Abstract
Deinococcus radiodurans (Drad) is the most radioresistant organism known. Although mechanisms that underlie the extreme radioresistance of Drad are incompletely defined, resistance to UV irradiation-induced killing was found to be greatly attenuated in an NO synthase (NOS) knockout strain of Drad (Δnos). We now show that endogenous NO production is also critical for protection of Drad against γ-irradiation (3000 Gy), a result of accelerated growth recovery, not protection against killing. NO-donor treatment rescued radiosensitization in Δnos Drad but did not influence radiosensitivity in wild type Drad. To discover molecular mechanisms by which endogenous NO confers radioresistance, metabolite profiling studies were performed. Untargeted LC-MS-based metabolite profiling in Drad quantified relative abundances of 1,425 molecules and levels of 294 of these were altered by >5-fold (p< 0.01). Unexpectedly, these studies identified a dramatic perturbation in carotenoid biosynthetic intermediates in Δnos Drad, including a reciprocal switch in the pathway end-products from deoxydeinoxanthin to deinoxanthin. NO supplementation rescued these nos deletion-associated changes in carotenoid biosynthesis, and fully-restored radioresistance to wildtype levels. Because carotenoids were shown to be important contributors to radioprotection in Drad, our findings suggest that endogenously-produced NO serves to maintain a spectrum of carotenoids critical for Drad’s ability to withstand radiation insult.
INTRODUCTION
D. radiodurans (Drad) is an extremeophilic bacterium that is remarkable for its capacity to withstand exposure to extreme environmental stress, including desiccation, oxidants, ultraviolet and ionizing radiation [1–5] This non-pathogenic and non-photosynthetic bacterium has gained particular notoriety as the most radioresistant organism known, able to withstand >10,000 Gy of ionizing radiation [2,5–7]. The extreme radioresistance of Drad is thought to arise from a synergy of multiple cellular defense mechanisms, including an extremely efficient system for repairing double-strand DNA breaks, high antioxidant activity, unusual cell envelope protective structure, and mechanisms that evolved to preserve protein functions.
Radiation insult can damage DNA, proteins, lipids and other macromolecules directly, and also via secondary radiation-induced reactive oxygen species (ROS) such as the hydroxyl radical [8,9]. Irradiation insult and secondary ROS cause single-and double-strand DNA breaks that, if repaired improperly or left unrepaired, can lead to mutation, genomic instability, and cell death [9–12]. Drad has highly efficient enzymatic DNA repair processes that allow for the rapid and unusually error free reassembly of DNA fragments caused by double strand DNA breaks [13–15]. However, the efficacy of these repair processes is contingent upon the preservation of enzymatic activities. Thus, protection of proteins from oxidation is a major determinant of radioresistance in Drad, and ROS-scavenging mechanisms additionally play a vital role in response to various environmental stressors. [16–18]. Consequently, Drad maintains powerful antioxidant mechanisms that prevent oxidation of proteins and thereby preserves the activity of DNA repair enzymes [16,18,19]. These mechanisms include efficient enzymatic ROS scavenging systems as well as small molecule antioxidants [20–22]. Indeed, Drad has developed powerful enzymatic mechanisms capable of detoxifying reactive species, mediated by scavenging enzymes such as superoxide dismutase, catalase, and peroxidase [19,23,24]. Exposure to radiation has been shown to induces expression of the above enzymes in Drad, and mutation of their cognate genes can result in increased sensitivity to radiation insult [20].
Surprisingly, incubation with ultrafiltered, protein-free Drad cell extract was shown to prevent oxidation of proteins in E. coli following exposure to extreme doses of ionizing radiation [22]. This latter finding suggests that small molecule antioxidants also comprise an element of Drad’s radio-defense systems. Notably, Drad contains C40 carotenoid pigments that give the bacterium its characteristic reddish-pink color and some of these carotenoid molecules are unique to Drad [25–27]. These long-chain, unsaturated terpenoids exhibit strong antioxidant properties in Drad, scavenging ROS and likely contributing significantly to radioresistance. Carotenoids are responsible for many of the colors of plants, animals, and microorganisms, functioning as accessory pigments in photosynthetic systems and playing vital roles in photoprotection that contributes to membrane fluidity and antioxidant defenses [28,29]. Drad synthesizes the unique carotenoid deinoxanthin from isoprenoid units, via a series of reactions catalyzed by carotenoid biosynthesis (Crt) enzymes [26]. It is notable that Drad mutants that are colorless due to a carotenoid synthesis deficiency exhibit enhanced sensitivity to ionizing radiation and ROS-induced oxidative damage, highlighting the importance of these membrane-localized pigments as potential contributors to Drad radioresistance mechanisms [21,30–33]. Deinoxanthin in particular has potent ROS-scavenging activity, as demonstrated by its efficient ability to quench singlet oxygen and hydroxyl radicals (Lemee et al 1997, Ji 2010, Carbanou 1989) and accordingly has been implicated as a major determinant of radioresistance in Drad. However, the deinoxanthin biosynthesis pathway in Drad and other carotenoid intermediates are incompletely characterized in terms of molecular identity and contribution to ROS scavenging efficiency. Moreover, regulatory mechanisms that control carotenoid biosynthesis in Drad are undefined.
In sum, the current dogma regarding the extreme radioresistance of Drad is that multiple passive and enzymatic antioxidant mechanisms serve to mitigate radiation-induced oxidative damage, preserve enzyme activities, and both efficiently and precisely promote the repair of radiation-induced DNA damage [14,16,17,22,34,35]. Yet, despite recent advances in our understanding of the physiological mechanisms underlying the enormous capacity of Drad to withstand radiation insult, underlying molecular mechanisms and regulatory processes await definition.
Despite 2 billion years of evolutionary separation from mammals, Drad is unusual for its expression of a nitric oxide synthase (NOS) hemi-enzyme that is highly homologous in sequence and structure to the oxygenase domain found in mammalian NOS isoforms [36,37]. Surprisingly, knock out of Drad nos (Δnos,) was found to sensitize Drad to UV irradiation-induced toxicity [36,37]. Indeed, following exposure to UV radiation (30m/cm2), Δnos Drad recover and replicate markedly slower and to a lesser extent, as compared with wild type Drad. In accord with the possibility that NO may also promote radioresistance in mammals, NOS inhibitor treatment has been shown to sensitization some mammalian tumors to the cytotoxic effects of ionizing radiation [38,39]. Together, these findings suggest that endogenously produced NO in both bacteria and mammals can afford protection against radiation-induced cell damage.
In attempt to uncover cytoprotective mechanisms in Drad, we employed an untargeted metabolite profiling to assess how loss of endogenous NO production impacts the small molecule composition of this bacterium. Notably, metabolomics offers a powerful “-Omics” approach for discovering differences in the expression of small molecules in complex biological matrices [40–42]. We reasoned that observed changes in Drad metabolite abundances that arise after loss of endogenous NO biosynthesis, can shed new light on the molecular basis for NO-mediated radioresistance.
RESULTS
Deletion of nos increases susceptibility of Drad to extreme ionizing radiation and this effect is rescued by exogenous NO supplementation
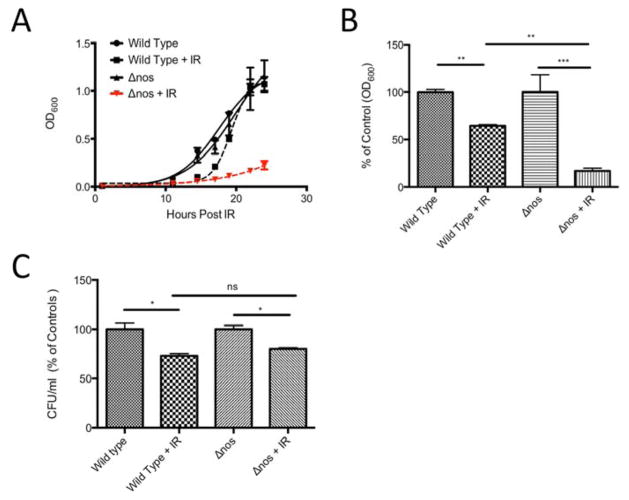
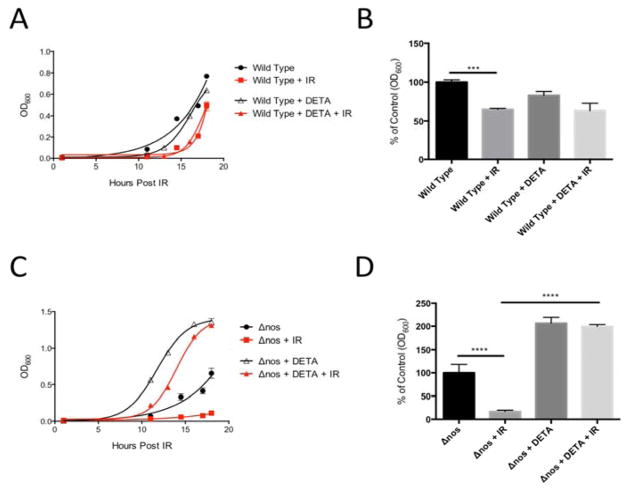
Following an 18 h exposure to extreme ionizing radiation from a γ-emitter (3000 Gy total dose), Δnos viable Drad decreased significantly, relative to wild type Drad (Fig 1A). Indeed, cell density measurements (OD600nm) showed that irradiated Δnos Drad decreased in abundance by over 83% post-irradiation, compared to non-irradiated controls and this recovery was more pronounced than in radiation-exposed wild type nos Drad, where cell density was reduced by only 35% (Fig. 1B). In contrast, the percentage of viable cells remaining after irradiation, measured as colony forming units (CFU), was not significantly different between wild type and Δnos strains (Fig. 1C). Thus, nos gene deletion does not appear to influence the extent of cell death in response to γ-irradiation, but rather suppresses the ability of Drad to recover after an γ-radiation insult, as previously shown for UV-irradiation [37]. Remarkably, exogenous NO supplementation with the NO-donating chemical DETA NONOate (25 μM) restored post-irradiation growth of Δnos to a degree that was equal to or greater than observed in non-irradiated controls (Fig. 2). In contrast, NO supplementation in nos-intact Drad did not influence growth recovery following irradiation-insult (Fig. 2). Together, these findings suggest that endogenous NO production protects against γ-irradiation-induced growth arrest in Drad, and the level of endogenous NO synthesis in wildtype Drad is sufficient for maximal radioprotection.
