SUMMARY
Influenza-virus antigenicity evolves to escape host immune protection. Antibody lineages within individuals evolve in turn to increase affinity and hence potency. Strategies for a “universal” influenza vaccine to elicit lineages that escape this evolutionary arms race and protect against seasonal variation and novel, pandemic viruses will require directing B-cell ontogeny to focus the humoral response on conserved epitopes on the viral hemagglutinin (HA). The unmutated common ancestors (UCAs) of six distinct, broadly-neutralizing antibody lineages from one individual bind the HA of a virus circulating at the time the participant was born. HAs of viruses circulating more than five years later no longer bind the UCAs, but mature antibodies in the lineages bind strains from the entire 18-year lifetime of the participant. The analysis shows how immunological memory shaped the response to subsequent influenza exposures and suggests that early imprinting by a suitable influenza antigen may enhance likelihood of later breadth.
INTRODUCTION
Influenza virus in humans evolves in response to pressure from immunity in the susceptible population, leading to progressive variation of viral antigenicity. Introduction of a new strain of influenza A from birds or swine (“antigenic shift”) initiates a cycle of antibody generation and viral escape (“antigenic drift”), the latter largely through mutation of surface residues on the viral hemagglutinin (HA) but secondarily through variation of antigenic determinants on the neuraminidase (NA). Detailed antigenic analysis of annual HA variation in H1 and H3 subtypes shows a punctuated evolutionary trajectory, with a shift in “antigenic cluster” (defined by reactivity with standard panels of ferret immune sera) every few years (Smith et al., 2004; Fonville et al., 2014). Strong selective pressure from widespread immunity in the human population thus appears to require more than one seasonal cycle.
The humoral response within individuals also evolves, through immune memory and B-cell affinity maturation. When stimulated by a new exposure (infection or vaccination), memory cells can re-enter germinal centers and undergo new rounds of somatic hypermutation and selection (Victora and Nussenzweig, 2012; De Silva and Klein, 2015). The net effect of this ongoing selection across the entire population exposed to the virus is a virus-immunity “arms race”. Mutated HA with reduced affinity for a particular antibody can in principle select for mutations in the latter that restore strong binding. We can study this evolutionary process by detecting B-cells descended from the same common ancestor and determining the sequences of their rearranged variable-domain genes (Moody et al., 2011).
Antigenic variation requires an annual revision of vaccine components. A more effective vaccine strategy would protect against many rounds of this seasonal variation and ideally against introduction of new serotypes from viruses circulating in animal reservoirs (a so-called “universal” influenza vaccine (Burton et al., 2012; Krammer and Palese, 2015). Broad protection will probably come from a humoral response to conserved sites on the viral HA. The two relatively invariant epitopes so far recognized are the receptor binding site (RBS) on the HA “head” and a surface along the HA “stem” (Knossow et al., 2002; Ekiert et al., 2009; Sui et al., 2009; Corti et al., 2011; Whittle et al., 2011; Corti and Lanzavecchia, 2013). Study of over 100 influenza (subtype H1) receptor binding site (RBS)-directed antibodies from three individuals, all of whom received the trivalent influenza vaccine in 2008 (Moody et al, 2011), has shown that antibodies engage the RBS through contacts that recapitulate many of those made by the viral receptor, sialic acid (Weis et al., 1988; Whittle et al., 2011; Schmidt et al., 2015). The key interactions come from a critical dipeptide (valine-aspartic-acid or a related sequence) at the tip of the third heavy-chain complementarity determining loop (CDR H3). This class of antibodies is nearly unrestricted in VH and VL gene usage; moreover, the lineages show that distinct affinity maturation pathways can lead from a single germline precursor (the unmutated common ancestor: UCA) to functionally similar outcomes. Many of these antibodies came from one individual (designated TIV01); they defined various clonal lineages, each with a unique germline precursor. A suitable set of three or four such antibodies would have in common only contacts with conserved, receptor-interacting amino-acid residues. We proposed that this sort of polyclonal response would approximate the broad immunity to H1 subtypes that a universal vaccine should elicit.
We have chosen six lineages of H1 RBS-directed antibodies from TIV01 and studied the binding of their UCAs and intermediates with members of a panel of HAs from viruses that circulated since that individual was born. We find that the UCAs of all six lineages bind the RBS of an H1 virus circulating in the year of TIV01’s birth (1990), but not the RBS of viruses circulating more than five years later. Certain early intermediates bind the initial strain more tightly, consistent with affinity maturation during a primary response; affinities of later intermediates and the “mature” antibodies from plasmacytes seven days post-vaccination indicate affinity maturation during a secondary response. The lineages gained breadth as they developed, and antibodies from the plasma cells stimulated by the vaccine had not lost affinity for the 1990 strain. The results show that we can study the virus-immunity arms race in humans by sampling an appropriate cohort of individuals and that we can reconstruct patterns of B-cell affinity maturation in those individuals. They further suggest that very early exposure to viral antigens may bias subsequent responses.
