Abstract
BACE1, a major therapeutic target for treatment of Alzheimer’s disease, functions within a narrow pH range. Despite tremendous effort and progress in the development of BACE1 inhibitors, details of the underlying pH-dependent regulatory mechanism remain unclear. Here we elucidate the pH-dependent conformational mechanism that regulates BACE1 activity using continuous constant pH molecular dynamics. The simulations reveal that BACE1 mainly occupies three conformational states, however, the relative population of the states shifts according to pH. At intermediate pH when the catalytic dyad is mono-protonated, a binding-competent state is highly populated, while at low and high pH a Tyr-inhibited state is dominant. Furthermore, our data provide strong evidence supporting conformational selection as a major mechanism for substrate and peptide-inhibitor binding. These new insights, while consistent with experiment, greatly extend the knowledge of BACE1 and have implications for further optimization of inhibitors and understanding potential side effects of targeting BACE1. Finally, the work highlights the importance of properly modeling protonation states in molecular dynamics simulations.
The β-site amyloid precursor protein (APP) cleavage enzyme (β-secretase or BACE1) is an aspartyl protease that catalyzes the cleavage of APP to generate β-amyloid (Aβ) peptides.1,2,3 Subsequent oligomerization and aggregation of Aβ are linked to the onset of Alzheimer’s disease (AD).4,5 While a complete understanding of the physiological roles of BACE1 has yet to be achieved6, BACE1 deficient mice demonstrate significantly reduced level of Aβ and little phenotypical abnormalities.7,8 Thus, blocking the enzymatic activity of BACE1 has emerged as a major avenue in the drug development efforts towards the treatment of AD.9
Over the past fifteen years since the discovery of BACE1, hundreds of inhibitors have been synthesized, and dozens have entered clinical trials including Merck’s MK8931 and AstraZeneca’s AZD3293 that are currently in Phase II/III trials.10 Despite the enormous efforts and progress in the development of inhibitors, the fundamental biology of BACE1 such as its physiological substrates and regulatory mechanism is not fully understood.6 Such knowledge, however, has implications in the prediction of side effects and further optimization of BACE1 inhibitors and the discovery of new AD targets.
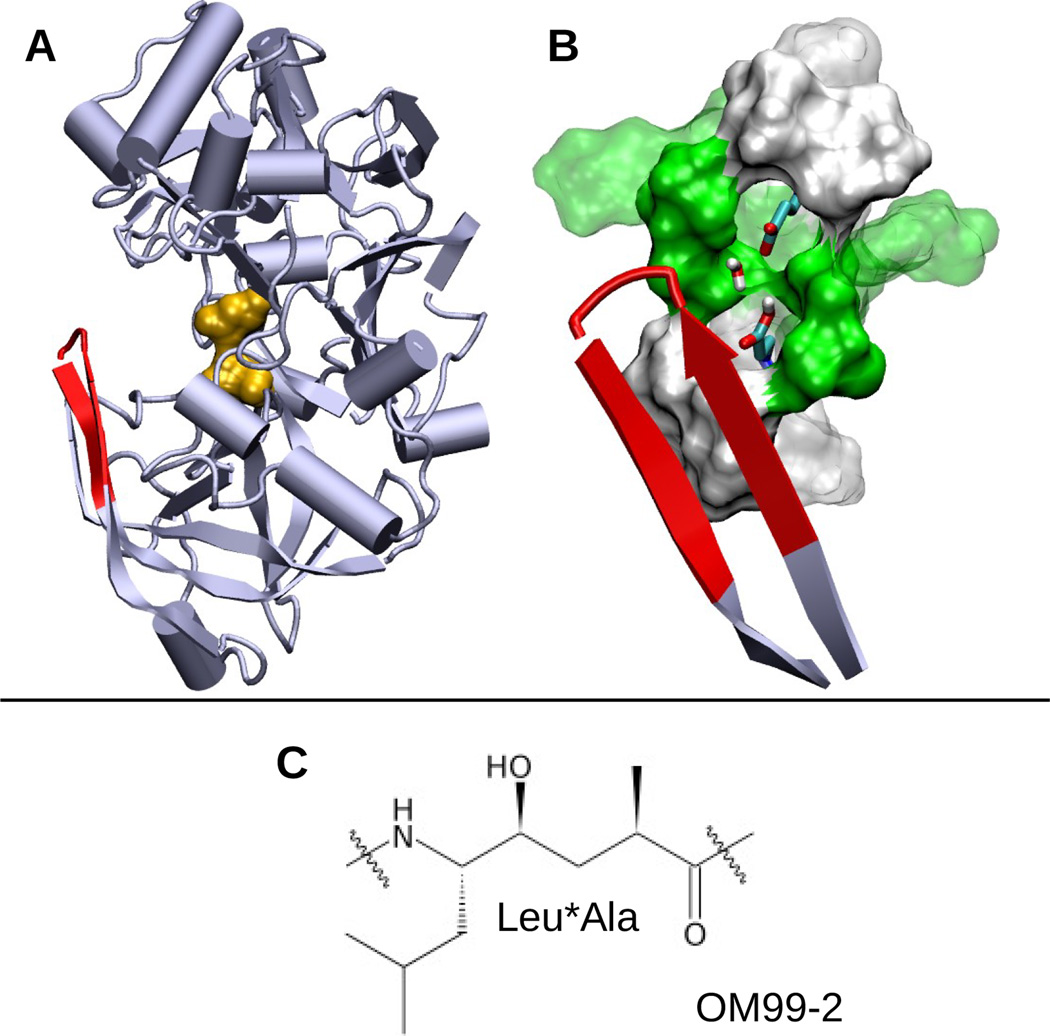