Figure 1. Δnos is more susceptible to ionizing radiation than wild type Drad.
Panel A: Growth curves (monitored by OD600) are depicted. Dashed lines indicate growth of wild type nos and Δnos Drad following exposure to extreme ionizing radiation (γ-radiation, 3000 Gy). Cultures were grown to OD 0.8–1.0 and irradiated followed by 1:100 dilution in fresh TGY media and monitoring of growth. Panel B: Relative growth of Δnos and wild type nos Drad with and without 18hrs post-irradiation. Panel C: The percentage of viable cells in post-irradiated samples from Panel B was determined based on colony forming assays (CFU/ml) and compared to controls. Data are plotted as mean +/− SD of three biological replicates. Statistical significance was assessed using one-way ANOVA and Tukey post-hoc testing. (*p<0.05, **p<0.01, ***p<0.001).
Figure 2. Exogenous NO rescues irradiation-induced growth defects in Δnos Drad.
Panel A and B: Following exposure to ionizing radiation (γ-radiation, 3000 Gy), supplementation of wildtype Drad with exogenous NO (treatment with 25 μM DETA NONOate) had no significant effect on growth recovery. Panel C and D: In contrast, exogenous NO treatment increased Δnos Drad growth to a level greater than that of non-irradiated controls and opposed the growth-suppressing action of ionizing radiation. Data are plotted as mean +/− SD of three biological replicates. The relative growth of Δnos and wild type nos Drad compared to controls in panels B and D was evaluated statistically at 18hrs post-irradiation using one-way ANOVA and Tukey post-hoc testing. (*p<0.05, **p<0.01, ***p<0.001).
Comparative metabolite profiling of wild type vs. Δnos Drad
Untargeted metabolite profiling was performed to assess the metabolomic consequences of nos gene deletion in Drad. Toward this end, untargeted metabolite profiling was performed essentially as described previously [40–42], employing C18 reverse phase (RP) chromatography for efficient resolution of hydrophobic molecules that were detected using ESI-MS (see metabolomic workflow in Fig. 3). This analytical strategy allowed separation, detection and relative quantification of diverse small molecule metabolites (50–1000 Da) for downstream analysis using chemometric analytical methods. These studies quantified relative abundances of 1,648 molecular features across all wild type nos and Δnos Drad extracts (Fig. 4). Considering only those features that could be detected in at least 80% of samples from at least one experimental group limited this dataset to 1,425 features, among the 1,648 total features observed across all samples (Fig. 4A). These molecular species were further considered for differential expression using chemometric profiling tools and algorithms (MassHunter Profinder in combination with MassProfiler Professional; Agilent Technologies, Santa Clara, CA).
Figure 3. Untargeted metabolite profiling workflow.
Δnos and wild type nos Drad cultures were harvested and subjected to metabolite profiling as depicted. Upon isolation, metabolism was rapidly quenched and metabolites were extracted by immersion in −70°C methanol (100%), followed by bead-assisted homogenization. Metabolite extracts were vacuum concentrated and stored at −80°C until LC-MS analysis. Residual tissue pellets were retained for protein assay and protein levels were used to normalize LC-MS injected metabolite extracts. Dried down extracts were resuspended in 100% methanol + 0.1% formic acid and an equivalent of 6.0 μg protein was injected for metabolite profiling and chemometric analysis, as described in Materials & Methods. Chemometric profiling, multivariate analysis, and pathway analysis were conducted using MassProfiler Professional B12.0 software (Agilent Technologies).
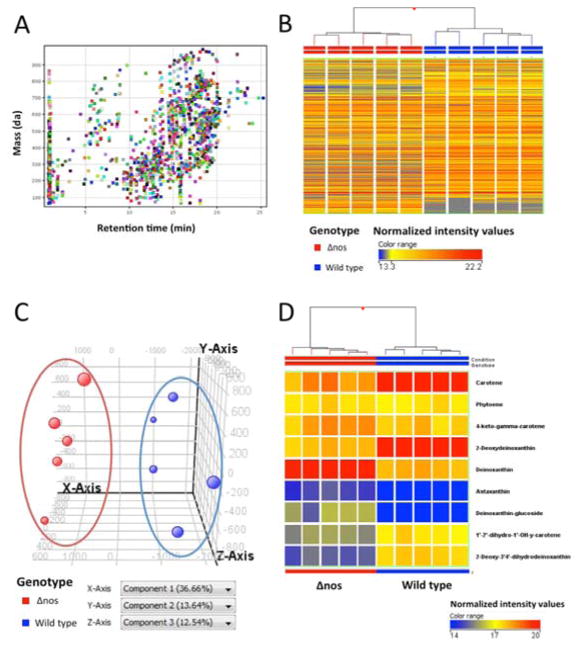
Figure 4. Untargeted metabolite profiling comparison of Δnos and wild type nos Drad.
Depicted LC-MS data were acquired using reverse phase (RP) chromatography and positive ion MS detection. Metabolite extracts of 5 independent cultures from each of wild type nos and Drad Δnos were analyzed. Panel A: Plot of retention time vs. neutral mass depicting 1,425 distinct features that were observed in at least 80% of samples from at least one group. Color coding of features is arbitrary, serving only to aid in visual discrimination. Panel B: Unsupervised Hierarchical Clustering Analysis (HCA), with samples color-coded by genotype, displays the expression pattern and clustering of these 1,425 metabolites quantified. Each column represents a single sample with horizontal hash marks representing metabolites and their relative abundances, color-coded by normalized expression values with red greatest and blue least. HCA was clustered according to a Euclidean distance metric and Ward’s linkage rule. Panel C: Principal Component Analysis (PCA) plot showing 3D visualization of similarities and differences in the metabolite composition of each Drad sample. Each data point corresponds with a single sample and is colored by genotype, representing the projection of 1,425 features considered in the first three principal components (PC). Principal component 1 (X-axis) accounted for over 36.66% of the variability between metabolite samples and principal component 2 (y-axis) and 3 accounted for 13.64% and 12.54%, respectively. As depicted by HCA and PCA, Δnos clustered independently of wild type samples. Panel D. Unsupervised HCA considering only molecules tentatively identified as carotenoids, with samples color-coded by genotype. Notably, 7 carotenoids in Drad’s biosynthesis pathway were found to be differentially-expressed between Δnos and wild type nos Drad, as assessed by unpaired t-test (p<0.01, see Table 1).
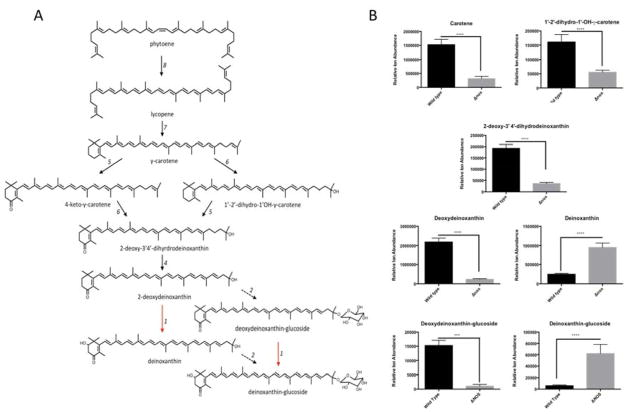
The 1,425 observed features were further assessed by unsupervised pattern recognition and statistical analyses. Unsupervised Hierarchical Clustering analysis (HCA) and Principal Component Analysis (PCA) were performed to test for group differences in metabolite profiles. As seen in the HCA dendrogram (Fig. 4B), and 3-dimensional PCA plot (Fig. 4C), samples from Δnos Drad clustered closely - yet were distinct from wild type nos expressing Drad. By investigating loading scores for the PCA plot, one can identify metabolites that contribute to the greatest extent in separating the groups along each principal component axis. Applying this strategy, we observed differential expression of many tetraterpenoids (including C40 carotenoids) dominantly contributed to the ability of PCA to distinguish Δnos from wild type Drad. Loading scores for PC1 (x-axis) and PC2 (y-axis) indicated that molecules tentatively identified as carotene (C40H56), deoxydeinoxanthin (C40H54O2), deinoxanthin (C40H54O3), and deinoxanthin-glucoside (C46H64O8) were among the features contributing most to the separation between Drad genotype differences (Fig. S1). Unsupervised HCA, considering only known carotenoid biosynthesis pathway metabolites, further indicated global perturbations in carotenoid composition associated with nos-deletion (Fig. 4D). Notably, we also observed alterations in the abundance of molecules whose formulae matched metabolic intermediates that have been reported as unique to the carotenogenesis pathway of Drad [26], including, 2-deoxy-3′,4′-dihydrodeinoxanthin (C40H56O2), deoxydeinoxanthin (C40H54O3), and deinoxanthin (C40H54O3) (see Fig. 5).
Figure 5. Carotenoid biosynthesis pathway in wildtype Drad and apparent perturbation in.
Δnos Drad.
Panel A: Solid black arrows refer to carotenoid biosynthesis (Crt) enzymes and biosynthesis steps that have been experimentally confirmed; adapted from (Tian and Hua, 2010). Red arrows indicate the hypothetical reactions catalyzed by a putative 2-hydroxylase that is presumably responsible for the conversion of deoxydeinoxanthin to deinoxanthin. Dashed black arrows refer to a glucosyltransferase enzyme in Drad whose function and metabolic products was described in this work, but which has yet to be fully characterized. 1: Putative 2-Hydroxylase; 2: enzyme with homology to glucosyltransferase in Chlorobium tepidum (CruC; DR_0089); 3: enzyme with homology to acetyltransferase in Chlorobium tepidum (CruD; DR_0090); 4: C-3′,4′ desaturase (CrtD; DR_2250); 5: Carotene ketolase (CrtO: DR_0093); 6: C-1′-2′ hydratase (CruF: DR_0091); 7: Lycopene cyclase (CrtLm: DR_0801); 8: Phytoene desaturase (CrtL; DR_0861). Panels B & C: Relative abundances of carotenoids known to be involved in D. radiodurans carotenoid biosynthesis. Notably, deoxydeinoxanthin was the predominant carotenoid species observed in wild type, whereas deinoxanthin was the predominant species in Δnos Drad. Panel D: Unpaired t-test results are depicted for carotenoid metabolites from Table 1 that were found to have statistically significant differences in abundance in nos vs. wild type Δnos Drad. Error bars are mean ion counts +/− SD, compared by unpaired t-test (* p<0.05, **p<0.01, ***p<0.001).
