Abstract
The retinal pigmented epithelium (RPE) is critically important to retinal homeostasis, in part due to its very active processes of phagocytosis and autophagy. Both of these processes depend upon the normal functioning of lysosomes, organelles which must fuse with (auto)phagosomes to deliver the hydrolases that effect degradation of cargo. It has become clear that signaling through mTOR complex 1 (mTORC1), is very important in the regulation of lysosomal function. This signaling pathway is becoming a target for therapeutic intervention in diseases, including age-related macular degeneration (AMD), where lysosomal function is defective. In addition, our laboratory has been studying animal models in which the gene (Cryba1) for βA3/A1-crystallin is deficient. These animals exhibit impaired lysosomal clearance in the RPE and pathological signs that are similar to some of those seen in AMD patients. The data demonstrate that βA3/A1-crystallin localizes to lysosomes in the RPE and that it is a binding partner of V-ATPase, the proton pump that acidifies the lysosomal lumen. This suggests that βA3/A1-crystallin may also be a potential target for therapeutic intervention in AMD. In this review, we focus on effector molecules that impact the lysosomal-autophagic pathway in RPE cells.
Keywords: AMD, Autophagy, βA3/A1-crystallin, Lysosome, mTORC1, Oxidative stress, RPE, V-ATPase
Lysosomes are cellular organelles that modulate various processes such as autophagy and heterophagy, plasma membrane repair, cholesterol homeostasis and cell death (Xu and Ren, 2015). The number, size and content of lysosomes vary in different cell types. The distribution of lysosomes within the cell is determined by the nutrient sensing machinery at the lysosomal membrane, and is an important factor in lysosomal catabolic function. In this review, we focus on the effector molecules present in retinal pigmented epithelial (RPE) cells that impact the lysosomal-autophagic pathway.
Retinal Pigmented Epithelium (RPE)
The RPE is a single layer of cells interposed between the neurosensory retina and Bruch's membrane (Strauss, 2005). En face, RPE cells assume a hexagonal, cobblestone-like appearance. The cells are highly polarized and contain abundant melanin granules that absorb scattered light, thereby reducing photo-oxidative stress on the retina (Beatty et al., 1999). In addition, the RPE has several other functions that are crucial to the retina's functional integrity. Perhaps its most important function is the phagocytosis of shed photoreceptor outer segments (POS) and the subsequent degradation and recycling of their molecular components for re-use in the visual cycle (Young and Bok, 1969 and Bok, 1993). Apical microvilli of the RPE extend around the POS and ingest shed rod and cone outer segment discs into the RPE as membrane bound phagosomes. These phagosomes fuse with lysosomes to form phagolysosomes. The acid hydrolases from the lysosomes digest the outer segment material, critical components of which are returned to the photoreceptors for re-use. In a related process, called autophagy, damaged intra-cellular components including organelles, protein aggregates, and membranes are packaged into autophagosomes, which like phagosomes, fuse with lysosomes to effect cargo degradation.
Lysosomes and Autophagy
Much is now known about the molecular mechanisms of autophagosome formation (Mizushima and Kamatsu, 2011, Yang and Klionsky, 2010 and Rubinsztein et al., 2012), however, we know less about the end stages of macroautophagy, particularly the role of lysosomes in the degradation of autophagosome contents (Shen and Mizushima, 2014). The process is different from microautophagy and chaperone-mediated autophagy, where cellular materials to be degraded are directly delivered to the lysosomes, independent of autophagosomes (Kaushik and Cuervo, 2012). Therefore, lysosomes are indispensible in the degradation and recycling processes of all three major autophagy types.
Lysosomes are the major digestive organelle in eukaryotic cells (Saftig, 2006). They have a lipid bilayer membrane with an acidic lumen containing over 60 acidic hydrolases, each capable of degrading specific substrates (Settembre et al., 2013). The acidification of lysosomes is established by vacuolar-type H+-ATPases (V-ATPase) (Sun-Wada et al., 2003 and Mindell, 2012) which are multi-subunit complexes, composed of a peripheral V1 domain that hydrolyzes ATP and an integral V0 domain, that translocates protons from the cytoplasm to the lumen (Toei et al., 2010).
Lysosomal dysfunction may result from abnormal functioning of any of the myriad of proteins required for maintaining lysosomal homeostasis. However, in each case, the disease phenotype and tissue (s) affected can be different. Therefore, the mechanisms by which lysosomal function is regulated in the RPE may be unique. RPE cells are not only among the most active phagocytic cells in the body, continuously phagocytosing shed POS, but also are post-mitotic cells with high metabolic activity, where a high rate of autophagy would be expected. Therefore, lysosomal-mediated removal of waste products in the RPE is essential to insure functional integrity of the neural retina. The lysosomal degradation pathway declines with age in the human brain, contributing to the pathogenesis of neurodegenerative diseases (Cuervo and Dice, 2000 and Nixon, 2013). While RPE lysosomal dysfunction is now thought to be a significant risk factor for age-related macular degeneration (AMD), our knowledge of how such abnormalities contribute to the disease process remains limited (Kaarniranta et al., 2013). In 1 year old rats with a spontaneous mutation in the Cryba1 gene (encoding for βA3/A1-crystallin) (Sinha et al., 2008), electron microscopy (EM) showed large aggregates of lipofuscin-like material (arrows in Figure 1A) and large vacuoles containing many degenerated cellular organelles (arrowheads in Figure 1A) indicative of inefficient lysosomal clearance (Zigler et al., 2011). Interestingly, similar structures are also seen in EM sections of the fovea from a 95-year old male patient with geographic atrophy (Figure 1B). Therefore, understanding the lysosomal-mediated clearance mechanisms in the RPE may help to understand the pathophysiology of AMD.