RESULTS
TIV01 immune history and RBS-directed lineages
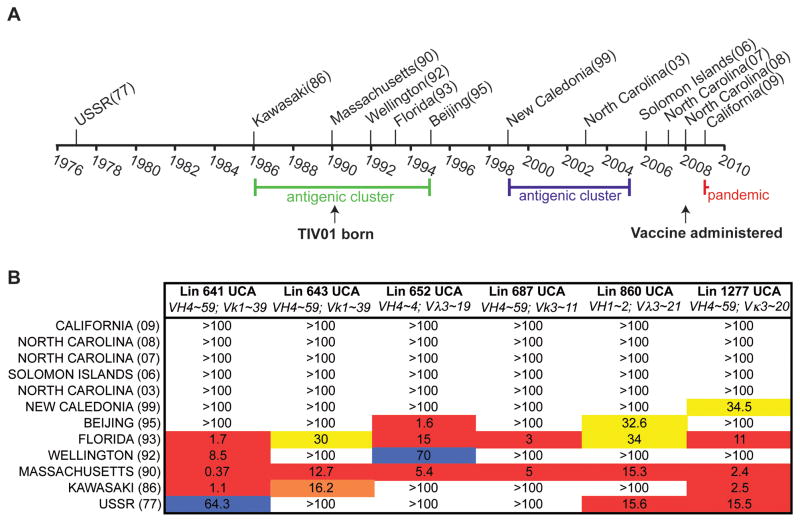
TIV01 was 18 at the time of the 2008 study and reported no previous vaccination history. We prepared a panel of HA1 head domains from H1 strains circulating in various years since 1990 and examined their affinities for the UCAs of antibodies in six lineages of RBS-directed antibodies (Figure 1A and Figure S1). The average VH-gene mutational frequency of all the RBS-directed antibody lineages isolated from TIV01 was ~5%, substantially greater than the likely level of somatic hypermutation during a primary response (Figure S2A, B). We infer that the antibodies identified in the day 7 samples represent a recall response. Mutation could have occurred because of an influenza-virus infection some time between 1990 and 2006 and of course during the response to the vaccine. The initial screen had no bias for HA binding: of the 404 paired sequences recovered from the entire cohort of five individuals at day 7, 252 were positive (by ELISA) for at least one HA in the panel tested (Moody et al., 2011), and Figure S2A, B shows that at least 90% of the 174 HA positive antibodies that came from TIV01 had somatic mutation levels greater than 2.5%. Thus, not only were the lineages we analyzed part of a secondary response, but also the vast majority of all antibodies with detectable HA affinity. A similar analysis of lineages from other individuals in the same cohort also shows an almost exclusively secondary response (Figure S2C). The six chosen lineages represent three different VH genes and five different VL genes (3 Vκ and 2 Vλ) and a total of 56 mature antibodies (Figure 1B). Among the lineages we analyzed is the broadly neutralizing antibody lineage 860, which includes the well-characterized antibodies, CH65 and CH67 (Whittle et al., 2011; Schmidt et al., 2013; Xu et al., 2014; Schmidt et al., 2015).
Figure 1. Timeline of seasonal H1 influenzas and lineage UCA reactivity.
(A) H1 influenza strains from 1977–2010 used in this study. TIV01 birth and administration of the vaccine are marked. (B) Six RBS-directed lineages (Lin) with their VH and VL listed. The seasonal strains and year of isolation (in parenthesis) are on the left. KD values for the Fab binding to monomeric HA heads are in μM. The “heat map” color scheme is arbitrary, as an aid in visualization; warm colors are high affinity interactions (e.g. values <1μM-15μM in red) and lower affinity interactions in cool colors (e.g. values 60–75μM in blue); color changes represent 15μM intervals; affinities marked as >100μM are those beyond the limit of detection. See also Figure S1 and Figure S3.
