator at 65 °C and plated on TTH plates with 30 μg ml−1 kanamycin. Cells were repetitively streaked on non-selective TTH plates and grown in non-selective TTH medium until kanamycinR was lost. Genomic DNA of kanamycinS cells was purified using the FastDNA SPIN Kit for Soil (MP Biomedicals) and loss of the ago gene was confirmed by PCR amplification of genomic DNA and sequencing of the target region. This strain is named HB27Δago, or knockout strain.
The genes encoding Strep(II)-tagged TtAgo protein and kanamycinR marker with upstream pSLPa promoter were PCR amplified from pWUR627 and pMK184 (ref. 28), respectively (Extended Data Table 5). PCR products were cloned into pWUR676 as indicated in Extended Data Table 5. HindIII-linearized pWUR676 was used to transform strain HB27Δago as described earlier. This strain is named HB27Δago::sago (Fig. 1). Genomic DNA was purified using the FastDNA SPIN Kit for Soil (MP biomedicals) and insertion of the sago-kanamycinR cassette was confirmed by PCR amplification from genomic DNA and sequencing of the target region.
Transformations
T. thermophilus strains were cultivated in TTH medium in a 65 °C shaker incubator until an OD600nm of 0.4 was reached. The culture was diluted 1:1 in pre-warmed TTH medium and incubated for another hour at 65 °C. 0.5 ml of the culture was transferred to a new tube, which was incubated at 65 °C for 30 min. One-hundred nanograms of plasmid pMK184 or pMHPnqosGFP was added and the mixture was incubated for 4 h at 65 °C without shaking, after which it was serial diluted and plated on TTH plates (TTH medium solidified with 1.5% agar) and on selective TTH plates (TTH plates supplied with 50 μg ml−1 kanamycin or 100 μg ml−1 hygromycin). After 48 h of incubation at 65 °C, colonies were counted. Competence was determined as the amount of kanamycinR or hygromycinR colony-forming units (c.f.u.; counted on selective plates) per μg DNA, divided by total c.f.u. (counted on non-selective plates). To show relative competence, HB27 wild-type transformation efficiency was set to 1, with the competences of other strains normalized against this number.
DNA purification
For plasmid purification, T. thermophilus HB27 and HB27Δago cultures were cultivated in triplicates in TTH medium supplied with 30 ng μl−1 kanamycin and 100 ng μl−1 hygromycin. Five OD600nm units of each overnight culture were harvested and plasmids were isolated with the Fermentas GeneJET plasmid Miniprep Kit (Thermo Scientific) according to the manual provided by the manufacturer and quantified using a NanoDrop ND1000 spectrophotometer. For complete DNA (containing both genomic and plasmid DNA) purification, T. thermophilus HB27 and HB27Δago cultures were cultivated in triplicates to an OD600nm of 0.500. One OD600nm unit was harvested and complete DNA was isolated using the JGI ‘bacterial genomic DNA isolation using CTAB’ protocol. 2.5 μg DNA of each purification was resolved on 0.8% agarose gels and stained with SYBR Safe Nucleic Acid Stain (Invitrogen), visualized using a G:BOX Chemi imager and analysed using GeneTools analysis software (Syngene).
RNA sequencing
Triplicate T. thermophilus strains were cultivated in 20 ml TTH medium in a 65 °C shaker incubator overnight. Cultures were diluted 1/100 and grown to an OD600nm of 0.5, after which cells were harvested by centrifugation. After harvesting, RNA was purified using the mirVana RNA isolation kit (Ambion) according to the instructions provided by the manufacturer. Biological triplicates of purified RNA were sequenced by BaseClear BV by Illumina sequencing. Reads were mapped on genomes and plasmid using Rockhopper29, but rather than using the programs calculated expression rates and significance, the percentage of raw counts mapped on each gene were normalized against the total number of raw counts mapped on the genome. Variance in expression was calculated by dividing the average of the triplicate normalized counts mapped on single genes in strain HB27 by the average of the triplicate normalized counts mapped on the same gene in strain HB27Δago.
TtAgo expression and purification from E. coli KRX
The ago gene was PCR amplified from T. thermophilus (ATCC 27634) genomic DNA (gene TTHB0068, base positions on pTT27: 61573–59516), and directionally cloned into a pET-52b(+) expression vector as indicated in Extended Data Table 5 (pWUR627). By introduction of mutations according to the QuikChange Site-Directed Mutagenesis Kit instruction manual (Stratagene), pWUR642 was generated (Extended Data Table 5). The inserts of pWUR627 and pWUR642 were PCR amplified and ligated into pCDF-1b as indicated in Extended Data Table 5 (pWUR702 and pWUR703). These plasmids were transformed into E. coli KRX (Promega) simultaneously with pRARE (Novagen), purified from E. coli Rosetta DE3 (Novagen). Strains were cultivated in LB medium containing the corresponding antibiotics (50 ng ml−1 streptomycin, 34 μg ml−1 chloramphenicol) in a shaker incubator at 37 °C. When the culture reached an OD600nm of 0.7–0.8, cells were cold-shocked by incubation in an ice bath for 15 min. Expression was induced by adding isopropyl-β-d-thiogalactoside (IPTG) and l-Rhamnose to a final concentration of 1 mM and 0.1% (w/v), respectively, and expression was continued for 16 h in a shaker incubator at 20 °C. Cells were harvested by centrifugation. For plasmid quantification, plasmids were isolated from 5 OD600nm units of harvested cells using the Fermentas GeneJET plasmid Miniprep Kit (Thermo Scientific) according to the manual provided by the manufacturer and quantified using a NanoDrop ND1000 spectrophotometer. For TtAgo purification, harvested cells were resuspended in Buffer I (20 mM Tris-HCl pH 8, 1 M NaCl, supplied with either 2 mM MnCl2 or 2 mM MgCl2), and disrupted using a French pressure cell. Expressed proteins have an N-terminal Strep(II)-tag and were isolated using Strep-Tactin affinity chromatography (IBA) with an adapted protocol. Before loading of the cell-free extract, columns were equilibrated in Buffer I. After loading, columns were washed with 9 column volumes of Buffer I and with 9 column volumes of Buffer II (20 mM Tris-HCl pH 8, 0.5 M NaCl, supplied with 2 mM MnCl2). Proteins were eluted in Buffer III (Buffer II supplemented with 2.5 mM d-Desthiobiotin (Sigma-Aldrich)). For purification of TtAgo used in Mn/Mg gradient experiments, no Mn or Mg was added to purification buffers. For other activity assays, MnCl2 or MgCl2 was added to all buffers at a final concentration of 0.5 mM.
