Abstract
The treatment of glioblastoma (GBM) remains challenging in part due to the presence of stem-like tumor-propagating cells that are resistant to standard therapies consisting of radiation and temozolomide. Among the novel and targeted agents under evaluation for the treatment of GBM are BRAF/MAPK inhibitors, but their effects on tumor-propagating cells are unclear. Here, we characterized the behaviors of CD133+ tumor-propagating cells isolated from primary GBM cell lines. We show that CD133+ cells exhibited decreased sensitivity to the anti-proliferative effects of BRAF/MAPK inhibition compared to CD133− cells. Furthermore, CD133+ cells exhibited an extended G2/M phase and increased polarized asymmetric cell divisions. At the molecular level, we observed that polo-like kinase (PLK) 1 activity was elevated in CD133+ cells, prompting our investigation of BRAF/PLK1 combination treatment effects in an orthotopic GBM xenograft model. Combined inhibition of BRAF and PLK1 resulted in significantly greater anti-proliferative and pro-apoptotic effects beyond those achieved by monotherapy (p<0.05). We propose that PLK1 activity controls a polarity checkpoint and compensates for BRAF/MAPK inhibition in CD133+ cells, suggesting the need for concurrent PLK1 inhibition to improve antitumor activity against a therapy-resistant cell compartment.
Introduction
Patients with glioblastoma multiforme (GBM), the most common and malignant type of brain tumor in adults, have a poor prognosis despite aggressive first line treatment, which consists of resection followed by radiotherapy with concurrent and adjuvant temozolomide (1). The genetic and phenotypic heterogeneity of GBM, poses a major hurdle for the effective treatment of these tumors. Transcriptomic subclassification analyses have revealed discrete molecular subgroups among series of GBM (2,3), and single-cell RNA sequencing has further demonstrated the presence of multiple molecular subgroups in different cells within a single tumor (4). The intra-tumoral heterogeneity further manifests as mosaic expression of receptor tyrosine kinases (RTKs) (5,6), gene copy number variation (7), the presence of multiple genetically distinct clones (8), and the existence of phenotypically distinct tumor-propagating cells (TPCs), as highlighted by studies examining the tumorigenicity of xeno-transplanted cells sorted from GBM surgical specimen (9,10). One TPC population of particular interest expresses the cell surface antigen CD133, and CD133+ TPCs were shown to exhibit elevated resistance to standard therapy (11–16). In contrast, NG2 positivity, that is associated with oligodendrocyte progenitor cells (OPCs), has been shown to identify TPCs that respond well to chemotherapy (17,18).
With increasingly routine tumor molecular profiling and the ongoing movement towards the use of targeted therapeutics, it is anticipated that molecular-informed therapeutic decision-making will improve the survival of patients with GBM. Differences between stem and progenitor-like TPCs and other GBM cells could lead to distinct, insufficient responses to those recently emerging targeted therapies and need to be investigated.
NSC (neural stem cells), OPCs, and TPCs share the ability to undergo asymmetric cell division (ACD). Cells acquiring polarity and as a result segregating cell fate determinants unequally between daughter cells at cytokinesis define ACD. Changes in ACD have been associated with tumor initiation for several cancer types, including GBM (19–21). ACD regulation requires the coordinated activity of a network of polarity regulators and mitotic kinases. This network is well characterized in invertebrate stem cells, and has been shown to include polo kinase (19). However, for normal mammalian stem and progenitor cells and TPCs, the extent to which polo-like kinase 1 (PLK1; 22), the mammalian homologue of polo kinase, affects ACD is unknown.
Here, we have used human GBM models, to examine ACD in CD133+ versus CD133−NG2+ cell populations, and to study their response to BRAF/MAPK pathway inhibition. In a subset of malignant astrocytoma the gene encoding Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A) is deleted, and RAS-RAF-MAPK signaling is constitutively activated due to the BRAFV600E mutation (23). BRAFV600E astrocytoma have been reported to show transient sensitivity to the anti-tumor effects of the mutant-selective inhibitor vemurafenib, in preclinical (24,25) and clinical settings (26). Based on clinical data from vemurafenib treatment of BRAFV600E-melanoma (27), combination therapies are needed to achieve a more durable anti-tumor effect than BRAFV600E/MAPK pathway inhibition alone.
Our data show that CD133+ TPCs display reduced basal proliferation that is less affected by BRAF/MAPK inhibition, in relation to CD133−NG2+ cells. The lower sensitivity to BRAF/MAPK inhibition is accompanied by increased PLK1 activity and higher rates of polarity and ACD, which can be suppressed by PLK1 inhibition. Combined use of PLK1 and BRAF/MAPK inhibitors increases the anti-proliferative and pro-apoptotic response, especially towards CD133+ cells, and reduces growth of an intracranial BRAFV600E mutant xenograft more effectively than single agent treatment. The data support the existence of a mitotic polarity/ACD checkpoint in CD133+ TPCs, which is susceptible to PLK1 inhibition. We conclude that investigations into the mechanism of ACD aid in the identification of approaches that support a more durable BRAF/MAPK inhibitor anti-tumor effect.
Materials and Methods
Immunofluorescence (IF)
Tissue sections were obtained from 8 GBM specimens that were collected from consenting patients and distributed anonymously by the brain tumor research core at UCSF. Tissue sections from primary GBM were cut at 5μm from frozen tissue samples and fixed with 4% paraformaldehyde in PBS (PFA) for 10 minutes at room temperature. Tissue sections from PFA-perfused mouse brains were cut and stained as free-floating 30μm sections or as 12μm sections. Cells were pulsed with thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) for 30 min at indicated time points, then fixed for 10 minutes with 4% PFA at room temperature. EdU was detected using the Click-it EdU detection kit according to manufacturer’s instructions (Life Technologies).
All samples for IF were washed with PBS, blocked with PBS+5% normal goat serum for 1 hour at room temperature, and incubated with primary antibody overnight at 4°C (Supplemental Table 1). Samples were washed with PBS and incubated with secondary antibodies for 1 hour at room temperature, washed with PBS+0.2μg/ml DAPI (to stain nuclei, shown in blue), then with PBS, and mounted with Vectashield (Vecta Labs #H-1000). Quantification of cell counts and immunostainings in tumor sections were performed manually for CD133, NG2, and activated cleaved caspase 3 (CC3), and the number of Ki67+ cells and total cell numbers (DAPI) were counted automatically using ImageJ.
Proliferation assays
Cells from culture or following FACS were plated into black Cell-Bind 96-well plates (Corning #3340) at 500 cells/well in 100ul of appropriate media ± inhibitors as indicated in figures and legends, with a minimum of 3 technical replicates per condition. At 24, 72 and 120 hours, Alamar Blue (1:10) was added and fluorescence measured (excitation 560nm, emission 580nm) after 1 and 3 hours. Fold change was calculated from the vehicle reading taken at 24 hours post-plating.
Cell pair & polarity assays
Primary and adherent cell lines were dissociated to single cells, passed through a 70μm cell strainer, and plated at low density (2000 cells per 1.9cm2 well) onto poly-L-lysine coated glass coverslips. Cells were allowed to attach or divide (polarity assays; 24h, cell pair assays 36–48h), inhibitors were added as indicated in figures and figure legends, and cells were fixed and processed for IF. For polarity analysis, the sum of the grey values in each of 24×15° segments was measured using the Oval Profile plugin for ImageJ, expressed as a percentage of the grey value for each cell, aligned with the maximum at 0° using R, and values averaged across >30 cells/experiment. Cell pairs were scored for asymmetric and symmetric protein distribution as previously described (20).