Germline precursors bind an early seasonal influenza
All six UCAs bound A/Massachusetts/1/1990, which was isolated in the year TIV01 was born (Figure 1B). They also bound A/Florida/2/1993, but most had low affinities for HAs from viruses isolated after 1995, and all failed to bind HAs from 1999 onwards. In no case did the UCA of the lineages bind the H1 A/Solomon Islands/03/2006 included in the vaccine. The loss of affinity corresponds accurately to the “antigenic cartography” of H1N1 strains (Bedford et al., 2014). Viruses between 1986 and 1995 belonged to the same “antigenic cluster”; A/Beijing/262/1995 was a transition to the 1999–2005 cluster that included A/New Caledonia/20/1999. The concordance suggests that during the course of the decade, seasonal influenza viruses had evolved resistance to similar antibodies present in the general population. This conclusion is consistent with an analysis of H3N2 antigenicity from 1968 to 2003, which showed that substitutions around the rim of the RBS determined the principal antigenic changes during that time period (Koel et al., 2013) and hence that antibodies binding in or near the RBS were widespread enough to exert strong selective pressure.
Viral resistance to RBS-directed germline precursors
The one difference between all HAs that bind these UCAs and those that do not is the loss, in 1995, of residue K133a at the edge of the receptor-binding pocket. Deletion of K133a from A/Massachusetts/1/1990 eliminates detectable binding by all six UCAs; insertion of K133a into A/Solomon Islands/03/2006 generates moderate to high affinity for four of the six (Figure S3). We conclude that viruses that had lost K133a escaped infectivity neutralization by antibodies such as those studied here and that the presence of this residue was a principal determinant of neutralization susceptibility.
The germline origins of the six antibody lineages have in common only JH6, which encodes a string of tyrosines immediately C-terminal to the critical dipeptide at the tip of CDR H3. Hydrogen bonds between K133a and the phenolic hydroxyls are probably present in many of the UCA-bound complexes. While the UCAs appear dependent on K133a, subsequent affinity maturation to an immunogen during a secondary exposure has apparently made the antibody response independent of its presence.
Affinity maturation of RBS-directed clonal lineages
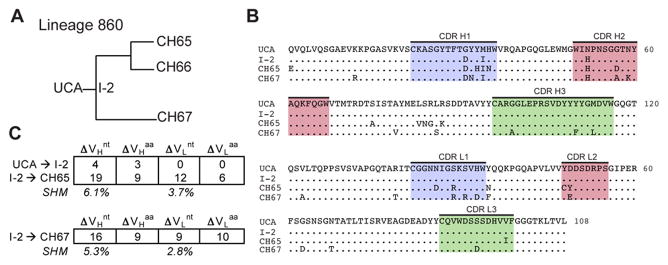
We examined initial steps of affinity maturation in three of the lineages (Figures 2–4) by measuring binding of early intermediates to the 1990 A/Massachusetts and 1993 A/Florida HAs (Figure 5). In lineage 860, somatic mutations encoding three amino-acid residues, all in the heavy chain, accumulated between the germline response and intermediate antibody I-2. Because neither Fab had detectable affinity for A/Solomon Islands/03/2006, we concluded in a previous paper that fixation of the three mutations must have come from enhanced affinity of I-2 for the virus that induced the primary response (Schmidt et al., 2013). Indeed, the data in Figure 5 show that I-2 binds the putative primary antigen (i.e., some strain between 1990 and 1993) substantially more tightly than does the UCA. The limited number of mutations between UCA and I-2 did not, however, impart increased breadth, which presumably came from mutations fixed by exposure to later strains (Figure S4).
Figure 2. Properties of Clonal Lineage 860.
(A) Phylogenetic tree of Lineage 860. (B) Sequence alignment of VH and VL domains for clonal members. CDR 1 (blue), 2 (pink) and 3 (green) are highlighted; (.) denotes conservation in reference to the UCA. (C) Summary of nucleotide (nt) and amino acid (aa) changes in VH and VL. The percent somatic hypermutation (SHM) for the mature antibodies are listed. See also Figure S2 and Figure S4.
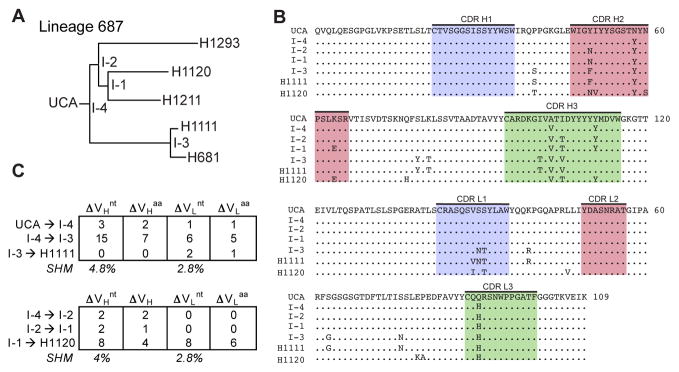
Figure 4. Properties of Clonal Lineage 687.
