Abstract
The proteasome system of Mycobacterium tuberculosis is required for causing disease. Proteasomes are multi-subunit chambered proteases and, until recently, were only known to participate in adenosine triphosphate (ATP)-dependent proteolysis in bacteria. In this review, we discuss the latest advances in understanding how both ATP-dependent and ATP-independent proteasome-regulated pathways contribute to M. tuberculosis virulence.
Keywords: Mycobacterium tuberculosis, proteasome, pupylation, pathogenesis
Tuberculosis Infections
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB), which affects nearly one-third of the world's population. If active TB is not properly treated, the disease can be fatal, making TB one of the greatest killers on earth. The latest report from the World Health Organization reported that in 2013, about nine million people developed TB and 1.5 million died from the disease [1]. Drug-resistant M. tuberculosis strains are on the rise, including the emergence of multi-drug resistant and extensively-drug resistant (XDR) resistant strains that are untreatable with the two most powerful anti-TB drugs (isoniazid and rifampicin), in addition to fluoroquinolones and at least one of three injectable second-line drugs. To date, cases of XDR-TB have been reported in at least 100 countries [2]. Thus, there is an urgent need to develop new drugs to battle the TB pandemic.
TB is contracted by the inhalation of air-borne droplets containing M. tuberculosis bacilli released from an infected individual (reviewed in [3]). Bacteria most commonly infect the lungs and reside in alveolar macrophages. If the host cannot control the infection, M. tuberculosis growth will result in the destruction of lung tissue and eventually the death of the host. Despite the high global mortality caused by TB, 90% of immunocompetent individuals harboring the tubercle bacillus are able to suppress bacterial growth, and often do not progress to active disease. Vertebrate hosts have several effective mechanisms to control microbial growth; in mammals, these include the production of reactive oxygen and reactive nitrogen species (ROS and RNS, respectively) by phagocytes (reviewed in [4, 5]). Nitric oxide (NO) is an antimicrobial free radical produced by the inducible NO synthase (iNOS or NOS2) in activated macrophages (reviewed in [6]). Among its potential activities, NO can penetrate bacterial membranes and combine with ROS such as superoxide to generate peroxynitrite, a highly toxic molecule. RNS and ROS can damage nucleic acids and proteins, and cause lipid peroxidation [7, 8]. Mice lacking iNOS are highly susceptible to M. tuberculosis, demonstrating a critical function for NO during infections [9]. While a role for NO in human tuberculosis is controversial, there is abundant indirect evidence suggesting NO participates in controlling bacterial growth in humans [10–15]. Relevant to this review, the investigation of NO resistance ultimately led to the identification of a bacterial proteasome as an important regulator of M. tuberculosis pathogenesis.
The M. tuberculosis Proteasome: Guardian of the Cell
For many years, Carl Nathan hypothesized that M. tuberculosis encodes gene products that allow it to resist NO-mediated killing in animals. To test this hypothesis, his team took a forward genetic approach and screened for M. tuberculosis mutants that were sensitive to NO in vitro, with the idea that these mutants would be attenuated in mice producing NO, but fully virulent in mice lacking iNOS. Darwin et al. [16] identified several NO-hypersensitive mutants with transposon insertions in genes predicted to encode components of a bacterial proteasome. The NO-hypersensitivity phenotype of these mutant strains was mimicked in wild-type (WT) M. tuberculosis treated with eukaryotic proteasome inhibitors, further supporting a role for proteasome-dependent proteolysis in NO resistance [16].
Proteasomes are responsible for the majority of cytosolic and nuclear proteolysis in eukaryotes [17]. In particular, the 26S proteasome is a multi-subunit, barrel-shaped protease composed of two functionally distinct sub-complexes, the 20S core particle (CP) and the 19S regulatory particle (RP). The 20S CP is composed of four stacked hetero-heptameric rings forming a cylinder, whereas the 19S RP is a complex of at least 19 proteins located at one end or both ends of the CPs (reviewed in [18]). RPs include a ring of six different ATPases that unfold and translocate substrates into CPs as well as non-ATPase subunits that function in various aspects of substrate recognition and processing. Before being targeted to the CP, most substrates need to be covalently modified with a 76-amino acid protein called ubiquitin (reviewed in [19]). Ubiquitylated proteins are recognized by the RP, and unfolded by the hexameric ATPase ring in order to deliver the protein into the 20S CP for degradation.