Cell culture and drug treatment
Primary human GBM cell lines (SF7996, SF8565, proneural subtype) were isolated from surgical specimens obtained through the UCSF Brain Tumor Research Center by Dr. Joseph Costello’s laboratory at UCSF (29). Primary cell lines were maintained in ultra low attachment plates (Corning) in Neurobasal-A media (1x B27-A, 1x N2, Penicillin/Streptomycin, L-glutamine, 10ng/ml EGF and 10ng/ml bFGF2). Primary cells were passaged using Accutase #AM105. DBTRG-05MG and SF188 were obtained from the Nicolaides and Pieper labs at UCSF and were cultured in 10cm plates (Corning) in DMEM (4.5g/ml glucose, pen/strep, 10% FBS) and passaged using 0.25% Trypsin. DBTRG cells were also kept in primary cell conditions to increase the content of CD133+ cells. Cell lines were authenticated by DNA fingerprinting. Cells were treated for 2h with vehicle (DMSO) or 5μM of the actin-organization inhibitor Latrunculin-A (LatA). For Alamar Blue assays, cells were incubated with vehicle, 1μM PD901, 5nM BI2536, or a combination of 1μM PD901 and 5nM BI2536 for five days. DBTRG-05MG cells were treated with vehicle, 1μM PLX4720, 10nM BI2536 or a combination of 1μM PLX4720 and 10nM BI2536. For desensitization of DBTRG-05M, cells were treated every three days with escalating dosage of PLX4720 up to 6μM.
Flow cytometry and fluorescence activated cell sorting (FACS)
Cells were analyzed by flow cytometry using the FACS Calibur. Cell sorting was performed using a BD Biosciences ARIA3 cytometer. For flow cytometry, cells from culture were dissociated by incubating with PBS (Ca2+, MG2+ -free) + 0.2mM EDTA for 10 minutes at 37°C, followed by mechanical dissociation in PBS+0.2mM EDTA+0.5% BSA (FACS buffer) until a single cell suspension was achieved. Cells from tumor were dissociated in 20U/ml papain (Worthington Biochem), followed by antigen recovery by incubation in neurosphere media for 3 hours at 37°C. Alternatively, tumors were dissociated using the neural stem cell dissociation kit (P) (Miltenyi). Live dissociated cells were stained with CD133 and NG2 for 15 minutes on ice, washed in FACS buffer, and stained with secondary antibodies. For phospho-PLK1 (pT120-PLK1) and phospho-ERK (pERK) detection, cells were fixed in 2% PFA and permeabilized using 100% methanol. For FACS, cells were re-suspended in FACS buffer containing 0.2ug/ml DAPI.
Statistics and analysis
Appropriate statistical analyses were performed as indicated in figure legends using GraphPad Prism 5.0, Microsoft Excel, or TreeStar FlowJo.
Cell cycle analysis
Cultured cells were pulsed with EdU (10μM) for 30 minutes prior to dissociation and processing for FACS. For ex vivo analysis of tumor cells, mice were injected with 100mg/kg EdU 30 minutes to two hrs before tumor isolation. DAPI (1μg/ml) was added to cell suspensions 30 minutes before analysis to measure DNA content.
RNA isolation and qPCR
Total RNA was isolated from FACS-enriched cells or tumor tissue using Trizol reagent. RNA was reverse transcribed (Life Technologies #4368814), and quantitative real time PCR performed using Power SYBR qPCR mix (Life Technologies) using an Applied Biosystems 7900HT thermal cycler, with primer sets indicated in Supplemental Table 2. Fold changes were calculated using the ΔΔCt method (30).
Xenograft models and preclinical treatment
For orthotopic tumor models, 6 week old athymic mice were implanted with luciferase-expressing DBTRG-05MG cells (3×105 cells/mouse) at 1mm anterior, 2mm lateral, and 3mm deep (from Bregma). For flank xenografts, 3×107 cells from previous generation flank tumors were harvested and implanted as previously described (25). Tumor growth was measured by bioluminescence imaging and expressed as normalized bioluminescence (fold-change from the start of treatment). Treatment was started at 7–21 days post implantation, and continued for up to 9 days; PLX4720 was injected I.P at 20mg/kg daily, whereas BI2536 was injected I.P. at 50mg/kg twice a week.
Results
CD133 and NG2 identify functionally distinct subpopulations in human GBM
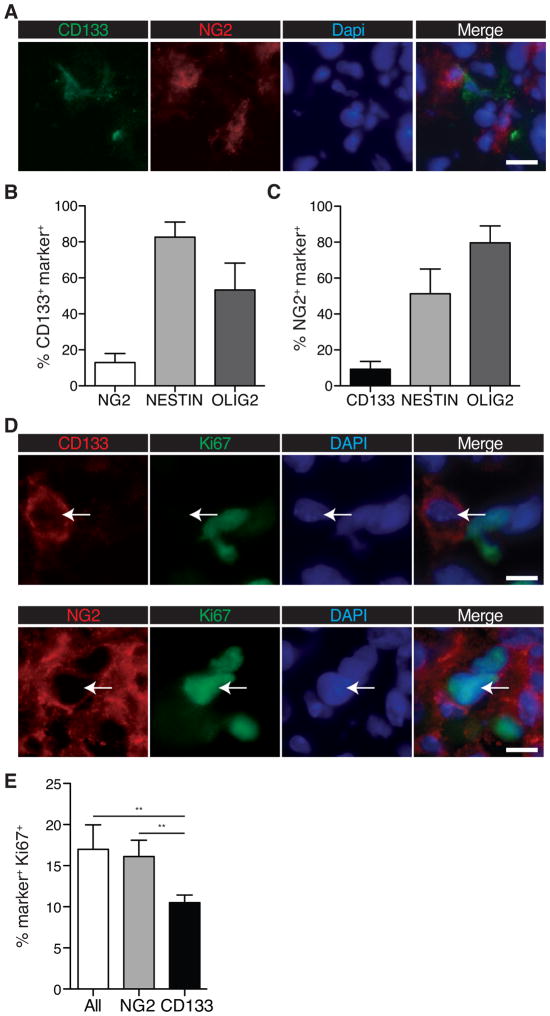
To examine the proportion of CD133 and NG2 positive cells in GBM, we performed co-immunofluorescence on human GBM surgical specimens using CD133 and NG2 antibodies (Fig. 1A). We found the frequencies of CD133+ and NG2+ cells to be variable between patient samples, with CD133 antibody labeling 2–20% of tumor cells, and NG2 antibody labeling 4–23% of the cells (Fig. S1A). Less than 20% of CD133+ cells displayed NG2 staining, with NG2+ cells showing a similar low frequency of CD133 co-staining, indicating that the markers identify largely non-overlapping CD133+ and CD133−NG2+ populations (Fig. 1B, C). In contrast, co-staining with NSC-marker NESTIN and the OPC-marker OLIG2 revealed 83±8% NESTIN positivity of CD133+ cells, and 80±9% OLIG2 positivity of NG2+ cells (Fig. 1B, C). To assess proliferative differences between the two CD133+ and NG2+ populations, we stained specimens for the proliferation marker Ki67 (Fig. 1D), and quantified the proportion of actively cycling cells in each subpopulation. We found that the frequency of actively cycling NG2+ cells was not significantly different from that of bulk tumor cells. We also discovered a significantly lower frequency of Ki67 positivity among CD133+ cells than for NG2+ cells: 11±1% vs. 16±2%, respectively (**p ≤ 0.005; Fig. 1E).