BACE1 is a monomeric protein primarily localized in the endosome and trans-Golgi apparatus. The catalytic domain of BACE1 contains about 400 residues among which two catalytic aspartates are located between the N- and C-terminal domains (Figure 1A). The aspartyl dyad hydrolyzes peptide bonds through a general acid-base mechanism, in which Asp228 acts as a base to activate a bridging catalytic water while Asp32 acts as an acid to protonate the substrate carbonyl group.11,12 Lying directly over the catalytic site is a β-hairpin loop spanning residues Tyr68 to Glu77, commonly known as the flap, which controls substrate access (Figure 1A and B). Both crystal structures and molecular dynamics simulations suggest that the flap opens and closes at room temperature13,14.
Figure 1.
Structures of BACE1 and the inhibitor OM99-2. A. Cartoon representation of apo BACE1 (PDB ID: 1SGZ) with the catalytic dyad (Asp32 and Asp228) colored orange and the flap (β-hairpin loop, residues 68–77) colored red. B. Active site of BACE1. Asp32, Asp228 and bridging water are shown in stick model. Polar (green) and hydrophobic (white) residues within 5 Å and 7 Å of Asp32/Asp228 are depicted as solid and transparent surfaces, respectively. C. The central hydroxyethylene group (Leu*Ala) of the peptidomimetic inhibitor OM99-2. The entire sequence is Glu-Val-Asn-Leu*Ala-Ala-Glu-Phe.
A unique aspect of the activation and inhibition of BACE1 is the delicate pH dependence. Fluorescence experiments showed that the peptide cleavage activity of BACE1 occurs in a very narrow pH range, peaking at pH 4.5 and sharply declining below pH 4 and above pH 5.15,14 Surface plasmon resonance experiments revealed that the binding of the peptidomimetic inhibitor OM99-2 (Figure 1C) decreases as pH is increased from 3 to 5 and is completely abolished at pH 6 and 7.14 These observations, in conjunction with the crystal structures resolved at several pH, led to the following hypothesis14: at pH 5 a conformational switch occurs preventing substrate/inhibitor binding at high pH, while at pH 4 the active site loses the bridging water deactivating BACE1. However, since these crystal structures have relatively low resolution (2.35–2.7 Å) and differences between them are insignificant (Table S1), the detailed mechanism of the pH-regulated activity and binding of BACE1 remains unclear.
Here we report an atomically-detailed mechanism underlying the pH-dependent enzymatic activity and inhibitor binding of BACE1 using a state-of-the-art molecular dynamics technique, continuous constant pH molecular dynamics (CpHMD)16,17 in explicit solvent with pH replica-exchange18. The CpHMD method allows simultaneous titration of all ionizable sites in response to conformational dynamics of the protein at a specified pH, thereby offering more accurate pKa predictions than the static-structure based methods, and most importantly, the atomistic details of pH-coupled conformational events (see a recent review19).
