Abstract
Leucine is a proteogenic amino acid that also regulates many aspects of mammalian physiology, in large part by activating the mTOR complex 1 (mTORC1) protein kinase, a master growth controller. Amino acids signal to mTORC1 through the Rag guanine triphosphatases (GTPases). Several factors regulate the Rags, including GATOR1, a GTPase activating protein (GAP); GATOR2, a positive regulator of unknown function; and Sestrin2, a GATOR2-interacting protein that inhibits mTORC1 signaling. We find that leucine, but not arginine, disrupts the Sestrin2-GATOR2 interaction by binding to Sestrin2 with a Kd of 20 µM, which is the leucine concentration that half-maximally activates mTORC1. The leucine-binding capacity of Sestrin2 is required for leucine to activate mTORC1 in cells. These results indicate that Sestrin2 is a leucine sensor for the mTORC1 pathway.
It has long been appreciated that in addition to being a proteogenic amino acid, leucine is also a signaling molecule that directly regulates animal physiology, including satiety (1), insulin secretion (2), and skeletal muscle anabolism (3, 4). Because the liver has a low capacity to metabolize leucine, its blood concentrations fluctuate in accord with its consumption so that dietary leucine can directly impact physiology (5–7). A key mediator of the effects of leucine is the mTORC1 protein kinase (8, 9), which regulates growth by controlling processes like protein and lipid synthesis as well as autophagy.
Many environmental signals besides leucine regulate the mTORC1 pathway, including other amino acids like arginine, as well as glucose and various growth factors and forms of stress (10, 11). How mTORC1 senses and integrates these diverse inputs is not well understood, but it is clear that the Rheb and Rag guanine triphosphatases (GTPases) have necessary but distinct roles. Rheb is a monomeric GTP binding protein and the Rags function as obligate heterodimers of RagA or RagB bound to RagC or RagD (12–14). Both the Rheb and Rag GTPases localize, at least in part, to the lysosomal surface (15–18), which is an important site of mTORC1 regulation (19). In a Rag-dependent manner amino acids promote the translocation of mTORC1 to the lysosome where Rheb, if bound to GTP, stimulates its kinase activity. Growth factors trigger the GTP-loading of Rheb by driving its GTPase activating protein (GAP), the tuberous sclerosis (TSC) complex, off the lysosomal surface (18).
Regulation of the Rag GTPases by amino acids is complex, and many distinct factors have important roles (20). A lysosome-associated super-complex containing Ragulator, SLC38A9, and the vacuolar adenosine triphosphase (v-ATPase) interacts with the Rag GTPases and is necessary for the activation of mTORC1 by amino acids (21–24). Ragulator anchors the Rag heterodimers to the lysosome and has nucleotide exchange activity for RagA and RagB (21, 25). SLC38A9 is an amino acid transporter and a potential lysosomal arginine sensor (23), but the function of the v-ATPase in mTORC1 activation is unclear. Two GAP complexes stimulate GTP hydrolysis by the Rag GTPases, with GATOR1 acting on RagA and RagB (26) and Folliculin (FLCN)-Folliculin interacting protein 2 (FNIP2) on RagC and RagD (27). The separate GATOR2 complex negatively regulates GATOR1 through an unknown mechanism and is necessary for mTORC1 activation (26). Lastly, the Sestrins are GATOR2-interacting proteins that inhibit mTORC1 signaling but whose molecular function is not known (28, 29).
The amino acids sensors upstream of mTORC1 have been elusive for many years. While SLC38A9 is a strong candidate for sensing arginine at lysosomes (23), the long-sought sensor of leucine was unknown. We demonstrate that Sestrin2 is a leucine sensor for the mTORC1 pathway.