Figure 1. Proliferation rates and compartment overlap of CD133+ and NG2+ GBMs.
(A) IF image of patient GBM specimen stained with antibodies against CD133 (green) and NG2 (red); DAPI was used to stain DNA (blue). (N=8; error bars represent SEM) Quantification of (B) CD133 and (C) NG2 co-expression with NESTIN and OLIG2. A minimum of n=77 cells were counted per tumor, for 8 specimens total. Error bars represent SEM.
(D) Co-IF of a primary human GBM specimen for CD133 (red, top panels, arrow), NG2 (red, bottom panels, arrow) and the proliferative marker Ki67 (green).
(E) Quantification of Ki67 expression in CD133+ and NG2+ cells of primary human GBM specimens. (N≥107 CD133+/NG2+ cells counted per tumor from each of 6 specimens; 2-tailed t-test; Error bars represent SEM. Scale bars represent 20μM.)
GBM cell cultures maintain discrete populations of CD133+ and NG2+ cells
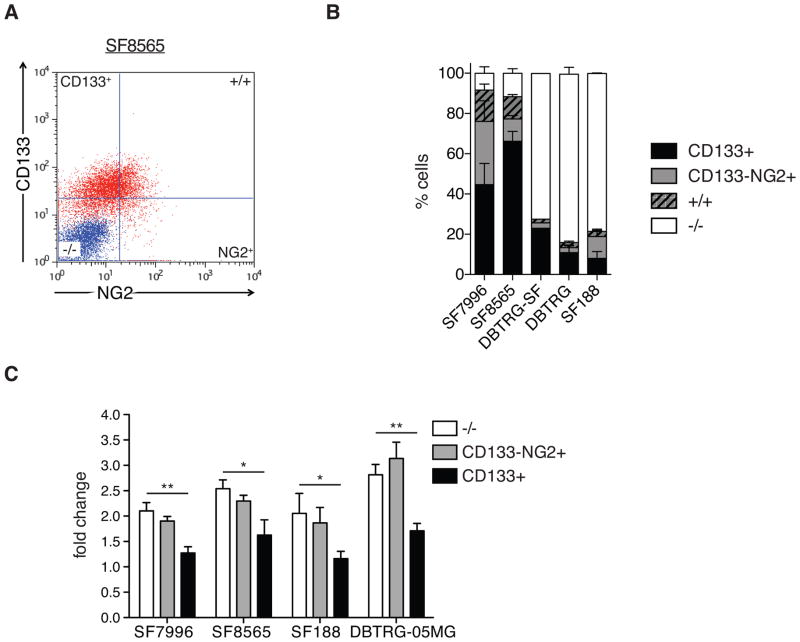
We next examined if the proliferative differences between CD133+ and NG2+ cells remain evident in established GBM cell lines (DBTRG-05MG, aka DBTRG; SF188) and primary GBM cell cultures (SF8565 and SF7996). We used flow cytometry to quantify the frequencies of CD133+NG2− (aka CD133+) and CD133−NG2+ cells. Primary SF7996 and SF8565 cell lines contain a higher proportion of CD133+ cells than established DBTRG and SF188 cell lines. We detected a CD133+ population that was larger than the CD133+NG2+ population in all cell lines (Fig. 2A, 2B, S2A and S2B), consistent with results from the analysis of patient tumors.
Figure 2. GBM cell cultures maintain discrete populations of CD133+ and NG2+ cells.
(A) Flow cytometry plot showing CD133 and NG2 expression in SF8565 cell line. The isotype control sample is labeled in blue and antibody-stained samples are in red.
(B) Quantification of frequency of CD133+NG2− (CD133), CD133−NG2+ (NG2), double-positive (+/+) and marker-negative (−/−) cells in SF8565, SF7996, SF188 and DBTRG-05MG cell lines. Error bars represent SEM.
(C) Alamar Blue viability assay of FACS-enriched CD133+ and CD133−/NG2+ populations from SF7996, SF8565, SF188, and DBTRG-05MG cells. Fold changes in fluorescence after five days of growth demonstrates the lower viability of CD133+ cells compared with CD133−/NG2+ cells (n=3 individual experiments; 1-way ANOVA with Tukey post-hoc test, Error bars represent SEM; *p≤0.005; **p≤0.01).
To further characterize the CD133+ and CD133−NG2+ cells, we examined FACS-enriched populations as well as unsorted cells for the expression of NSC genes NESTIN, SOX2 and MUSASHI1; the OPC-specific transcription factor SOX10; and glial differentiation markers MBP and GFAP. We found that NSC gene expression is enriched in the CD133+ cells compared with NG2+ cells and unsorted cells, while the SOX10 expression is enriched in the NG2+ population. Glial differentiation marker expression shows no enrichment in CD133+ cells while in NG2+ cells MBP expression is elevated (Fig. S2C).
To investigate CD133+ cell characteristics in vitro, we subjected FACS-enriched CD133+, CD133−NG2+ cells, and marker-negative cells to Alamar Blue assays. CD133+ cells showed significantly less viability than CD133−NG2+ and marker-negative cells, the latter two of which were very similar (Fig. 2C).
ACD is elevated in CD133+ GBM cells, in relation to NG2+ cells
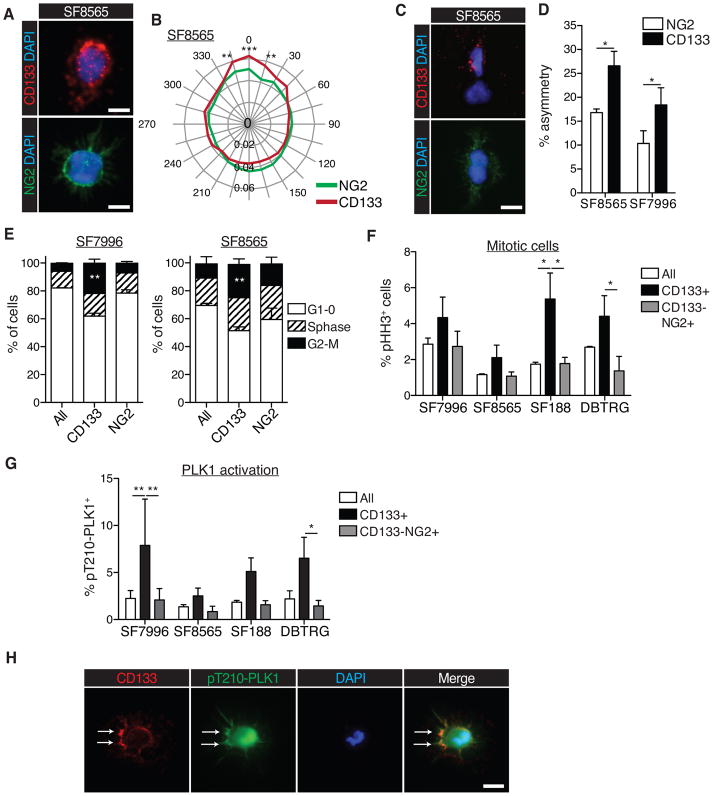
Next we examined asymmetric cell divisions (ACD) in CD133+ vs. CD133−NG2+ cells. Immunocytochemistry detecting the subcellular localization of CD133 and NG2 and associated radial profile plots (Fig. 3A, 3B and S3A) revealed that CD133 protein in CD133+ cells exhibited a more polarized or asymmetric subcellular distribution than NG2 protein in NG2+ cells, which is consistent with the more unequal partitioning of CD133 into daughter cells, as determined by pair assays (Fig. 3C–D and Fig. S3B, C) (21). Taken together, CD133+ cells consistently exhibited higher rates of polarity and ACD than CD133−NG2+ cells.