In bacteria, proteasomes are found in members of the order Actinomycetales [20–23]. Similar to eukaryotic proteasomes, bacterial 20S CPs are also barrel-shaped, with four stacked seven–subunit rings (two homo-heptameric β-subunit rings in between two homo-heptameric α-subunit rings), and are tightly gated to prevent unregulated proteolysis (reviewed in detail elsewhere [24]). Unlike their eukaryotic counterparts, complex RPs have not been identified with bacterial 20S CPs; however, ATPases similar to those found in eukaryotic RPs are found in all proteasome-bearing bacteria. Interestingly, robust interactions have not been detected between bacterial ATPases and 20S CPs in vitro.
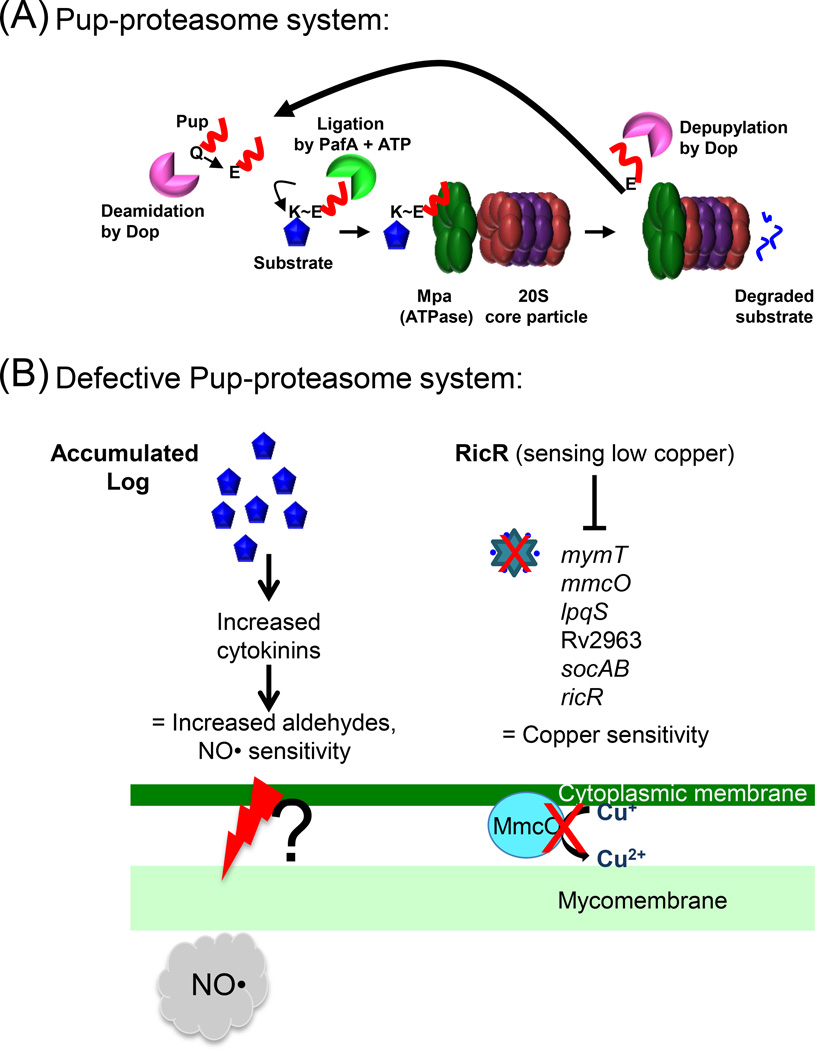
Over the past 10 years, we and other groups have characterized the biochemistry of proteasomal degradation in M. tuberculosis. Most critically, it was determined that many proteins destined for proteasomal degradation are post-translationally modified with the small protein Pup (prokaryotic ubiquitin-like protein), which functions like ubiquitin [25, 26]. Unlike ubiquitin, Pup is an intrinsically disordered protein [27, 28]. Pup is encoded upstream of the proteasome 20S CP genes, prcBA, and is translated with a carboxyl-terminal glutamine that must be converted to glutamate by the enzyme Dop (deamidase of Pup). Proteasome accessory factor A (PafA) then catalyzes the formation of an isopeptide bond between Pup and a lysine of a target protein [26, 29, 30]. Thereafter, Pup interacts with the mycobacterial proteasome ATPase Mpa for unfolding and translocation of the pupylated substrate into the 20S CP [26, 29, 31]. Additionally, Dop can remove Pup from substrates prior to degradation, and Mpa appears to facilitate this process [32–36] (Figure 1A). Over 60 confirmed targets of pupylation have been identified in M. tuberculosis, all of which require PafA for modification [37]. Unlike eukaryotes that have hundreds if not over a thousand ubiquitin ligases, PafA incredibly appears to be the only Pup ligase in bacteria.