Differentially expressed metabolites in Δnos Drad
Unpaired t-tests, corrected for multiple hypothesis testing, confirmed diverse and significant nos deletion-associated alterations in the abundance of carotenoids. Indeed, these studies recognized 294 features, defined by chromatographic retention times, accurate mass and ion abundances, that were significantly altered in expression level by >5-fold, with p< 0.01. These differentially-abundant molecules comprised many membrane components such as glycerophospholipids, phosphoethanolamines and carotenoid pigments. Among these 294 features with significant differences between groups, 9 molecules were tentatively identified as elements in the proposed carotenoid biosynthesis pathway of Drad (Unpaired t-tests, p < 0.01; see Table 1).
Table 1.
Differentially expressed carotenoid molecules in Δnos Drad compared to wild type Drad.
| Compound | Altered | Retention | ||||
|---|---|---|---|---|---|---|
| p (Corr) | in Δnos | Fold Change | Time | Mass | Formula | |
| Phytoene | 2.88E-01 | N.S.* | 1.09 | 18.78 | 544.501 | C40 H56 |
| Carotene | 3.53E-06 | down | −4.97 | 18.54 | 536.447 | C40 H56 |
| 4-keto-γ-carotene | 4.94E-02 | N.S. | 1.41 | 19.01 | 550.413 | C40 H54 O |
| 1′-2′-dihydro-1′-OH-y-carotene | 3.53E-06 | down | −2.90 | 17.97 | 554.457 | C40 H58 O |
| 2-Deoxy-3′, 4′-dihydrodeinoxanthin | 5.94E-08 | down | −5.44 | 18.07 | 566.415 | C40 H56 O2 |
| 2-Deoxydeinoxanthin | 1.29E-08 | down | −9.90 | 18.07 | 566.415 | C40 H54 O2 |
| Deinoxanthin | 1.77E-07 | up | 3.94 | 16.92 | 582.410 | C40 H54 O3 |
| Astaxanthin | 2.42E-06 | up | 13.98 | 15.95 | 596.396 | C40 H52 O4 |
| Deinoxanthin-glucoside | 1.75E-06 | up | 10.16 | 15.33 | 744.471 | C46 H64 08 |
not significant
Carotenoid biosynthetic pathways are depicted in Fig. 5A. Remarkably, marked differences in the abundance of 7 of these carotenoid pathway intermediates was evident in wild type nos vs. Δnos expressing Drad strains (Fig. 5B). Unpaired t-test results showed that levels of carotene (C40H56), 2-deoxy-3′,4′-dihydrodeinoxanthin (C40H56O2), deoxydeinoxanthin (C40H54O2), and deinoxanthin (C40H54O3) were significantly altered by nos-deletion (p < 0.001). Unexpectedly, these findings revealed that deoxydeinoxanthin is the predominant carotenoid in the wild type Drad, whereas, deinoxanthin is the majority carotenoid constituent in Δnos Drad. Indeed, according to these relative abundance values, wild type Drad had a deoxydeinoxanthin: deinoxanthin ratio of 11:1 and this ratio was inverted to 1:7 in Δnos Drad. Additionally, unpaired t-tests identified a compound corresponding in mass to the glucoside derivative of deoxydeinoxanthin (C46H64O7) and this species was significantly less in Δnos Drad (p < 0.001). Instead, deinoxanthin-glucoside (C46H64O8) was found to be the predominant glucoside species in nos-deleted Drad (p < 0.001, Figure 5B). With the exception of deinoxanthin (C40H52O4), and deinoxanthin-glucoside, the relative abundances of all other observed carotenoid molecules were significantly lower or unchanged in Δnos vs. wild type Drad (see Table 1 for comparative statistics).
Confirmation of carotenoid structure and identity by MS/MS analysis
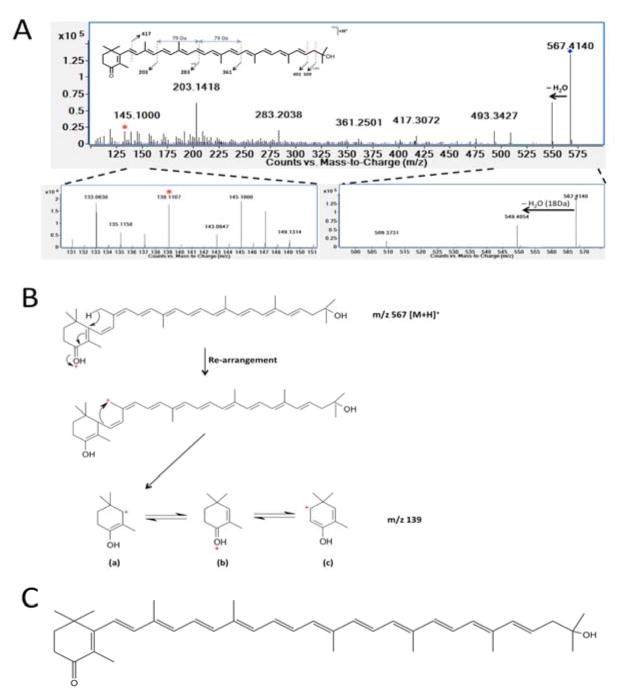
LC-MS/MS analyses were conducted on metabolite extracts in attempt to validate the inferred structural identity of molecules that were altered by nos deletion in Drad. Using a quadrupole-TOF (Q-TOF), putative carotenoid species were fragmented in a collision cell and the ascribed molecular structures were assessed. AS shown in Fig. 6A–C, fragmentation of the molecule inferred to be deoxydeinoxanthin (m/z 567.414, [M + H]+) yielded daughter ions characteristic of a single hydroxyl group loss (m/z 549.405) as well as daughter ion that was indicative of a non-hydroxylated, but ketolated carotenoid β-ring fragment (m/z 139.111 (Figure 6A–C). The structure and identity of deinoxanthin (C40H56O3, m/z 583.409, [M + H]+) was similarly validated in Δnos metabolite extracts (Fig. S2). Fragmentation of m/z 583.409 [M + H]+ yielded daughter ions characteristic of two hydroxyl group losses (m/z 565.395, m/z 547.393) as well as fragment ions indicative of a hydroxylated and ketolated carotenoid β-ring (m/z 155.106 & 137.096). The structures of deoxydeinoxanthin-glucoside (C46H64O7, m/z 729.463, [M + H]+) and deinoxanthin-glucoside (C46H64O8, m/z 745.459, [M + H]+) were also validated in wild type and Δnos Drad, respectively (Figs S3 and S4). These compounds yielded spectral peaks indicative of loss of glucose (−180 Da) as well as MS spectra that corresponded to fragments of deoxydeinoxanthin and deinoxanthin.
Figure 6. Fragmentation and identification of deoxydeinoxanthin (m/z 567.414 [M + H]+).
MS/MS fragmentation confirmed the structure and identity of deoxydeinoxanthin in wild type Drad. Panel A: MS/MS fragmentation spectra of deoxydeinoxanthin at 10eV collision induced dissociation (CID). Fragmentation yielded a daughter ion characteristic of a hydroxyl group loss (m/z 549.405 ([M+H]+ - 18), as well as a unique β-ring fragment at m/z 139.111. Panel B: Schematic describing inferred deoxydeinoxanthin fragmentation and structural rearrangement of the 139.111 ion unique to this compound. Panel C: Proposed structure of deoxydeinoxanthin.
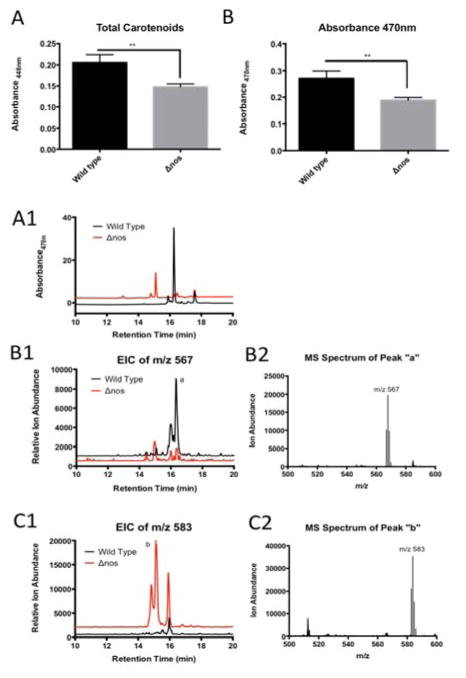
Optical spectroscopy alone and with LC-MS confirms a decrease in total carotenoid abundance in Δnos vs. wild type nos Drad and an altered distribution of carotenoid species
The total carotenoid content of wild type and Δnos Drad was determined by photodiode array detection (PDA). PDA analysis of crude MeOH extracts of carotenoid pigments revealed significant decreases in total levels of carotenoids in Δnos, compared to wildtype nos Drad, based on absorbance at 448 and 470 nm (Fig. 7A and 7B, respectively). Rigorous absolute quantitative assessment of the carotenoid composition of wildtype and Δnos Drad was conducted by diverting a fraction of the reverse phase LC flow used for MS detection to a PDA, allowing additional spectral characterization. This instrument configuration allowed simultaneous assessment of both absorbance and MS for each chromatographically-resolved carotenoid peaks of interest in Drad extracts. Absorbance at 470 nm reconfirmed a marked perturbation in the distribution of carotenoid species in Δnos Drad, compared to carotenoids in Drad expressing wild type nos (Fig. 7A1, 7B1 and 7C1). Notably, extracted ion chromatographs (EICs) and MS spectra from these parallel analyses confirmed a reciprocal switch in the predominance of deoxydeinoxanthin (m/z 567.414, [M + H]+) in wild tye Drad to deinoxanthin (m/z 583.409, [M + H]+) in nos-deleted Drad (Fig. 7B2 and 7C2).