Figure 1. Effects of Lysosomal dysfunction on RPE cell ultrastructure.
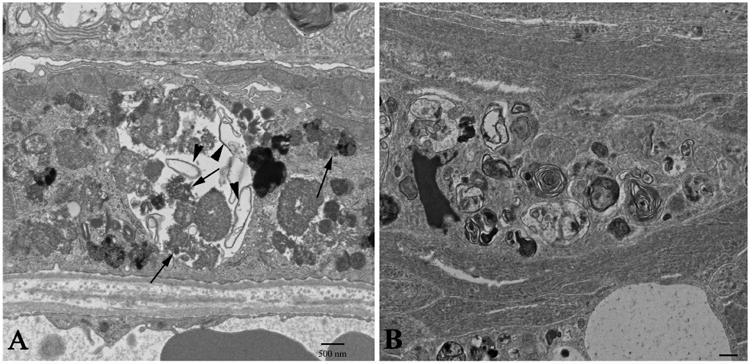
Transmission electron microscopy (TEM) was used to compare the cellular ultrastructure of the RPE in the Nuc1 rat (A) and a 95-year old human subject with geographic atrophy (B). Nuc1 is a spontaneous mutation in Cryba1, the gene encoding βA3/A1-crystallin, a lysosomal protein in RPE cells that participates in lysosomal-mediated clearance. The Nuc1 RPE at 1 year of age shows a large vacuole containing both partially degraded cellular organelles (arrowheads) and lipofuscin-like aggregates (arrows). The RPE from the foveal region of a 95-year old geographic atrophy subject (B) shows similar changes in the fibro-cellular formation located above Bruch's membrane near the area of atrophy. Scale bar= 500nm.
In the RPE, lysosomes degrade both extracellular (POS) and intracellular (autophagy) material. Recently, it has become very clear that lysosomes and mTORC1 signaling are interconnected (Bar-Peled and Sabatini, 2014, Betz and Hall, 2013 and Puertollano, 2014). An elegant study demonstrated that lysosomal positioning within the cell regulates mTORC1 signaling (Korolchuk et al., 2011) while another showed that long starvation periods lead to mTORC1 reactivation and, thereby, formation of proto-lysosomes that develop into mature lysosomes (Yu et al., 2010).
mTOR Signaling and Autophagy
The mammalian target of rapamycin (mTOR), now officially known as the mechanistic TOR, is an atypical serine/threonine kinase that has been conserved throughout evolution. It interacts with many other proteins to form at least two distinct multiprotein complexes, namely mTORC1 and mTORC2 (Laplante and Sabatini, 2013). The mTOR complexes have different upstream inputs and downstream outputs (Zoncu et al., 2011). mTORC1 integrates multiple signals either to promote cellular growth when growth factors, nutrients and energy are available, or to induce catabolic processes during stress. Active mTORC1 has a number of downstream biological effects, including suppression of autophagy (Zoncu et al., 2011).
Several studies have shown that inhibition of mTORC1 activity is crucially important for autophagy induction in eukaryotic cells subjected to nutrient deprivation (Yang and Klionsky, 2010 and Laplante and Sabatini, 2012). Although mTORC1 is inhibited by both glucose/growth factor and amino acid deprivation, the signalling mechanisms involved are different. In the presence of glucose and growth factors, the TSC1/2 (tuberous sclerosis complex), a heterodimeric complex, which is a negative regulator of mTORC1 is phosphorylated and inactivated by several growth factor effector kinases such as Akt/PKB (protein kinase B) and ERK1/2 (extracellular-signal-regulated kinase 1/2). This leads to activation of mTORC1 (Wullschleger et al., 2006), and inhibition of de novo autophagosome formation (Kim et al., 2011 and Ganley et al., 2009). In contrast, glucose starvation activates AMPK (5′-AMP-activated protein kinase), which inhibits mTORC1 by phosphorylation and activation of its negative regulator, TSC1/2 (Inoki et al., 2003b).
Recent studies showed that amino acid- mediated activation of mTORC1 is dependent on formation of a four component super complex with V-ATPase, Ragulator and members of the Rag family of GTPases (Sancak et al., 2010 and Efeyan et al., 2013). V-ATPase is crucial to this process, functioning as a sensing device that responds to the lysosomal amino acid content by activating the Rag family GTPases. Upon activation, the Rag GTPases regulate the translocation and activation of mTORC1 on the lysosomal surface (Settembre et al., 2012).