CpHMD simulations with pH replica-exchange were performed on BACE1 and its complex with inhibitor OM99-2 starting from the respective crystal structures (PDB ID: 1SGZ and 1FKN) using CHARMM (version C37b)20. The particular CpHMD used in the study employs a hybrid-solvent scheme that combines the accuracy of conformational sampling in explicit solvent with the efficiency of evaluating solvation forces on the titration coordinates by the generalized-Born model18. apo BACE1 was simulated using 24 pH replicas in the pH range 1–8, while the holo form was simulated using 32 pH replicas at pH −0.5–9.0. The apo and holo proteins were simulated respectively for 21 and 11 ns per replica under NPT conditions, resulting in the aggregated sampling time of 504 and 352 ns, respectively. In each simulation, all Asp, Glu, and His sidechains were allowed to titrate, while all Arg and Lys sidechains were modeled in the charged state. The first 1 ns per replica was excluded in analysis. Detailed simulation protocols and parameters are provided in Supporting Information (SI). Convergence tests for sampling protonation and conformational states are presented in Figure S1–S7.
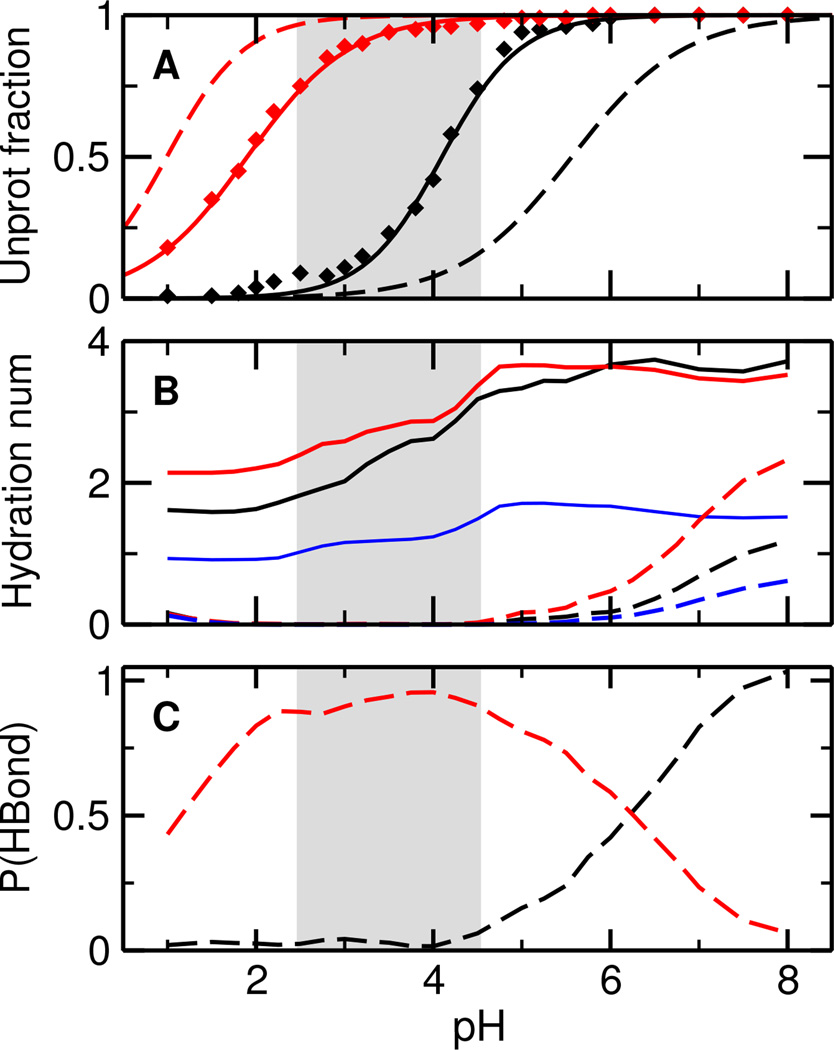
To elucidate the catalytic roles of Asp32 and Asp228, we examine their titration and solvent environment. Asp32 and Asp228 display very different titration behavior, with the calculated pKa’s of 4.1 and 1.9, respectively (Figure 2A, solid lines). The first solvation shell of both Asp (total hydration number) contains approximately two water molecules at pH below 3, one of which is a bridging water (Figure 2B, solid lines). As pH increases to 3–5.5, coinciding with the titration range of Asp32, the total hydration number increases to just above 3, while the number of bridging water remains constant. The increase in hydration is due to the deprotonation of Asp32, which induces a conformational rearrangement of the active site and consequently water entrance, a phenomenon previously observed for other proteins such as staphylococcal nuclease in both experimental21 and simulation studies22. As pH further increases, there is no change in the hydration number, as expected, since both Asp remain deprotonated.