Figure 3. ACD is elevated and cell cycle dynamics are different in CD133+ cells, in relation to NG2+ cells.
(A) Representative image of polarized CD133 localization (red; top), unpolarized NG2 localization (green; bottom), and DAPI (blue; top and bottom), in SF8565 cells (scale bars represent 10μm).
(B) Radial plot showing quantification of CD133 and NG2 polarization. Quantification was performed using the Oval Profile plugin for ImageJ and expressed as a percentage of the grey value for each cell, aligned with the maximum at 0°; values are averaged across ≥30 cells/experiment (2-way ANOVA with Bonferroni post-hoc test; *p≤0.05; **p≤0.01; ***p≤0.001; error bars represent SEM)
(C) Pair assays visualizing the asymmetric distribution of CD133 (red; top panel), symmetric distribution of NG2 (green; bottom panel) and DAPI (blue; top and bottom), in SF8565 cells (scale bars represent 10μm).
(D) Quantification of asymmetric division frequency of CD133+ and NG2+ cells from two different cell lines (n=4 individual experiments per cell line, ≥30 cell pairs were scored per experiment; 2-way paired t-test; error bars represent SEM; *p≤0.05).
(E) Cell cycle analysis of two different GBM cell lines, stratified by expression of CD133 and NG2, and showing an increase in CD133+ cells in G2/M phase. NG2=CD133−NG2+ cells (n=3 individual experiments per cell line, 2-way ANOVA with Bonferroni post-hoc test; error bars represent SEM; **p<0.005).
(F) Quantification of flow cytometry analyses to detect pHH3 in cell lines stratified by expression of CD133 and NG2 (n=2 individual experiments; 2-way ANOVA with Bonferroni post-hoc test, error bars represent SEM; *p<0.05).
(G) Flow cytometry analysis to detect activated phospho-PLK1 (pT210-PLK1) in cell lines stratified by expression of CD133 and NG2. CD133+ cells show elevated levels of activated PLK1 (n=3 individual experiments; 2-way ANOVA with Bonferroni post-hoc test, error bars represent SEM; *p≤0.05; **p≤0.01).
(H) Immunocytochemistry to detect phosphorylated form of PLK at T120 residue (pT210-PLK; green), CD133 (red), and DAPI staining (blue). Arrows depict enriched localization of activated PLK1 and co-localization with CD133 at the cortex (scale bar represents 10μm).
CD133+ cells have differential regulation of cell cycle dynamics
Since ACD and cell cycle control are tightly linked, we next examined if CD133+ and CD133−NG2+ cells exhibit distinct cell cycle dynamics. Rates at which CD133+ and CD133−NG2+ cells incorporated EdU were compared, with results indicating that CD133+ cells have an extended G2/M phase in relation to CD133−NG2+ and unsorted cells (Figs 3E and S3D). To distinguish M phase cells we stained with the mitotic marker phospho-histone H3 (pHH3). CD133+ cells showed increased pHH3 positivity, in relation to CD133−NG2+ cells (Fig. 3F). However, the fraction of CD133+ cells that stained for pHH3 did not exceed 5% for any cell line, indicating that most G2/M phase cells, as indicated by EdU incorporation, are in G2 phase. It is important to note that the higher proportion of G2/M cells in the CD133+ population merely reflects the prolonged duration of these cell cycle phases in this population and does not imply overall increased rates of proliferation.
Progression from G2 to M phase occurs, in part, as a result of PLK1 activity (31). PLK1 activation requires Threonine210 (T210) phosphorylation, and pT210-PLK1-directed flow cytometry is used surrogate for assessing PLK1 activation. To understand the molecular differences associated with extended G2/M phase in CD133+ subpopulations, the frequency of pT210-PLK1+ CD133+ and NG2+ cells was determined by flow cytometry and found to be higher in the CD133+ fraction than in the CD133− NG2+ or bulk tumor fraction (Fig. 3G). To determine whether PLK1 activation is associated with CD133 polarization, we co-stained for pT210-PLK1 and CD133. In addition to substantiating PLK1 localization to the centrosome in mitosis (Fig. S3E; 22), we detected active PLK1 at the cell periphery, overlapping with CD133 staining (Fig. 3H). Based on the totality of this data, we concluded that CD133+ cells differ from autologous CD133−NG2+ cells by having increased rates of polarized, asymmetric divisions, an extended G2/M phase and elevated levels of PLK1 activation.
A reciprocal relationship between polarity and PLK1 activity regulates CD133+ cell entry into mitosis
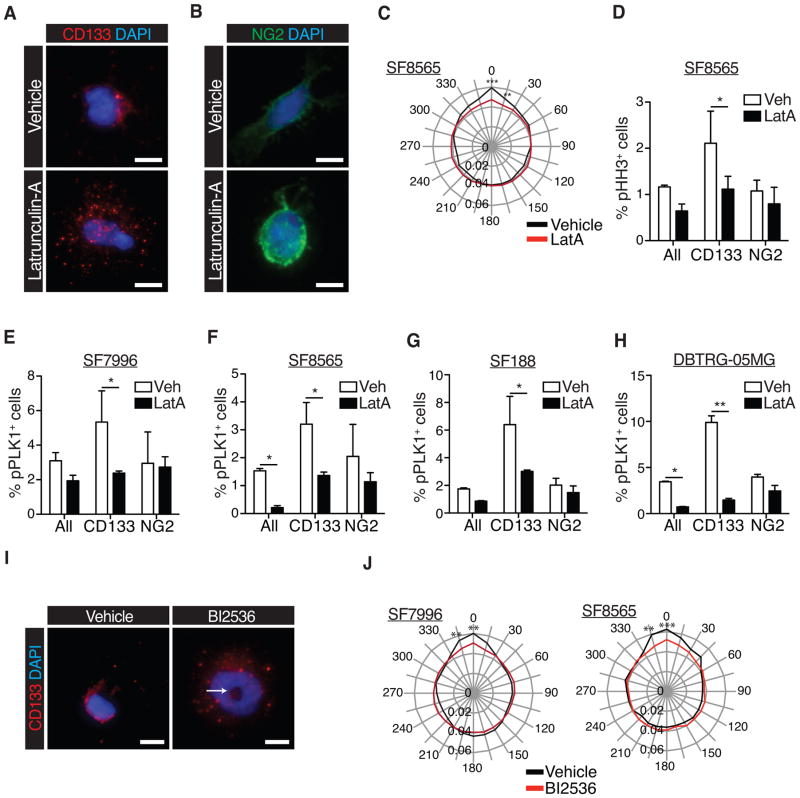
The polarized co-localization of active PLK1 and CD133, as well as elevated PLK1 activity in CD133+ cells and the cell cycle data, suggests a link between polarity, PLK1 activation and passage through G/M phase. To test this hypothesis, we treated cells with the Latrunculin-A (LatA), which is known to disrupt cell polarization by inhibiting actin dynamics (20). We first determined the cellular distribution of CD133 by immunocytochemistry. The effect of LatA was evident by disrupted cellular morphologies with fewer projections and a more rounded appearance. In contrast to control treated cells, which displayed polarized, membrane-bound CD133, LatA treated cells showed CD133+ staining as uniformly distributed dots, such that the polarization of CD133 was significantly decreased (Fig. 4A, C, S4A). The localization of NG2, which was largely uniform around the cortex in control cells, showed no significant changes following LatA treatment, except a stronger enrichment at the cortex (Fig. 4B).