(A) Phylogenetic tree of Lineage 687. (B) Sequence alignment of VH and VL domains for clonal members. CDR 1 (blue), 2 (pink) and 3 (green) are highlighted; (.) denotes conservation in reference to the UCA. (C) Summary of nucleotide (nt) and amino acid (aa) changes in VH and VL. The percent somatic hypermutation (SHM) for the mature antibodies are listed. See also Figure S2.
Figure 5. Affinity maturation of clonal lineages.
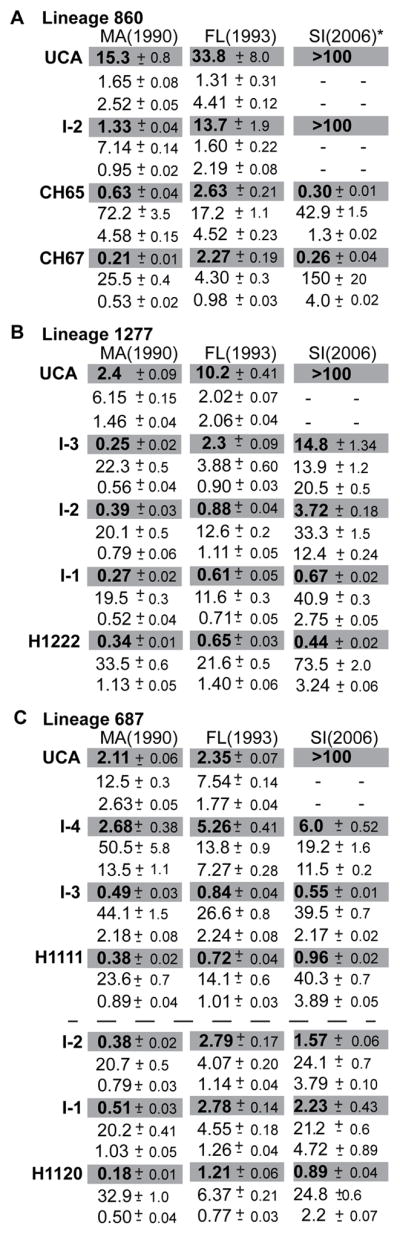
Biolayer interferometric analysis of binding by members of Lineage 860 (A) Lineage 1277 (B) and Lineage 687 (C) to three H1 seasonal isolates—Massachusetts (MA) (1990), Florida (FL) (1993) and Solomon Islands (SI) (2006). KD values (μM) are highlighted in gray; immediately below are the ka (M−1ms−1) and koff (10−2 s−1) values, respectively, that we used to obtain KD. See also Figure S3.
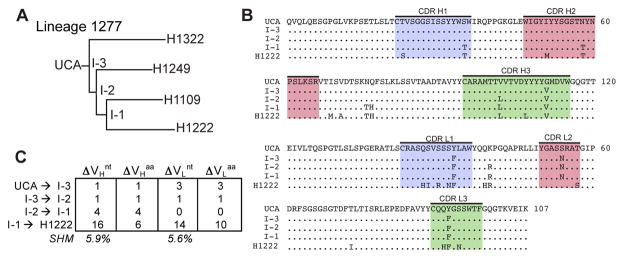
In lineage 1277, increased affinity for A/Massachusetts/1/1990 occurred at the first assignable intermediate, with little affinity gain through two additional intermediates to the contemporary antibody (Figures 3 and 5). Those later intermediates did have successively greater affinities for A/Solomon Islands/03/2006, however, perhaps driven by a post-1995 strain that had lost K133a. The earliest intermediates in lineages 1277 and 687 both bind A/Solomon Islands/03/2006 with moderate affinity, and hence each lineage has acquired some degree of breadth even with only 4 and 3 amino-acid residue changes, respectively (Figures 3 and 4). We have drawn similar conclusions from the more detailed structural and biophysical analyses we have carried out on lineages 641, 643 and 652, which will be the subjects of a separate paper.
Figure 3. Properties of Clonal Lineage 1277.
(A) Phylogenetic tree of Lineage 1277. (B) Sequence alignment of VH and VL domains for the clonal members. CDR 1 (blue), 2 (pink) and 3 (green) are highlighted; (.) denotes conservation in reference to the UCA. (C) Summary of nucleotide (nt) and amino acid (aa) changes in VH and VL. The percent somatic hypermutation (SHM) for the mature antibodies are listed. See also Figure S2.