Although the mTOR signaling pathway is highly conserved and ubiquitously expressed, its regulation is cell and tissue specific. RPE cells express both mTORC1 and mTORC2 complexes that are functionally active (Chen et al., 2010). Increased mTORC1 activation in senescent RPE cells leads to age-related decline in RPE cell function and rapamycin-mediated inhibition of mTORC1 prevents replicative senescence in cultured RPE cells (Yu et al., 2014 and Chen et al., 2010). In mouse models of retinal degeneration, rapamycin treatment prevented photoreceptor dysfunction (Zhao et al., 2011). In our Cryba1 cKO (conditional knockout) mouse model, where Cryba1 is knocked out specifically in the RPE, we have recently shown that mTORC1 activation leads to impaired lysosomal function and decreased autophagy in the RPE (Valapala et al., 2014). We demonstrated that βA3/A1-crystallin regulates lysosome-mediated degradation in the RPE by modulating V-ATPase via the AKT/mTORC1 signaling cascade. We also reported that βA3/A1-crystallin binds to the V0 domain of V-ATPase, the first such binding partner in a mammalian system (Valapala et al., 2014). V-ATPase is a master regulator for amino acid sensing in lysosomes and for translocation of amino acids into the lumen, a requirement for mTORC1 activation (Zoncu et al., 2011). These findings suggest that βA3/A1-crystallin is essential for mTORC1 signaling in the lysosomes of RPE (Figure 2). Our mouse models, both Cryba1 cKO (Valapala et al., 2014b) and Cryba1 KO develop a slowly progressive AMD-like pathology that is associated with inefficient lysosomal clearance (Figure 3).
Figure 2. A schematic diagram showing activation of mTORC1 and a possible role for βA3/A1-crystallin.
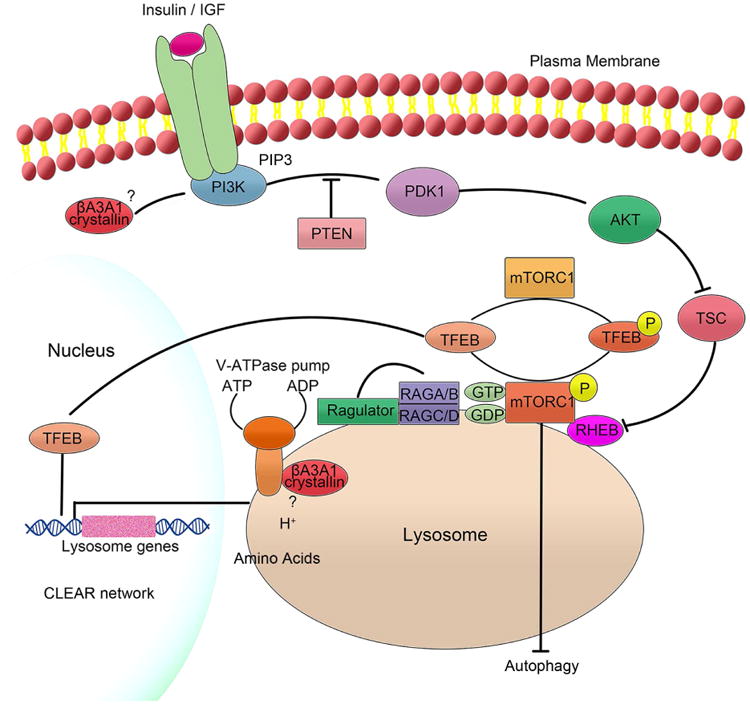
mTORC1 has been shown to integrate inputs from extracellular signal proteins, such as growth factors as well as amino acids and other nutrients. It is now known that V-ATPase interacts in an amino acid sensitive manner with pentameric Ragulator, a scaffolding complex that anchors the heterodimeric Rag GTPases to the lysosomes. This leads to the translocation of the inactive mTORC1 to the lysosomal surface. Once mTORC1 is on the surface of the lysosomes, it is activated by Rheb that is also localized to the lysosomal surface. It has been postulated that amino acids are probably translocated to the lysosomal lumen by V-ATPase and that amino acid signaling from the lysosomal lumen plays an important role in the complex process of recruiting mTORC1 to the lysosomal surface and activating it. We have recently shown that βA3/A1-crystallin is localized to the lysosomal lumen of RPE cells and is a binding partner of V-ATPase. It is also known that in the presence of growth factors or insulin, Akt inhibits Tsc, which releases its inhibitory activity on Rheb, thus allowing the activation of mTORC1. We have previously shown that βA3/A1-crystallin regulates cell survival in astrocytes through PI3K/Akt/mTOR. Further, following autophagy induction both in vivo and in vitro, phospho-Akt and phospho-Raptor decrease, while phospho mTOR increases in RPE cells, inhibiting autophagy and Akt/mTORC1. mTORC1 also regulates lysosomal function by directly preventing autophagy and Transcription factor EB (TFEB) activation. Unphosphorylated TFEB accumulates in the nucleus, where it activates genes in the Coordinated Lysosomal Expression and Regulation (CLEAR) network (such as V-ATPase) that act to support lysosomal function. Once TFEB is phosphorylated by mTORC1, TFEB transiently binds to the lysosomal surface and is also retained in the cytoplasm. It is possible that upstream inputs, such as from βA3/A1-crystallin to mTORC1 can contribute to novel regulation of TFEB in RPE cells. βA3/A1-crystalin produces two closely related proteins, βA3 and βA1, differing only in 17 amino acids in their amino termini from a single Cryba1 mRNA using an alternative translation by leaky scanning. It is possible that βA3- and βA1-crystallins exert their functions independently in the activation of mTORC1 in RPE, e.g. via modulation of V-ATPase/mTOR and PI3K/Akt/mTOR.