TtAgo purification from T. thermophilus
HB27Δago::sago was cultivated in TTH medium supplemented with 30 ng ml−1 kanamycin at 65 °C. After overnight growth, cells were harvested and TtAgo was purified as described earlier. After purification, elution fractions were resolved on SDS–PAGE gels and purified proteins were stained using Coomassie brilliant blue stain. A band corresponding to the region with the molecular weight of Argonaute (75–80 kDa) was excised from the gel and subjected to in-gel digestion using a Perkin Elmer Janus Automated Workstation. Peptide mixtures were injected onto a nanoACQUITY UPLC (Waters Corporation) coupled to a LTQ-Orbitap XL (Thermo Fisher Scientific) via an Advion Biosciences Nanomate. Peptides were eluted over a 30 min gradient (5–40% ACN). MaxQuant (v.1.4.1.2) and its embedded Andromeda search engine were used to search the data against a database containing T. thermophilus sequences extracted from Uniprot (8 August 2013). Methionine oxidation was used as a variable modification and a maximum of two missed trypsin cleavages were allowed. Peptide and protein posterior error probabilities (PEP) were calculated using a target-decoy search using the revert scheme. The light version of intensity-based absolute quantification (iBAQ) was used to rank the identified proteins by estimated relative abundance.
Guide co-purification and sequencing
Proteinase K (Ambion) and CaCl2 (final concentration, 5 mM) were added to purified proteins and samples were incubated for 1 h at 37 °C. Nucleic acids were separated from protein content using Roti phenol/chloroform/isoamyl alcohol pH 7.5–8.0 (Carl Roth GmbH) and further purified by ethanol precipitation. Precipitation was performed overnight at −20 °C in the presence of linear polymerized acrylamide as carrier.
Purified nucleic acids were [γ-32P]ATP labelled with T4 PNK (Fermentas) in exchange- or forward-labelling reactions and thereafter separated from free [γ-32P] ATP using a Sephadex G-25 column (GE). Labelled nucleic acids were incubated with nucleases (DNase-free RNase A (Fermentas),RQ1 RNase-free DNaseI (Promega) or P1 nuclease (Sigma)) for 1 h at 37 °C. After nuclease treatment, samples were mixed with Loading Buffer (95% (deionized) formamide, 5 mM EDTA, 0.025% SDS, 0.025% bromophenol blue and 0.025% xylene cyanol), heated for 5 min at 95 °C and resolved on 15% or 20% denaturing polyacrylamide gels. Radioactivity was captured from gels using phosphor screens.
Nucleic acids were purified from TtAgo and treated with RNaseA, as described earlier. The small 5′-phosphorylated DNA molecules were poly-adenylated at their 3′ end using recombinant terminal deoxynucleotidyl transferase (TdT, Invitrogen), according to the instructions of the manufacturer. After purification of the product using the QIAquick nucleotide removal kit (Qiagen), 5′-phosphorylated and 3′-polyadenylated products were ligated to the 3′ end of oligonucleotide BG4409 (Extended Data Table 4b) using T4 RNA ligase (Ambion), according to the instructions of the manufacturer. After purification of the product using the QIAquick nucleotide removal kit (Qiagen), the product was PCR amplified using primers BG4409 and BG4436 (anchored poly-T primer (partially degenerate); Extended Data Table 4b). The PCR amplification product was gel purified using the GeneJET gel extraction kit (Fermentas) and sent for sequencing by Imagif, Plateforme de Séquençage à Haut Débit by Illumina sequencing with an adapted RNA-seq protocol. Sequences were analysed with FastQC software (Babraham Bioinformatics). After mapping on genome and plasmids, duplicate reads were removed using SAMtools software26, to exclude a bias for preferentially PCR amplified reads in downstream analysis. Unique read data sets were re-analysed with FastQC software and remapped on genome and plasmid DNA using Tablet software (James Hutton Institute)28.
DNA guides and targets
The sequence of guide BG3466 is based on let-7 miRNA, and has been used before in experiments performed with TtAgo8,12,13, whereas the sequence of guide BG4017 is based on the reverse complementary sequence of let-7 miRNA (Extended Data Table 4b). Both guides have a 5′ phosphate, are 21-nucleotides long and have been PAGE purified after synthesis. Oligonucleotides BG4262–BG4265 (Extended Data Table 4b) were used in activity assays as an ssDNA target or mixed together with 2× STE buffer (20 mM Tris-HCl pH 8, 100 mM NaCl, 2 mM EDTA) in a 1:1:2 ratio (BG4262:BG4263:2× STE or BG4264:BG4265:2× STE) and incubated at 95 °C for 5 min. Samples were cooled down to room temperature (20 °C). Annealed oligonucleotides were used as inserts for plasmid pWUR677 (generated from pFU98)29 to generate pWUR704 and pWUR705. For experiments with nicked and linearized targets, pWUR704 and pWUR705 were treated with Nb.BsmI or SpeI, respectively. Plasmid pWUR708 was generated as pWUR704 and pWUR705 but with annealed BG3467 and BG3468 oligonucleotides as insert.