Figure 7. Photodiode array (PDA) analysis demonstrates that total carotenoid abundance is significantly decreased and the major species differs in Δnos vs. wildtype nos Drad.
Panel A: Total carotenoids abundance, measured by PDA at absorbance 448 nm shows significant decreases in total carotenoid content in Δnos compared to wild type nos Drad (unpaired t-test). Panel B: Absorbance at 470 nm, further confirms a significant decrease in carotenoid abundance in Δnos Drad (*p<0.05, **p<0.01, ***p<0.001). Error bars represent mean ion counts +/− standard deviation of the means. Panel A1: LC-resolved carotenoid profile of wild type (black trace) and Δnos (red trace) Drad, measured by PDA at absorbance 470 nm. A reverse phase C18 column and gradient elution was performed for resolution of carotenoids, as described in Methods. Panel B1/B2: Extracted ion chromatographs (EIC) for deoxydeinoxanthin (m/z 567.414, [M + H]+) indicate that deoxydeinoxanthin is the major carotenoid species in wild type and is significantly depleted in Δnos Drad. Panel C1/C2: EIC for deinoxanthin (m/z 583.409, [M + H]+) shows that deinoxanthin is the predominant species in Δnos Drad.
The carotenoid content in wild type and Δnos Drad was further quantified using an isocratic LC platform. Toward this end, metabolite extracts were normalized to absorbance at 470 nm and an equivalent quantity of each sample was analyzed using LC-PDA-MS. Results confirm a clear disturbance in carotenoid composition in Δnos vs. wild type Drad, characterized by an inversion in relative expression levels of deoxydeinoxanthin vs. deinoxanthin (Fig. S5).
Δnos and wildtype nos Drad express indistinguishable levels of known carotenoid gene (Crt) transcripts
Expression levels of mRNA transcripts for enzymes in the putative Deinococcus carotenoid biosynthesis (Crt) pathway were examined by quantitative real time PCR (q-RT-PCR) using primers for known Crt genes (see Table S1). Expression levels of CrtE (DR1395), CrtI (DR0861), CrtF (DR0091), CrtD (DR2250), and CrtO (DR0093) were quantified relative to gapdh (DR1343) expression levels. These qRT-PCR results revealed no significant changes in gene expression for any of the above listed carotenoid biosynthesis genes (Fig. S6). Primers for CrtB (DR0862), and CrtLm (DR0801) could not be effectively optimized after several attempts - hence relative expression levels of cognate transcripts in Drad were not quantified.
Exogenous NO application restores carotenoid biosynthesis in Δnos Drad
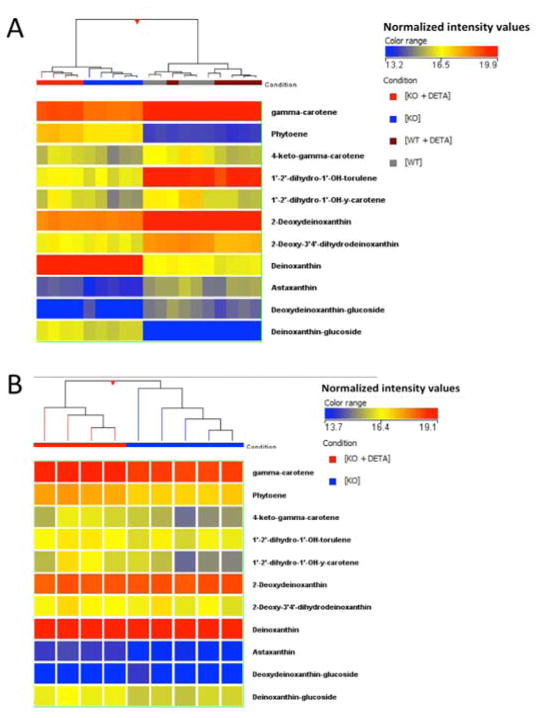
Pharmacological complementation with the slow-releasing NO-donor molecule DETA NONOate demonstrated that NO supplementation restores an apparent wildtype carotenoid profile in Δnos Drad. As shown by unsupervised HCA heatmaps (Fig. 8A and 8B), NO supplementation of Δnos Drad restored carotenoid precursors toward the levels observed in wild type Drad (i.e., complemented the defect elicited by nos deletion). Notably, restored species included carotene and phytoene. Additionally, DETA NONOate significantly increased the abundance of the terminal carotenoid product deinoxanthin and astaxanthin (a bi-ring, keto-carotenoid), that was otherwise markedly decreased by nos deletion in Drad. Treatment with exogenous NO had no discernable effects on abundances of observed carotenoids in wild type Drad (Fig. 8A and 8B; p > 0.05)
Figure 8. Exogenous NO exposure alters carotenoid biosynthesis in Δnos but not wildtype nos Drad.
Panel A: Unsupervised HCA, comparing relative levels of carotenoids and precursor molecules in Drad with and without nos, before and after NO exposure following treatment with DETA NONOate (DETA, 25 μM). Results show a clear separation of Δnos + DETA from untreated Δnos and that NO treatment elicited a restoration of carotenoid profile to more closely resemble wild type Drad. Panel B: Expanded view of carotenoid profile in Δnos Drad, with and without DETA treatment. Of 35 observed carotenoids, 12 were found to be differentially-expressed between Δnos and Δnos + DETA by unpaired t-testing (p< 0.01, Table 4.2). Note that exogenous NO supplementation had no significant effects on carotenoid abundances in wild type nos Drad (p>0.05).
Wild Type and Δnos Drad have similar antioxidant capacities
To determine the effect of nos deletion on the total antioxidant capacity of Drad, antioxidant capacity assays were performed on whole Drad extracts (Fig. S7A) as well as pigment-enriched DMSO extracts of Drad strains (Fig. S7B). Results demonstrated no significant difference in antioxidant capacity of Δnos vs. wild type Drad strains, considering both whole-cell lysates and pigment-enriched Drad extracts.
DISCUSSION
Deletion of nos significantly sensitized Drad to cytostasis following exposure to extreme ionizing γ-irradiation (3000 Gy), pointing to a fundamental role of endogenous NO production as a contributor to Drad’s extraordinary radioresistance (Fig. 1). This radioprotective effect of NO did not arise from diminished radiation-induced cytostasis cell killing since the number of colony forming units (CFUs) after irradiation was unchanged in Δnos vs. wildtype Drad, instead nos was found to be required for rapid growth recovery. The radiosensitizing effect of nos deletion can be specifically ascribed to a depletion of NO levels, because repletion with exogenous NO fully complemented the growth defect in Δnos, but had no affect on the growth of wild type Drad following irradiation (Fig. 2). We subsequently employed an untargeted metabolite profiling approach in attempt to uncover potential mechanism(s) of endogenous NO-conferred radiation resistance in Drad.
Our metabolomic data revealed that nos deletion profoundly affects carotenoid biosynthesis and composition in Drad, which is known to contain various C40 carotenoid species in its multilayered cell envelope [25,43]. Notably, nos deletion resulted in significant alterations in the expression of carotenoid pigments unique to Drad [26,27,33]. These long-chain, unsaturated tetraterpenoid molecules have been reported to be essential elements of the bacterium’s antioxidant and radiation defense mechanisms [21,30–33]. Indeed, Drad mutants deficient in carotenoid biosynthesis are highly sensitized to radiation and oxidative insults, presumably due to marked free-radical scavenging abilities of carotenoids and necessity for protecting against protein and DNA structural damage after radiation insult [21,30,32,33,44]. As determined by untargeted metabolite profiling, nos deletion significantly affected the expression of major carotenoids including carotene, deinoxanthin, and deoxydeinoxanthin (Fig. 5 and Table 1). These compounds represent upstream precursors (carotene) as well as the terminal products (deinoxanthin/deoxydeinoxanthin) in Drad’s proposed carotenoid biosynthetic pathway (Figure 4) [26]. Additionally, novel deoxydeinoxanthin-and deinoxanthin-glucoside conjugates were detected in Δnos and wild type nos bacteria, respectively.
Deinoxanthin, not deoxydeinoxanthin, had previously been identified as the major carotenoid in wild type R1 Drad [27,30,31]. In order to reconcile the differences between our findings and previous reports, we conducted MS/MS fragmentation to determine the structure and identity of Drad carotenoids. MS/MS analyses of pigment extracts confirmed that deoxydeinoxanthin and deinoxanthin are indeed the predominant carotenoids in wild type and Δnos, respectively (Fig. 6 and S2). Furthermore, these findings are consistent with a recent report that questioned the original findings of Lemme et al., [27] and support of our finding that carotenoids other than deinoxanthin can significantly predominate in wild type Drad [45].
Optical absorbance measurements on crude pigment extracts supported our MS-based finding of a significant decrease in total carotenoid abundance in Δnos and prompted a more rigorous combined MS-and optical absorbance-based analysis of pigment extracts. Results from the combined analytical platform confirmed that nos deletion elicits a profound increase in deinoxanthin and decrease in deoxydeinoxanthin (Fig. 7). These results further substantiated findings of untargeted metabolite profiling experiments; nos deletion decreases total carotenoid abundance, but also increases the ratio of deinoxanthin to deoxydeinoxanthin.
Pharmacological complementation experiments, using the long-lasting NO donor DETA NONOate, support the hypothesis that NO mediates carotenoid biosynthesis in Drad [46]. Application of exogenous NO to Δnos caused significant changes in carotenoid expression that were not observed in exogenous NO-treated wild type bacteria (Fig. 8). NO-induced changes in carotenoid expression caused Δnos to more closely resemble wild type bacteria in terms of carotenoid profile, increasing overall carotenoid abundance. The broad-spectrum effect of NO on carotenoid abundance suggests that NO may regulate carotenoid biosynthesis by a multiplicity of actions in Drad.
Quantitative real-time PCR (qRT-PCR) analyses were performed to investigate the possibility that NO was regulating the transcription of carotenoid biosynthesis (Crt) genes as a basis for alterations in carotenoid abundance. However, nos deletion was not found to be associated with significant differences in expression of Crt gene levels (Fig. S6).