Figure 3. Lysosomal dysfunction inhibits organelle clearance by selective autophagy.
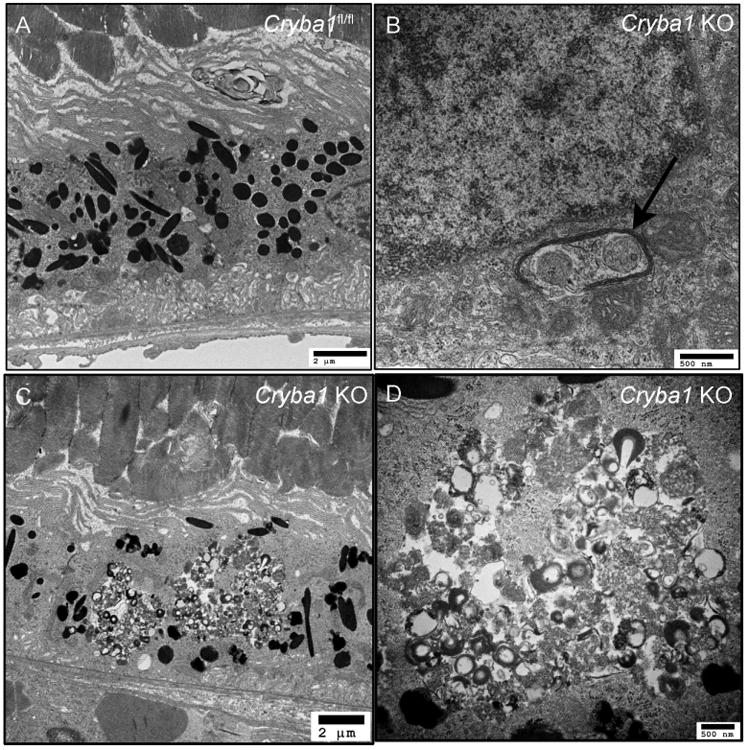
Transmission electron microscopy showing RPE in a 20 month old Cryba1 floxed (wild type) mouse (A). The Cryba1 knockout mouse at the same age shows degenerative changes in the RPE (C), including accumulation of undigested material (D is higher magnification of C). Inefficient lysosomal clearance affects mitophagy as seen in (B). Damaged mitochondria are enclosed by an autophagosome (arrow), but not cleared.
Oxidative Stress and Autophagy
Postmitotic RPE cells in the macula are constantly exposed to a high metabolic and oxidative stress environment (Bok 1993 and Decanini et al., 2007). During RPE cell aging, the capacity to neutralize mitochondrial-derived ROS diminishes due to decreased anti-oxidant production, reduced ability to repair DNA or protein damage, and disturbed proteolysis (Kaarniranta et al., 2009 and Blasiak et al., 2013). The inadequately neutralized ROS damage cellular proteins, leading to detrimental protein aggregation. Lipofuscin is one consequence of this aggregation because oxidized PUFAs are not efficiently digested in lysosomes of aged RPE cells (Schutt et al., 2002, Bergman et al., 2004, Vives-Bauza et al., 2008, Krohne et al., 2010 and Valapala et al., 2014). Lipofuscin is an autofluorescent heterogeneous mixture of lipid–protein aggregates, which sensitizes RPE cells to light induced oxidative stress, ultimately evoking further protein misfolding (Figure 4). In all cells, the heat-shock protein (Hsp) stress response is capable of refolding misfolded proteins, thereby improving cellular survival under oxidative stress (Ryhänen et al., 2009). Upregulation of Hsps has been detected in RPE homogenates isolated from human donor AMD samples (Schutt et al., 2002 and Decanini et al., 2007). This is an indication of stressed RPE cells, but importantly, it also reveals dysfunction in proteasomal clearance (Kapphahn et al., 2007, Li et al., 2008, Fernandes et al., 2008 and Ryhänen et al., 2009). Once Hsp repair capacity is exceeded, individual polypeptides can be degraded by either the proteasome or by chaperone-mediated autophagy, while aggregates are degraded by selective macroautophagy, also called aggrephagy (Hyttinen et al., 2014). In aggrephagy, cellular organelles and protein aggregates are encapsulated from the cytoplasm into autophagosomes, which then fuse with lysosomal vesicles for degradation (Lamark and Johansen, 2012). However, autophagy activity decreases with aging in RPE cells (Rodríguez-Muela et al., 2013, Viiri et al., 2013, Ferguson and Green, 2014 and Toops et al., 2015). One explanation for this decreased autophagy might be the accumulation of lipofuscin, a hallmark of aging, because it suppresses lysosomal function and autophagic clearance in RPE cells (Ryhänen et al., 2009, Krohne et al., 2010, Viiri et al., 2013, Mitter et al., 2014 and Valapala et al., 2014). Lipofuscin components have also been shown to inhibit V-ATPase, thereby elevating lysosomal pH and impairing the digestion of phagocytosed POS (Finnemann et al., 2002, Bergmann et al., 2004, Lamb and Simon 2004, Vives-Bauza et al., 2008 and Guha et al., 2014).
Figure 4. Multifunctional p62 in the regulation of proteolysis.