Activity assays
Purified TtAgo, ssDNA or ssRNA guides, and ssDNA targets (Extended Data Tables 4b, 5) were mixed in 5:1:1 ratio (TtAgo:guide:target) in 2× Reaction Buffer (20 mM Tris-HCl pH 8, 250 mM NaCl supplied with varying concentrations of MnCl2 or MgCl2). Reaction mixtures were incubated for 1 h at 75 °C. Reactions were stopped by the addition of Loading Buffer and heated for 5 min at 95 °C before the samples were resolved on 15% or 20% denaturing polyacrylamide gels. Gels were stained using SYBR gold Nucleic Acid Gel Stain (Invitrogen) and nucleic acids were visualized using a G:BOX Chemi imager (Syngene). Because DNA-guided cleavage of ssDNA is observed in the presence of 5–10 μM Mn2+ (Extended Data Fig. 5j), but comparable cleavage levels are observed in the presence of Mg2+ only at tenfold higher concentrations (Extended Data Fig. 5j), all activity assays were performed in the presence of 0.5 mM MnCl2.
Purified TtAgo, ssDNA guides and plasmid targets were mixed in a 25:5:1 ratio (TtAgo:guide:target) in 2× Reaction Buffer supplemented with 0.5 mM MnCl2. Samples were incubated for 16 h at 75 °C. Reactions were stopped by adding Proteinase K solution (Ambion) and CaCl2 (final concentration, 5 mM) and samples were incubated for 1 h at 65 °C. Samples were mixed with 6× loading dye (Fermentas) before they were resolved on 0.8% agarose gels. Agarose gels were stained with SYBR safe or SYBR gold Nucleic Acid Gel Stain (Invitrogen) and nucleic acids were visualized using a G:BOX Chemi imager (Syngene).
Plasmid pWUR704 was linearized with TtAgo–siDNA complexes as described earlier. The DNA was purified from the activity assay sample by PCI extraction followed by ethanol precipitation. Purified DNA was cut either by XbaI or by NheI. Restriction site overhangs were filled in with Klenow Fragment (Thermo Scientific) according to the manual provided by the manufacturer. Blunt-end linear plasmid was closed by T4 ligase ligation according to the manual provided by the manufacturer (Thermo Scientific). Ligated plasmids were treated with HindIII (in the case of the XbaI-treated plasmids) or SalI (in the case of NheI-treated plasmids) to eliminate the possible background of the original plasmid. Plasmids were transformed to NEB 5-α E. coli competent cells (New England Biolabs) according to the manual provided by the manufacturer. Colonies were picked, grown overnight in LB medium at 37 °C and miniprepped with the Fermentas GeneJET Plasmid Miniprep Kit (Thermo Scientific). Purified plasmids were sent to GATC Biotech (Germany) for target site sequencing.
Statistical analysis
All P values stated in this manuscript are calculated by a two-tailed distributed two-sample t-test assuming equal variances. For the calculation of P values of the transformation efficiencies, competence (calculated as described earlier) from biological duplicates of each strain was used as the input. For the calculation of P values of plasmid purification, plasmid DNA yield of biological triplicates of each strain were used as the input. For the calculation of P values of plasmid DNA content of complete DNA purification, plasmid DNA content of biological triplicates of each strain were used as the input. For the calculation of P values of differences in expression levels of specific genes, normalized raw mapped counts of biological triplicates of each strain were used as the input. All P values calculated are considered to be significant, as for all calculations P < 0.02).
Extended Data
Extended Data Figure 1. Analyses of TtAgo in T. thermophilus and E.coli.
a, TtAgo decreases plasmid transformation efficiency of T. thermophilus. Transformation efficiency of different ago mutant strains relative to the transformation efficiency of wild-type strain HB27. HB27EC is an HB27 mutant selected for high competence, and HB27Δago is an ago gene knockout strain (Fig. 1a). Strains were transformed with plasmid pMK184 (Extended Data Table 5). Transformations were performed in biological duplicates for each strain. Error bars indicate standard deviations. b, Effect on TtAgo expression on plasmid content in E. coli KRX. TtAgo and TtAgoDM were expressed in E. coli KRX from plasmid pWUR702 and pWUR703. Plasmids were purified from biological triplicate cultures in which expression was induced (+) or not induced (−). Compared with TtAgoDM expression, TtAgo expression in E. coli KRX does not lead to reduced plasmid content. Changes in plasmid yield between induced and not induced cultures probably originate from protein expression energy costs. Error bars indicate standard deviations. c, 10–150-nucleotide (nt) RNA with 5′-OH group co-purifies with TtAgo. 15% denaturing polyacrylamide gels with nucleic acids co-purified with TtAgo and TtAgoDM. Nucleic acids are phosphorylated in a T4 PNK forward reaction (5′-OH groups, and to a lesser extend 5′-P groups, are labelled) using [γ-32P] ATP, and resolved on 15% denaturing polyacrylamide gels. Nucleic acids were not treated (lane 1, 5), RNaseA treated (lanes 2, 6), DNaseI treated (lane 3, 7) or Nuclease P1 treated (lane 4, 8).