The UCAs of the six lineages studied had no detectable affinity for the year 2008 trivalent inactivated vaccine HA immunogens, but they all bound tightly with HAs likely to be close relatives of the eliciting viral antigen. Thus, the affinity increase inferred from binding measurements with only the vaccine immunogen overestimates the gain in response to any single exposure, when the course of affinity maturation has involved succession of mutated immunogens. We note that the affinities measured here are for monomeric Fabs and that dimeric IgGs (or IgMs) will show stronger differential effects.
Breadth of RBS-directed clonal lineages
The UCAs of the six lineages bind HAs only from H1 strains isolated around 1990, but a representative, affinity-matured antibody (or late intermediate) from each of the six lineages binds with high affinity to nearly all seasonal H1 HAs in a panel covering the period from 1977 to 2008 (Figure 6). These lineages illustrate how an initially restricted response can broaden after encountering an antigenically distinct HA, if there is a shared, conserved epitope such as the RBS. The enhanced breadth extends even to H1 viruses circulating before 1990: matured antibodies in all six lineages bind more tightly to the 1977 and 1986 strains than do their respective UCAs.
Figure 6. Breadth in RBS-directed clonal lineages.
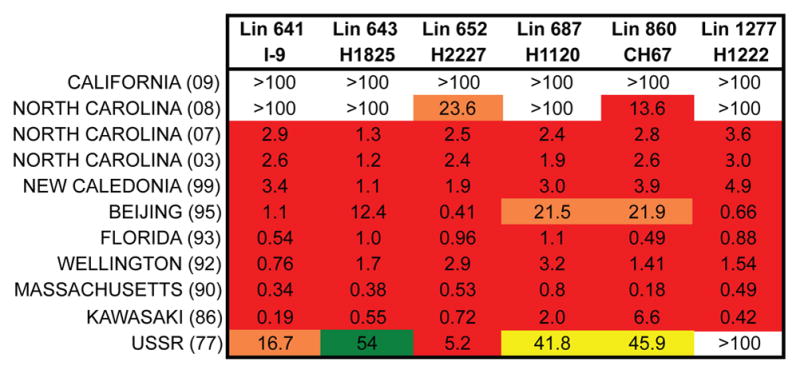
Antibodies from clonal lineages assayed for binding to H1 influenzas. KD values are for Fabs and expressed in μM. The “heat map” color scheme is arbitrary: see legend to Figure 1.
DISCUSSION
The changing antigenicity of circulating influenza viruses and the spectra of human serological responses have been the subjects of an extensive literature since the 1930s (Davenport et al., 1953; Jensen et al., 1956; Kim et al., 2009; Li et al., 2012; Li et al., 2013; Hensley, 2014). By 1960 T. Francis, Jr., could write, based on serological observations, that “the antibody which is first established continues to characterize… [an age] cohort of the population throughout its life” and designated the phenomenon by the colorful phrase “original antigenic sin” (OAS). Subsequent observations of a strong serum response to the most recent exposure appeared at odds with this interpretation, but a contemporary analysis has shown that the highest serological titers in a large, unvaccinated, Vietnamese cohort were indeed for viruses that circulated when the person was young (mean age about 6 years), the likely time frame of a first infection, although that the increases in antibody titers immediately following a new infection were for recent strains (Fonville et al., 2014). Thus, early exposure dominated the steady-state profile, while recent infection determined the immediate response.
Lineage analysis, structures, and affinities of clonally related human antibodies, lineage intermediates, and their UCAs with HAs of viruses circulating during the lifetime of the donor have now allowed us to relate the dynamics of memory, recall, and somatic hypermutation to the previous serological characterizations. The results of this “immuno-viral archeology” show that RBS-directed antibodies from TIV01 lineages amplified by the short-term (7-day) response to vaccination bound strongly to HAs from most of the potential previous exposures but that their UCAs dated to the earliest exposure and lost affinity as the virus mutated. That is, even the immediate post-exposure response, although it had highest affinity for the most recent strains, descended from the first encounter. A large majority of the antibodies from the TIV cohort, including antibodies with epitopes other than the RBS, had somatic mutation levels high enough to indicate a recall rather than primary response (Figure S2 B, C), consistent with the published results of several related studies (Wrammert et al, 2008; Jackson et al, 2014; Tan et al, 2014). The antigenic distance, irrespective of epitope, between influenza virus isolates from well separated time points thus appears to be within a range that strongly favors recall, consistent with less direct inference from serological data (Fonville et al, 2014). Thus, successive influenza-virus infections or vaccinations “update” an existing repertoire, at least for relatively conserved epitopes such as the RBS, but do not appear to add many new components.