Figure 2.
Simulated titration and solvent accessibility of the catalytic dyad in BACE1. A. Unprotonated fractions of Asp32 (black) and Asp228 (red) in the apo (solid) and holo state (dashed) at different pH. The curves are the best fits to the Hill equation. B. Hydration number of Asp32 (black), Asp228 (red), and the number of bridging water (blue). Dashed lines represent the holo state. Hydration number refers to the number of water molecules within the first solvation shell of Asp32/Asp228, which is defined as any water (oxygen) within 3.5 Å from Asp32/Asp228 (either carboxylate oxygen). C. Occupancy of a hydrogen bond between the hydroxyl group of Leu*Ala in OM99-2 and Asp32 (black) or Asp232 (red). A hydrogen bond is considered present if the donor-acceptor distance is below 3.5 Å and the acceptor-donor-hydrogen angle is less than 30°. The gray area indicates the simulated active pH range.
For holo BACE1, the titration curve of Asp32 is shifted to higher pH, while that of Asp228 is shifted to lower pH (Figure 2A, dashed lines). The resulting pKa’s are 5.6 and 1.0, 1.5 higher and 0.9 lower than the respective apo-state values due to hydrogen bonding with the hydroxyl group of Leu*Ala (see below). The presence of the inhibitor precludes water from entering the active site when the catalytic dyad is in the mono-protonated state (Figure 2B, dashed lines). However, as Asp32 begins to deprotonate, water starts to enter, analogous to the scenario for the apo protein. Interestingly, in lieu of the bridging water, the hydroxyl group of Leu*Ala donates a hydrogen bond to either Asp32 or Asp228 (Figure 2C). At pH below 4, Leu*Ala exclusively forms hydrogen bond with Asp228. As pH increases, Asp32 starts to deprotonate resulting in the hydrogen bond being shared between the two Asp. Finally, as pH is above 7, Asp32 becomes fully deprotonated and the exclusive hydrogen bond acceptor.
Our calculated site-specific pKa’s are in agreement with the consensus that Asp32 and Asp228 are the respective acid and base in the enzymatic reaction,11,14 although the macroscopic pKa’s of 4.1 and 1.8 (Figure S8) are 1.1 and 1.7 units lower than the respective values (5.2 and 3.5) inferred from a kinetic experiment.11 The discrepancy can be attributed to the implicit-solvent model which underestimates desolvation free energies.23 Thus, the active pH range, characterized by a mono-protonated catalytic dyad, is approximately 2.5–4.5 (Figure 2, gray area), one pH unit below the experimentally observed range of 3.5–5.5.15,14 We will keep the shift in mind in the discussion of conformational changes induced by protonation-state change of the catalytic dyad. Additionally, our data show that a bridging water is always present regardless of pH, thus supporting the hypothesis that the active site contains a bridging water which acts as the attacking water in the catalytic reaction11, but contradicting the hypothesis that the active site may be dehydrated at highly acidic pH14.