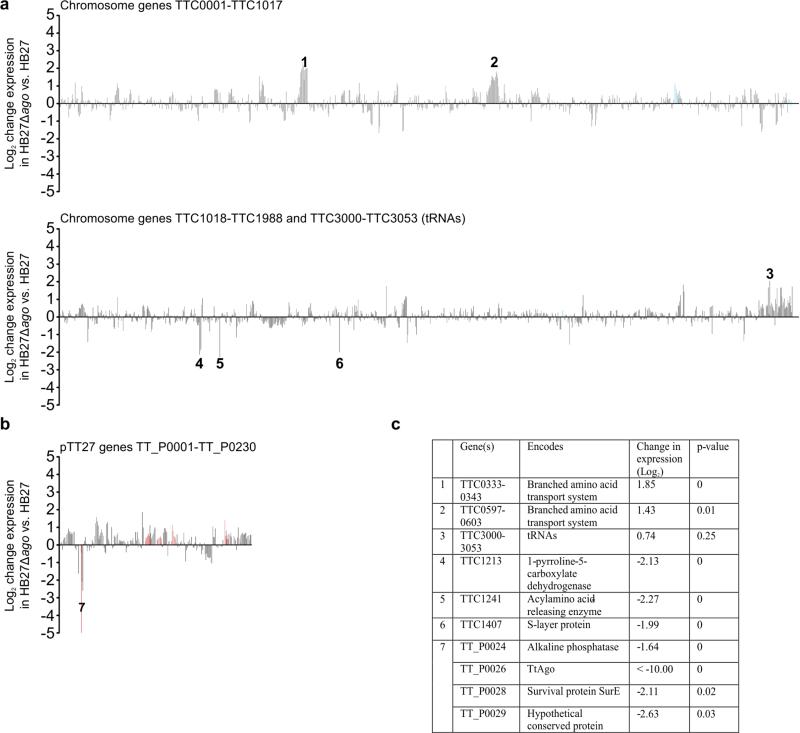
Extended Data Figure 2. Change in transcription of T. thermophilus genes after ago gene knockout.
a, b, RNA-seq analysis was performed on biological triplicates for each strain. Change in gene expression of genes encoded on the chromosome (a) or on the megaplasmid (b) is shown as the log2 of the fold difference in expression of the average of normalized mapped reads on that gene in HB27Δago compared with the average of normalized mapped reads on that gene in HB27. Peaks corresponding to genes involved in host defence are coloured red, whereas peaks corresponding to genes involved in competence are coloured blue. c, Genes or operons containing genes with a log2 expression change greater than 2 or −2.
Extended Data Figure 3. TtAgo cleaves ssDNA using ssDNA guides.
a, 21-nucleotide (nt) DNA and RNA guides are complementary to the 45-nucleotide DNA targets. Predicted cleavage positions are indicated with a black triangle. b, 20% denaturing polyacrylamide gel loaded with samples in which TtAgo and TtAgoDM were provided with an RNA or an DNA guide to cleave a 45-nucleotide ssDNA target. c, 21-nucleotide RV and FW DNA guides are complementary to the 98-nucleotide ssDNA targets. Predicted cleavage positions are indicated with a black triangle. d, 98-nucleotide ssDNA targets are incubated with TtAgo and TtAgoDM, provided with complementary and non-complementary DNA guides, and resolved on 15% denaturing polyacrylamide gels.
Extended Data Figure 4. Effect of variation of the 5′-end deoxynucleoside of the siDNA and effect of the temperature on TtAgo cleavage efficiency.
a–d, Cleavage of 98-nucleotide ssDNA target (Extended Data Fig. 3c) by TtAgo loaded with complementary siDNAs containing a different 5′ deoxynucleoside, as shown in red. The concentrations of each siDNA were varied (indicated on top of the gels). Products of the reaction were resolved on 15% denaturing polyacrylamide gels. e, TtAgo expression plasmid pWUR702 (no guides added) incubated with TtAgo and TtAgoDM at different temperatures. f, pWUR708 plasmid (FW and RV guides added; Fig. 4b) incubated with TtAgo and TtAgoDM at different temperatures, resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); OC, open circular; SC, supercoiled. g, 98-nucleotide RV target cleavage (FW guide added) incubated with TtAgo and TtAgoDM at different temperatures, resolved on a 15% denaturing acrylamide gel. M2, O'RangeRuler 5 bp DNA Ladder (Thermo Scientific).
Extended Data Figure 5. Activity analyses of TtAgo.