NO is a bioactive signaling molecule widely recognized for its ability to affect protein activity via post-translational modifications (PTMs), such as S-nitrosylation of cysteine residues and nitrosylation of transition metals in proteins. Our finding that Crt gene transcript levels remained stable, while carotenoid levels profoundly changed, is in accord with the notion that NO regulates carotenoid biosynthesis through post-translational modification of Crt gene(s) rather than by influencing gene expression. Instead, we hypothesize that NO addition to catalytic domains of Crt enzymes provides a mechanistic basis for control of carotenoid biosynthesis. This may occur by outcompeting the binding of oxygen to non-heme iron containing catalytic domains of Crt enzymes or by S-nitrosylation of active-site thiols that interfere with or otherwise modulate the activity of multiple Crt enzymes [47]. This type of broad-spectrum modulation of Crt enzyme activities may explain the seemingly diverse effects of endogenously produced NO on carotenoid biosynthesis in Drad. Notably, the most profound and reproducible effect of nos deletion was a decrease in the ratio of deoxydeinoxanthin to deinoxanthin (Figs. 5 and S5). A putative hydroxylase that is as yet unidentified, is thought to covert deoxydeinoxanthin to deinoxanthin and we speculate that NO interferes with catalytic activity of this enzyme. Notaby, s constitutive inhibitory action of NO on deoxydeinoxanthin hydroxylase would explain the apparent lack of deinoxanthin accumulation in wild type Drad and build-up as a result of nos-delection.
Carotenoids are notable for their potent antioxidant properties and have been implicated as critical elements in Drad’ s defense against oxidative insult. Proper carotenoid biosynthesis has previously been shown to be necessary for the maintenance of antioxidant activity and radioresistance to ionizing radiation in Drad [32,33,44,48]. Carotenoids have high potential for ROS-scavenging as demonstrated by their ability to quench singlet oxygen and oxidative free radicals such as the hydroxyl species [27,30,31]. Although there was a significant decrease in overall carotenoid abundance in Δnos (Fig. 7), as well as a decrease in growth recovery following extreme (3000 Gy) ionizing radiation (Fig. 1), the antioxidant capacity of Δnos was similar to wild type (Fig. S7). Together, these findings suggest that carotenoids contribute to radioresistance in Drad via mechanisms that do not depend on increased antioxidant activity.
This study identified novel and unreported deoxydeinoxanthin-and deinoxanthin-glucosides in Drad (Fig. S3 and S4). Glucoside derivatives of carotenoids stabilize the membrane in thermophilic bacteria, and were reported to contribute to their resistance to heat-induced cell death [29,49,50]. Although carotenoid-glucosides were not previously identified in Drad, carbohydrate-containing lipids have been shown and implicated as a determinant of radioresistance in Drad [51,52]. Interestingly, a gene with high homology to a carotenoid glucosyltransferase of the thermophilic bacterium Chlorobium tepidum was previously identified in Drad [26,53], it was not known to be functional. We now hypothesize that the carotenoid glucosyltransferase of Drad plays an important functional role in carotenogenesis. Indeed, wild type Drad carotenoid-glucosides were comprised of deoxydeinoxanthin conjugated to a glucose moiety, whereas Δnos Drad contained deinoxanthin-conjugated to glucose. Deoxydeinoxanthin and deinoxanthin differ only by a hydroxyl group on the cyclized β-ring (C2) of the molecule. The presence or absence of this functional group affects hydrophobicity and would likely promote incorporation into the cell envelope in different orientations, altering the characteristics of the membrane. This type of membrane misincorporation may extend to the respective novel carotenoid-glucosides, altering the membrane properties of Drad and decreasing resistance to environmental stressors such as heat and radiation.
Taken together, our studies affirm the utility of untargeted metabolite profiling to identify alterations in metabolism that are associated with gene deletion and dampened radioresistance in Drad. Our data indicate that endogenous NO generation plays critical roles in the maintenance of radioresistance, in association with altered patterns of carotenoid biosynthesis and diminished total carotenoid levels. These findings are mutually supportive of a model in which NO regulates Drad carotenoid biosynthesis which, in turn, facilitates the bacterium’s ability to withstand ionizing radiation. Further, we identified novel deinoxanthin-and deoxydeinoxanthin-glucosides in Drad, whose relative abundances were also profoundly and reciprocally altered by nos deletion. Carotenoid-glucosides, previously unreported in Drad, may have vital yet unappreciated roles in the maintenance of radioresistance. Indeed, membrane fluidity has been suggested as a determinant of radioresistance in Drad, and carotenoid-glucosides are known to play a vital membrane stabilization role in thermophilic bacterium [49,50]. We propose that nos deletion-induced alterations in carotenoid composition alter the fluidity and photoprotective properties of Drad’s membrane, decreasing the ability of this bacterial master of radioresistance to withstand and recover from radiation insult.
MATERIALS AND METHODS
Bacterial strains and growth
Bacterial wild-type strain Deinococcus radiodurans (Drad) and nitric oxide synthase mutant (Δnos) were generously donated from Dr. Brian Crane (Cornell University, Ithaca, NY)[37]. Strains were cultured aerobically in TGY media (0.5% tryptone, 0.3% yeast extract, and 0.1% glucose; pH 7) at 32°C to an optical density (OD600) of 0.8–1.0, and harvested for metabolomic analysis. To avoid light-induced changes in gene expression and metabolic activity, cells were grown in a light-shielded shaker-incubator set to 225RPM.
Cell density (growth curve) and colony forming unit (CFU) assays
Bacteria were diluted 1:100 in fresh TGY and grown under the conditions described above. Cell density as measured by OD600 was recorded as a function of time to construct growth curves. CFU assays were also conducted to determine the percentage of viable cells in each culture. Cells were plated on LB-agar plates (Sigma-Aldrich) and colonies were counted at 72hrs and compared to controls.
Gamma irradiation
Drad bacterial strains were grown as noted above to an optical density (OD600) of 0.8–1.0. For irradiation-only studies, these cultures were harvested by centrifugation, resuspended in fresh TGY media, and exposed to 3000Gy of ionizing radiation (137Cs source) at room temperature using the Memorial Sloan Kettering Cancer Center irradiator. Following irradiation, an aliquot of each culture was used to dilute cells 1:100 in fresh TGY. Cell density, measured at OD600, was recorded as a function of time to construct growth curves. CFU assays were also conducted to determine the post-irradiation percentage of viable cells in each culture. Toward this end, an aliquot of each irradiated culture was plated on LB-agar plates (Sigma-Aldrich) and colonies were counted at 72 h post-irradiation (CFU assay) to quantify the percentage of viable cells in irradiated samples vs. controls. For irradiation with simultaneous NO-supplementation experiments, detaNONOate (Cayman Chemical Company, Ann Arbor, MI) was added to a final concentration of 25μM to TGY both 1-hour before and immediately following irradiation.
Metabolite extraction
Bacterial cultures were harvested by centrifugation and washed in ice-cold ddH2O (Millipore Direct-Q UV3 filtration system). Methanol (MeOH; −70°C) was used to rapidly quench metabolism and extract small molecule metabolites. Bacterial pellets were homogenized using a stainless steel bead beater (TissueLyser II; Qiagen, Valencia, CA). The homogenate was then pelleted by centrifugation (14,000 RPM, 4°C) and the supernatant was collected. Successive methanolic extraction steps were performed until the debris pellet was colorless, with homogenization and centrifugation after each volume. Methanolic supernatants were pooled and vacuum centrifuged until dry (3 hours) using a vacuum concentrator (Eppendorf Vacufuge, Eppendorf, Germany), and stored at −80°C until the day of LC-MS metabolomic analysis. Vacuum concentration was conducted in the dark to avoid degradation of light-sensitive metabolites and pigments.
Normalization of metabolites to total protein
To account for differences in starting material between samples, a normalization based on total protein was conducted on the debris pellet that remained after methanolic extraction. 0.2 M NaOH was added to each tissue pellet and these samples were heated for 20 min at 95°C with frequent vortexing. The dissolved proteinaceous solution was spun down at 10,000 RPM for 5 min to remove debris. A DC protein assay (Bio-Rad, Hercules, CA) was conducted on the supernatant. Total protein (μg) was calculated and recorded for each sample so that metabolite extracts could later be resuspended to equivalent concentrations.
Sample preparation for metabolomic analysis
Vacuum concentrated metabolite extracts were resuspended in 100% MeOH w/0.1% formic acid to their appropriate concentrations based on protein content from normalization steps. These resuspensions were spun at 15,000 RPM for 10 min to remove insoluble debris. The metabolite-containing supernatants from these samples were then transferred to 250 μL conical polypropylene vials, capped, and loaded into the LC autosampler for subsequent LC-MS analysis. MeOH-diluted samples, at the desired concentration of protein equivalent, were injected into the LC-MS platform for chromatographic separation and MS detection.
Reverse phase (RP) LC-MS analytical platform
Metabolite extracts were analyzed on an Agilent model 1200/6230 Liquid Chromatography/Accurate-Mass Time-Of-Flight mass spectrometry (LC-MS) system (Agilent Technologies, Santa Clara, CA) equipped with dual electrospray ionization source, using positive and negative ion-monitoring with reverse phase (RP) chromatographic separation. The LC system comprised a C18 Zorbax SB-AQ™ column (2.1 × 100mm) with 1.8 μm particle size (Agilent Technologies, Santa Clara, CA). A 0.5 μM guard column was placed in front of the RP column to prolong column lifetime (Upchurch Scientific, Oak Harbor, WA). The chromatography platform included a binary pump, on-line degasser, thermostat-regulated dual 54-well plate autosampler, and a temperature controlled column compartment set to 8°C. LC parameters for RP chromatographic separation were as follows: 4 μl injection volume, 60°C column temperature, and a 0.4 ml/min mobile phase flow rate. The mobile phase solvents consisted ultrapure ddH2O w/0.1% formic acid (solvent A) and 100% acetonitrile w/0.1% formic acid (solvent B) with gradient steps as follows: 0–2 min, 10% B; 2.1–17 min, to 95% B; 17.1–27 min, hold at 95% B; 27.1, drop to 10% B; 27.1–34.0 hold at 10% B.