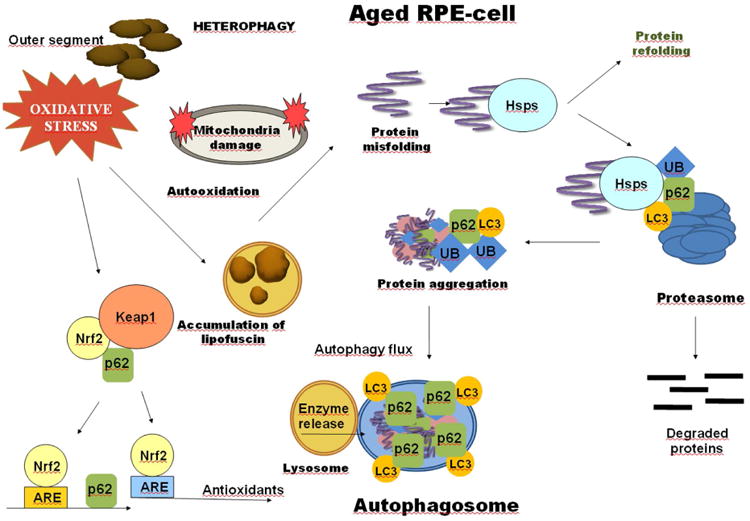
RPE cells in the macula are constantly exposed to the daily heterophagy. The capacity to defend against oxidative stress decreases in aged RPE cells. The simultaneous oxidative stress and impaired defense systems damage cellular proteins and evoke detrimental protein aggregation. Prior to aggregation, heat-shock proteins (Hsps) attempt to refold misfolded proteins. Once Hsp repair capacity is exceeded, individual polypeptides can be degraded by the ubiquitin (Ub) targeted proteasome, while aggregates are degraded by autophagy. p62 sorts proteins between proteasomal and autophagic clearance pathways. It binds to Ub cargoes and to LC3. p62 interacts with the Nrf2/ARE by disrupting the cytoplasmic Nrf2-Keap1 complex and thereby regulates antioxidant production. A functional ARE element is located in the regulatory region of the p62 gene.
The p62 protein sorts proteins between the proteasomal and autophagic clearance pathways (Kirkin et al., 2009). In this process, p62 selectively targets ubiquitinated protein aggregates for autophagic degradation. First, it binds to the perinuclear protein aggregates and undergoes autophagic clearance, making it a useful biomarker of autophagy activity (Bjørkøy et al., 2006, Clausen et al., 2010, Larsen et al., 2010 and Viri et al., 2013). Its accumulation in macular RPE cells rather than in the cells of the periphery suggests that autophagy activity declines in AMD (Viiri et al., 2013 and Valapala et al., 2014). Second, p62 interacts with the Nrf2/ARE (nuclear factor-erythroid 2-related factor-2/antioxidant response element) pathway by disrupting the cytoplasmic Nrf2-Keap1 complex to regulate antioxidant production (Jain et al., 2010 and Wang et al., 2014). A functional ARE element is located in the p62 gene promoter (Jain et al., 2010 and Hirotsu et al., 2012). Nrf2 and p62 create a regulatory loop where Nrf2 activates p62 expression, while Nrf2 nuclear localization is facilitated by p62 (Lau 2010). In addition, Keap1 elimination is processed by p62 dependent autophagy (Taguchi et al., 2012). Nrf2 signaling dysfunction plays an important role in the oxidative stress response (Sachdeva et al., 2014) of RPE cells, and been found to decline in the RPE of AMD samples (Wang et al, 2014). With decreased Nrf2 signaling, p62 can decrease and thus, impair aggrephagy during AMD.
Autophagy in Retinal Diseases
Autophagy clearly plays a protective role against disease in the retina and RPE. It has recently been found that the retina, and in particular the photoreceptors and RPE, of wild-type mice have constitutive autophagic events and that light exposure induces an additional autophagic response (Chen et al., 2013). Mice deficient in Beclin 1 or Atg7 develop severe retinal degeneration upon light exposure, indicating that autophagy is important for maintaining retinal homeostasis. Furthermore, impaired mitophagy, with Park2 deficiency, results in mitochondrial dysfunction and retinal degeneration. Given the highly abundant mitochondria in photoreceptors, mitophagy in addition to macroautophagy, plays an essential role in photoreceptor homeostasis. Since autophagy in photoreceptors is covered in another review in this issue, we will not discuss it further here.
In general, autophagy decreases with aging (Cuervo and Dice, 2000); further, decreased autophagy with aging in retinas of C57BL6 mice has recently been described (Rodriguez-Muela et al, 2013). The aging retinas did not have an increase in autophagy related compartments, suggesting that the defect occurs during autophagosomal formation and not during degradation. The decline in macroautophagy was partially compensated by chaperone-mediated autophagy, where several rate-limiting components were upregulated, such as Lamp2A and Hsc70. Similarly, in mice deficient of Atg5 specifically in rod photoreceptors, retinas displayed increased TUNEL positive rods, coincident with decreased scotopic vision. Because the decreased rod mediated vision with impaired autophagy mirrors that of age-related vision loss, the authors speculated that impaired autophagy contributes to decreased vision with aging. The reduction in retinoids from impaired autophagy is perhaps a specific aspect of autophagy that explains the decreased vision during aging (Kim et al., 2013), especially because the visual function can be recovered to some extent, with vitamin A supplementation (Owsley et al, 2006).