Leucine directly regulates the Sestrin2-GATOR2 interaction
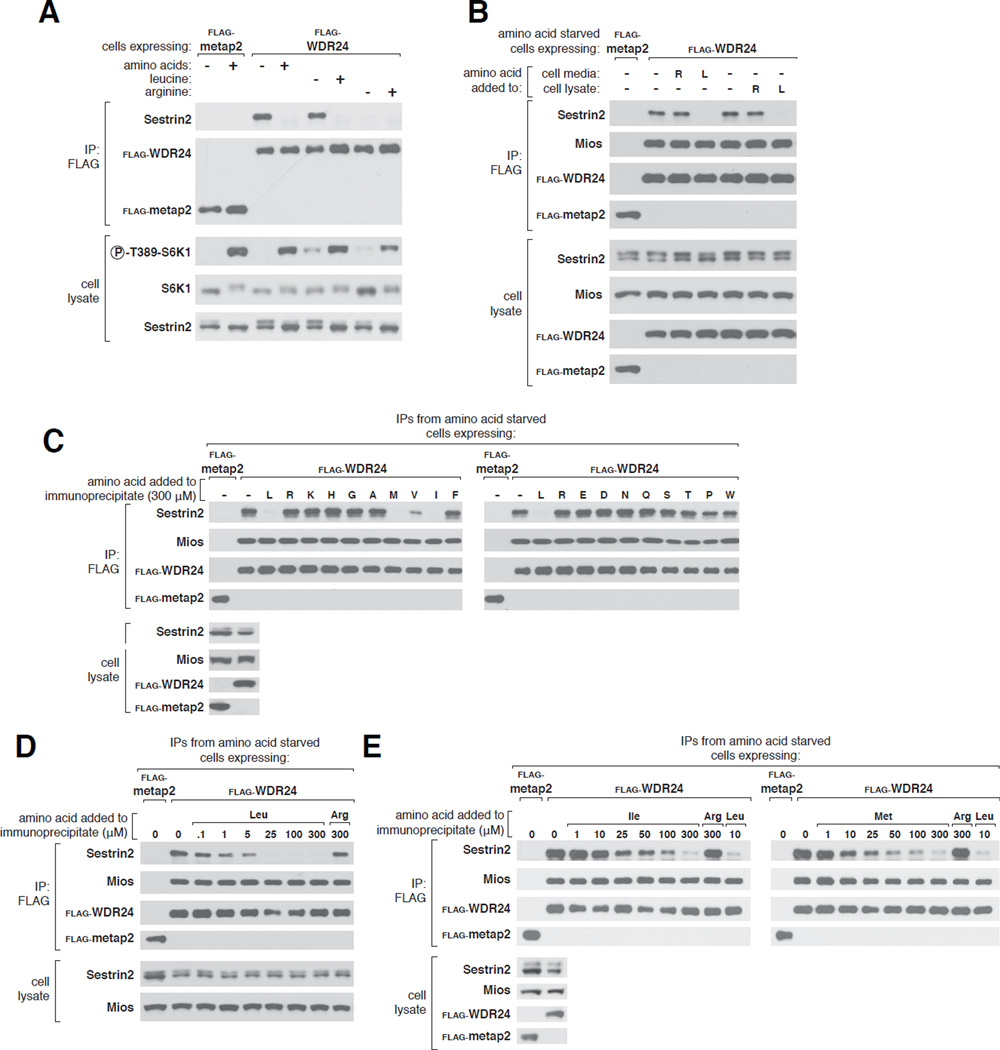
Activation of mTORC1 by amino acids requires the pentameric GATOR2 complex (26). Although its molecular function is unknown, epistasis-like experiments suggest that GATOR2 suppresses GATOR1, the GAP for and inhibitor of RagA and RagB (26). Within cells Sestrin2 binds to GATOR2 in an amino acid-sensitive manner (28, 29), with removal and re-addition of all amino acids from the culture media inducing and reversing the interaction, respectively (28). Although several amino acids can regulate mTORC1 signaling, arginine and leucine are the best established and deprivation of either strongly inhibits mTORC1 in various cell types (8, 30, 31). In human embryonic kidney-293T (HEK-293T) cells removal of either leucine or arginine from the cell medium inhibited mTORC1 signaling, as read out by ribosomal protein S6 kinase 1 (S6K1) phosphorylation, to similar extents. Strikingly, however, only leucine depletion caused Sestrin2 to bind to GATOR2, inducing the interaction as effectively as complete amino acid starvation (Fig. 1A). Leucine re-addition rapidly reversed the binding and amino acids did not affect the interaction between WDR24 and Mios, two core components of GATOR2 (Fig. 1A, and S1A).
Figure 1. Leucine, but not arginine, disrupts the Sestrin2-GATOR2 interaction in cells and in vitro.
A) Binding of Sestrin2 to GATOR2 in HEK-293T cells stably expressing FLAG-WDR24 (a component of GATOR2). Cells were deprived of leucine, arginine, or all amino acids for 50 minutes. Where indicated, cells were re-stimulated with leucine, arginine, or all amino acids for 10 minutes and FLAG immunoprecipitates prepared from cell lysates. Immunoprecipitates and lysates were analyzed by immunoblotting for the indicated proteins. FLAG-metap2 served as a negative control.
B) Effects of leucine and arginine on the Sestrin2-GATOR2 interaction in ice-cold detergents lysates of amino acid-starved cells. HEK-293T cells stably expressing FLAG-metap2 or FLAG-WDR24 were deprived of all amino acids for 50 minutes. Leucine or arginine was added to the culture media or cell lysates and FLAG immunoprecipitates prepared and analyzed as in (A).
C) Effects of individual amino acids on the purified Sestrin2-GATOR2 complex. FLAG immunoprecipitates were prepared from HEK-293T cells stably expressing FLAG-metap2 or FLAG-WDR24 and deprived of all amino acids for 50 minutes. Indicated amino acids (300 µM) were added directly to the immunoprecipitates, which, after re-washing, were analyzed as in (A).
D) Disruption of the purified Sestrin2-GATOR2 complex by leucine. Experiment was performed and analyzed as in (C) except that indicated concentrations of leucine or arginine were used.