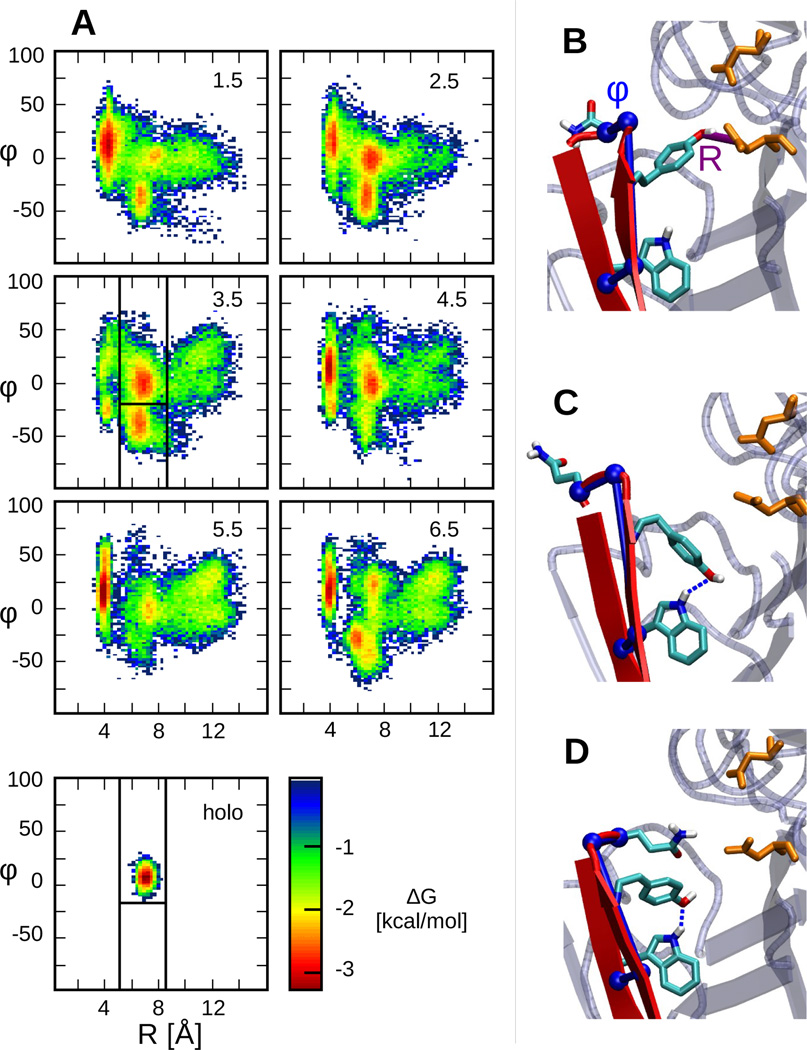
Next, we examine the conformational dynamics of BACE1 at different pH (Figure 3). Following previous simulation work by others13,24, we use two order parameters, R and φ, to characterize the flap conformation. R is defined as the distance between Tyr71:OH and Asp32:Cγ, while φ is a pseudo dihedral angle formed by Trp76:C-Val69:N-Thr72: CA-Gln73:CA. Tyr71 is an important residue on the flap as it is conserved in all pepsin-like aspartic proteases, and its orientation was found to modulate the flap conformation in BACE124. φ describes the twist of the flap, where a positive twist orients the flap toward the active site and a negative twist presents a more open active site24.
Figure 3.
pH-dependent conformational states of BACE1. A. Free energy surface of apo BACE1 as a function of R and φ at various pH conditions indicated on the upper right corner. The lower left panel presents the FES of holo BACE1 at pH 4.5. The free energy is calculated using −kTlnP(R;φ), where k is the Boltzmann constant and T is the temperature. Vertical and horizontal lines in the panel for pH 3.5 define the various states. B. Representative snapshot of the Tyr-inhibited state. The order parameters, R and φ defined in the main text, are shown in purple and blue, respectively. C. Representative snapshot of the binding-competent state. D. Representative snapshot of the Gln-inhibited state.
Remarkably, the free energy surface (FES) as a function of R and φ reveals three local free energy minima (Figure 3A). We refer to these three states as the Tyr-inhibited, binding-competent and Gln-inhibited states. In the Tyr-inhibited state, corresponding to the minimum located at R<5 Å, Tyr71 interacts with Asp32, causing the flap to cover the active site, preventing ligand binding (Figure 3B). Similar self-inhibited states have been reported for aspartic proteinases25,26 as well as BACE113,24. Separated from the Tyr-inhibited state by a barrier of about 3 kcal/mol are two states with the free energy minima located between 5 and 8.5 Å of R, one below and one above −18°. In the state with φ >−18°, Tyr71 points towards the base of the flap allowing the formation of a hydrogen bond with the indole group of Trp76 (Figure 3C). Importantly, this state is exclusively sampled in the holo simulations. Thus, we call it the binding-competent state. In the state with φ <−18°, Tyr71 maintains the same orientation, however, due to the twist of the flap (change in φ), the sidechain of Gln73 occludes the active site (Figure 3D). Thus, we call this state the Gln-inhibited state. Note, unlike Tyr71, the distance between Gln73 and Asp32 rarely becomes short enough to induce the formation of a hydrogen bond (Figure S9). The free energy barrier separating the Gln-inhibited and binding-competent states is very small (<1 kcal/mol). Finally, we refer to the regions in the FES with R > 8.5 Å and no visible free energy minimum as the diffuse states of BACE1.