Detecting the likely eliciting HA (or a close relative) resolves a puzzle in our previous work on these and other influenza antibody lineages from various subjects. The low affinity of the UCAs for the vaccine immunogen has seemed to imply a weak threshold for the binding needed to activate the originating, naive B-cell. We recognized that the extent of somatic hypermutation indicated a recall response, but we have not had until now an estimate for the HA affinity required to initiate a germinal-center reaction. The dissociation constants for the Fab-HA head interactions reported here correspond, of course, to lower affinities than those expected for a trimeric HA with a dimeric B-cell receptor (IgM or IgG). For example, in cases for which we can compare the dissociation constant (Kd) for Fab-head binding with an avidity estimate from ELISA measurements for the corresponding IgGs and immobilized HAs, we find half-maximal binding from the latter experiments that are lower than the monomer-monomer Kds by between two and three orders of magnitude. The binding of an Fab to a monomeric head is a direct way to relate affinity changes to the structural consequences of mutations in the Fab, since avidity effects, which can depend on the details of the measurement, are difficult to normalize. We have in practice found an excellent correlation between avidity, as measured by ELISA with HA or with head, and affinity of Fab for head, as determined here (Kuraoka et al, submitted).
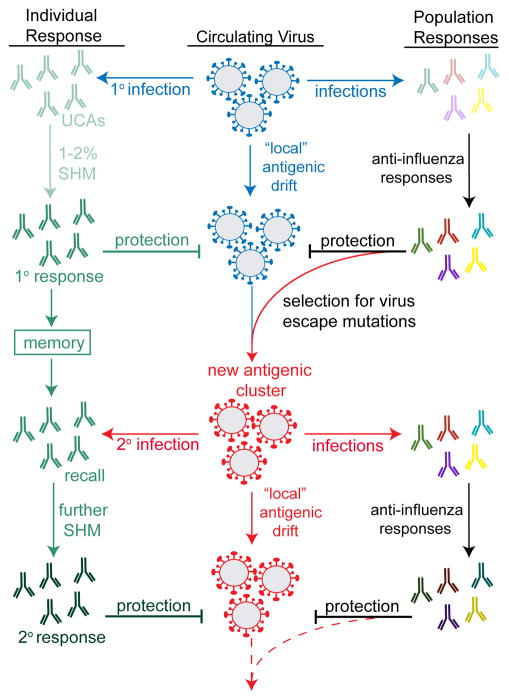
Somatic hypermutation and fixation of mutations over a period of 18 years, presumably by exposure to viruses that had evolved resistance to one or more of the earlier antibodies in the lineages, follows the course expected for co-evolution of influenza virus and responding human antibodies. Determination of single-genome HIV glycoprotein sequences from infected individuals participating in longitudinal studies and concomitant B-cell lineage analyses have enabled studies of the HIV-antibody arms race as it plays out in a single person (Liao et al., 2013; Fera et al., 2014). A more complex relationship dictates influenza virus and human humoral immunity co-evolution, which proceeds by episodic exposure (from vaccination or occasional infection) of an individual immune system to antigen and continuous exposure of the circulating virus to herd immunity (Figure 7). Nonetheless, our results suggest that it should be possible to carry out studies of the arms race between influenza virus and the collective immune response of the sampled population.
Figure 7. Co-evolution of virus and antibody response.
Left column: Episodic exposure to influenza virus governs evolution of the antibody response (eliciting naive response and maturing that response by affinity maturation in germinal centers). Center column: Essentially continuous selection for mutations that escape host immunity governs evolution of viral epitopes targeted by protective antibodies in the population as a whole. Right column: collective immunity is an aggregate of many individual responses. The data presented in this paper suggest that a small number of individual responses may be enough to give a representative sample of the population response. Punctuated evolution of influenza HA antigenicity, documented for both H1 and H3 influenzas (Smith et. al., 2004, Bedford et. al., 2014), defines chronologically successive antigenic clusters; the color change in the “virus” icon from blue to red symbolizes a shift from one cluster to the next. Increasing color intensity of the antibody icons represents increased affinity. SHM: somatic hypermutation.