E) Disruption of the Sestrin2-GATOR2 interaction by isoleucine and methionine. Experiment was performed and analyzed as in (C) except that the indicated concentrations of isoleucine, methionine, leucine, or arginine were used.
Sestrin2 is homologous to two other proteins, Sestrin1 and Sestrin3 (32–34), and when overexpressed all three can interact with GATOR2 (28). As with Sestrin2, leucine starvation and stimulation strongly regulated the interaction of endogenous Sestrin1 with GATOR2 (Fig. S1B). In contrast, endogenous Sestrin3 bound to GATOR2 irrespective of leucine concentrations (Fig. S1B), suggesting that this interaction is constitutive or regulated by signals that remain to be defined.
While enzymatic events triggered by leucine might mediate the effects of leucine on the Sestrin2-GATOR2 interaction, it was tempting to consider that leucine might act directly on the complex. Consistent with this possibility, the addition of leucine, but not arginine, to ice-cold detergent lysates of cells deprived of all amino acids abrogated the interaction to the same extent as leucine-stimulation of live cells (Fig. 1B). Even more intriguingly, leucine also disrupted the interaction when added directly to immunopurified Sestrin2-GATOR2 complexes isolated from amino acid-deprived cells. Of the 18 amino acids tested at 300 µM each, only those most similar to leucine –methionine, isoleucine, and valine– had any effect on the Sestrin2-GATOR2 interaction in vitro (Fig. 1C).
When added to the purified complexes, leucine dose-dependently disrupted the Sestrin2- GATOR2 complex, with the half maximal effect at about 1 µM (Fig. 1D). Methionine and isoleucine were considerably less potent, acting at concentrations approximately 10- and 25-fold greater than leucine, respectively (Fig. 1E). These values reflect only the relative potencies of these amino acids as equilibrium conditions were not attained because the large assay volume precluded Sestrin2 from rebinding to GATOR2 once dissociated.
Sestrin2 binds leucine with a Kd of 20 µM
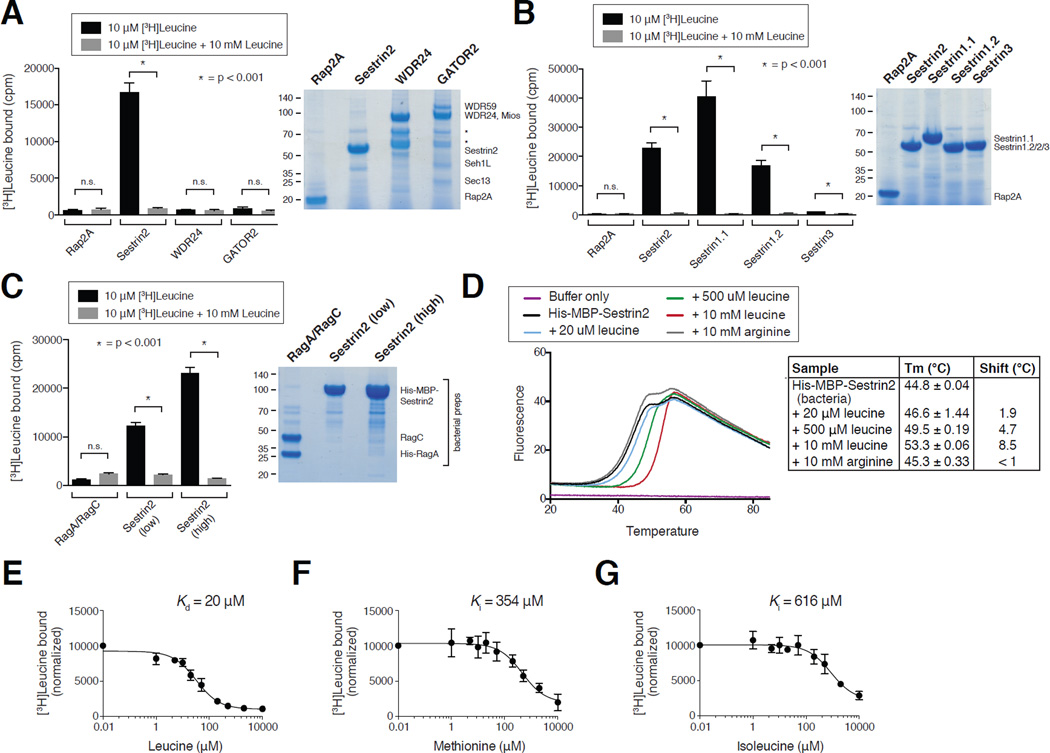
Given that leucine disrupts the purified complex, we reasoned that leucine might directly bind to Sestrin2 or GATOR2. To test this, we developed an equilibrium binding assay in which purified proteins immobilized on agarose beads were incubated with radioactive amino acids, and the bound amino acids were quantified after washing. Radiolabeled leucine bound to Sestrin2, but not WDR24, the GATOR2 complex, or the control protein Rap2A, in a manner that was fully competed by excess non-radiolabeled leucine (Fig. 2A). In contrast, arginine did not bind to either Sestrin2 or Rap2A (Fig. S2A). Consistent with the differential sensitivities of the Sestrin1- and Sestrin3-GATOR2 complexes to leucine, Sestrin1 bound leucine to a similar extent as did Sestrin2, whereas Sestrin3 bound very weakly (Fig. 2B and S2A). Drosophila dSestrin (CG11299-PD) also bound leucine, albeit at lower amounts than the human protein (Fig. S2B and C).