Post-column LC flow was analyzed by an Agilent model 6230 Accurate Mass time-of-flight (TOF) mass spectrometer, equipped with dual spray electrospray ionization (ESI) source. A separate isocratic pump was set to continuously deliver an internal reference mass solution (m/z 121.0509 and 922.0093 in positive ion mode, m/z 60.0172 and 966.0007 in negative ion mode) to the second ESI source, enabling continuous and automatic mass calibration during sample analysis. Positive and negative mass spectra were acquired in 2 GHz (extended dynamic range) mode with 1.41 spectra/sec sampled over a mass/charge (m/z) range of 50–1000 Daltons. The TOF capillary voltage was set at 3500 V, the fragmentor at 175V, the nebulizer pressure was 45 psi, and the nitrogen drying gas at 300°C, delivered at a flow rate of 10 l/min. Data were saved in centroid mode using Agilent MassHunter Workstation Data acquisition Software (B05.00).
Chemometric and multivariate analysis of untargeted LC-MS data and molecular feature extraction (MFE)
Raw LC-MS data files from all experiments were processed with Agilent MassHunter Qualitative Analysis Software (B05.00; Agilent Technologies) for downstream comparative data analysis using MassProfiler Professional (Agilent, B12.01) and MassHunter Profinder (B06.00), as described elsewhere [40,41]. The algorithm used for molecular feature extraction (MFE) treats all of the mass spectral data from the experiment as a large, three-dimensional array of values (i.e., retention time, m/z, and ion abundance). It then searches for and aligns features (ions) with a common elution profile (e.g. identical m/z values and similar retention times). Aligned features are further grouped into distinct “compounds” based on natural abundance isotope patterns as well as their detection as dimers, common adducts, including those with salts, and neutral loss of H2O. Recursive analysis was also conducted to re-mine the raw data for missed features in order to eliminate false-positives and false-negatives.
MFE was restricted to the m/z range of 50–1000 Da and completed within the time-span of the LC gradient. Singly charged ions with two or more ion species (including an M+1 isotope peak or adduct ion) and a minimum peak height of 250 ion counts were selected for MFE. The allowed adduct ions included H+, H−, Na+, & NH4+. The continuously injected reference ions (m/z 121.0509 and 922.0093 in positive ionization mode as well as m/z 60.0172 and 966.0007 in negative ionization mode) were excluded from analysis. Compounds with a minimum absolute peak height of 1000 counts were further filtered for between-sample alignment and statistical analysis.
Ion Alignment, Chemometric Profiling, and Multivariate analysis
MassProfiler Professional (Agilent, B12.01) was used to perform multivariate analysis on data sets. This chemometric software package employs an algorithm for data alignment across multiple groups and treats all imported files as a single dataset. Each aligned compound is associated with a neutral mass, retention time, and ion intensity. Between sample alignment of ions based on retention time (RT) and mass measurements was performed at this point as well. Alignment parameters allowed for a retention time window of 0.4 min and a mass tolerance of 10 mDa. Following feature alignment, unsupervised pattern recognition algorithm analysis was utilized to examine data sets for similarly changing features, expected and unexpected clusters, and the presence of outlying samples. All data considered in multivariate analysis were preprocessed by limiting the species considered to only singly-charged ions that were observed at 80% frequency or more in at least one treatment group. T-tests and ANOVA statistical analysis (with alpha error rate set to 5% or lower) were also conducted to determine what metabolites were significantly different between experimental groups. A Benjamini Hochberg correction was applied to these analyses to adjust for false-positive discovery arising from multiple hypothesis testing. MassHunter Profinder (B06.00) was used to examine raw data chromatograms for accuracy and confirm trends seen in statistical analyses conducted by MassProfiler Professional.
Metabolite identification
As a first step in the structural identification of unknown metabolites, compounds were assigned tentative formulae by utilizing a molecular formula generator (MFG) algorithm. The MFG algorithm was used to generate and score molecular formula by calculating goodness-of-fit to accurate monoisotopic mass, isotope abundance ratios, and spacing between predicted isotope peaks. A putative compound formula was assigned based on the MFG result and further comparison to databases of known metabolites (both in-house and web-based) provided tentative metabolite identifications. An in-house metabolite database (discussed below) and several web-based, metabolomics-specific databases were utilized for metabolite identification.
Web-based metabolomics databases are available for identification of small molecule metabolites based on observed accurate mass and, in some cases, retention time. Databases utilized for identification in these experiments include (1) general mass spectral databases (METLIN), (2) large-scale molecular datasets of genes and small molecules (Kyoto Encyclopedia of Genes, KEGG; and Genomes BioCyc) and (3) biochemical databases (Chemspider) [54–56]. Searching unknown masses against any or all of these databases with mass error set to less than 10 ppm allows for confident assignment of molecular identities to ions.
In-house metabolite Database
Validation of tentatively assigned compound IDs was accomplished by attempting to match observed chromatographic retention times and/or fragmentation patterns with pure molecule standards. Notably, our laboratory generated an in-house metabolite database comprised of m/z and retention times for numerous common metabolites. This database is unique to our metabolomic platform, chromatographic separation techniques, and MS instrument parameters. Currently, over 150 compounds have been analyzed by LC-MS and annotated in our database. Searching untargeted data against this database can give confident molecular identifications and provide insight as to what molecules and metabolic pathways to more rigorously examine.
Molecules that remained unidentified after all database search options were exhausted were assigned tentative molecular formulae based on observed accurate mass and MFG scoring. Processed metabolomics datasets were then considered against relevant biochemical pathways in available databases (ex. KEGG & BioCyc) or literature to pinpoint the location of any possible metabolic dysfunction(s).
MS/MS fragmentation
MS/MS fragmentation was preformed to further characterize the molecular structure and identity of compounds of interest. The reverse phase LC system utilized in untargeted profiling experiments was coupled to an Agilent model 6550A IFunnel Accurate Mass quadrupole time-of-flight (Q-TOF) mass spectrometer for fragmentation of target ions. LC parameters were maintained as described above. Mass spectra were acquired in 2 GHz (extended dynamic range) mode with 2.0 spectra/sec sampled over a mass/charge (m/z) range of 50–1000 Daltons. The Q-TOF capillary voltage was set at 3500V, the fragmentor at 140V, the nebulizer pressure was 35 psi, and the nitrogen drying gas was 200°C, delivered at a flow rate of 14 l/min. Collision induced dissociation (CID) of target molecules was accomplished with a collision energy of 10eV. Data were saved in centroid mode using Agilent MassHunter Workstation Data acquisition Software (B.06.00).
Photdiode Array (PDA) detection
PDA analyses were performed on freshly extracted and non-vacuum concentrated methanolic metabolite extracts. A Hewlett Packard 8453 Diode array spectrophotometer was utilized to conduct a multi-wavelength absorbance scan (190–1100nm) of crude Drad extracts. The machine was blanked with the appropriate solvent sample and then crude pigment extract samples were analyzed. Absorption spectra were profiled between 190 and 1100nm. Total carotenoid content was determined based on absorbance at 448nm, applying the protocol established by Donaldson [57]. Absorbance was also measured at 470nm to assess carotenoid content in bacterial strains.
LC-PDA-MS analysis
LC-PDA-MS examination of samples was accomplished by coupling the LC-MS metabolomics platform to an Agilent 1200 series PDA detector. The LC gradient employed in untargeted profiling analysis was again utilized for chromatographic separation. Absorbance at 470 nm ad 280nm was measured by PDA. Bypassing the PDA allowed for column effluent to be analyzed by MS. EICs of carotenoids were extracted from MS data and ascribed to absorbance peaks from PDA analysis. An isocratic LC elution with ACN:methanol:isopropanol (4:5:1) modified from [48] was also utilized as an alternative and complementary carotenoid separation technique.
qRT-PCR of D. radiodurans’ carotenoid biosynthesis enzymes
The expression levels of mRNA transcripts for enzymes in the putative Deinococcus radiodurans carotenoid biosynthesis (Crt) pathway were examined by quantitative real time PCR (q-RT-PCR). Designed primers are listed in Table S2. An Eppendorf Realplex2 Mastercycler was used for the qPCR reaction and monitoring. Expression values were normalized to GAPDH and statistical comparisons were quantified using Prism 6 (GraphPad Software Inc.). The specificity of primers listed in Table S2 were confirmed by gel electrophoresis and Sanger sequencing (DNA Sequencing Core Facility at Cornell University, Ithaca, NY).
NO-supplementation/scavenging
For studies involving exogenous NO supplementation, the long lasting NO donor DETA NONOate (Cayman Chemical Company, Ann Arbor, MI) was added to a final concentration of 25 μM to TGY. Bacterial cultures were grown in NO-supplemented or scavenged media, as described above, to an optical density of 0.8–1.0 and harvested for metabolite profiling experiments.
Antioxidant capacity assays
Antioxidant capacity assays (Caymen Chemical Company, Ann Arbor, MI) were conducted on both whole cell extracts as well as pigment-enriched methanolic extracts from liquid Drad cultures. Fresh cell lysates or metabolite extracts resuspended in DMSO (50 μL) were spun down for 15 min at 7,500 RPM to remove insoluble cellular debris. This antioxidant assay relies on the principal that antioxidants in the sample inhibit the oxidation of ABTS (2,2′-Azino-di-[3-ethylbenzthiazoline sulphonate]) to ABTS•+ by a ferryl myoglobin radical formed from the reaction of metmyoglobin and hydrogen peroxide. ABTS•+ is a soluble chromogen that is green in color and concentration can be determined based on absorbance at 750 nm, where A750 decreases proportionally as the concentration of antioxidants in a given sample increases. Trolox, a water-soluble vitamin E analog, served as a reference antioxidant for generation of a standard curve. Assays were conducted according to the manufacture’s instructions, with the antioxidant capacity of each sample being calculated as molar equivalents of Trolox antioxidant standard.