Figure 1. The Pup-proteasome System (PPS) of M. tuberculosis.
(A) ATP-dependent degradation by the proteasome. 20S core particle subunits are colored in red (α-subunit, PrcA) and purple (β-subunit, PrcB). Pup (red line) is an intrinsically disordered protein. In M. tuberculosis, the last amino acid of Pup is glutamine (Gln, Q), which must be deamidated on its side chain by Dop to convert it to glutamate (Glu, E). The Glu is the phosphorylated by PafA, making it prime for attack by the amino group on the side chain of a substrate lysine (Lys, K). Although Pup is largely disordered, it forms helices upon interacting with amino-terminal coiled-coil domains of Mpa. This interaction is essential for targeting pupylated proteins for degradation. In M. tuberculosis, Pup appears to be recycled by Dop in an Mpa-dependent manner, suggesting depupylation can occur just before proteins are threaded into the proteasome core particle. See text for additional details and references. (B) Disrupted proteasomal degradation results in several dysregulated pathways. Left: Accumulation of the enzyme Lonely Guy (Rv1205, Log) results in the hyperproduction of cytokinins, which can break down into aldehydes. Aldehydes synergize with NO by an unknown mechanism to kill mycobacteria. A mechanism of aldehyde-mediated toxicity might include damage (as represented by the red lightening bolt) to the mycobacterial membranes. This damage may be exacerbated in the presence of host-produced NO. Right: The RicR regulon is partially repressed in mpa and pafA mutants, leading to reduced copper (Cu) resistance. Red Xs indicate loss of MymT (Cu metallothionein) and MmcO (mycobacterial multi-copper oxidase) activities.
In addition to observations that mutations in Pup-proteasome system (PPS) genes result in increased NO susceptibility in vitro, Nathan and colleagues also determined that PPS mutants are highly attenuated in vivo [16, 38]. PPS mutant strains do not grow to high numbers in the lungs or spleens of mice, and in the case of prcBA mutants, the bacteria are eventually cleared [39, 40]. Interestingly, an mpa mutant in the CDC1551 M. tuberculosis background has a conspicuous in vitro growth defect [41], which is not observed in the H37Rv M. tuberculosis background [16]. In addition, Bishai and co-workers showed that besides reduced numbers of bacteria in the lungs of the mpa mutant-infected mice, there are also reduced levels of interferon-γ (IFN-γ) production in the infected animals [41]. It is unclear if this is simply due to lower bacterial numbers, or if disruption of the PPS alters the immune response in another way.
While it remains to be determined if all pupylation leads to protein degradation, proteasome-dependent proteolysis is nonetheless an essential weapon in an arsenal that allows the robust growth and persistence of M. tuberculosis in animals.
NO: You Win or You Die
Although the Nathan lab determined that a link exists between proteasome activity and NO resistance over 10 years ago, it remained unknown how proteolysis protected against NO toxicity. There were two main hypotheses: the proteasome degrades damaged proteins that would otherwise misfold, aggregate and ultimately kill the bacteria; or, the accumulation of one or more specific proteasome substrates exacerbates NO toxicity. Recently, the question of why PPS mutants are more sensitive to NO than WT bacteria may have been at least partially answered. A suppressor screen for secondary mutations in an mpa mutant background that could revert its NO-hypersensitive phenotype to WT identified insertions in a previously uncharacterized gene, Rv1205. Remarkably, genetic disruption of Rv1205 not only restores WT NO resistance to an mpa mutant but also partially restores virulence in mice. Rv1205 encodes a proteasome substrate that accumulates in mpa, pafA, and prcBA mutants [42]. Based on structural homology searches and deep bioinformatics analysis, Rv1205 was determined to be homologous to members of the LONELY GUY (LOG) family of plant enzymes that catalyze the last step of cytokinin biosynthesis [43].
Cytokinins are plant hormones that stimulate transcriptional responses via two-component regulatory systems to shape plant development (reviewed in [44, 45]). In order to produce a cytokinin, a precursor molecule, which is an adenosine monophosphate carrying either an isoprene or aromatic group at the position six nitrogen (N6) of the adenine base, must be processed by LOG to remove the phosphoribose group; the liberated modified adenine base is the bioactive cytokinin. Because Rv1205 has phosphoribohydrolase activity like plant LOGs, Rv1205 is referred to as "Log" [42].