Figure 2. Sestrin2 binds leucine with a Kd of 20 µM.
A) Binding of radiolabeled leucine to Sestrin2, but not WDR24, GATOR2, or the control protein Rap2A. FLAG immunoprecipitates prepared from HEK-293T cells transiently expressing indicated proteins or complexes were used in binding assays with [3H]Leucine as described in the methods. Unlabeled leucine was added where indicated. Values are Mean ± SD for 3 technical replicates from one representative experiment. SDS-PAGE followed by Coomasie blue staining was used to analyze immunoprecipitates prepared in parallel to those included in the binding assays. Asterisks indicate breakdown products in the WDR24 and GATOR2 purifications.
B) Leucine-binding capacities of Sestrin1 (two isoforms), Sestrin2, and Sestrin3. FLAG immunoprecipitates were prepared and binding assays performed and analyzed as in (A).
C) Leucine binds to bacterially-produced Sestrin2, but not the RagA/RagC heterodimer. Leucine binding assays were performed as described in the methods and analyzed as in (A) with His- MBP-Sestrin2 or His-RagA/RagC bound to the Ni-NTA resin.
D) Effects of leucine and arginine on the melting temperature of bacterially-produced Sestrin2 in a thermal shift assay. His-MBP-Sestrin2 was incubated with the Sypro orange dye with or without leucine or arginine. Upon heating the sample the change in fluorescence was captured and used to calculate melting temperatures (Tm) under the indicated conditions. Values are Mean ± SD from 3 replicates.
E) Sestrin2 binds leucine with a Kd of 20 µM. FLAG-Sestrin2 immunoprecipitates prepared as in (A) were used in binding assays with 10 µM or 20 µM [3H]Leucine and indicated concentrations of unlabeled leucine. In the representative graph shown each point represents the normalized mean ± SD for n = 3 in an assay with 10 µM [3H]Leucine. The Kd was calculated from the results of six experiments (three with 10 µM and three with 20 µM [3H]Leucine).
F) Methionine can compete the binding of leucine to Sestrin2. FLAG-Sestrin2 immunoprecipitates prepared as in (A) were used in binding assays with 10 µM [3H]Leucine and indicated concentrations of unlabeled methionine. In the graph shown each point represents the normalized mean ± SD for n = 3. The Ki was calculated using data from the three experiments.
G) Isoleucine can compete the binding of leucine to Sestrin2. FLAG-Sestrin2 immunoprecipitates prepared as in (A) were used in binding assays with 10 µM [3H]Leucine and indicated concentrations of unlabeled isoleucine. In the graph shown each point represents the normalized mean ± SD for n = 3. The Ki was calculated using data from the three experiments.
As all of these proteins were expressed in and purified from human HEK-293T cells, it remained formally possible that an unidentified protein that co-purifies with Sestrin2 (and Sestrin1) is the actual receptor for leucine. To address this possibility, we prepared human Sestrin2 in bacteria, a heterologous system that does not encode a Sestrin homologue or even a TOR pathway. Consistent with the results obtained with Sestrin2 prepared in human cells, radiolabelled leucine bound to bacterially-produced Sestrin2, but not the RagA-RagC heterodimer, which was used as a control (Fig. 2C). Furthermore, in a thermal shift assay, leucine, but not arginine, shifted the melting temperature by up to 8.5°C of bacterially-produced Sestrin2, but not of two control proteins (Fig. 2D and S2E–F). Collectively, these data strongly argue that leucine binds directly to Sestrin2.
While the thermal shift assay is valuable for assessing the capacity of a protein to bind a ligand, this method is not suitable for obtaining an accurate Kd (35). Therefore, we used a competition binding assay with increasing amounts of unlabeled leucine to determine that leucine has a Kd for Sestrin2 of 20 ± 5 µM (Fig. 2E). In comparison, methionine and isoleucine competed leucine binding with inhibitory constants (Ki) of 354 ± 118 µM and 616 ± 273 µM, respectively (Fig. 2F and G). These values are approximately one eighteenth and one thirtieth the affinity of leucine for Sestrin2, and correlate well with the relative potencies of leucine, methionine, and isoleucine in disrupting the Sestrin2-GATOR2 interaction in vitro (Fig. 1D and E).