Figure 4. PLK1 activity controls mitotic entry and cell polarity in CD133+ cells.
(A) Immunocytochemistry to detect CD133 in GBM cells treated with vehicle (DMSO) or Latrunculin-A (LatA; CD133 is in red, DAPI in blue; scale bars represent 10μm).
(B) Immunocytochemistry to detect NG2 in GBM cells treated with vehicle (DMSO) or Latrunculin-A (LatA; NG2 is in green, DAPI in blue; scale bars represent 10μm).
(C) Radial plot showing quantification of polarized CD133 and NG2 distribution in the SF8565 GBM cell line after treatment with vehicle or LatA (n=2 individual experiments per cell line, ≥30 cells were scored per experiment, 2-way ANOVA with Bonferroni post-hoc test; **p≤0.01; ***p≤0.001).
(D) Flow cytometry analyses quantifying the frequency of pHH3+, mitotic cells in a primary GBM cell line. CD133= CD133+NG2− cells, NG2+=CD133−NG2+ cells (n=3 individual experiments per cell line; 2-way ANOVA with Bonferroni post-hoc test; error bars represent SEM).
(E–H) Flow cytometry analyses quantifying the frequency of pT210-PLK1+ cells (%pPLK1+ cells) in four different cell lines, following treatment with LatA (n=4 individual experiments per cell line; 2-way ANOVA with Bonferroni post-hoc test; error bars represent SEM). (*p≤0.05 in C–H)
(I) Immunocytochemistry for CD133 (red) and DAPI staining (blue) in GBM cells treated with Vehicle or 1nM BI2536 for 2hrs. Arrow depicts the typical polo arrest phenotype.
(J) Quantification of CD133 and NG2 polarization in vehicle and BI2536–treated (1nM, 2h) GBM cell lines (n=2 individual experiments per cell line, ≥30 cells scored per experiment; 2-way ANOVA with Bonferroni post-hoc test; **p≤0.01; ***p≤0.001).
We next determined if LatA treatment alters mitotic entry and PLK activity. To this end, we subjected LatA treated cells to flow cytometric analysis for pHH3, pT210-PLK1, CD133 or NG2 expression. A decrease in mitotic CD133+ cells, but not CD133−NG2+ or unsorted cells, was detected following LatA treatment (Fig. 4D, S4B). Depending on the cell line, the frequency of CD133+pT210-PLK1+ cells was reduced two to ten-fold by LatA treatment (Fig. 4E–H).
Lastly, we tested whether PLK1 activity is required for cell polarity by plating cells at clonal densities in the presence of vehicle or the PLK1 inhibitor BI2536 (31). Treated cells were fixed and stained for CD133 (Fig. 4I). Quantification of CD133 polarization revealed that PLK1 inhibition with BI2536 significantly reduced CD133 polarization (Fig. 4J). We therefore concluded that PLK1 activity, mitotic entry and cell polarization are linked.
CD133+ cells are less responsive to targeted inhibition of MAPK signaling pathway
We next determined whether CD133+ cells show differential response to MAPK pathway inhibition. First, Alamar Blue assays were performed on all cell lines to determine an effective dosage for the anti-proliferative effect of the MAPK inhibitor PD0325901 (PD901 (Fig. S5A–C, E). Similarly, we determined the response of DBTRG cells carrying the BRAFV600E mutation to increasing doses of PLX4720, the Vemurafenib tool compound (Fig. S5D).
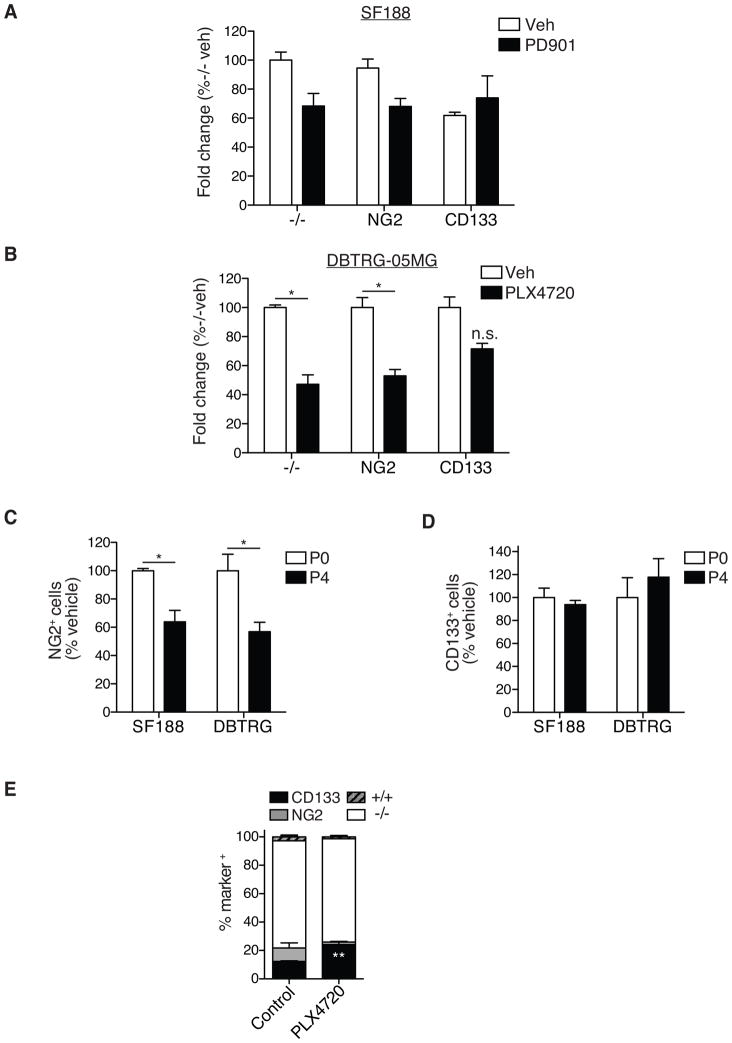
Next we subjected FACS-enriched CD133+, CD133−NG2+, and marker-negative cells to a viability assay after five-day treatments with PD901 or PLX4720. PD901 and PLX4720 reduced the viability of CD133−NG2+ and marker-negative cells, respectively, but exerted no effect on CD133+ cells (Fig. 5A, B). We then assessed the effects of extended drug treatment by propagating cells in the continuous presence of PD901 or PLX4720, and quantifying the frequency of CD133+ and NG2+ cells by flow cytometry at each passage, for up to four passages. During the course of two weeks of treatment, the proportion of NG2+ cells decreased by 36% (SF188) and by 43% (DBTRG) (Fig. 5C), respectively, while the proportion of CD133+ cells remained unchanged (Fig. 5D).