a, b, AT-rich (17% GC) insert of pWUR704 (a) and GC-rich insert (59% GC) of pWUR705 (b). The target sequence is boxed. Restriction sites HindIII and BsmI are indicated in grey. Sequences are displayed in the 3′–5′ direction to allow comparison with Fig. 4b, which shows guide base pairing to this sequence. c, d, SpeI-linearized plasmid pWUR704 (c) and pWUR705 (d) incubated with TtAgo–siDNA and TtAgoDM–siDNA complexes targeting both strands of the plasmid, and resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); M2, open circular and linearized pWUR704 (c), or open circular and linearized pWUR705 (d); OC, open circular. FW guide: BG3466. RV guide: BG4017. High salt concentration (250 mM NaCl) in the reaction buffer cause the TtAgo-treated samples to run higher in the gel than M1 and M2. e, Two-step plasmid cleavage. Target pWUR704 was first nicked by a TtAgo–siDNA complex targeting the first strand (FW guide, lane 1), after which a TtAgo–siDNA complex targeting the other strand was added (RV guide, lane 2). FW guide is still present, and its presence is therefore indicated as (+). LIN, linear; M1, 1 kb Generuler marker (Fermentas); OC, open circular; SC, supercoiled. f, g, Nb.BsmI-nicked plasmid pWUR704 (f) and pWUR705 (g) incubated with TtAgo–siDNA and TtAgoDM–siDNA complexes targeting the un-nicked strands of the plasmid (33 bp away from the nicking site), and resolved on 0.8% agarose gels. LIN, linear; M1, 1 kb Generuler marker (Fermentas); M2, open circular and linearized pWUR704 (a), or open circular and linearized pWUR705 (b); OC, open circular. High salt concentrations (250 mM NaCl) in the reaction buffer cause the TtAgo-treated samples to run higher in the gel than M1 and M2. h, TtAgo dsDNA cleavage site analysis. (i) Plasmid pWUR704 with TtAgo–siDNA target sequences. Predicted cleavage sites are indicated with black triangles. (ii) pWUR704 was linearized using TtAgo–siDNA complexes targeting the plasmid on both strands. (iii) The linearized plasmid was cleaved using either NheI (as shown) or XbaI (not shown). (iv) Restriction site overhangs and possible overhangs resulting from TtAgo–siDNA cleavage were filled using Klenow fragment polymerase (Fermentas). (v) Blunt-end DNA was ligated using T4 DNA ligase (Fermentas), after which the plasmid could be transformed and later sequenced to determine the cleavage site. Sequences revealed that TtAgo–siDNA complexes indeed cleaved the target at the predicted locations (as shown in a), and are shown in more detail in Fig. 4b and Extended Data Fig. 5a, b. Note that in this picture target sequences are displayed in reversed order compared with Fig. 4b and Extended Data Fig. 5a, b. j, TtAgo prefers Mn2+ over Mg2+ as a divalent cation for cleavage. (i) 21-nucleotide DNA guide and 98-nucleotide ssDNA target used. The predicted cleavage site is indicated with a black triangle. (ii) 98-nucleotide ssDNA target cleavage reaction with TtAgo loaded with a 21-nucleotide siDNA in the presence of increasing concentrations of Mg2+, as indicated on top of the gel. (iii) 98-nucleotide ssDNA target cleavage reaction with TtAgo loaded with a 21-nucleotide siDNA in the presence of increasing concentrations of Mn2+, as indicated. Samples were resolved on 15% denaturing polyacrylamide gels.
Extended Data Table 1.
Expression profile of T. thermophilus genes involved in competence and host-defence
| a | ||
|---|---|---|
| Genes involved in competence | ||
| Gene | Encoded protein | Log2 fold change (P-value) |
| TTC1603 | ComEC | 0.09 (0.49) |
| TTC1602 | ComEA | −0.05 (0.74) |
| TTC1873 | DprA | 0.22 (0.19) |
| TTC0854 | PilA1 | 1.15 (0.02) |
| TTC0855 | PilA2 | 0.97 (<0.01) |
| TTC0856 | PilA3 | 0.82 (<0.01) |
| TTC0858 | PilA4 | 0.45 (0.17) |
| TTC1716 | PilD | 0.40 (0.11) |
| TTC1622 | PilF | 0.10(0.11) |
| TTC0440 | PilC | 0.43 (<0.01) |
| TTC1017 | PilQ | 0.15 (0.30) |
| TTC0857 | ComZ | 0.61 (0.05) |
| TTC1013 | PilM | 0.25 (0.06) |
| TTC1014 | PilN | 0.21 (0.05) |
| TTC1015 | PilO | 0.20 (0.43) |
| TTC1016 | PilW | −0.17 (0.12) |
| TT_P0190 | PilA | −0.01 (0.93) |
| b | ||
|---|---|---|
| Genes involved in host-defence | ||
| Gene | Encoded protein | Log2 fold change (P-value) |
| TT_P0026 | TtAgo | <−10 (<0.01) |
| TTC1926 | Cas2 | 0.16 (0.03) |
| TTC1927 | Cas6 | 0.02 (0.88) |
| TT_P0101 | Cas2 | 0.18 (0.61) |
| TT_P0102 | Csm1 | 0.39 (0.04) |
| TT_P0103 | Csm2 | 0.46 (<0.01) |
| TT_P0104 | Csm3 | 0.54 (<0.01) |
| TT_P0105 | Csm4 | 0.64 (0.01) |
| TT_P0106 | Csm5 | 0.52 (<0.01) |
| TT_P0107 | Csx1 | 0.28 (0.07) |
| TT_P0115 | Cmr2 | 0.32 (0.06) |
| TT_P0116 | Cmr3 | 0.30 (0.16) |
| TT_P0117 | Cmr1 | 0.43 (0.01) |
| TT_P0118 | Cmr4 | 0.41 (<0.01) |
| TT_P0119 | Cmr5 | 0.48 (<0.01) |
| TT_P0120 | RAMP | 0.38 (0.03) |
| TT_P0132 | Cas3 | 0.22 (0.20) |
| TT_P0133 | Cas4 | 1.14 (<0.01) |
| TT_P0134 | Cas8C | 0.80 (<0.01) |
| TT_P0135 | Cas7 | 0.50 (<0.01) |
| TT_P0136 | Cas4 | 0.48 (0.01) |
| TT_P0195 | Cas2 | 1.42 (0.15) |
| TT_P0196 | Cas1 | 0.52 (0.20) |
| TT_P0197 | Cas4 | 0.34 (0.44) |
| TT_P0204 | Cas6 | 0.73 (0.01) |
| TT_P0215 | Cas1 | 0.16 (0.42) |
Expression values are given as log2 values of fold expression levels of the gene in strain HB27Δago relative to strain HB27, and P values (t-test) are indicated in brackets. Changes in expression are considered substantial if the log2 value > 2 and P < 0.02 (Extended Data Fig. 2).
Extended Data Table 2.