The FES reveals that apo BACE1 samples distinct conformational states in a pH-dependent manner, and upon inhibitor binding only the binding-competent state is visited (Figure 3A, bottom plot). At pH below 2 or above 5, when the catalytic dyad is di-protonated or di-deprotonated, the Tyr-inhibited state is dominant (Figure 3A, pH 1.5, 5.5 and 6.5). At pH 2.5–4.5, the simulated active pH range when the catalytic dyad is in the mono-protonated form, the binding-competent state becomes more populated (Figure 3A, pH 2.5, 3.5 and 4.5). More quantitative analysis will be given later. Interestingly, in the FES at pH 3.5 the Tyr-inhibited state is significantly diminished, suggesting that it is the most active pH in our simulation. This is consistent with the experimental observation that the most active pH is about 0.8 units below the pKa of Asp32. In contrast to the FES of apo BACE1, the FES of the holo protein under all pH conditions shows only one free energy minimum located in the same region as the binding-competent state of the apo BACE1. This is consistent with the fact that the crystal structures of BACE1 bound to OM99-2 obtained at pH 5 and pH 7.4 are nearly identical and supports the hypothesis that OM99-2 binding locks BACE1 into a single conformation14.
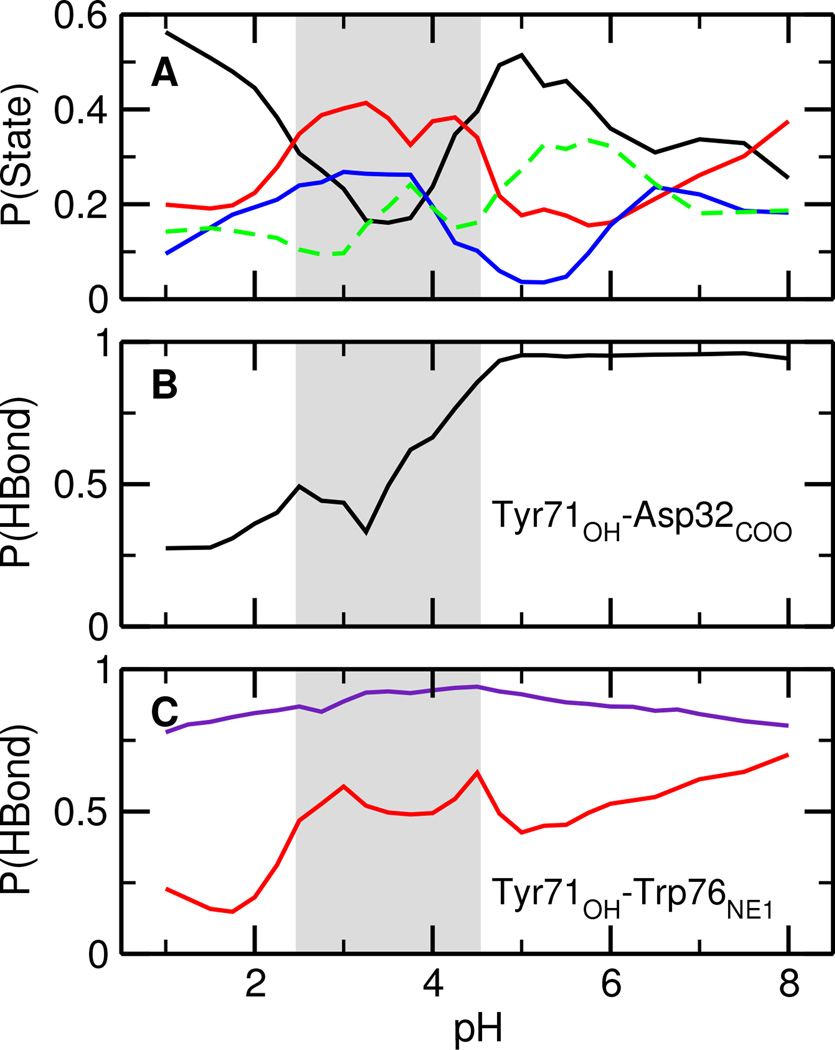
To quantify the extent of the conformational switch induced by pH, the occupancies of the aforementioned states were calculated as a function of pH (Figure 4A). The occupancy of the diffuse states remains nearly constant (about 20%) in the entire pH range, whereas the occupancies of the other three states vary dramatically. In particular, the occupancy profile of the Tyr-inhibited state has a complementary shape as the binding-competent/Gln-inhibited states. Thus, pH shifts the relative population between the states. Since the Gln-inhibited state has a low population (below 25% in the entire pH range) and readily converts to the binding-competent state, we will focus on the latter in the discussion below.
Figure 4.