Supplementary Material
Table S1. qRT-PCR primers designed against Deinococus radiodurans carotenoid biosynthetic enzymes
Principal component 1 (depicted on the x-axis) accounted for over 36.66% of the variability between D. radiodurans metabolite samples and principal component 2 (y-axis) accounted for 13.64%. Metabolite features from these data were assigned tentative identifications by searching accurate mass and calculated formulae against a custom database of small molecule metabolites (METLIN, KEGG & personal database). The differential expression of many C40 compounds allowed PCA to distinguish Δnos from wild type. Loading scores for PC1 and PC2 showed that molecules tentatively identified as carotene (C40H56, m/z 537.448, [M + H]+, deoxydeinoxanthin (C40H54O2, m/z 567.414, [M + H]+), deinoxanthin (C40H54O3, m/z 583.409, [M + H]+), and deinoxanthin-glucoside (C46H64O8, m/z 745.459, [M + H]+) are among features contributing to the greatest separation between genotype groups.
MS/MS fragmentation confirmed the structure and identity of deinoxanthin in Δnos Drad. Panel A: MS/MS fragmentation spectra of deinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of two hydroxyl group losses (m/z 565.395; ([M+H]+ - 18) and m/z 547.392; ([M + H]+ - 36), as well as a unique β-ring fragments identified as m/z 137.096 and m/z 155.106. These fragments are indicative of hydroxylated and ketolated β-rings, respectively. Panel B: Proposed structure of deinoxanthin.
MS/MS fragmentation confirmed the structure and identity of deoxydeinoxanthin-glucoside in wild type D. radiodurans. Panel A: MS/MS fragmentation spectra of deoxydeinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of a carbohydrate sugar loss (m/z 567.414; [M + H]+ – 162), hydroxyl group loss (m/z 549.405; [M + H]+ – 180), as well as a β-ring fragment unique to deoxydeinoxanthin (m/z 139.111). Panel B: Proposed structure and fragmentation pattern of deoxydeinoxanthin-glucoside.
MS/MS fragmentation confirmed structure and identity of deinoxanthin-glucoside in Δnos D. radiodurans. Panel A: MS/MS fragmentation spectra of deinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of a carbohydrate sugar loss (m/z 583.409; [M + H]+ – 162), hydroxyl group loss (m/z 565.395; ([M + H]+ – 180), as well as β-ring fragments unique to deinoxanthin (m/z 137.092 & m/z 155.101). Panel B: Proposed structure and fragmentation pattern of deinoxanthin-glucoside.
Panel A: LC-resolved carotenoid profile of wild type (black trace) and Δnos (red trace), measured by PDA at absorbance 470 nm. A reverse phase C18 column and isocratic elution were employed. The mobile phase consisted of acetonitrile:methanol:isopropanol (4:5:1). Absorbance units are plotted on the y-axis and retention time (RT) in min on the x-axis. Panel B: Extracted ion chromatographs (EIC) for deoxydeinoxanthin (m/z 567.414, [M + H]+) indicate that deoxydeinoxanthin is the major carotenoid species in wild type Δnos Drad and is significantly depleted in nos Drad. Panel C: EIC for Deinoxanthin (m/z 583.409, [M + H]+) shows that deinoxanthin is the predominant species in Δnos Drad.
Quantitative real-time PCR (qRT-PCR) was used to determine gene expression levels of known carotenoid biosynthesis enzymes in Δnos and wild type Drad. As determined by unpaired t-tests, no significant differences in Crt gene expression were observed in these analyses. Primers for CrtB (DR0862), and CrtLm (DR0801) could not be optimized after several attempts. qRT-PCR also confirmed deletion of nos. (*p<0.05, **p<0.01, ***p<0.001). Error bars represent mean ion counts +/− SD.
Cell lysates (Panel A) and DMSO extracts (Panel B) from Δnos vs. wild type nos Drad were analyzed for antioxidant capacity (Caymen Chemical Company, Ann Arbor, MI). Antioxidant capacity was calculated as molar equivalents of Trolox antioxidant standard. Notably, nos deletion had no significant influence on Drad antioxidant capacity as assessed by unpaired t-test (p> 0.05).
NO production by D. radiodurans (Drad) is required for its extreme resistance to γ-irradiation
Untargeted metabolite profiling uncovered broad metabolic effects of nos deletion in Drad
NO was revealed to be required for physiological carotenoid biosynthesis in Drad
NO supplementation of nos-null Drad restored both carotenoid biosynthesis and radioresistance
Carotenoids are contributors to radioresistance in Drad and may mediate radioprotection by NO
Results demonstrate the power of untargeted metabolite profiling for biological discovery
Acknowledgments
This work was supported by grants from NICHD and NHLBI to SSG. (PO1 HD67244 and R37 HL87062), as well as a NIH T32 training grant to AH (T32 GM073546 to AH). We thank Dr. Brian Crane (Cornell University) for the generous gift of D. radiodurans strains studied in this investigation.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Duggan DE, Anderson AW, Elliker PR, Cain RF. ULTRAVIOLET EXPOSURE STUDIES ON A GAMMA RADIATION RESISTANT MICROCOCCUS ISOLATED FROM FOODa,b,c. Journal of Food Science. 1959;24:376–382. [Google Scholar]
- 4.Duggan DE, Anderson AW, Elliker PR. Inactivation of the Radiation-Resistant Spoilage Bacterium Micrococcus Radiodurans. I. Radiation Inactivation Rates in Three Meat Substrates and in Buffer. Appl Microbiol. 1963;11:398–403. doi: 10.1128/am.11.5.398-403.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson AW, Nordon HC, Cain RF, Parrish G, Duggan DE. Studies on a radioresistant micrococcus: I. Isolation, Morphology, Cultural Characteristics and Resistance to Gamma Radiation. Food Technology. 1956;10:575–578. [Google Scholar]
- 6.Cox MM, Battista JR. Deinococcus radiodurans - the consummate survivor. Nat Rev Microbiol. 2005;3:882–892. doi: 10.1038/nrmicro1264. [DOI] [PubMed] [Google Scholar]
- 7.Thornley MJ, Horne RW, Glauert AM. The fine structure of Micrococcus radiodurans. Arch Mikrobiol. 1965;51:267–289. doi: 10.1007/BF00408143. [DOI] [PubMed] [Google Scholar]
- 8.Hagen U. Mechanisms of induction and repair of DNA double-strand breaks by ionizing radiation: some contradictions. Radiat Environ Biophys. 1994;33:45–61. doi: 10.1007/BF01255273. [DOI] [PubMed] [Google Scholar]
- 9.Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 10.Chandra J, Samali A, Orrenius S. Triggering and modulation of apoptosis by oxidative stress. Free Radic Biol Med. 2000;29:323–333. doi: 10.1016/s0891-5849(00)00302-6. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington: American Society for Microbiology (ASM); 1995. p. xvii.p. 698. [Google Scholar]
- 12.Bruge F, Tiano L, Cacciamani T, Principi F, Littarru GP. Effect of UV-C mediated oxidative stress in leukemia cell lines and its relation to ubiquinone content. Biofactors. 2003;18:51–63. doi: 10.1002/biof.5520180207. [DOI] [PubMed] [Google Scholar]
- 13.Minton KW. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 14.Minton KW. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 15.Blasius M, Sommer S, Hubscher U. Deinococcus radiodurans: what belongs to the survival kit? Crit Rev Biochem Mol Biol. 2008;43:221–238. doi: 10.1080/10409230802122274. [DOI] [PubMed] [Google Scholar]
- 16.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly MJ. Death by protein damage in irradiated cells. DNA Repair (Amst) 2012;11:12–21. doi: 10.1016/j.dnarep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- 20.Markillie LM, Varnum SM, Hradecky P, Wong KK. Targeted mutagenesis by duplication insertion in the radioresistant bacterium Deinococcus radiodurans: radiation sensitivities of catalase (katA) and superoxide dismutase (sodA) mutants. J Bacteriol. 1999;181:666–669. doi: 10.1128/jb.181.2.666-669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian B, Sun Z, Shen S, Wang H, Jiao J, et al. Effects of carotenoids from Deinococcus radiodurans on protein oxidation. Lett Appl Microbiol. 2009;49:689–694. doi: 10.1111/j.1472-765X.2009.02727.x. [DOI] [PubMed] [Google Scholar]
- 22.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One. 2010;5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, Wu Y, Sheng D, Zheng Z, Gao G, et al. Chemiluminescence assay for reactive oxygen species scavenging activities and inhibition on oxidative damage of DNA in Deinococcus radiodurans. Luminescence. 2004;19:78–84. doi: 10.1002/bio.761. [DOI] [PubMed] [Google Scholar]
- 24.Omelchenko MV, Wolf YI, Gaidamakova EK, Matrosova VY, Vasilenko A, et al. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol Biol. 2005;5:57. doi: 10.1186/1471-2148-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bamji MS, Krinsky NI. The carotenoid pigments of a radiation-resistant Micrococcus species. Biochim Biophys Acta. 1966;115:276–284. doi: 10.1016/0304-4165(66)90426-0. [DOI] [PubMed] [Google Scholar]
- 26.Tian B, Hua Y. Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 2010;18:512–520. doi: 10.1016/j.tim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Lemee L, Peuchant E, Clerc M, Brunner M, Pfander H. Deinoxanthin: A new carotenoid isolated from Deinococcus radiodurans. Tetrahedron. 1997;53:919–926. [Google Scholar]
- 28.Umeno D, Tobias AV, Arnold FH. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol Mol Biol Rev. 2005;69:51–78. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara M, Yuan H, Yang Q, Hoshino T, Yokoyama A, et al. Stabilization of liposomal membranes by thermozeaxanthins: carotenoid-glucoside esters. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1999;1461:147–154. doi: 10.1016/s0005-2736(99)00173-x. [DOI] [PubMed] [Google Scholar]
- 30.Carbonneau MA, Melin AM, Perromat A, Clerc M. The action of free radicals on Deinococcus radiodurans carotenoids. Arch Biochem Biophys. 1989;275:244–251. doi: 10.1016/0003-9861(89)90370-6. [DOI] [PubMed] [Google Scholar]