From these studies, it was found that WT M. tuberculosis secretes cytokinins. Importantly, an mpa mutant produces cytokinins in strikingly greater abundance, correlating with the accumulation of Log in this strain, whereas a log mutant shows dramatically reduced cytokinin production. Interestingly, the accumulation of cytokinins is not directly accountable for sensitizing M. tuberculosis to NO. A metabolomics analysis revealed that an mpa mutant accumulates high concentrations of a cytokinin breakdown product, para-hydroxybenzaldehyde (pHBA). Cytokinins can be broken down into adenine and aldehydes (reviewed in [46]) and while pHBA alone is not toxic to M. tuberculosis, the addition of NO with pHBA or another cytokinin aldehyde results in robust bacterial killing in vitro [42]. It is unknown how pHBA and NO synergize to kill; it is possible that the aldehydes react with proteins or other molecules in the mycobacterial membrane, which sensitizes it to NO or other RNS (Figure 1B). Taken together, the tight regulation of Log levels by the PPS is essential to prevent unintentional NO hypersensitivity during infections.
What is the function of Log and cytokinins in M. tuberculosis? This question has yet to be answered, but we speculate that cytokinins have a signaling role during infections. In addition to plants using cytokinins as signal transducers, microbial pathogens and symbionts can subvert plant development by producing cytokinins; this can include tumor formation, which promotes microbial growth in plant tissues (reviewed in [47]). Thus, it is tempting to speculate that M. tuberculosis, which is never found naturally outside of humans, uses cytokinins to signal transcriptional changes in mammalian cells to favor its growth in vivo. Alternatively, cytokinins could be used among the bacteria to signal specific messages during infection to alter the bacterial transcriptome and promote their survival.
It is worth discussing here a previously reported and somewhat curious observation: the complementation of an M. tuberculosis prcBA deletion-disruption mutant (ΔprcBA::hyg) with a prcBA allele encoding a catalytically inactive 20S CP can restore NO resistance to WT levels in vitro, but not WT persistence in mice [39]. Recent studies have shown that the production of inactive 20S CPs in M. tuberculosis is highly effective in trapping known proteasome substrates [64]. Thus, it is possible that catalytically inactive 20S CPs are able to trap substrates and reduce the amount of free or active protein in the bacterial cytosol. For example, trapping and inactivating Log in this way might be sufficient to prevent the accumulation of aldehydes in vitro. In contrast, long-term infections of M. tuberculosis likely requires the proteasomal degradation of numerous substrates affecting various pathways required for successful growth in a mammalian host. Proteasomal degradation might also simply be necessary to recycle amino acids or liberate other nutrient sources such as metal co-factors during an infection.
The Metal Throne
In addition to providing resistance to host-derived NO, the proteasome regulates other pathways important for pathogenesis. As with other chambered proteases that degrade transcription factors (reviewed in [48–50]), it was hypothesized that the M. tuberculosis proteasome could affect gene expression. A transcriptomic analysis of mpa and pafA mutants revealed the dysregulation of several genes regulated by zinc and copper [51]. Members of the Zur (zinc uptake regulator) regulon are up regulated in mpa and pafA mutants compared to WT bacteria. Zur is a zinc-sensing transcriptional repressor, which under low zinc concentrations is released from the operators of eight M. tuberculosis promoters that control the expression of 24 genes [52]. Among them is the esx-3 operon, which encodes an essential type VII secretion system involved in iron and zinc uptake in M. tuberculosis [53–56]. While we do not know how the increased expression of the Zur regulon affects pathogenesis (if at all), it seems likely that the perturbation of zinc and iron homeostasis would disrupt the physiology of M. tuberculosis during infections.
The transcriptomic analysis of PPS mutants also identified a copper-responsive regulon unique to pathogenic mycobacteria. The RicR (regulated in copper repressor) regulon includes five loci distributed throughout the M. tuberculosis chromosome and is repressed in both mpa and pafA mutants. RicR is a paralog of CsoR (copper sensitive operon repressor), which is released from its operators when bound to copper [51, 57, 58]. When the concentration of intracellular copper is low, RicR represses transcription of ricR itself, mymT (encoding a copper metallothionein), lpqS (encoding a putative lipoprotein), Rv2963 (encoding a putative permease), socAB (small ORF induced by copper A and B), and mmcO (mycobacterial multi-copper oxidase) [51]. Metallothioneins are small, typically cysteine-rich proteins that sequester metals to protect against metal-catalyzed toxicity. MymT binds up to six copper (Cu+1) ions, and a mymT mutant is hypersensitive to copper in vitro [59]. Similarly, a mutant deficient in the multi-copper oxidase MmcO, which oxidizes Cu+1 to Cu+2, is sensitive to copper in vitro [60, 61]. While single mutants of mymT, mmcO, or any other RicR-regulated gene do not have virulence defects in mice [59, 61], constitutive repression of the entire RicR regulon results in a strong growth defect in vivo [61]. Similarly, mpa and pafA mutants, in which the entire ricR regulon is partially repressed, are also hypersensitive to copper [61] (Figure 1B).