Figure 5. Decreased sensitivity of CD133+ subpopulations to MAPK pathway inhibition.
Alamar Blue viability assay using FACS-enriched CD133+, NG2+ and marker-negative (−/−) cells from SF188 GBM cell lines (A) treated for 5 consecutive days with 0.5μM PD901 (MAPK inhibitor) and vehicle (VEH) (B) DBTRG-05MG treated for 5 consecutive days with 1μM PLX4720 and vehicle (VEH) (n=3 individual experiments per cell line; 2-way ANOVA with Bonferroni post-hoc test; error bars represent SEM).
Flow cytometry analyses of (C) CD133−NG2+ (NG2) and (D) CD133+NG2− (CD133) cell frequency in SF188 and DBTRG-05MG GBM cell lines following treatment with 0.5μM PD901 (SF188) or 1μM PLX4720 (DBTRG-05MG) for four passages (P0=passage 0; P4= passage 4; n=2 individual experiments per cell line, 1-way ANOVA with Tukey post-hoc test; error bars represent SEM; *p≤0.05 in B and C).
(E) Flow cytometry analyses of CD133+ and CD133−NG2+ (NG2) cell frequency in subcutaneous DBTRG-05MG xenografts; established tumors were transplanted into new hosts on average every 6 weeks and received 186-days of continuous treatment with vehicle or PLX4720 (10mg/kg/day). (N=4 individual tumor specimen; 2-way ANOVA with Bonferroni post-hoc test; error bars represent SEM; **p≤0.01).
To examine the response of CD133+ cells to MAPK-pathway inhibition in vivo, subcutaneous DBTRG xenograft tumors were serially propagated in athymic mice that received PLX4720 at 10mg/kg/day (186 days total treatment). Flow cytometry analysis of disaggregated tumor cells revealed a change in the cellular composition of tumors following extended BRAFV600E inhibition. Whereas CD133−NG2+ cells were effectively eliminated, the frequency of CD133+ cells was significantly increased (Fig. 5E).
Based on these data, we concluded that short-term, intermediate and chronic BRAF/MAPK inhibition fails to diminish the viability of CD133+ cells.
Targeting PLK1 in combination with BRAF/MAPK inhibition reduces GBM cell growth
Next we assessed if elevated PLK1 activity provides a point of susceptibility in CD133+ cells, and furthermore tested whether combined PLK1 and BRAF/MAPK inhibition is more effective than BRAF/MAPK inhibition alone at inhibiting tumor cell viability in general and/or in specific cell populations. By itself, PLK1 inhibitor BI2536 (Fig. S6A–D) showed anti-viability activity against unsorted cells in the nanomolar range.
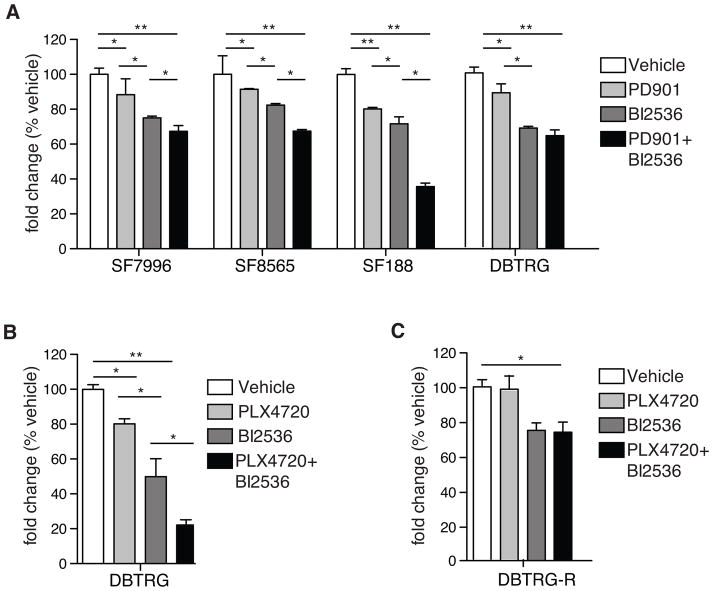
Next, we performed Alamar Blue viability assays on GBM cell lines treated for 5 days with PD901 or PLX4702 alone, or in combination with BI2536. BI2536 decreased viability more substantially than PD901 and PLX4720, in comparing single agent effects. In all cases, combination treatments inhibited proliferation to a greater extent than any single agent treatment (Fig. 6A, B). Treatment of DBTRG cells with PLX4720, for an extended length of time and with increasing concentrations of inhibitor, resulted in tumor cell adaptation to inhibitor, such that cell viability in the presence of inhibitor became similar to that of untreated cells. In contrast, BI2536 single agent treatment of DBTRG showed sustained adverse effect on cell viability (Fig. 6C).
Figure 6. Targeting PLK1 in combination with BRAF/MAPK inhibition reduces GBM cell viability.
(A) Alamar Blue viability assay of four GBM cell lines treated with vehicle (VEH), 1μM PD901, 5nM BI2536, or a combination of PD901 and BI2536 for five days (n=4 individual experiments/cell line; error bars represent SEM).
(B) Growth assay of DBTRG-05MG cells treated with vehicle, PLX4720, BI2536 or a combination of PLX4720 and BI2536 (n=3 individual experiments/cell line; 1-way ANOVA with Tukey post-hoc tests for A and B; error bars represent SEM).
(C) Growth assay of DBTRG-05MG cells pre-treated with an escalating dosage of PLX4720, treated with vehicle, PLX4720, BI2536 or a combination of PLX4720 and BI2536 (n=3 individual experiments/cell line; 1-way ANOVA with Tukey post-hoc tests for A and B; error bars represent SEM). *P≤0.05; **p≤0.01 in A, B.