Autophagy appears to have a biphasic response in AMD. Autophagy is increased in the RPE in aging and early AMD to compensate for oxidative stress and damaged organelles (Mitter et al., 2014). In two AMD mouse models and human AMD samples, LC3, ATG7 and ATG9 were increased in the RPE and retinal layers. Likewise, Wang et al. reported that Atg12 immunolabeling and Atg12-Atg5 and LC3 proteins were increased in the RPE/Bruch's membrane of elderly mice (Wang et al., 2009). However, LC3, ATG7, and ATG9 are decreased in advanced AMD samples, suggesting that autophagy failure contributes to late disease (Mitter et al., 2014).
Dysregulated inflammation contributes to AMD pathology. In addition to genetic variants in multiple complement factors being associated with AMD risk (Edwards et al., 2005, Haines et al., 2005, Klein et al., 2005, Yates et al., 2007, Maller et al., 2007 and Kondo et al., 2010), the NLRP3 inflammasome has been implicated in geographic atrophy development (Tarallo et al, 2012). A decrease in Dicer causes an increase in Alu RNAs, oxidative stress, mitochondrial dysfunction, oxidized mitochondrial DNA, and lysosomal permeability, all of which can activate the inflammasome and are relevant stimulants in AMD (Halle et al., 2008, Hornung et al., 2008, Tschopp et al., 2010, Zhou et al., 2011, Kauppinen et al., 2012 and Shimada et al., 2012). Autophagy controls NLRP3 inflammasome activation by degrading inflammasome components and effector molecules (Shi et al., 2012 and Harris et al., 2011). With an autophagy decline, inflammasome control can be compromised, resulting in excessive activation and potential tissue injury (Zhou et al., 2011 and Nakahira et al., 2011). Given the role of the inflammasome in AMD, impaired inflammasome control by decreased autophagy in the RPE may contribute to an exaggerated inflammatory response during late AMD.
Perspective
Lysosomes are a heterogeneous collection of distinct organelles, specialized for intracellular digestion. mTORC1 regulates the biogenesis, distribution, and activity of lysosomes. In neurodegenerative diseases, such as Alzheimer's and Parkinson's, several studies suggest that defective lysosomal clearance is involved in disease pathogenesis (Bergamini et al., 2004, Keller, 2004 and Shintani and Klionsky, 2004). We believe that prolonged impairment of lysosomal clearance in the RPE, as seen in our Cryba1 genetic animal models, can lead to pathological changes reminiscent of AMD. The mTORC1 pathway regulates many major cellular processes and is implicated in an increasing number of pathological conditions, including cancer, obesity, type 2 diabetes and neurodegeneration (Efeyan et al., 2012). A multicenter study (Interventions Testing Program) conducted by the National Institute of Aging, reported that mTOR inhibition with rapamycin extends the life span of mice (Harrison et al., 2009). While rapamycin or rapalogs (Lamming et al., 2013) have shown therapeutic efficacy for age-related pathologies in animal models, significant side effects limit their use in humans. Therefore, selective targeting of the mTORC1 signaling pathway may offer a safe mode for the treatment of age-related diseases, such as AMD. A better understanding of the functions of the mTOR interacting proteins would allow for the development of novel modulators of mTOR complexes that perturb their function in specific ways. The mTORC1 signaling pathway in the lysosome is becoming a legitimate target for developing therapeutic approaches for human diseases, such as AMD, where dysfunction of the lysosomal–autophagic pathway is apparent. We are optimistic that βA3/A1-crystallin represents a potential avenue of targeting the autophagic-lysosomal process in RPE in an effort to restore or maintain normal lysosomal function in human AMD disease.
In the RPE, lysosomes modulate both heterophagy and autophagy to maintain retinal homeostasis.
The mTORC1 signaling pathway regulates the biogenesis, distribution and activity of lysosomes.
βA3/A1-crystallin is a novel target for restoring normal lysosome function in human AMD disease.