Several studies suggest that either host-restricted access to metals (e.g. iron, zinc, and manganese) or host-imposed metals (e.g. copper and zinc) can play a role in immunity against pathogens (reviewed in [62, 63]). It would thus be unsurprising if defects in metal homeostasis were at least partially responsible for the attenuated phenotype of M. tuberculosis PPS mutants in mice. We do not yet understand how the PPS is connected to the regulation of the Zur and RicR regulons, and there are currently no data to suggest either regulator is a substrate of the PPS. One possibility is there is an accumulation of proteasome substrates in PPS mutants that are metal binding proteins, resulting in the increased sequestration of metals. The accumulation of a zinc- or copper-binding protein could potentially result in the induction of the Zur regulon or repression of the RicR regulon, respectively.
Different Roads Sometimes Lead to the Same 20S Proteasome
It was noted that defects in mpa and pafA result in a subtle growth defect in vitro, whereas a prcBA mutant has a dramatic growth defect in liquid and, in particular, on solid medium [39, 40]. It was thus speculated that there were additional ways for proteins to be targeted to the 20S proteasome in M. tuberculosis. This hypothesis was finally tested when a new proteasome activator was discovered that promotes the degradation of peptides or unfolded proteins by the mycobacterial proteasome without using ATP or Pup [64, 65]. This protein, PafE (proteasome accessory factor E; also known as Bpa for bacterial proteasome activator), forms dodecamers and is functionally homologous to several eukaryotic ATP-independent proteasome activators (reviewed in [66]). Similar to other proteasome activators, PafE caps the ends of 20S CPs and enhances proteasomal degradation, presumably by opening the entrance to the 20S CP to allow proteins to enter [64, 65].
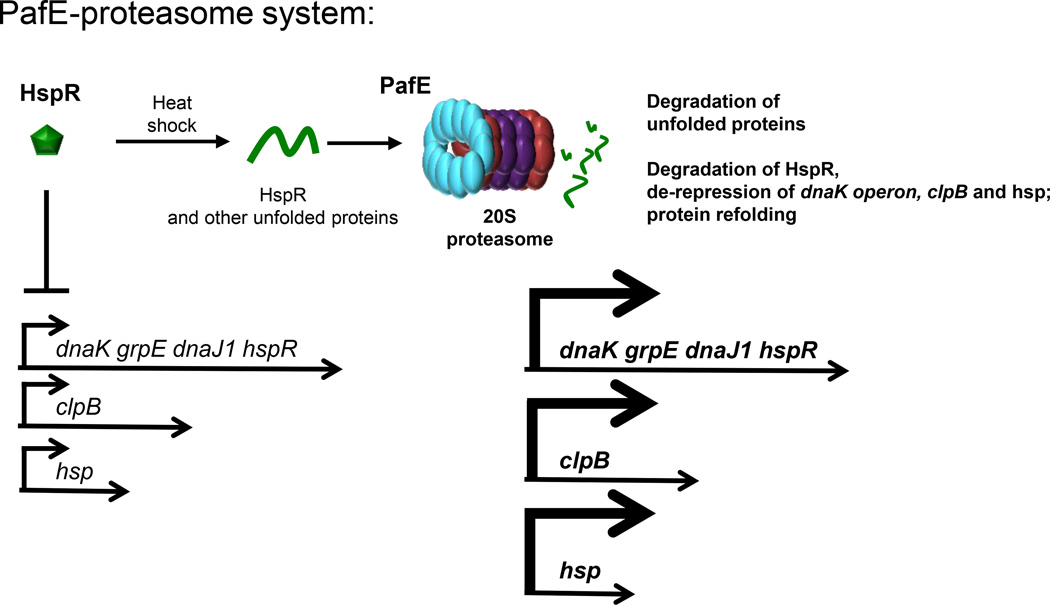
A proteomic search for PafE-dependent proteasome substrates revealed a distinct class of proteins that so far do not appear to be targets of pupylation [64]. Among the most abundant proteins to accumulate in a pafE mutant is the heat shock protein repressor (HspR). In M. tuberculosis, HspR is encoded by the last gene of the dnaK (or hsp70) operon and represses expression from the dnaK promoter, as well as the promoters of two other heat-shock responsive protein chaperone genes (Figure 2). The dnaK operon encodes a chaperone system that is essential for protein quality control during heat shock (reviewed in [67]). Upon heat shock, it is hypothesized that HspR partially unfolds in order to detach from its promoter to allow expression of the dnaK operon. When first described, it was unknown if HspR itself was degraded or simply failed to bind DNA during heat shock [68]. Jastrab et al. showed that PafE stimulates proteasomal degradation of HspR, and at elevated temperatures HspR is even more rapidly degraded in vitro [68]. These data suggest that the denaturation of HspR functions to both release the repressor from the dnaK operator and facilitate its degradation by the PafE-proteasome (Figure 2).