Mass-spectrometry data of identified proteins after Strep(II)-tag affinity purification
| a | ||||||
|---|---|---|---|---|---|---|
| Proteins with most abundant peptides | ||||||
| Protein ID | Name | Peptides | Sequence coverage (%) | Mol. Weight (kDa) | PEP | iBAQ |
| Q72G73 | TT_C1975 | 31 | 42.8 | 76.8 | 1.64E-109 | 32756000 |
| P61490 | groL | 16 | 28.4 | 57.9 | 1.09E-36 | 704330 |
| Q746M7 | TtAgo | 14 | 19 | 76.7 | 1.95E-37 | 2642000 |
| Q72JL4 | TT_C0758 | 11 | 26.3 | 49.3 | 1.72E-41 | 2238900 |
| Q72J15 | TT_C0966 | 9 | 25.3 | 41.6 | 3.57E-29 | 1088200 |
| Q72H68 | TT_C1627 | 7 | 16.1 | 48.1 | 1.06E-19 | 340990 |
| Q72HX7 | TT_C1355 | 7 | 8.7 | 82.5 | 1.05E-19 | 272740 |
| Q72GH6 | TT_C1872 | 5 | 15.9 | 33.3 | 4.09E-17 | 612660 |
| Q72K98 | TT_C0549 | 5 | 16.3 | 35.9 | 3.04E-13 | 585590 |
| Q72GW4 | tuf1 | 5 | 12.3 | 44.8 | 1.97E-14 | 497870 |
| b | |||
|---|---|---|---|
| Peptides matched against TtAgo | |||
| Sequence | Mass (Da) | Protein | PEP |
| AFGASGASLR | 935.48248 | TtAgo | 0.00054319 |
| AQETALALLR | 1084.6241 | TtAgo | 5.97E-06 |
| AVSKPADALR | 1026.5822 | TtAgo | 0.0052086 |
| EGIAYDLVSVR | 1220.6401 | TtAgo | 0.0012876 |
| EIASWIGR | 930.49232 | TtAgo | 0.0065071 |
| LADGLYVPLEDK | 1331.6973 | TtAgo | 0.00097635 |
| LGEEDPK | 786.37595 | TtAgo | 0.013635 |
| LGLGTPEAVR | 1011.5713 | TtAgo | 0.00069792 |
| LYPASGFAFPR | 1224.6291 | TtAgo | 0.021229 |
| MGQNYAYR | 1001.4389 | TtAgo | 0.0050656 |
| SVLSALAR | 815.4865 | TtAgo | 0.0013518 |
| TEVFLNR | 877.46577 | TtAgo | 0.0013027 |
| VAWVADPKDPR | 1252.6564 | TtAgo | 0.023399 |
| VYPVQGR | 817.44464 | TtAgo | 0.033518 |
a, Only proteins of which five or more peptides were discovered are shown. The ‘Peptides’ column shows how many peptides are matches against a certain protein. iBAQ, intensity-based absolute quantification; PEP, posterior error probability. b, Peptides identified that match TtAgo sequence.
Extended Data Table 3.
TtAgo preferentially acquires ssDNA guides from plasmid DNA
| DNA locus | Size | Copies per cell | Total DNA per cell | Reads aligned to DNA locus | Normalized reads* | Normalized reads* corrected for DNA per cell |
|---|---|---|---|---|---|---|
| E. coli K12 chromosome | 4.64 Mb | 1 | 4.64 Mb | 23*106 | 1 | 1 |
| pWUR702 | 5.6 kb | 20-40 | 0.17 Mb | 45*106 | 2 | 54 |
| pRARE | 4.7 kb | 10-12 | 52 kb | 2*106 | 0.1 | 8.8 |
Estimated relative quantities of guides complementary to plasmid and chromosome DNA per cell.
Reads are normalized against the number of reads mapped against the E. coli K12 chromosome.
Extended Data Table 4.
Strains and oligonucleotides
| a | |||
|---|---|---|---|
| Strain | Abbreviations | Description | Source, reference |
| Thermus thermophilus HB27 | HB27, wild type | ATCC BAA-163 / DSM 7039 / NBRC 101085 | DSMZ |
| T. thermophilus HB27EC | HB27EC | ago::agoISTth7 and multiple mutations, selected for enhanced competence | This study |
| T. thermophilus HB27 Δago | HB27Δago, knockout | Δ ago | This study |
| T. thermophilus HB27 Δago::strep(II)-ago | HB27Δago::sago | HB27Δago with strep(II)-tag-ago fusion and kanamycin marker insert | This study |
| b | |||
|---|---|---|---|
| Experiment | Primers | Sequence (5’-3’) | Description, restriction sites |
| Genomic mutants | BG3524 | AAAAAAAAGCTTCCTCAACGGGGAGGTTCCGGA | upstream region ago (fw), HindIII |
| BG3525 | AAAAAAGTCGACGCTCAGATTTGCATAGGAGCTGC | upstream region ago (rv), SalI | |
| BG3526 | AAAAAAGTCGACATGGCAAGCTGGAGCCACCCG | strep(II)-ago (fw), SalI | |
| BG3527 | AAAAAATCTAGACTAAACGAAGAAGAGCTTTTCCCG | strep(II)-ago (rv), XbaI | |
| BG3528 | AAAAAATCTAGATGCCCAAGCGGGGCGGAACC | downstream region ago (fw), XbaI | |
| BG3529 | AAAAAAGAATTCGGTCAATCCGCCCCGCTTCCA | downstream region ago (rv), EcoRI | |
| BG3563 | GGCCGTCTAGACCCGGGAGTATAACAGAAACCTT | PslpA-KanR-stop (fw), XbaI | |
| BG3564 | GCGCGTCTAGATCAAAATGGTATGCGTTTTGACAC | PslpA-KanR-stop (rv), XbaI | |
| Expression vectors TtAgo | AgoFW | GCGCGCGGTACCAGATGAACCACCTTGGAAAAACGG | T. thermophilus HB8 ago (fw), KpnI |
| AgoRV | GCGCGCGCGGCCGCGAATTCCTAAACGAAGAAGAGCTTTTCCC | T. thermophilus HB8 ago (rv), NotI | |
| BG4207 | GCGCGCACATGTCAAGCTGGAGCCACCCGCAG | strep(II)-ago (fw), PciI | |
| BG4208 | GCGCGCCCTAGGTTAATTAGTGGTGGTGATGG | strep(II)-ago (rv), AvrII | |
| Site directed mutagenesis of ago gene | BG3454 | GGCGGAGCTCGCCGTGGGCTTTGCCGCCGGCGGAAGGGAGTCCTTTCG | HB8 ago D478A (fw) |
| BG3455 | CGAAAGGACTCCCTTCCGCCGGCGGCAAAGCCCACGGCGAGCTCCGCC | HB8 ago D478A (rv) | |
| BG3456 | CCCGGGTCCTCCTCCTTCGGGCCGGCCGCGTGCCCCAGGACGAG | HB8 ago D546A (fw) | |
| BG3457 | CTCGTCCTGGGGCACGCGGCCGGCCCGAAGGAGGAGGACCCGGG | HB8 ago D546A (rv) | |
| Guide sequencing | BG4409 | GAGAGAGGATCCCGAATTGTGCAGCTGTCAATCAACC | 5’ Amplification primer, BamHI |