Acknowledgments
We would like to thank all members of Sinha, Kaarniranta, Handa and Lutty laboratories. DS is a recipient of the Carolyn K. McGillvray Memorial Award for Macular Degeneration Research from BrightFocus Foundation and the Sybil B. Harrington Special Scholar Award for Macular Degeneration from Research to Prevent Blindness. Dr. Handa is the Robert Bond Welch Professor. The authors would like to acknowledge funding support from National Institutes of Health: EY019037-S (DS), EY019904 (JTH) EY14005 (JTH), EY01765 (Wilmer Imaging Core); IPA from National Eye Institute (DS); RPB Senior Scientist Award (JTH); Thome Foundation (JTH); Research to Prevent Blindness (unrestricted grant to The Wilmer Eye Institute). We apologize to our colleagues for not being able to cite important contributions due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty S, Boulton M, Henson D, Koh H, Murray I. Macular pigment and age related macular degeneration. Br J Ophthalmol. 1999;83:867. doi: 10.1136/bjo.83.7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The role of macroautophagy in the ageing process, anti-ageing intervention and age-associated diseases. Int J Biochem Cell Biol. 2004;36:2392–2404. doi: 10.1016/j.biocel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Bergmann M, Schutt F, Holz FG, Kopitz J. Inhibition of the ATP-driven proton pump in RPE lysosomes by the major lipofuscin fluorophore A2-E may contribute to the pathogenesis of age-related macular degeneration. Faseb j. 2004;18:562–564. doi: 10.1096/fj.03-0289fje. [DOI] [PubMed] [Google Scholar]
- Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- Blasiak J, Glowacki S, Kauppinen A, Kaarniranta K. Mitochondrial and nuclear DNA damage and repair in age-related macular degeneration. Int J Mol Sci. 2013;14:2996–3010. doi: 10.3390/ijms14022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sawada O, Kohno H, Le YZ, Subauste C, Maeda T, Maeda A. Autophagy protects the retina from light-induced degeneration. J Biol Chem. 2013;288:7506–7518. doi: 10.1074/jbc.M112.439935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang J, Cai J, Sternberg P. Altered mTOR signaling in senescent retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2010;51:5314–5319. doi: 10.1167/iovs.10-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice DJ. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000 Oct 6;275(40):31505–13. doi: 10.1074/jbc.M002102200. 2000. J Biol Chem. 275, 31505. [DOI] [PubMed] [Google Scholar]
- Decanini A, Nordgaard CL, Feng X, Ferrington DA, Olsen TW. Changes in select redox proteins of the retinal pigment epithelium in age-related macular degeneration. Am J Ophthalmol. 2007;143:607–615. doi: 10.1016/j.ajo.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Ritter R, Abel K, Manning A, Panhuysen C, Farrer L. Complement factor H polymorphism and age-related macular degeneration. 2005;308:421. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson TA, Green DR. Autophagy and phagocytosis converge for better vision. Autophagy. 2014;10:165–167. doi: 10.4161/auto.26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AF, Zhou J, Zhang X, Bian Q, Sparrow J, Taylor A, Pereira P, Shang F. Oxidative inactivation of the proteasome in retinal pigment epithelial cells. A potential link between oxidative stress and up-regulation of interleukin-8. J Biol Chem. 2008;283:20745–20753. doi: 10.1074/jbc.M800268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann SC, Leung LW, Rodriguez-Boulan E. The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2002;99:3842–3847. doi: 10.1073/pnas.052025899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Coffey EE, Lu W, Lim JC, Beckel JM, Laties AM, Boesze-Battaglia K, Mitchell CH. Approaches for detecting lysosomal alkalinization and impaired degradation in fresh and cultured RPE cells: evidence for a role in retinal degenerations. Exp Eye Res. 2014;126:68–76. doi: 10.1016/j.exer.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JM, Amadio M, Viiri J, Pascale A, Salminen A, Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev. 2014;18:16–28. doi: 10.1016/j.arr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003b;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K, Sinha D, Blasiak J, Kauppinen A, Vereb Z, Salminen A, Boulton ME, Petrovski G. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy. 2013;9:973–984. doi: 10.4161/auto.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K, Salminen A, Eskelinen EL, Kopitz J. Heat shock proteins as gatekeepers of proteolytic pathways-Implications for age-related macular degeneration (AMD) Ageing Res Rev. 2009;8:128–139. doi: 10.1016/j.arr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Kapphahn RJ, Bigelow EJ, Ferrington DA. Age-dependent inhibition of proteasome chymotrypsin-like activity in the retina. Exp Eye Res. 2007;84:646–654. doi: 10.1016/j.exer.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen A, Niskanen H, Suuronen T, Kinnunen K, Salminen A, Kaarniranta K. Oxidative stress activates NLRP3 inflammasomes in ARPE-19 cells--implications for age-related macular degeneration (AMD) Immunol Lett. 2012;147:29–33. doi: 10.1016/j.imlet.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol. 2004;36:2376–2391. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, Ferguson TA. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- Klein R, Zeiss C, Chew E, Tsai J, Sackler R, Haynes C, Henning A, SanGiovanni J, Mane S, Mayne S, Bracken M, Ferris F, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Bessho H, Honda S, Negi A. Additional evidence to support the role of a common variant near the complement factor I gene in susceptibility to age-related macular degeneration. Eur J Hum Genet. 2010;18:634–635. doi: 10.1038/ejhg.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O'Kane CJ, Deretic V, Rubinsztein DC. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohne TU, Kaemmerer E, Holz FG, Kopitz J. Lipid peroxidation products reduce lysosomal protease activities in human retinal pigment epithelial cells via two different mechanisms of action. Exp Eye Res. 2010;90:261–266. doi: 10.1016/j.exer.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb L, Simon J. A2E: a component of ocular lipofuscin. Photochem Photobiol. 