Figure 2. The PafE-proteasome System of M. tuberculosis.
PafE (blue) forms dodecameric rings and caps 20S CPs. This allows the ATP-independent opening of 20S CPs to facilitate the degradation of peptides and denatured proteins as well as HspR. HspR represses expression of the dnaK operon, clpB and hsp (Rv0249c). See text for details.
The observation that the PafE-proteasome degrades unfolded proteins suggests it could help minimize proteotoxic stress during heat shock by at least two ways: to degrade any misfolded proteins before they aggregate and to increase production of the DnaK protein quality control system to minimize misfolding of newly translated proteins (Figure 2). While these may be the primary functions of PafE, future studies will determine if this road to degradation regulates other aspects of M. tuberculosis pathogenesis (see Outstanding Questions).
Finally, PafE and Mpa do not appear to work together on the same 20S CP. Delley et al. reported that PafE can prevent the degradation of a pupylated substrate by Mpa and 20S CPs in vitro, suggesting PafE competes with Mpa for 20S CP access [65]. Moreover, proteomics analysis of a M. tuberculosis ΔpafE::hyg mutant demonstrated that the pupylome differs from the ‘PafEylome’ [64]. Perhaps most relevantly, PafE is exclusively found in proteasome-bearing bacteria, unlike Mpa, further suggesting an independent role for PafE in proteolysis. While we cannot rule out that 20S CPs can be simultaneously capped with PafE and Mpa, it seems more likely that these two proteasome activators function independently and for different purposes.
Concluding Remarks and Future Directions
Proteolysis should never be dismissed as ‘housekeeping’. Regulated protein degradation by proteasomes and other proteases is critical for the normal function of all living organisms. For M. tuberculosis, proteasomal degradation affects numerous pathways that are essential for it to be a successful pathogen.
Part of the success of M. tuberculosis lies in its ability to survive within its host for a long or latent period without causing overt symptoms, and until waning immunity allows the bacteria to revive and cause disease. In addition to host immune effectors like RNS and ROS, bacteria face inhospitable conditions such as hypoxia and nutrient starvation during latency [69–71]. In eukaryotes, proteasomal degradation can have nutritional roles under starvation conditions through the recycling of amino acids [72, 73]. In M. smegmatis, a non-pathogenic, saprophytic Mycobacterium species, the PPS is needed for survival under conditions of nitrogen limitation and, to a lesser extent, during carbon starvation [74]. It is possible that nutrient limitation is one reason why a proteasome core protease mutant is unable to persist in mice [39, 40].
Is winter finally coming for TB? Because the proteasome is essential for M. tuberculosis to cause lethal infections, it is a highly attractive drug target. Considerable effort is being placed on the inhibition of the 20S CP of M. tuberculosis with the expectation that inhibitors specific to the M. tuberculosis proteasome, and ineffective against mammalian proteasomes, can be developed [75, 76]. In contrast to the 20S CP, other components of the PPS (i.e. Dop and PafA) and PafE are biochemically distinct from those of the eukaryotic proteasome system. As such, these enzymes may provide more specific targets for the development of TB drugs. In addition, a recent report identified peptide aptamers that could target Pup to prevent it from interacting with Mpa, thus inhibiting proteolysis of a pupylated protein [77]. Although this required the production of the peptide within mycobacteria, this study nonetheless showed that Pup, which has no enzymatic activity, could be targeted. In addition to providing hope for finding new TB therapeutics, the continued characterization of pathways regulated by the M. tuberculosis proteasome will undoubtedly reveal new insights into the physiology of one of the world's most successful pathogens.
Outstanding Questions.
What are the roles of cytokinins during tuberculosis infections?
What are the sources of copper and zinc during infections?