Sestrin2 regulates mTORC1 through GATOR2
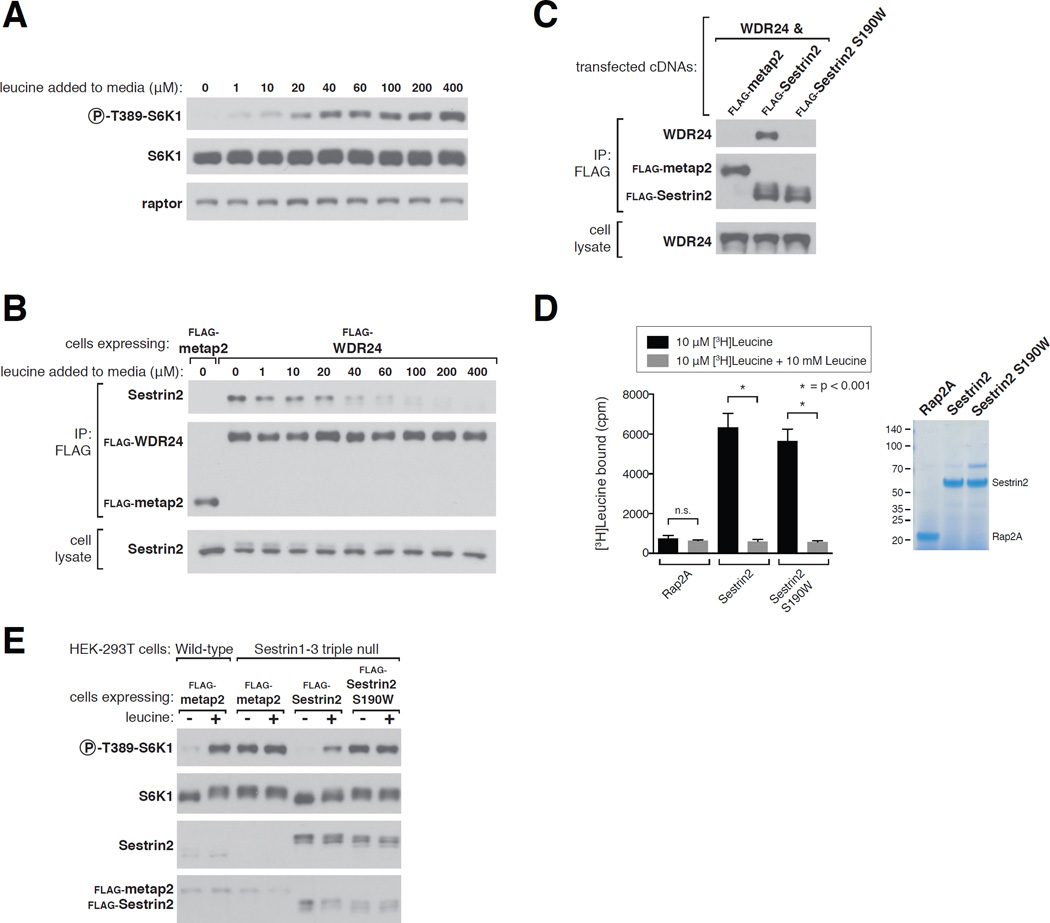
Consistent with leucine regulating mTORC1 by modulating the binding of Sestrin2 to GATOR2, 20–40 µM leucine had half-maximal effects on both the Sestrin2-GATOR2 interaction and mTORC1 activity in HEK-293T cells (Fig. 3A and B). This concentration range encompasses the Kd of leucine for Sestrin2, indicating that the affinity of Sestrin2 for leucine is physiologically relevant.
Figure 3. Sestrin2 regulates mTORC1 through GATOR2.
A) Effects of varying leucine concentrations on mTORC1 activity, as measured by the phosphorylation of S6K1. HEK-293T cells were deprived of leucine for 50 minutes and restimulated with leucine at the indicated concentrations for 10 minutes. Cell lysates were analyzed via immunoblotting for the indicated proteins and phosphorylation states.
B) Effects of varying leucine concentrations on the Sestrin2-GATOR2 interaction. HEK-293T cells stably expressing the indicated proteins were starved as in (A) and FLAG immunoprecipitates were collected. The immunopurified complexes were treated with the indicated concentrations of leucine and then analyzed as in Figure 1C.
C) Decreased GATOR2-binding capacity of the Sestrin2 S190W mutant. FLAG immunoprecipitates were prepared from HEK-293T cells transiently expressing the indicated proteins and were analyzed by immunoblotting for the indicated proteins.
D) Determination of leucine-binding capacity of Sestrin2 S190W. Assays were performed and immunoprecipitates analyzed as in Figure 2A.
E) In Sestrin1-3 triple null cells expressing Sestrin2 S190W the mTORC1 pathway cannot sense the absence of leucine. Wild-type HEK-293T cells and Sestrin1-3 triple null HEK-293T cells generated with the CRISPR/Cas9 system were used to express the indicated FLAG-tagged proteins. Cells were starved for leucine for 50 minutes and, where indicated, stimulated with leucine for 10 minutes and lysates analyzed via immunoblotting.
