Abstract
Optineurin is a cytosolic protein encoded by the OPTN gene. Mutations of OPTN are associated with normal tension glaucoma and amyotrophic lateral sclerosis. Autophagy is an intracellular degradation system that delivers cytoplasmic components to the lysosomes. It plays a wide variety of physiological and pathophysiological roles. The optineurin protein is a selective autophagy receptor (or adaptor), containing an ubiquitin binding domain with the ability to bind polyubiquitinated cargoes and bring them to autophagosomes via its microtubule-associated protein 1 light chain 3-interacting domain. It is involved in xenophagy, mitophagy, aggrephagy, and tumor suppression. Optineurin can also mediate the removal of protein aggregates through an ubiquitin-independent mechanism. This protein in addition can induce autophagy upon overexpression or mutation. When overexpressed or mutated, the optineurin protein also serves as a substrate for autophagic degradation. In the present review, the multiple connections of optineurin to autophagy are highlighted.
Keywords: Optineurin (OPTN), Autophagy, Ubiquitin–proteasome system (UPS), Normal tension glaucoma (NTG), Amyotrophic lateral sclerosis (ALS), Autophagy receptor, Autophagy inducer, Protein aggregate, Mitophagy, Tumor suppressor, Aggrephagy, Xenophagy
1. Introduction
Optineurin is a gene linked to normal tension glaucoma (NTG) and amyotrophic lateral sclerosis (ALS) (Kachaner et al., 2012b; Osawa et al., 2011; Ying and Yue, 2012). It is also associated with Paget's disease of the bone (Albagha et al., 2010). This gene encodes a cytosolic protein that interacts with a number of proteins and participates in basic cellular functions such as vesicle trafficking, maintenance of the Golgi apparatus, NF-κB pathway, anti-bacterial and antiviral signaling, cell division control, and autophagy. Mutation or level alteration of optineurin results in adverse consequences in the cells leading to diseases (Gao et al., 2014; Park et al., 2010; Turturro et al., 2014). The molecular mechanisms, however, are largely not understood.
The ubiquitin–proteasome system (UPS) and autophagy are two main systems by which the cell degrades cytoplasmic constituents. The UPS targets short-lived or abnormally folded proteins, while the autophagy targets long-lived macromolecular complexes and organelles (Glickman and Ciechanover, 2002; Kirkin et al., 2009).
Degradation of a protein via the UPS involves two successive steps: tagging of the substrate protein by covalent attachment of single or multiple ubiquitin molecules and the subsequent degradation of the tagged protein by the 26S proteasome. At the tagging or conjugation stage, ubiquitin is covalently attached to the protein substrate through a series of ATP-dependent enzymatic reactions by E1 (ubiquitin activating), E2 (ubiquitin conjugating) and E3 (ubiquitin ligating) enzymes (Glickman and Ciechanover, 2002; Pickart, 2004). This process renders the ubiquitinated protein to be recognized by the proteasome and be degraded to small peptides (Zheng et al., 2014).
Autophagy (cellular self-eating or self-digestion) is a basic catabolic mechanism that involves bulk cell degradation of cellular components (Klionsky, 2005). There are three forms of autophagy: macroautophagy, microautophagy (Mijaljica et al. 2011), and chaperone-mediated autophagy (Kaushik and Cuervo, 2012). Among them, macroautophagy (herein referred to as autophagy) is the major pathway to eradicate damaged cell organelles or unused proteins. It is initiated with the sequestration of cytoplasmic components such as the entire organelles, lipid vesicles, or protein aggregates within double-membrane vesicles (so-called autophagosomes). These vesicles are then fused with lysosomes to generate autolysosomes, in which the autophagic cargo is degraded by acidic hydrolases (Galluzzi et al., 2014; Klionsky, 2005). Autophagy can be relatively nonselective, virtually any portion of the cytoplasm can be targeted to lysosomal degradation, triggered by nutrient deprivation. It can also be highly selective, triggered by damaged organelle or intracellular pathogens (Mizushima and Komatsu, 2011). Defects in the autophagic machinery have been associated with diseases, including aging, cancer, neurodegenerative diseases, cardiovascular disorders, and infectious/inflammatory conditions.
The UPS and autophagy were regarded originally as independent and separate pathways, but were found more recently to be closely connected (Wojcik, 2013). The crosstalk between ubiquitination and autophagy is guided by autophagy receptors or adaptors such as multi-domain scaffold/adaptor protein p62/sequestosome-1 (p62/SQSTM-1) and nuclear domain 10 protein 52 (NDP52). These receptors/adaptors can bind both ubiquitin and autophagy-related gene 8 family members microtubule-associated protein 1A/1B-light chain 3/γ-aminobutyric acid receptor-associated protein (LC3/GABARAP) (Wilde et al., 2011) and act as a bridge recognizing selective ubiquitinated proteins or cargoes and bringing them into autophagosomes (Komatsu et al., 2007).
Similar to p62, optineurin has also been shown to be an autophagy receptor or adaptor. In addition, optineurin, upon upregulation or mutation, can induces autophagy and become a substrate for autophagic clearance. Accumulating evidence indicates that optineurin partakes in various cellular functions through autophagy. Herein, we briefly illustrate the multiple connections of optineurin with autophagy.
2. Optineurin gene and protein
2.1. Gene structure, and mutations in diseases
The human optineurin gene (OPTN) is located at chromosome 10p13 and spans about a 37 kb genomic region. Its mRNA contains a total of 16 exons. The first 3 exons (exons 1-3) are noncoding sequence and the remaining13 exons (exons 4-16) code for a 577-amino acid (aa) protein (Rezaie et al., 2005; Ying and Yue, 2012).