Concurrent inhibition of PLK1 and BRAFV600E has heightened adverse effect on cell viability
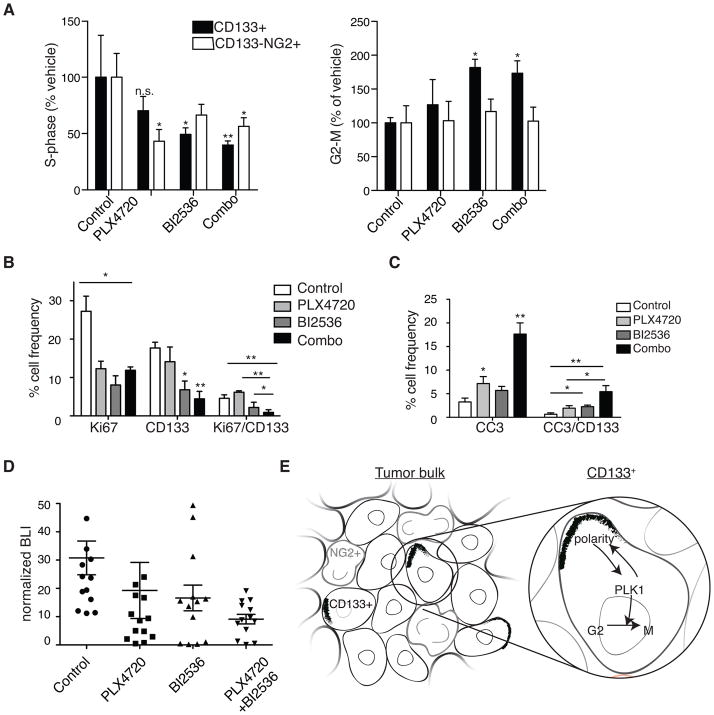
We next examined inhibitor effects on cell subpopulations in vivo. Mice with subcutaneous BRAFV600E mutant DBTRG xenograft tumors were treated with PLX4720 or BI2536 alone or in combination. After 5 days treatment mice were injected with EdU, then euthanized, with subcutaneous tumors immediately resected then disaggregated to create cell suspensions for flow sorting. CD133+ and CD133−NG2+ cells were analyzed for proliferation and cell cycle effects. Single agent and combination treatments reduced S phase cells in both CD133+ and CD133−NG2+ cell populations, with greatest S-phase effect observed for CD133+ cells in tumors exposed to both inhibitors (Fig. 7A). Combination treatment, as well as BI2536 only treatment, significantly increased the number of CD133+ cells in G2/M, indicative of M phase arrest (Fig. 7A). Further evidence that BI2536 effectively reaches tumor cells was provided by IF of tumor tissue for pHH3 which revealed increased numbers of mitotic cells with abnormally shaped nuclei typically associated with PLK1 inhibition (Fig. S7A) (31).
Figure 7. Effect of combined inhibition of BRAFV600E and PLK1 on CD133+ cells and tumor growth in vivo.
(A) Ex vivo flow cytometry analyses of subcutaneous DBTRG xenografts treated for five days with PLX4720 daily at 20mg/kg, BI2536 twice at 50mg/kg or a combination of both inhibitors (Combo). Dissociated tumor cells were analyzed for incorporation of EdU (S-phase) or G2/M following an EdU pulse 30 min before tumors were harvested. (N=5 individual experiments; 2-way ANOVA with Bonferroni post-hoc test, values compared to vehicle treated conditions for each cell type.)
(B) Quantification of Ki67+, CD133+ and CD133+Ki67+ double positive cells by IF performed on intracranial xenograft tumor samples of treated mice. (N ≥ 3 individual tumors from each treatment group; a minimum of three sections for each treatment group were counted.)
(C) Quantification of cleaved caspase 3 (CC3)+, CD133+ and CD133+CC3+ double-positive cells by IF performed on intracranial xenograft tumor samples of treated mice. (N ≥ 3 individual tumors from each treatment group; a minimum of three sections for each treatment group were counted) *P≤0.05; **p≤0.005 in A–C.
(D) Normalized bioluminescence (BLI) readings from intracranial DBTRG-05MG tumors at 7–9 days post-treatment initiation. Mice were either untreated (control) or treated with either daily PLX4720 at 20mg/kg, twice-weekly BI2536 at 50mg/kg, or a combination (n=12–13 animals per treatment group, 1-way ANOVA with Tukey post-hoc test).
(E) Model for a PLK1 inhibitor-sensitive polarity checkpoint in a heterogeneous GBM tumor where CD133+ cells co-exist with CD133−NG2+ (NG2+) cells. NG2+ cells are outlined in grey; polarized CD133 are black crescents.
Mice with intracranial xenografts were also treated with inhibitors, alone or in combination, for up to 9 days, to investigate whether an orthotopic microenvironment affects inhibitor activities against their corresponding targets. Dissociated DBTRG tumor cells were examined by flow cytometry for pT210-PLK1 and phospho-ERK (pERK) following PLX4720 and/or BI2536 treatments, with results showing the most substantial decrease in active PLK1 following animal subject treatment with PLK1 inhibitor, alone or with PLX4720 (Fig. S7B). In contrast, PLX4720 showed greater inhibitory effect against pERK, an indicator for BRAFV600E (24), than BI2536, with combination PLX4720 + BI2536 showing the most substantial effect in suppressing pERK (Fig. S7C). Similar results were obtained with MAPK inhibitor PD901 in cells (Fig. S7D). A qPCR analysis of CD133+ and OPC maker OLIG2 in agent-treated subcutaneous DBTRG xenografts further revealed reduced CD133+ expression only after PLK1 inhibition by BI2536 and PLX4720/BI2536 combination treatment (Fig. S7E).
Intracranial BRAFV600E xenografts were also examined for treatment effects on CD133+ cell proliferation by Ki67 staining and apoptosis by cleaved caspase-3 staining, with corresponding results showing combination treatment as producing the most significant anti-proliferative response (six-fold vs. control and PLX4720 treated and three-fold vs. BI2536 treated) towards CD133+ cells (Fig. 7B). The combination treatment also had a strong pro-apoptotic effect on tumor cells and CD133+ cells in particular (five-fold vs. control and two point five-fold vs. single agents; Fig. 7C). Anti-proliferative and pro-apoptotic effects were confirmed by bioluminescence imaging of luciferase-modified DBTRG intracranial xenografts, which showed combination therapy as having greatest activity in suppressing tumor growth (Fig. 7D).
Discussion
Recent reports have shown that TPCs are genetically and phenotypically heterogeneous, consisting of populations with distinct responsiveness to therapy (13,16). These observations have prompted us to perform side-by-side analyses of CD133+ and NG2+ TPC subpopulations for differential response to MAPK pathway inhibition. Consistent with the results of previous studies (32,33) we determined that autologous CD133+ cells show reduced proliferation in relation to unsorted tumor cells as well as in relation to CD133− tumor cells, whether in human tumor tissue or cell culture, and irrespective of cell culture conditions (34–36). In contrast, we found NG2+ cells as having very similar proliferation characteristics as bulk, unsorted tumor cells, and that were at higher rates than CD133+ cells.
In addition, our results show distinct cell cycle dynamics and ACD rates, with CD133+ cell characteristics that are reminiscent of the healthy mouse brain, and in which NSCs comprise a partly quiescent population that, when activated to proliferate, predominantly divides asymmetrically (37–39). NG2+ tumor cells, on the other hand, undergo fewer ACD than CD133+ cells and predominantly divide symmetrical, in line with previous data in oligodendroglioma (20). Our findings that CD133+ TPCs are less proliferative while maintaining a higher rate of ACD is consistent with a role for ACD in restricting proliferation.
Many ACD regulators, which have been extensively studied in invertebrates, are conserved in the mammalian genome (19). The activity of one these, PLK1, is known to increase in G2 phase of the cell cycle (40). The reciprocal relationship between PLK1 activity and cell polarity, reported in invertebrate model systems (41–43) and shown for the first time here in CD133+ TPCs, suggests a cell cycle checkpoint in which PLK1 activation and cell cycle G2-to-M progression occurs only when appropriate polarity is achieved (Fig. 7E).