To formally test whether Sestrin2 regulates mTORC1 by interacting with GATOR2, we identified a Sestrin2 mutant (S190W) that still binds leucine but has a severely decreased capacity to bind GATOR2 (Fig. 3C and D). In Sestrin1-3 triple null HEK-293T cells, mTORC1 signaling was active and unaffected by leucine deprivation (Fig. 3E). In these cells expression of wild-type Sestrin2 restored the leucine sensitivity of the mTORC1 pathway, but that of Sestrin2 S190W had no effect (Fig. 3E). Thus, Sestrin2 must be able to interact with GATOR2 for the mTORC1 pathway to sense the absence of leucine.
For leucine to activate mTORC1, Sestrin2 must be able to bind leucine
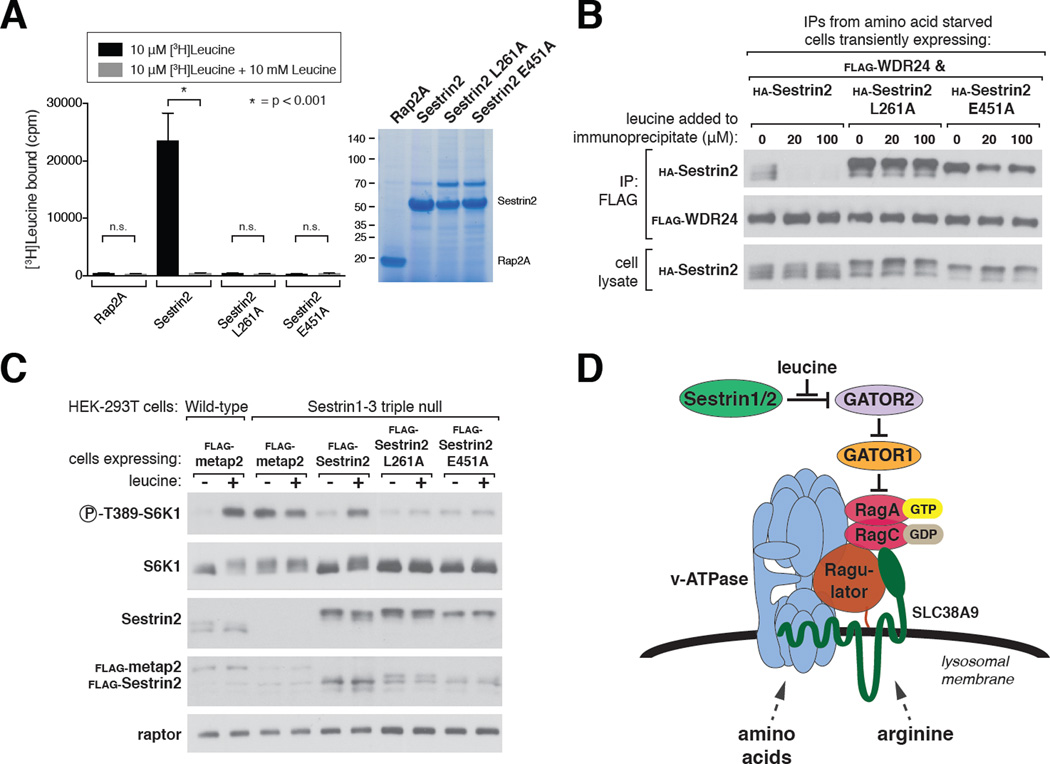
To test whether the leucine-binding capacity of Sestrin2 is necessary for mTORC1 to sense the presence of leucine, we identified two Sestrin2 mutants, L261A and E451A, which do not bind leucine to an appreciable degree (Fig. 4A). Leucine did not significantly affect the interaction of the mutants with GATOR2 in vitro, consistent with Sestrin2 mediating the effects of leucine on the Sestrin2-GATOR2 complex (Fig. 4B). Expression of wild-type Sestrin2 in the Sestrin1-3 triple null cells restored the leucine sensitivity of the mTORC1 pathway in these cells, but that of either mutant inhibited signaling and rendered it insensitive to leucine (Fig. 4C and S3A). Furthermore, in Sestrin1-3 triple null cells expressing the mutants, the localization of mTOR to lysosomes in the presence of leucine was decreased, while that of RagC was not affected (Fig. S4A–D). Thus, activation of mTORC1 by leucine requires the binding of leucine to Sestrin2.
Figure 4. The capacity of Sestrin2 to bind leucine is required for the mTORC1 pathway to sense leucine.
A) The Sestrin2 L261A and E451A mutants do not bind leucine. Binding assays were performed and immunoprecipitates analyzed as in Figure 2A.
B) Leucine-insensitivity of the interactions of Sestrin2 L261A or E451A with GATOR2. FLAG immunoprecipitates were prepared from cells transiently expressing the indicated proteins. The immunoprecipitates were treated with the indicated concentrations of leucine and analyzed as in Figure 1C.