Characterization of the Tyr-inhibited and binding-competent states of BACE1. A. Occupancy of different conformational states of apo BACE1. The Tyr-inhibited, Gln-inhibited, binding-competent, and diffuse states are shown in black, blue, red, and green, respectively. B. Occupancy of the hydrogen bond Tyr71:OH⋯Asp32:COO in the Tyr-inhibited state of apo BACE1. C. Occupancy of the hydrogen bond Tyr71:OH⋯Trp76:NE1 in the binding-competent population (red) and holo BACE1 (purple). The gray area indicates the simulated active pH range.
At pH below 2 the Tyr-inhibited state has the highest occupancy, accounting for about 50% of the total population. This data offers an explanation for the inactivity of BACE1 at low pH when the catalytic dyad is in the di-protonated state. As pH increases from 2 to 3.5, the occupancy of the Tyr-inhibited state steadily decreases whereas that of the binding-competent state steadily increases. The former reaches a minimum at about pH 3.5 while the latter reaches a broad maximum in the pH range 2.5–4.5 where it becomes the most occupied state, accounting for approximately 40% of the total population. This data provides quantitative support for the active pH range of 2.5–4.5, which corresponds to the mono-protonated state of the catalytic dyad. As pH increases from 3.5 to 5, Asp32 is becoming deprotonated, the occupancy of the Tyr-inhibited state increases, and that of the binding-competent state decreases, further confirming that BACE1 is most active in the mono-protonated state and explaining why the inhibitor cannot bind at pH 6 and 7.14
Curiously, the occupancy analysis does not resolve which state dominates the conditions of pH above 5. To understand it, we consider the occupancy of a hydrogen bond between Tyr71 and Asp32 in the Tyr-inhibited state, which closes the active site. Remarkably, at pH 5 and above, the hydrogen bond is nearly 100% present (Figure 4B), indicating that at high pH when Asp32 is deprotonated, the active site can be fully occluded. Once this happens, the probability for converting to the binding-competent state is very low due to the high energy barrier (Figure 3A, pH 5.5 and 6.5). Thus, the data supports the hypothesis that the self-inhibited conformation is the most stable state at high pH.13,14 Moreover, at high pH, Asp32 is fully deprotonated, therefore unable to donate a proton to the substrate required by catalysis.
Finally, we examine the binding-competent states by focusing on a hydrogen bond between Tyr71 and Trp76 which is nearly always present when OM99-2 is bound (Figure 4C, purple). In apo BACE1, the occupancy of this hydrogen bond reaches maximum at pH 2.5–4, coinciding with the pH range for the maximum occupancy of the binding-competent state (Figure 4C, red). This observation is consistent with the finding that apo BACE1 readily assumes the binding-competent state in the active pH range, thus suggesting that peptide-inhibitor/substrate binding takes place, at least in part, via the conformational selection mechanism. We note that unlike OM99-2, which resembles the natural substrate, entrance of many small-molecule inhibitors requires the flap to further open leading to the loss of the Tyr71–Trp76 hydrogen bond.
In summary, constant pH molecular dynamics simulations revealed a detailed mechanism of the pH-regulated enzymatic activity and peptide-inhibitor binding of BACE1 (Figure 5), a major pharmaceutical target for treatment of AD. At room temperature, due to the mobility of the flap, apo BACE1 mainly samples two distinct states, Tyr-inhibited and binding-competent. pH shifts the relative population between the two such that at intermediate pH where the catalytic aspartates are in the mono-deprotonated form, the binding-competent state is the most occupied. However, at both low and high pH, the Tyr-inhibited state is thermodynamically more stable and large free barriers separate the states. This mechanism is consistent with the experimental observations and previous simulation studies but it is somewhat different from the the mechanism proposed by Shimizu et al.14 Specifically, instead of dehydration of the active site at low pH, our simulations demonstrate that the active site remains hydrated and adopts a Tyr-inhibited state. Thus, our simulations suggest that mutation of Tyr71 to a sidechain (such as Phe) incapable of hydrogen bonding with Asp32 may significantly increase the enzyme activity, especially at high pH. More importantly, our simulations offer an unprecedented, detailed view of the interplay between pH, protonation state, hydrogen bonding and conformational dynamics. The resemblance between the binding-competent state of apo BACE1 and the peptide inhibitor-bound state suggests a conformational selection mechanism for binding. Finally, the present work demonstrates the utility of constant pH molecular dynamics in gaining novel insights into the intricacy of pH-dependent enzyme catalysis and inhibition.
Figure 5.
Schematic representation of the pH-dependent mechanism of BACE1 activity and inhibition. The active pH range is shifted by one unit to match experiment.