How is the proteasome linked to the Zur and RicR regulons?
Does proteasome-dependent proteolysis help generate nutrients during infections?
What are the other functions of Mpa-dependent and PafE-dependent proteasomes?
Trends.
The proteasome is a highly regulated protease that is required for the pathogenesis of Mycobacterium tuberculosis.
Both ATP-dependent and ATP-independent pathways to the proteasome are needed to promote bacterial virulence.
The M. tuberculosis proteasome degrades an enzyme that produces cytokinins, which break down into aldehydes that synergize with nitric oxide to kill bacteria.
Defects in proteasomal degradation perturb metal homeostasis, including zinc and copper regulated genes required for pathogenesis.
Proteasome function is required for a robust heat shock response.
Acknowledgments
We thank Samuel Becker and Jordan Jastrab for critical review of a draft version of this manuscript, and the anonymous reviewers for excellent suggestions. Tuberculosis proteasome research in the K.H. Darwin lab is supported by NIH grants HL92774 and AI088075. K.H.D. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. M.I.S. received support from the Jan T. Vilcek Endowed Fellowship fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. World Health Organization; 2014. Tuberculosis fact sheet 2014. [Google Scholar]
- 2.WHO. 2014 Update. World Health Organization; 2014. Multidrug-Resistance Tuberculosis (MDR-TB) [Google Scholar]
- 3.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 4.Bowman LA, McLean S, Poole RK, Fukuto JM. The diversity of microbial responses to nitric oxide and agents of nitrosative stress close cousins but not identical twins. Adv Microb Physiol. 2011;59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 5.Fang FC. Antimicrobial actions of reactive oxygen species. MBio. 2011;2 doi: 10.1128/mBio.00141-11. e00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 8.Szabo C. Multiple pathways of peroxynitrite cytotoxicity. Toxicol Lett. 2003;140–141:105–112. doi: 10.1016/s0378-4274(02)00507-6. [DOI] [PubMed] [Google Scholar]
- 9.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson S, Bonecini-Almeida Mda G, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, Ho JL. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–2302. [Google Scholar]
- 11.Wang CH, Liu CY, Lin HC, Yu CT, Chung KF, Kuo HP. Increased exhaled nitric oxide in active pulmonary tuberculosis due to inducible NO synthase upregulation in alveolar macrophages. Eur Respir J. 1998;11:809–815. doi: 10.1183/09031936.98.11040809. [DOI] [PubMed] [Google Scholar]
- 12.Choi HS, Rai PR, Chu HW, Cool C, Chan ED. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am J Respir Crit Care Med. 2002;166:178–186. doi: 10.1164/rccm.2201023. [DOI] [PubMed] [Google Scholar]
- 13.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Modlin RL. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 14.Rockett KA, Brookes R, Udalova I, Vidal V, Hill AV, Kwiatkowski D. 1,25-Dihydroxyvitamin D3 induces nitric oxide synthase and suppresses growth of Mycobacterium tuberculosis in a human macrophage-like cell line. Infect Immun. 1998;66:5314–5321. doi: 10.1128/iai.66.11.5314-5321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung JY, Madan-Lala R, Georgieva M, Rengarajan J, Sohaskey CD, Bange FC, Robinson CM. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect Immun. 2013;81:3198–3209. doi: 10.1128/IAI.00611-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 17.Arrigo AP, Tanaka K, Goldberg AL, Welch WJ. Identity of the 19S 'prosome' particle with the large multifunctional protease complex of mammalian cells (the proteasome) Nature. 1988;331:192–194. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- 18.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 20.Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Baumeister W. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 21.Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 22.Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pouch MN, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol Microbiol. 2000;35:368–377. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 24.Samanovic MI, Li H, Darwin KH. The Pup-Proteasome System of Mycobacterium tuberculosis. Subcell Biochem. 2013;66:267–295. doi: 10.1007/978-94-007-5940-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 29.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 30.Guth E, Thommen M, Weber-Ban E. Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated pup intermediate. J Biol Chem. 2011;286:4412–4419. doi: 10.1074/jbc.M110.189282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol. 2010;17:1352–1357. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH. "Depupylation" of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol Cell. 2010;39:821–827. doi: 10.1016/j.molcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol Microbiol. 2010;77:1123–1135. doi: 10.1111/j.1365-2958.2010.07276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delley CL, Striebel F, Heydenreich FM, Ozcelik D, Weber-Ban E. Activity of the mycobacterial proteasomal ATPase Mpa is reversibly regulated by pupylation. J Biol Chem. 2012;287:7907–7914. doi: 10.1074/jbc.M111.331124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2010;75:744–754. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- 36.Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol. 2010;192:2933–2935. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected] PLoS One. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 39.Gandotra S, Lebron MB, Ehrt S. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6:e1001040. doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamichhane G, Raghunand TR, Morrison NE, Woolwine SC, Tyagi S, Kandavelou K, Bishai WR. Deletion of a Mycobacterium tuberculosis proteasomal ATPase homologue gene produces a slow-growing strain that persists in host tissues. J Infect Dis. 2006;194:1233–1240. doi: 10.1086/508288. [DOI] [PubMed] [Google Scholar]