C) In Sestrin1-3 triple null cells expressing Sestrin2 L261A or E451A the mTORC1 pathway cannot sense the presence of leucine. Cells were generated and analyzed as in Figure 3E.
D) Model showing how amino acid inputs arising from multiple sensors in distinct compartments impinge on the Rag GTPases to control mTORC1 activity.
Conclusions
Sestrin2 has several properties consistent with it being a leucine sensor for the mTORC1 pathway: (1) it binds leucine at affinities consistent with the concentrations at which leucine is sensed; (2) Sestrin2 mutants that do not bind leucine cannot signal the presence of leucine to mTORC1; and (3) loss of Sestrin2 and its homologs renders the mTORC1 pathway insensitive to the absence of leucine. Although we have not investigated Sestrin1 as thoroughly, it appears to behave similarly to Sestrin2, so we propose that Sestrin1 and Sestrin2 are leucine sensors upstream of mTORC1.
Given that Sestrin2 has appreciable affinity for methionine, it would not be surprising if in contexts where leucine concentrations are low and those of methionine are high, Sestrin2 may serve as a methionine sensor for the mTORC1 pathway.
Sestrin2 binds to and likely inhibits GATOR2, but how this leads to suppression of mTORC1 awaits elucidation of the molecular function of GATOR2. In addition, structural studies are needed to understand how the binding of leucine to Sestrin2 disrupts its interaction with GATOR2 and why leucine binds very poorly to Sestrin3.
As Sestrin1 and Sestrin2 are soluble proteins, it is likely that they sense free leucine in the cytosol. Although these concentrations are unknown, the Km of the human leucyl-tRNA synthetase (LRS) for leucine has been reported to be 45 µM (36), which is similar to the affinity of Sestrin2 for leucine, suggesting that cytosolic free leucine concentrations are within this range. Like Sestrin2, LRS can bind isoleucine and methionine at lower affinities than leucine (about 30 fold less in the case of LRS) (36). The similarities between the amino acid binding characteristics of Sestrin2 and LRS support the notion that the affinity and specificity of Sestrin2 for leucine are sufficient for it to serve as a leucine sensor.
Our work suggests a model in which signals emerging from distinct amino acid sensors in different cellular compartments converge on the Rag GTPases at the lysosomal surface to regulate mTORC1 activity (Fig. 4D). The putative arginine sensor SLC38A9 likely monitors lysosomal contents and Sestrin2 is almost certainly a cytosolic sensor. There must also be an amino acid sensor upstream of the GAP for RagC and RagD, the FLCN-FNIP2 complex, but its identity and cellular localization are unknown. A future challenge is to elucidate how the Rag GTPases integrate the inputs coming from the different sensors, which will likely require a much better understanding of the function of each Rag in the heterodimer. Moreover, in vivo characterization of the different sensors will be needed to comprehend how specific tissues adapt the amino acid sensing pathway to their particular needs.
Given that Sestrin2 (and Sestrin1) are likely to have leucine-binding pockets, these proteins may be targets for developing small molecule modulators of the mTORC1 pathway. Leucine attenuates the decrease in skeletal muscle protein synthesis that occurs in the elderly and stimulates satiety (1, 37). Thus, small molecules that potently mimic the effects of leucine on Sestrin2 could have therapeutic value. Furthermore, caloric restriction (CR) inhibits mTORC1 signaling (38, 39) and is associated with increases in healthspan and lifespan in multiple organisms (40, 41). Thus, small molecules that antagonize the effects of leucine on Sestrin2 might have CR-mimicking properties.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Sabatini Lab for helpful insights, in particular Shuyu Wang for experimental advice; Olesya Levsh from the lab of Jing-Ke Weng for generously providing the control proteins used in the thermal shift assays; Navitor Pharmaceuticals for providing the His-MBP-TEV-Sestrin2 pMAL6H-C5XT plasmid; and Cell Signaling Technology (CST) for many antibodies. This work was supported by grants from the NIH (R01 CA103866 and AI47389) and Department of Defense (W81XWH-07-0448) to D.M.S., and fellowship support from the NIH to R.L.W. (T32 GM007753 and F30 CA189333) and L.C. (F31 CA180271) and from the Paul Gray UROP Fund to S.M.S. (3143900). K.S. is a Pfizer fellow of the Life Sciences Research Foundation. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
REFERENCES AND NOTES
- 1.Potier M, Darcel N, Tomé D. Protein, amino acids and the control of food intake. Current Opinion in Clinical Nutrition and Metabolic Care. 2009;12:54–58. doi: 10.1097/MCO.0b013e32831b9e01. [DOI] [PubMed] [Google Scholar]
- 2.Panten U, Christians J, von Kriegstein E, Poser W, Hasselblatt A. Studies on the mechanism of L-leucine-and alpha-ketoisocaproic acid-induced insulin release from perifused isolated pancreatic islets. Diabetologia. 1974;10:149–154. doi: 10.1007/BF01219672. [DOI] [PubMed] [Google Scholar]
- 3.Greiwe JS, Kwon G, McDaniel ML, Semenkovich CF. Leucine and insulin activate p70 S6 kinase through different pathways in human skeletal muscle. American journal of physiology. Endocrinology and metabolism. 2001;281:E466–E471. doi: 10.1152/ajpendo.2001.281.3.E466. [DOI] [PubMed] [Google Scholar]