Supplementary Material
ACKNOWLEDGEMENTS
Financial support is provided by National Science Foundation (MCB1305560) and National Institutes of Health (GM098818).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Details of the computational protocols and additional analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G. Mol. Cell. Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 2.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson- Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 3.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis J-C, Collins F, Treanor J, Rogers G, Citron M. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 5.Bateman RJ, Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, Santacruz A, Buckles V, Oliver A, Moulder K, Aisen PS, Ghetti B, Klunk WE, McDade E, Martins RN, Masters CL, Mayeux R, Ringman JM, Rossor MN, Schofield PR, Sperling RA, Salloway S, Morris JC. N. Engl. J. Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandalepas PC, Vassar R. J. Neurochem. 2012;120(Suppl 1):55–61. doi: 10.1111/j.1471-4159.2011.07512.x. [DOI] [PubMed] [Google Scholar]
- 7.Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, Martin L, Louis J-C, Yan Q, Richards WG, Citron M, Vassar R. Nature Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 8.Nishitomi K, Sakaguchi G, Horikoshi Y, Gray AJ, Maeda M, Hirata-Fukae H-F, Becker AG, Hosono M, Sakaguchi I, Minami SS, Nakajima Y, Li H-F, Takeyama C, Kihara T, Ota A, Wong PC, Aisen PS, Kato A, Kinoshita N, Matsuoka Y. J. Neurochem. 2006;99:1555–1563. doi: 10.1111/j.1471-4159.2006.04178.x. [DOI] [PubMed] [Google Scholar]
- 9.Yuan J, Venkatraman S, Zheng Y, McKeever BM, Dillard LW, Singh SB. J. Med. Chem. 2013;56:4156–4180. doi: 10.1021/jm301659n. [DOI] [PubMed] [Google Scholar]
- 10.Vassar R. Alzheimer’s Res. Ther. 2014;6:89. doi: 10.1186/s13195-014-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toulokhonova L, Metzler WJ, Witmer MR, Copeland RA, Marcinkeviciene J. J. Biol. Chem. 2003;278:4582–4589. doi: 10.1074/jbc.M210471200. [DOI] [PubMed] [Google Scholar]
- 12.Hong L, Tang J. Biochemistry. 2004;43:4689–4695. doi: 10.1021/bi0498252. [DOI] [PubMed] [Google Scholar]
- 13.Gorfe AA, Caflisch A. Structure. 2005;13:1487–1498. doi: 10.1016/j.str.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu H, Tosaki A, Kaneko K, Hisano T, Sakurai T, Nukina N. Mol. Cell. Biol. 2008;28:3663–3671. doi: 10.1128/MCB.02185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grüninger-Leitch F, Schlatter D, Küng E, Nelböck P, Döbeli H. J. Biol. Chem. 2002;277:4687–4693. doi: 10.1074/jbc.M109266200. [DOI] [PubMed] [Google Scholar]
- 16.Lee MS, Salsbury FR, Jr, Brooks CL., III Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 17.Khandogin J, Brooks CL., III Biophys. J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace JA, Shen JK. J. Chem. Theory Comput. 2011;7:2617–2629. doi: 10.1021/ct200146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Morrow BH, Shi C, Shen JK. Mol. Simulat. 2014;40:830–838. doi: 10.1080/08927022.2014.907492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartles C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damjanović A, Brooks BR, García-Moreno EB. J. Phys. Chem. A. 2011;115:4042–4053. doi: 10.1021/jp110373f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi C, Wallace JA, Shen JK. Biophys. J. 2012;102:1590–1597. doi: 10.1016/j.bpj.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace JA, Wang Y, Shi C, Pastoor KJ, Nguyen B-L, Xia K, Shen JK. Proteins. 2011;79:3364–3373. doi: 10.1002/prot.23080. [DOI] [PubMed] [Google Scholar]
- 24.Spronk SA, Carlson HA. Proteins. 2011;79:2247–2259. doi: 10.1002/prot.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreeva N, Dill J, Gilliland GL. Biochem. Biophys. Res. Commun. 1992;184:1074–1081. doi: 10.1016/0006-291x(92)90701-l. [DOI] [PubMed] [Google Scholar]
- 26.Gustchina A, Li M, Phylip LH, Lees WE, Kay J, Wlodawer A. Biochem. Biophys. Res. Commun. 2002;295:1020–1026. doi: 10.1016/s0006-291x(02)00742-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.