- 31.Ji HF. Insight into the strong antioxidant activity of deinoxanthin, a unique carotenoid in deinococcus radiodurans. Int J Mol Sci. 2010;11:4506–4510. doi: 10.3390/ijms11114506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B, Xu Z, Sun Z, Lin J, Hua Y. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim Biophys Acta. 2007;1770:902–911. doi: 10.1016/j.bbagen.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Yang Q, Luo X, Fang C, Zhang Q, et al. Knockout of crtB or crtI gene blocks the carotenoid biosynthetic pathway in Deinococcus radiodurans R1 and influences its resistance to oxidative DNA-damaging agents due to change of free radicals scavenging ability. Arch Microbiol. 2007;188:411–419. doi: 10.1007/s00203-007-0262-5. [DOI] [PubMed] [Google Scholar]
- 34.Shuryak I, Brenner DJ. A model of interactions between radiation-induced oxidative stress, protein and DNA damage in Deinococcus radiodurans. J Theor Biol. 2009;261:305–317. doi: 10.1016/j.jtbi.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Microbiol. 2009;7:237–245. doi: 10.1038/nrmicro2073. [DOI] [PubMed] [Google Scholar]
- 36.Adak S, Bilwes AM, Panda K, Hosfield D, Aulak KS, et al. Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc Natl Acad Sci U S A. 2002;99:107–112. doi: 10.1073/pnas.012470099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel BA, Moreau M, Widom J, Chen H, Yin L, et al. Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proc Natl Acad Sci U S A. 2009;106:18183–18188. doi: 10.1073/pnas.0907262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janakiram NB, Rao CV. iNOS-selective inhibitors for cancer prevention: promise and progress. Future Med Chem. 2012;4:2193–2204. doi: 10.4155/fmc.12.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikora AG, Gelbard A, Davies MA, Sano D, Ekmekcioglu S, et al. Targeted inhibition of inducible nitric oxide synthase inhibits growth of human melanoma in vivo and synergizes with chemotherapy. Clin Cancer Res. 2010;16:1834–1844. doi: 10.1158/1078-0432.CCR-09-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbott GW, Tai KK, Neverisky DL, Hansler A, Hu Z, et al. KCNQ1, KCNE2, and Na+-Coupled Solute Transporters Form Reciprocally Regulating Complexes That Affect Neuronal Excitability. Sci Signal. 2014:7ra22. doi: 10.1126/scisignal.2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Park HC, Goligorsky MS, Chander P, Fischer SM, et al. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One. 2012;7:e37149. doi: 10.1371/journal.pone.0037149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witherspoon M, Chen Q, Kopelovich L, Gross SS, Lipkin SM. Unbiased metabolite profiling indicates that a diminished thymidine pool is the underlying mechanism of colon cancer chemoprevention by alpha-difluoromethylornithine. Cancer Discov. 2013;3:1072–1081. doi: 10.1158/2159-8290.CD-12-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothfuss H, Lara JC, Schmid AK, Lidstrom ME. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology. 2006;152:2779–2787. doi: 10.1099/mic.0.28971-0. [DOI] [PubMed] [Google Scholar]
- 44.Luan H, Meng N, Fu J, Chen X, Xu X, et al. Genome-wide transcriptome and antioxidant analyses on gamma-irradiated phases of deinococcus radiodurans R1. PLoS One. 2014;9:e85649. doi: 10.1371/journal.pone.0085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lysenko VS, Chistyakov VA, Zimakov DV, Soier VG, Sazykina MA, et al. Separation and mass spectrometry identification of carotenoid complex from radioresistant bacteria Deinococcus radiodurans. Journal of Analytical Chemistry. 2011;66:1281–1284. [Google Scholar]
- 46.Kovacik J, Babula P, Klejdus B, Hedbavny J, Jarosova M. Unexpected behavior of some nitric oxide modulators under cadmium excess in plant tissue. PLoS One. 2014;9:e91685. doi: 10.1371/journal.pone.0091685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas DD, Miranda KM, Colton CA, Citrin D, Espey MG, et al. Heme proteins and nitric oxide (NO): the neglected, eloquent chemistry in NO redox signaling and regulation. Antioxid Redox Signal. 2003;5:307–317. doi: 10.1089/152308603322110887. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z, Tian B, Sun Z, Lin J, Hua Y. Identification and functional analysis of a phytoene desaturase gene from the extremely radioresistant bacterium Deinococcus radiodurans. Microbiology. 2007;153:1642–1652. doi: 10.1099/mic.0.2006/002857-0. [DOI] [PubMed] [Google Scholar]
- 49.Lutnaes BF, Strand Å, Pétursdóttir SK, Liaaen-Jensen S. Carotenoids of thermophilic bacteria—Rhodothermus marinus from submarine Icelandic hot springs. Biochemical Systematics and Ecology. 2004;32:455–468. [Google Scholar]
- 50.Burgess ML, Barrow KD, Gao C, Heard GM, Glenn D. Carotenoid glycoside esters from the thermophilic bacterium meiothermusruber. J Nat Prod. 1999;62:859–863. doi: 10.1021/np980573d. [DOI] [PubMed] [Google Scholar]
- 51.Montaudon D, Carbonneau MA, Melin AM, Rebeyrotte N. Lipid composition, lipid fluidity and radioresistance of Deinococcus radiodurans and two mutant strains. Biochimie. 1987;69:1243–1250. doi: 10.1016/0300-9084(87)90152-0. [DOI] [PubMed] [Google Scholar]
- 52.Melin AM, Carbonneau MA, Rebeyrotte N. Fatty acids and carbohydrate-containing lipids in four Micrococcaceae strains. Biochimie. 1986;68:1201–1209. doi: 10.1016/s0300-9084(86)80065-7. [DOI] [PubMed] [Google Scholar]
- 53.Maresca JA, Bryant DA. Two genes encoding new carotenoid-modifying enzymes in the green sulfur bacterium Chlorobium tepidum. J Bacteriol. 2006;188:6217–6223. doi: 10.1128/JB.00766-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 55.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donaldson MS. A carotenoid health index based on plasma carotenoids and health outcomes. Nutrients. 2011;3:1003–1022. doi: 10.3390/nu3121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. qRT-PCR primers designed against Deinococus radiodurans carotenoid biosynthetic enzymes
Principal component 1 (depicted on the x-axis) accounted for over 36.66% of the variability between D. radiodurans metabolite samples and principal component 2 (y-axis) accounted for 13.64%. Metabolite features from these data were assigned tentative identifications by searching accurate mass and calculated formulae against a custom database of small molecule metabolites (METLIN, KEGG & personal database). The differential expression of many C40 compounds allowed PCA to distinguish Δnos from wild type. Loading scores for PC1 and PC2 showed that molecules tentatively identified as carotene (C40H56, m/z 537.448, [M + H]+, deoxydeinoxanthin (C40H54O2, m/z 567.414, [M + H]+), deinoxanthin (C40H54O3, m/z 583.409, [M + H]+), and deinoxanthin-glucoside (C46H64O8, m/z 745.459, [M + H]+) are among features contributing to the greatest separation between genotype groups.
MS/MS fragmentation confirmed the structure and identity of deinoxanthin in Δnos Drad. Panel A: MS/MS fragmentation spectra of deinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of two hydroxyl group losses (m/z 565.395; ([M+H]+ - 18) and m/z 547.392; ([M + H]+ - 36), as well as a unique β-ring fragments identified as m/z 137.096 and m/z 155.106. These fragments are indicative of hydroxylated and ketolated β-rings, respectively. Panel B: Proposed structure of deinoxanthin.
MS/MS fragmentation confirmed the structure and identity of deoxydeinoxanthin-glucoside in wild type D. radiodurans. Panel A: MS/MS fragmentation spectra of deoxydeinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of a carbohydrate sugar loss (m/z 567.414; [M + H]+ – 162), hydroxyl group loss (m/z 549.405; [M + H]+ – 180), as well as a β-ring fragment unique to deoxydeinoxanthin (m/z 139.111). Panel B: Proposed structure and fragmentation pattern of deoxydeinoxanthin-glucoside.
MS/MS fragmentation confirmed structure and identity of deinoxanthin-glucoside in Δnos D. radiodurans. Panel A: MS/MS fragmentation spectra of deinoxanthin at 10eV CID. Fragmentation yielded a daughter ion characteristic of a carbohydrate sugar loss (m/z 583.409; [M + H]+ – 162), hydroxyl group loss (m/z 565.395; ([M + H]+ – 180), as well as β-ring fragments unique to deinoxanthin (m/z 137.092 & m/z 155.101). Panel B: Proposed structure and fragmentation pattern of deinoxanthin-glucoside.
Panel A: LC-resolved carotenoid profile of wild type (black trace) and Δnos (red trace), measured by PDA at absorbance 470 nm. A reverse phase C18 column and isocratic elution were employed. The mobile phase consisted of acetonitrile:methanol:isopropanol (4:5:1). Absorbance units are plotted on the y-axis and retention time (RT) in min on the x-axis. Panel B: Extracted ion chromatographs (EIC) for deoxydeinoxanthin (m/z 567.414, [M + H]+) indicate that deoxydeinoxanthin is the major carotenoid species in wild type Δnos Drad and is significantly depleted in nos Drad. Panel C: EIC for Deinoxanthin (m/z 583.409, [M + H]+) shows that deinoxanthin is the predominant species in Δnos Drad.
Quantitative real-time PCR (qRT-PCR) was used to determine gene expression levels of known carotenoid biosynthesis enzymes in Δnos and wild type Drad. As determined by unpaired t-tests, no significant differences in Crt gene expression were observed in these analyses. Primers for CrtB (DR0862), and CrtLm (DR0801) could not be optimized after several attempts. qRT-PCR also confirmed deletion of nos. (*p<0.05, **p<0.01, ***p<0.001). Error bars represent mean ion counts +/− SD.
Cell lysates (Panel A) and DMSO extracts (Panel B) from Δnos vs. wild type nos Drad were analyzed for antioxidant capacity (Caymen Chemical Company, Ann Arbor, MI). Antioxidant capacity was calculated as molar equivalents of Trolox antioxidant standard. Notably, nos deletion had no significant influence on Drad antioxidant capacity as assessed by unpaired t-test (p> 0.05).