- 4.Nair KS, Schwartz RG, Welle S. Leucine as a regulator of whole body and skeletal muscle protein metabolism in humans. American Journal of Physiology -- Legacy Content. 1992;263:E928–E934. doi: 10.1152/ajpendo.1992.263.5.E928. [DOI] [PubMed] [Google Scholar]
- 5.Layman DK, Walker DA. Potential Importance of Leucine in Treatment of Obesity and the Metabolic Syndrome. 2006 doi: 10.1093/jn/136.1.319S. [DOI] [PubMed] [Google Scholar]
- 6.Frame EG. Journal of Clinical Investigation. 1958;37:1710–1723. doi: 10.1172/JCI103763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper AE, Miller RH, Block KP. Annual Review of Nutrition. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 8.Fox HL, Pham PT, Kimball SR, Jefferson LS, Lynch CJ. Amino acid effects on translational repressor 4E-BP1 are mediated primarily by L-leucine in isolated adipocytes. American Journal of Physiology -- Legacy Content. 1998;275:C1232–C1238. doi: 10.1152/ajpcell.1998.275.5.C1232. [DOI] [PubMed] [Google Scholar]
- 9.Lynch CJ, Fox HL, Vary TC, Jefferson LS, Kimball SR. Regulation of amino acid-sensitive TOR signaling by leucine analogues in adipocytes. Journal of cellular biochemistry. 2000;77:234–251. doi: 10.1002/(sici)1097-4644(20000501)77:2<234::aid-jcb7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 10.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature Cell Biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efeyan A, Sabatini DM. Biochem. Soc. Trans. Vol. 41. Portland Press Ltd.; 2013. pp. 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. Journal of cell science. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 13.Schürmann A, Brauers A, Maβmann S, Becker W, Joost H-G. Cloning of a Novel Family of Mammalian GTP-binding Proteins (RagA, RagBs, RagB1) with Remote Similarity to the Ras-related GTPases. The Journal of biological chemistry. 1995;270:28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 14.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. The Journal of biological chemistry. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 15.Buerger C, DeVries B, Stambolic V. Biochemical and Biophysical Research Communications. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- 16.Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. Journal of biochemistry. 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 17.Sancak Y, et al. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon S, et al. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efeyan A, Zoncu R, Sabatini DM. Trends Mol Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chantranupong L, Wolfson RL, Sabatini DM. Cell. 2015;161:67–83. doi: 10.1016/j.cell.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoncu R, et al. mTORC1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H+-ATPase. Science Signaling. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebsamen M, et al. Nature. 2015 [Google Scholar]
- 25.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, et al. A Tumor Suppressor Complex with GAP Activity for the Rag GTPases That Signal Amino Acid Sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsun Z-Y, et al. The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1. Molecular cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chantranupong L, et al. The Sestrins Interact with GATOR2 to Negatively Regulate the Amino-Acid-Sensing Pathway Upstream of mTORC1. Cell Reports. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parmigiani A, et al. Sestrins Inhibit mTORC1 Kinase Activation through the GATOR Complex. Cell Reports. 2014;9:1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. The Journal of biological chemistry. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 31.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 32.Peeters H, et al. PA26 is a candidate gene for heterotaxia in humans: identification of a novel PA26-related gene family in human and mouse. Human genetics. 2003;112:573–580. doi: 10.1007/s00439-003-0917-5. [DOI] [PubMed] [Google Scholar]
- 33.Buckbinder L, Talbott R, Seizinger BR, Kley N. Gene regulation by temperature-sensitive p53 mutants: identification of p53 response genes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10640–10644. doi: 10.1073/pnas.91.22.10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budanov AV, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 35.Rogez-Florent T, et al. J. Mol. Recognit. 2014;27:46–56. doi: 10.1002/jmr.2330. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, et al. Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic acids research. 2011;39:235–247. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. American journal of physiology. Endocrinology and metabolism. 2006;291:E381–E387. doi: 10.1152/ajpendo.00488.2005. [DOI] [PubMed] [Google Scholar]
- 38.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochimica et biophysica acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallinetti J, Harputlugil E, Mitchell JR. Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochemical Journal. 2013;449:1–10. doi: 10.1042/BJ20121098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. 1989;5:155–171. discussion 172. [PubMed] [Google Scholar]
- 41.Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cellular and molecular life sciences : CMLS. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D-H, et al. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 43.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.