The use of chemotherapeutics that disrupt cell polarity could activate this checkpoint, which, in turn, could spare tumor cells from continued cell progression and potential mitotic catastrophe. Through a pharmacologic approach we have shown that inhibition of this checkpoint not only increases tumor cell apoptosis, especially in the CD133+ subpopulation, but as well limits the proliferation of this important class of TPC.
The discovery of a BRAFV600E mutation in a subset of GBM (23) and encouraging GBM response to BRAFV600E inhibition in pre-clinical models (24,25) has prompted ongoing clinical trials for using BRAF inhibitors to treat glioma patients (26,27). However, pre-existing and acquired resistance to BRAFV600E inhibitor is known to occur (28,44) and as a result several combination therapy approaches are being investigated for increasing the duration and extent of anti-tumor activity from inhibiting mutant BRAF.
PLK1 is highly expressed in gliomas and GBM (45) and has previously been shown to enhance radio-sensitivity in brain tumor models (46). Our discovery that simultaneous MAPK pathway and PLK1 inhibition is especially effective against CD133+ GBM cell subpopulations, that are known to be resistant to routinely used therapies for GBM, supports the need to further develop this treatment concept for clinical translation and evaluation in GBM patients.
Supplementary Material
Acknowledgments
The authors thank the Pieper, Nicolaides and Costello labs for cells, and Cynthia Cowdrey and the BTRC tissue core for providing tissue sections. For critical discussion we thank M. Oft and the members of the Petritsch lab, S. Mueller and D. Haas-Kogan. We thank Y. Yashida, B. Reicholff, N. Andor, M. Kastner, S. Elmes, T. Mazor, R. Santos, H. Collins and N. Murphy for technical support. The authors thank C. Crook and H. Oft for volunteering. C. Petritsch wishes to dedicate this work to Florian Petritsch. This work was supported by funding by the National Institute of Health/NCI (to C. Petritsch R01CA164746-04), NINDS (to C. Petritsch and C.D. James RO1NS080619), Childhood Brain Tumor Foundation (to C. Petritsch), Voices against Brain Cancer Foundation (to C. Petritsch), and RAP-Jr. Investigator grant CTSI-UCSF (to C. Petritsch).
Footnotes
The authors disclose no potential conflicts of interest
References
- 1.Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014 doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 2.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell Elsevier Inc. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccirillo SGM, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–11. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 8.Andor N, Harness JV, Mueller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 10.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 11.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 12.Jamal M, Rath B. The brain microenvironment preferentially enhances the radioresistance of CD133+ glioblastoma stem-like cells. … (New York, NY) 2012;16:6049–59. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature Nature Publishing Group. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal Differentiation Mediated by NF-κB Promotes Radiation Resistance in Glioblastoma. Cancer Cell. 2013;24:331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Aoyagi M, Wakimoto H, Ando N, Nariai T, Yamamoto M, et al. Accumulation of CD133-positive glioma cells after high-dose irradiation by Gamma Knife surgery plus external beam radiation. J Neurosurg. 2010;113:310–8. doi: 10.3171/2010.2.JNS091607. [DOI] [PubMed] [Google Scholar]
- 16.Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, et al. Transcriptional profiles of CD133+ and CD133− glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–40. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 17.Al-Mayhani MTF, Grenfell R, Narita M, Piccirillo S, Kenney-Herbert E, Fawcett JW, et al. NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro- …. 2011;13:830–45. doi: 10.1093/neuonc/nor088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persson AAI, Petritsch C, Swartling FFJ, Itsara M, Sim FJ, Auvergne R, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–82. doi: 10.1016/j.ccr.2010.10.033. 2010/12/16 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-López S, Lerner RG, Petritsch C. Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell Mol Life Sci. 2014;71:575–97. doi: 10.1007/s00018-013-1386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell Elsevier Inc. 2011;20:328–40. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji a, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis-cytokinesis, DNA damage response, and development. Curr Opin Cell Biol. 2008:650–6024. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley M-YCY, Yu M, et al. Oncogenic BRAF Mutation and CDKN2A Inactivation Is Characteristic of a Subset of Pediatric Malignant Astrocytomas. Cancer Res. 2010;70:512–9. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolaides TP, Li H, Solomon Da, Hariono S, Hashizume R, Barkovich K, et al. Targeted therapy for BRAFV600E malignant astrocytoma. Clin Cancer Res. 2011;17:7595–604. doi: 10.1158/1078-0432.CCR-11-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huillard E, Hashizume R. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. PNAS. 2012;109:8710–5. doi: 10.1073/pnas.1117255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson GW, Orr Ba, Gajjar A. Complete clinical regression of a BRAF V600E-mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer. 2014;14:258. doi: 10.1186/1471-2407-14-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. 2011/06/07 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fouse S, Nakamura JL, James CD, Chang S, Costello JF. Response of primary glioblastoma cells to therapy is patient specific and independent of cancer stem cell phenotype. Neuro Oncol. 2014;16:361–371. doi: 10.1093/neuonc/not223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–22. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 32.Deleyrolle LP, Harding A, Cato K, Siebzehnrubl FA, Rahman M, Azari H, et al. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 2011;134:1331–43. doi: 10.1093/brain/awr081. 2011/04/26 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richichi C, Brescia P, Alberizzi V, Fornasari L, Pelicci G. Marker-independent Method for Isolating Slow-Dividing Cancer Stem Cells in Human Glioblastoma. … (New York, NY) 2013;15:840–7. doi: 10.1593/neo.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 35.Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 36.Jin F, Gao C, Zhao L, Zhang H, Wang HT, Shao T, et al. Using CD133 positive U251 glioblastoma stem cells to establish nude mice model of transplanted tumor. Brain Res. 2011;1368:82–90. doi: 10.1016/j.brainres.2010.10.051. [DOI] [PubMed] [Google Scholar]
- 37.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–44. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 38.Ponti G, Obernier K, Guinto C, Jose L, Bonfanti L, Alvarez-Buylla A. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc Natl Acad Sci U S A. 2013;110:E1045–54. doi: 10.1073/pnas.1219563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calzolari F, Michel J, Baumgart EV, Theis F, Gotz M, Ninkovic J. Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat Neurosci. 2015;18:490–492. doi: 10.1038/nn.3963. [DOI] [PubMed] [Google Scholar]
- 40.Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends in Biochem Sciences. 2012;37:534–42. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Rivers DM, Moreno S, Abraham M, Ahringer J. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol. 2008;180:877–85. doi: 10.1083/jcb.200710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noatynska A, Panbianco C, Gotta M. SPAT-1/Bora acts with Polo-like kinase 1 to regulate PAR polarity and cell cycle progression. Development. 2010;137:3315–25. doi: 10.1242/dev.055293. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–67. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietzmann K, Kirches E, von B, Jachau K, Mawrin C, Von Bossanyi P, et al. Increased human polo-like kinase-1 expression in gliomas. J Neurooncol. 2001;53:1–11. doi: 10.1023/a:1011808200978. [DOI] [PubMed] [Google Scholar]
- 46.Tandle AT, Kramp T, Kil WJ, Halthore A, Gehlhaus K, Shankavaram U, et al. Inhibition of polo-like kinase 1 in glioblastoma multiforme induces mitotic catastrophe and enhances radiosensitisation. Eur J Cancer. 2013;49:3020–8. doi: 10.1016/j.ejca.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.