Exposure of TIV01 to an H1 influenza virus circulating during infancy (1990–1993) elicited B-cell memory that dominated the response to a trivalent vaccine administered in 2008 (Moody et al, 2011). More than 60% of the heavy-chain variable regions cloned from sorted B-cells taken seven days post-vaccination have the sequence hallmarks of RBS-directed antibodies, and 50% belong to one of the six lineages studied here. The large number of distinct lineages of RBS-directed antibodies found in TIV01 is unusual, however. While RBS-directed antibodies may be relatively common (Schmidt et al, 2015), other individuals probably have at most one or two, making antigenic drift toward resistance possible. Indeed, the data in Figure 6 show that by 2008, a circulating virus (A/North Carolina/AF1292/2008) no longer bound antibodies from four of the six lineages. One objective for an improved influenza vaccine might be to increase the frequency of multiple RBS-directed antibodies in a large enough fraction of a population to hinder accumulation of such mutant viruses.
The observations reported here are consistent with the notion that viral antigens seen very early in life, as an infant’s immunity develops, pre-condition subsequent responses by biasing the memory repertoire more strongly that do antigens first seen at later times. This outcome would be similar to imprinting of the B-cell response to bacterial antigens by the early intestinal microbiome, as seen in experiments with mice (Hooper et al., 2012). In a recently published study of a donor who received the 2011 trivalent vaccine, four different day 7 antibodies that neutralized the pdm2009 component of the vaccine also neutralized the A/USSR/90/1977 isolate, suggesting priming by a strain closely related to the 1977 virus (Huang et al, 2015). The epitope recognized by those antibodies is on the head, but in a surface patch well separated from the RBS. The A/USSR/90/197 virus, which represents influenza viruses circulating during the donor’s childhood, was also the first reappearance of a seasonal H1 after its displacement by H2 in 1957. It would have been the earliest H1 strain that the donor could have seen. The published study did not include analysis of the properties of the UCAs of the day 7 antibodies, as we have done here for the six RBS-directed lineages from TIV01, and any inference that the germline precursors of the antibodies in that work would recognize the 1977 strain is indirect. Nonetheless, its results suggest that our findings from TIV01 apply more broadly, as they come from an individual over a decade older, who received a different trivalent vaccine, and they involve a different epitope.
The concept of “B-cell lineage vaccine design” underlies much of current research on vaccines for HIV, influenza, and other rapidly evolving pathogens (Haynes et al., 2012; Jardine et al., 2015). This approach assumes (1) that dominant responses to exposed, variable epitopes outcompete broadly neutralizing, subdominant responses to more cryptic, conserved epitopes and (2) that within the naive B-cell repertoires of most individuals in a population are cells that can respond to those conserved epitopes when appropriately stimulated. A stimulus that might, in principle, increase the likelihood of eliciting an otherwise subdominant response is a modified immunogen that binds with high affinity to the UCA of a lineage with broadly neutralizing potential. In the lineages studied here, the apparent priming stimulus was infection with a seasonal strain during the year after the person was born, with potential boosts from subsequent exposures between 1993 and 2008 and a known one from the 2008 vaccine. The boost(s) led to further affinity maturation, with development of increased breadth; analysis of the intermediates in certain lineages suggests where affinity maturation to the primary exposure ceased and where the secondary exposure(s) took over. Our observation, that early exposure appears to have biased immunological memory in this individual, suggests that vaccination of infants with HA immunogens selectively exposing conserved epitopes might be a constructive step toward a vaccine that elicits long-lasting immunity.
EXPERIMENTAL PROCEDURES
Subjects
TIV01 was recruited at Duke University and given the trivalent inactivated seasonal influenza vaccine (TIV) 2007–2008 containing A/Solomon Islands/3/2006 (H1N1), A/Wisconsin/67/2005 (H3N2), and B/Malaysia/2506/2004, as described previously (Liao et al., 2009; Moody et al., 2011).
Lineage analysis
The lineages analyzed here have been described and characterized previously (Moody et al., 2011; Schmidt et al., 2015). The phylogenic tree for each clone was estimated using the dnaml program from Phylip 3.69 package, and the UCA was obtained by aligning the inferred sequence at the root to germline gene libraries. The tree was then recomputed with the UCA at the root, and the sequences of intermediates at branch-points in that tree were inferred as described (Kepler, 2013). The degree of somatic hypermutation was such that we found no ambiguity in assigning from the IMGT database the alleles and sequences for heavy-chain V, D and J and light-chain V and J. The only potential uncertainties were at the V-D and D-J boundaries in the heavy chain, where residues encoded by potential n nucleotides cannot be derived from genome sequences. Five of the six clones had so few mutations in CDR H3 that the inferred UCA sequences were unambiguous; for the sixth (641), we followed the analysis in Moody et al., 2011. If the UCA sequence (the root of the tree) and sequences of the day 7 antibodies (the twigs of the tree) are known, then the inferred sequences of intermediates are essentially unambiguous. Paired-chain sequenced used in the original study ensures correct assignment of the heavy- and light-chain combination.