| BG4436 | GAGAGAGGATCCTTTTTTTTTTTTTTTTTTTTTTTTTTVN | 3’ Poly-T primer with ‘VN’ anchor, BamHI | |
| Target sequences | BG4262 | GGCCATTTAATTAAATTAAAAGCTTGAATGCAATATTTATTTAAAAATTTATACGAGGTAGTAGGTTGTATAGTATATTAAATTATTTAAATATAAAG | Low GC-content (17%) target oligonucleotide ‘FW-target’ |
| BG4263 | TCGACTTTATATTTAAATAATTTAATATACTATACAACCTACTACCTCGTATAAATTTTTAAATAAATATTGCATTCAAGCTTTTAATTTAATTAAAT | Low GC-content (17%) target oligonucleotide ‘RV-target’ | |
| BG4264 | GGCCAGGTCCACCATGCGTAAGCTTGAATGCCGGCCAGCCCAAGGGCTCTGCACGAGGTAGTAGGTTGTATAGTTGCTGGCAGGCGTAGGTCTAAGCG | High GC-content (59%) target oligonucleotide ‘FW-target’ | |
| BG4265 | TCGACGCTTAGACCTACGCCTGCCAGCAACTATACAACCTACTACCTCGTGCAGAGCCCTTGGGCTGGCCGGCATTCAAGCTTACGCATGGTGGACCT | High GC-content (59%) target oligonucleotide ‘RV-target’ | |
| BG3467 | CTAGACGAGGTAGTAGGTTGTATAGTA | Target sequence insert, XbaI, HindIII | |
| BG3468 | AGCTTACTATACAACCTACTACCTCGT | Target sequence insert, XbaI, HindIII | |
| siDNA and siRNA sequences | BG3466 | P-TGAGGTAGTAGGTTGTATAGT | FW-guide, based on let-7 miRNA |
| BG4017 | P-TTATACAACCTACTACCTCGT | RV-guide, based on reverse complement of let-7 miRNA | |
| BG4500 | P-AGAGGTAGTAGGTTGTATAGT | FW-guide, based on let-7 miRNA, 5’-end deoxyadenosine | |
| BG4501 | P-GGAGGTAGTAGGTTGTATAGT | FW-guide, based on let-7 miRNA, 5’-end deoxyguanosine | |
| BG4502 | P-CGAGGTAGTAGGTTGTATAGT | FW-guide, based on let-7 miRNA, 5’-end deoxycytidine | |
| BG4503 | P-TGAGGTAGTAGGTTGTATAGT | FW-guide, based on let-7 miRNA, 5’-end deoxythymidine | |
| BG4508 | P-UGAGGUAGUAGGUUGUAUAGU | FW-guide, based on let-7 miRNA | |
a, T. thermophilus strains used in this study. b, Oligonucleotides used in this study; restriction sites are underlined.
Extended Data Table 5.
Plasmids
| Plasmid | Description | Restriction sites used | Primers | Source, reference |
|---|---|---|---|---|
| pRARE | E. coli Rosetta (DE3) plasmid, encodes rare tRNAs, CamR | Novagen | ||
| pET-52b(+) | T7 RNA polymerase based expression vector, AmpR | Novagen | ||
| pWUR627 | T. thermophilus HB8 ago with N-term. strep(II)-tag in pET-52b(+) expression vector for TtAgo | KpnI | AgoFW | This study |
| NotI | AgoRV | |||
| pWUR641 | pWUR627, ago active site residue substituted (D546A) | - | BG3456 BG3457 | This study |
| pWUR642 | pWUR641, ago active site residue substituted (D478A) | - | BG3454 | This study |
| Expression vector for TtAgoDM(D478A,D546A) | BG3455 | |||
| pCDF-1b | T7 RNA polymerase based expression vector, SmR | Novagen | ||
| pWUR702 | strep(II)-ago insert from pWUR627 inserted in pCDF-1b | AvrII | BG4207 | This study |
| Expression vector for TtAgo | NcoI | BG4208 | ||
| pWUR703 | strep(II)-agodm(D478A,D546A) insert from pWUR642 inserted in pCDF-1b | AvrII | BG4207 | This study |
| Expression vector for TtAgoDM(D478A,D546A) | NcoI | BG4208 | ||
| pUC18 | AmpR | Thermo scientific | ||
| pUC19 | AmpR | Thermo scientific | ||
| pWUR673 | 2.4kb downstream sequence of ago inserted in pUC18 | XbaI | BG3528 | This study |
| EcoRI | BG3529 | |||
| pWUR674 | 1kb upstream sequence of ago inserted in pWUR673 | HindIII | BG3524 | This study |
| SalI | BG3525 | |||
| pWUR675 | ago with N-terminal strep(II)-tag inserted in pWUR674 | SalI | BG3526 | This study |
| XbaI | BG3527 | |||
| pWUR676 | KanR marker with pSLPa promoter inserted in pWUR675 | XbaI | BG3563 | This study |
| XbaI | BG3564 | |||
| pK18 | Recombination vector | 27 | ||
| pWUR701 | Insert from pWUR674 transferred to pK18 | HindIII | This study | |
| EcoRI | ||||
| pMHPnqosGFP | E. coli / T. thermophilus shuttle vector, HygR, sGFP under control of Pnqo promoter | 30 | ||
| pMKPnqosGFP | E. coli / T. thermophilus shuttle vector, KanR, sGFP under control of Pnqo promoter | 30 | ||
| pMK184 | E. coli / T. thermophilus shuttle vector, KanR | 31 | ||
| pFU98 | pSC101 ori, rbs-luxCDABE, CamR | 29 | ||
| pWUR677 | pFU98, CamR marker replaced by HygR marker | SacI | BG3870 | This study |
| NheI | BG3871 | |||
| pWUR704 | pWUR677, rbs-luxCDABE replaced by annealed BG4262-BG4263 | NotI | BG4262 | This study |
| SalI | BG4263 | |||
| pWUR705 | pWUR677, rbs-luxCDABE replaced by annealed BG4264-BG4265 | NotI | BG4264 | This study |
| SalI | BG4265 | |||
| PWUR708 | pWUR677, rbs-luxCDABE replaced by annealed BG3467-3468 insert | XbaI | BG3467 | This study |
| HindIII | BG3468 | |||
Supplementary Material
Acknowledgements