2004;79:127. doi: 10.1562/0031-8655(2004)079<0127:aacool>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Sabatini DM, Baur JA. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Invest. 2013;123:980–989. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen KB, Lamark T, Overvatn A, Harneshaug I, Johansen T, Bjorkoy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010;6:784–793. doi: 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang YS, Shen XF, Hui YN, Han J, Zhao W, Zhu J. Alterations of activity and intracellular distribution of the 20S proteasome in ageing retinal pigment epithelial cells. Exp Gerontol. 2008;43:1114–1122. doi: 10.1016/j.exger.2008.08.052. [DOI] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol. 2012;74:69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- Mitter SK, Song C, Qi X, Mao H, Rao H, Akin D, Lewin A, Grant M, Dunn W, Jr, Ding J, Bowes Rickman C, Boulton M. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014;10:1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Owsley C, McGwin G, Jackson GR, Heimburger DC, Piyathilake CJ, Klein R, White MF, Kallies K. Effect of short-term, high-dose retinol on dark adaptation in aging and early age-related maculopathy. Invest Ophthalmol Vis Sci. 2006;47:1310–1318. doi: 10.1167/iovs.05-1292. [DOI] [PubMed] [Google Scholar]
- Puertollano R. mTOR and lysosome regulation. F1000Prime Rep. 2014;6:52–52. doi: 10.12703/P6-52. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;12:478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Ryhanen T, Hyttinen JM, Kopitz J, Rilla K, Kuusisto E, Mannermaa E, Viiri J, Holmberg CI, Immonen I, Meri S, Parkkinen J, Eskelinen EL, Uusitalo H, Salminen A, Kaarniranta K. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med. 2009;13:3616–3631. doi: 10.1111/j.1582-4934.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva MM, Cano M, Handa JT. Nrf2 signaling is impaired in the aging RPE given an oxidative insult. Exp Eye Res. 2014;119:111–114. doi: 10.1016/j.exer.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P. Physiology of the lysosome. Oxford PharmaGenesis; 2006. [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt F, Bergmann M, Holz FG, Kopitz J. Isolation of intact lysosomes from human RPE cells and effects of A2-E on the integrity of the lysosomal and other cellular membranes. Graefes Arch Clin Exp Ophthalmol. 2002;240:983–988. doi: 10.1007/s00417-002-0558-8. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo j. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Klise A, Sergeev Y, Hose S, Bhutto IA, Hackler L, Jr, Malpic-Llanos T, Samtani S, Grebe R, Goldberg MF, Hejtmancik JF, Nath A, Zack DJ, Fariss RN, McLeod DS, Sundin O, Broman KW, Lutty GA, Zigler JS., Jr betaA3/A1-crystallin in astroglial cells regulates retinal vascular remodeling during development. Mol Cell Neurosci. 2008;37:85–95. doi: 10.1016/j.mcn.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Sun-Wada GH, Wada Y, Futai M. Lysosome and lysosome-related organelles responsible for specialized functions in higher organisms, with special emphasis on vacuolar-type proton ATPase. Cell Struct Funct. 2003;28:455–463. doi: 10.1247/csf.28.455. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJ, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, Ogura Y, Yoo JW, Lee DK, Provost P, Hinton DR, Nunez G, Baffi JZ, Kleinman ME, Ambati J. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toops KA, Tan LX, Jiang Z, Radu RA, Lakkaraju A. Cholesterol-mediated activation of acid sphingomyelinase disrupts autophagy in the retinal pigment epithelium. Mol Biol Cell. 2015;26:1–14. doi: 10.1091/mbc.E14-05-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- Valapala M, Edwards M, Hose S, Grebe R, Bhutto IA, Cano M, Berger T, Mak TW, Wawrousek E, Handa JT, Lutty GA, Samuel Zigler J, Jr, Sinha D. Increased Lipocalin-2 in the retinal pigment epithelium of Cryba1 cKO mice is associated with a chronic inflammatory response. Aging Cell. 2014b;13:1091–1094. doi: 10.1111/acel.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valapala M, Wilson C, Hose S, Bhutto IA, Grebe R, Dong A, Greenbaum S, Gu L, Sengupta S, Cano M, Hackett S, Xu G, Lutty GA, Dong L, Sergeev Y, Handa JT, Campochiaro P, Wawrousek E, Zigler JS, Jr, Sinha D. Lysosomal-mediated waste clearance in retinal pigment epithelial cells is regulated by CRYBA1/betaA3/A1-crystallin via V-ATPase-MTORC1 signaling. Autophagy. 2014;10:480–496. doi: 10.4161/auto.27292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D'Agostino VG, Govoni S, Pascale A, Agostini H, Petrovski G, Salminen A, Kaarniranta K. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013;8:e69563. doi: 10.1371/journal.pone.0069563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Anand M, Shiraz AK, Magrane J, Gao J, Vollmer-Snarr HR, Manfredi G, Finnemann SC. The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J Biol Chem. 2008;283:24770–24780. doi: 10.1074/jbc.M800706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cano M, Handa JT. p62 provides dual cytoprotection against oxidative stress in the retinal pigment epithelium. Biochim Biophys Acta. 2014;1843:1248–1258. doi: 10.1016/j.bbamcr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT Genetic Factors in AMD Study Group. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Young RW, Bok D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu P, Zhao Z, Cai J, Sternberg P, Chen Y. Subcellular distribution and activity of mechanistic target of rapamycin in aged retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2014;55:8638–8650. doi: 10.1167/iovs.14-14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Yasumura D, Li X, Matthes M, Lloyd M, Nielsen G, Ahern K, Snyder M, Bok D, Dunaief JL, LaVail MM, Vollrath D. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi A, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- Zigler JS, Jr, Zhang C, Grebe R, Sehrawat G, Hackler L, Jr, Adhya S, Hose S, McLeod DS, Bhutto I, Barbour W, Parthasarathy G, Zack DJ, Sergeev Y, Lutty GA, Handa JT, Sinha D. Mutation in the betaA3/A1-crystallin gene impairs phagosome degradation in the retinal pigmented epithelium of the rat. J Cell Sci. 2011;124:523–531. doi: 10.1242/jcs.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]