- 42.Samanovic MI, Tu S, Novak O, Iyer LM, McAllister FE, Aravind L, Darwin KH. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell. 2015;57:984–994. doi: 10.1016/j.molcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 44.Argueso CT, Ferreira FJ, Kieber JJ. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009;32:1147–1160. doi: 10.1111/j.1365-3040.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 45.Muller B. Generic signal-specific responses: cytokinin and context-dependent cellular responses. J Exp Bot. 2011;62:3273–3288. doi: 10.1093/jxb/erq420. [DOI] [PubMed] [Google Scholar]
- 46.Frebort I, Kowalska M, Hluska T, Frebortova J, Galuszka P. Evolution of cytokinin biosynthesis and degradation. J Exp Bot. 2011;62:2431–2452. doi: 10.1093/jxb/err004. [DOI] [PubMed] [Google Scholar]
- 47.Naseem M, Wolfling M, Dandekar T. Cytokinins for immunity beyond growth, galls and green islands. Trends Plant Sci. 2014;19:481–484. doi: 10.1016/j.tplants.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 50.Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 51.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Darwin KH. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Manganelli R. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serafini A, Pisu D, Palu G, Rodriguez GM, Manganelli R. The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One. 2013;8:e78351. doi: 10.1371/journal.pone.0078351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegrist MS, Steigedal M, Ahmad R, Mehra A, Dragset MS, Schuster BM, Rubin EJ. Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio. 2014;5:e01073–e01014. doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 58.Tan BG, Vijgenboom E, Worrall JA. Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucleic Acids Res. 2014;42:1326–1340. doi: 10.1093/nar/gkt902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, Nathan C. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol. 2008;4:609–616. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowland JL, Niederweis M. A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J Bacteriol. 2013;195:3724–3733. doi: 10.1128/JB.00546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi X, Festa RA, Ioerger TR, Butler-Wu S, Sacchettini JC, Darwin KH, Samanovic MI. The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence. MBio. 2014;5 doi: 10.1128/mBio.00876-13. e00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jastrab JB, Wang T, Murphy JP, Bai L, Hu K, Merkx R, Darwin KH. An adenosine triphosphate-independent proteasome activator contributes to the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2015;112:E1763–E1772. doi: 10.1073/pnas.1423319112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delley CL, Laederach J, Ziemski M, Bolten M, Boehringer D, Weber-Ban E. Bacterial proteasome activator Bpa (Rv3780) is a novel ring-shaped interactor of the mycobacterial proteasome. PLoS One. 2014;9:e114348. doi: 10.1371/journal.pone.0114348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Das Gupta T, Bandyopadhyay B, Das Gupta SK. Modulation of DNA-binding activity of Mycobacterium tuberculosis HspR by chaperones. Microbiology. 2008;154:484–490. doi: 10.1099/mic.0.2007/012294-0. [DOI] [PubMed] [Google Scholar]
- 69.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 70.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 71.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 2009;11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 72.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 73.Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elharar Y, Roth Z, Hermelin I, Moon A, Peretz G, Shenkerman Y, Gur E. Survival of mycobacteria depends on proteasome-mediated amino acid recycling under nutrient limitation. EMBO J. 2014;33:1802–1814. doi: 10.15252/embj.201387076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Nathan C. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461:621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin G, Li D, Chidawanyika T, Nathan C, Li H. Fellutamide B is a potent inhibitor of the Mycobacterium tuberculosis proteasome. Arch Biochem Biophys. 2010;501:214–220. doi: 10.1016/j.abb.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cobbert JD, DeMott C, Majumder S, Smith EA, Reverdatto S, Burz DS, Shekhtman A. Caught in action: selecting peptide aptamers against intrinsically disordered proteins in live cells. Sci Rep. 2015;5:9402. doi: 10.1038/srep09402. [DOI] [PMC free article] [PubMed] [Google Scholar]