We want to thank A. Hidalgo, C. E. César, M. Davids and R. H. J. Staals for advice on experimental procedures. Furthermore, we would like to thank R. Engelhart, B. van Genugten, G. Göertz and R. Stolk for experimental contributions. This work was financially supported by grants from the Netherlands Organization of Scientific Research (NWO) to J.O. (NWO-TOP, 854.10.003), and to S.J.J.B. (NWO Vidi , 864.11.005), and by project BIO2010-18875 from the Spanish Ministry of Science and Innovation, and an Institutional Grant from the Fundación Ramón Areces to CBMSO (J.B.).
Footnotes
Author Contributions M.M.J. and J.H.J. made genomic T. thermophilus mutants under the supervision of J.v.d.O. T. thermophilus experiments were performed by D.C.S., M.M.J. and J.H.J. under the supervision of J.B., S.J.J.B. and J.v.d.O. D.C.S. and E.R.W. purified RNA for RNA-seq, and D.C.S. analysed RNA-seq data under the supervision of S.J.J.B. and J.v.d.O. D.C.S. and A.P.S. performed experiments in which TtAgo expression in T. thermophilus was shown using mass spectrometry. D.C.S., M.M.J. and J.H.J. made all plasmid constructs under the supervision of S.J.J.B., J.B. and J.v.d.O. D.C.S., E.R.W. and Y.Z. purified and analysed TtAgo guides. In vitro activity assays were designed and analysed by D.C.S., S.J.J.B., Y.W., D.J.P. and J.v.d.O., and performed by D.C.S. and Y.Z. under the supervision of S.J.J.B. and J.v.d.O. All authors read and approved the submitted manuscript.
Author Information The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus under accession number GSE52738. The siDNA sequence data discussed in this publication have been deposited in NCBI s BioSample database and are accessible under accession number SAMN02593821. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Online Content Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
References
- 1.Ketting RF. microRNA biogenesis and function: an overview. Adv. Exp. Med. Biol. 2011;700:1–14. doi: 10.1007/978-1-4419-7823-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb. Perspect. Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova KS, Wolf YI, van der Oost J, Koonin EV. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Averhoff B. Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 2009;33:611–626. doi: 10.1111/j.1574-6976.2008.00160.x. [DOI] [PubMed] [Google Scholar]
- 6.Gregory ST, Dahlberg AE. Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Microbiol. Lett. 2008;289:187–192. doi: 10.1111/j.1574-6968.2008.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containingargonaute silencingcomplex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368––374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 15.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MIDdomains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012;31:3588–3595. doi: 10.1038/emboj.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collin RG, Morgan HW, Musgrave DR, Daniel RM. Distribution of reverse gyrase in representative species of eubacteria and archaebacteria. FEMS Microbiol. Lett. 1988;55:235–240. [Google Scholar]
- 17.Charbonnier F, Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J. Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duguet M. The helical repeat of DNA at high temperature. Nucleic Acids Res. 1993;21:463–468. doi: 10.1093/nar/21.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates AD, Maxwell A. DNA Topology. 2nd edn Oxford Univ. Press; 2005. [Google Scholar]
- 21.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomyof siRNAs for mediating efficient RNAi in Drosophila melanogasterembryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng G, et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl Acad. Sci. USA. 2013;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travers A, Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nature Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 25.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 26.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laptenko O, et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25:2131–2141. doi: 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne I, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 29.Uliczka F, et al. Monitoring of gene expression in bacteria during infections using an adaptable set of bioluminescent, fluorescent and colorigenic fusion vectors. PLoS ONE. 2011;6:e20425. doi: 10.1371/journal.pone.0020425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cava F, et al. Expression and use of superfolder green fluorescent protein at high temperatures in vivo: a tool to study extreme thermophile biology. Environ. Microbiol. 2008;10:605–613. doi: 10.1111/j.1462-2920.2007.01482.x. [DOI] [PubMed] [Google Scholar]
- 31.Cava F, et al. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 2007;64:630–646. doi: 10.1111/j.1365-2958.2007.05687.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.