Expression and purification of HA
All rHA “head” constructs were cloned into pFastBac vector for insect cell expression. All contained a cleavable C-terminal His6X tag and were purified from supernatants by passage over Co-NTA agarose (Clontech) followed by gel filtration chromatography on Superdex 200 (GE Healthcare) in 10 mM Tris-HCl, 150 mM NaCl at pH 7.5. The tag was removed using PreScission protease (MolBioTech, ThermoScientific) and the protein repurified using Co-NTA agarose to remove the protease, tag and non-cleaved protein. All mutations were made using QuikChange Mutagenesis (Agilent).
Fab expression and purification
The genes for the heavy- and light-chain variable domains were synthesized and codon optimized by Integrated DNA Technologies or GenScript and subcloned into protein expression vectors containing human heavy- and light-chain constant domains. All heavy-chain constructs contained a non-cleavable His6X tag. Fabs were produced by transient transfection in HEK 293T cells using Lipofecatamine 2000 (Invitrogen). Supernatants were harvested five days later, clarified by centrifugation and the Fabs were purified using Co-NTA agarose (Clontech) followed by gel filtration chromatography on Superdex 200 (GE Healthcare). The buffer purification buffer was 10 mM Tris-HCl, 150 mM NaCl at pH 7.5. All constructs were confirmed by DNA sequencing at the DNA Sequencing Core Facility at Dana Farber Cancer Institute.
Interferometry and binding experiments
Interferometry experiments were performed using a BLItz instrument (forteBIO, Pall Corporation). Purified Fab was immobilized on a Ni-NTA biosensor, and cleaved rHA heads were titrated to obtain binding kinetics and affinities. KD values were obtained by applying a 1:1 binding isotherm using vendor-supplied software under the “Advanced Kinetics” program. UCA binding was tested at an initially high protein concentration of rHA head (75μM); for detectable binding, rHA was then titrated at three different concentrations (chosen depending on the apparent KD from the high concentration); a global fit to the titration curves was applied to obtain the KD. For mature antibodies, a single high concentration (25μM) was used to obtain an approximate KD value. All experiments were performed in 10 mM Tris-HCl, 150 mM NaCl at pH 7.5 and at room temperature.
Supplementary Material
Acknowledgments
We thank members of the Harrison Laboratory for discussions. The work at Harvard Medical School and Boston Children’s Hospital was supported by NIH Grant P01AI089618. SCH is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions A.G.S. and S.C.H. designed research; A.G.S., K.T.D., K.R.M., T.B.K., H.X.L, M.A.M., performed research; A.G.S., K.T.D., K.R.M., T.B.K., H.X.L, M.A.M., B.F.H., and S.C.H. analyzed data; A.G.S. and S.C.H. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, Rambaut A. Integrating influenza antigenic dynamics with molecular evolution. eLife. 2014;3:e01914. doi: 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annual review of immunology. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Davenport FM, Hennessy AV, Francis T., Jr Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. The Journal of experimental medicine. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nature reviews Immunology. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NM, Pham QT, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature biotechnology. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Current opinion in virology. 2014;8:85–89. doi: 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KY, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, Daniels RS, Townsend AR. Focused antibody response to influenza linked to antigenic drift. Journal of clinical investigation. 2015;125:2631–2645. doi: 10.1172/JCI81104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Mooy MA, Haynes BF, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host and Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KE, Davenport FM, Hennessy AV, Francis T., Jr Characterization of influenza antibodies by serum absorption. The Journal of experimental medicine. 1956;104:199–209. doi: 10.1084/jem.104.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Research. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol. 2009;183:3294–3301. doi: 10.4049/jimmunol.0900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M, Gaudier M, Douglas A, Barrere B, Bizebard T, Barbey C, Gigant B, Skehel JJ. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology. 2002;302:294–298. doi: 10.1006/viro.2002.1625. [DOI] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nature reviews Drug discovery. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. B-cell selection in germinal centers elicited by complex antigens. 2015 doi: 10.1016/j.immuni.2016.02.010. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. The Journal of experimental medicine. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PloS one. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YC, Blum LK, Kongpachith S, Ju CH, Cal X, Lindstrom TM, Sokolove J, Robinson WH. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clinical Immunology. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Schmidt AG, O’Donnell T, Therkelsen MD, Kepler TB, Moody MA, Haynes BF, Liao H, Harrison SC, Shaw DE. Key mutations stabilize antigen-binding conformation during affinity maturation of a broadly neutralizing influenza antibody lineage. Proteins. 2014 doi: 10.1002/prot.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.