The OPTN gene was found by Rezaie et al. in 2002 to be a disease-causing gene in NTG, a subtype of primary open angle glaucoma (POAG). Four mutations in OPTN, Glu50→Lys (E50K), Met98→Lys (M98K), Arg545→Gln (R545Q), and 691_692insAG (2-bp “AG” insertion), were detected from 54 families with adult-onset POAG in which most displayed normal intraocular pressure (Rezaie et al., 2002). Among the mutations, E50K is seen associated with a more progressive and severe disease (Aung et al., 2005; Hauser et al., 2006). Other OPTN alterations observed include Lys322→Glu (E322K), His26→Asp (H26D), Glu103→Asp (E103D), Val148→Val (V148V), intron IVS7+24G→A, and His486→Arg (H486R) (Leung et al., 2003; Willoughby et al., 2004).
OPTN mutations were also reported in patients with ALS. Maruyama et al. (2010) identified a homozygous deletion of exon 5, a homozygous nonsense Gln398→stop (Q398X) and a heterozygous missense Glu478→Gly (E478G) mutations in Japanese ALS patients. Besides exon 5 deletion, exon 1-5 and 3-5 heterozygous OPTN deletions were also detected, indicating that the exon might be a hotspot for OPTN deletion in ALS and that OPTN deletions are ALS-specific events (Iida et al., 2012). Other OPTN alterations reported in ALS include 382_383insAG (691_692insAG or 2-bp “AG” insertion), Arg96→Leu (R96L), Gln165→stop (Q165X), Gln454→Glu (Q454E), and a heterozygous truncating mutation p.Lys440→Asnfs*8 (c.1320delA) that causes a frameshift and a premature stop codon ((Weishaupt et al., 2013). The 382_383insAG or 691_692insAG mutation has been previously described in familial POAG (Rezaie et al., 2002). Additionally, 3 POAG alterations (M98K, E322K, and R545Q) were also noted in ALS (Weishaupt et al., 2013).
2.2. Protein structure and function
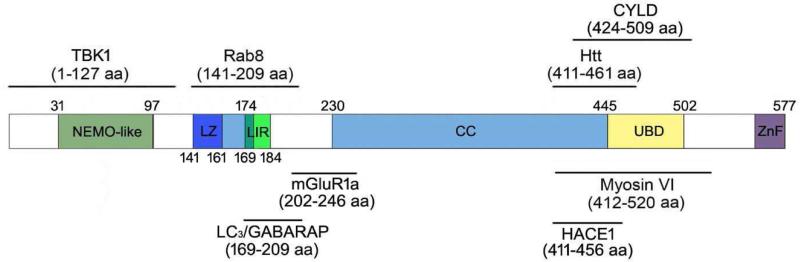
The optineurin protein is expressed in many tissues including the heart, brain, skeletal muscle, liver, and the eye (Li et al., 1998; Rezaie et al., 2005). The protein contains a NF-κB-essential molecule (NEMO)-like domain, leucine zipper and coiled-coil motifs, an ubiquitin-binding domain (UBD), a LC3-interacting region (LIR), and a carboxyl (C)-terminal C2H2 type of zinc finger (Ying and Yue, 2012). The endogenous or ectopically expressed optineurin has been shown to interact with itself to localize in foci and form high molecular weight protein complexes (homo-oligomers) in cells (Gao et al., 2014). It also binds with Ras-related protein 8, huntingtin, myosin VI, transferrin receptor, metabotropic glutamate receptor, transcription factor IIIA, serine/threonine kinase receptor-interacting protein 1, CYLD that is a product of a familial cylindromatosis tumor-suppressor gene (Nagabhushana et al., 2011), LC3/GABARAP (Ying and Yue, 2012), polo-like kinase 1 (Kachaner et al., 2012a), TANK (TRAF-associated NF-kB activator) binding kinase 1 (TBK1), and HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1; Liu et al., 2014). These interactions, shown in Figure 1, depict basic optineurin functions or participation in cellular processes (Kachaner et al., 2012b; Ying and Yue, 2012) such as vesicle trafficking, maintenance of the Golgi apparatus, NF-κB pathway, anti-bacterial and antiviral signaling, cell division control (Kachaner et al., 2012a), and autophagy (Chalasani et al., 2014; Liu et al., 2014; Shen et al., 2011; Wong and Holzbaur, 2014). Optineurin has in addition been found to be phosphorylated, which is essential for its functions in mitosis (Kachaner et al., 2012a) and autophagic signaling pathway (Korac et al., 2013; Rogov et al., 2013; Sorbara and Girardin, 2014; Weidberg and Elazar, 2011; Wild et al., 2011; Wong and Holzbaur, 2014).
Figure 1.
Schematic representation of human optineurin protein domains and the binding sites of optineurin-interacting proteins. CC, coiled-coil; CYLD, a protein encoded by a familial cylindromatosis tumor-suppressor gene; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; Htt, huntingtin; LZ, leucine zipper domain; LC3/GABARAP, microtubule-associated protein 1A/1B-light chain 3/γ-aminobutyric acid receptor-associated protein; LIR, LC3-interacting region; NEMO-like, NF-κB-essential molecule-like domain; mGluR1a, metabotropic glutamate receptor 1a; Rab8, Ras-related protein; TBK1, TANK (TRAF-associated NF-kB activator) binding kinase 1; UBD, ubiquitin-binding domain; ZnF, zinc finger; aa, amino acid.
The endogenous optineurin has been demonstrated to be a short-lived protein with a half-life of approximately 8 h (Shen et al., 2011). In a normal homeostatic situation, the turnover of endogenous optineurin involves mainly UPS (Shen et al., 2011).
3. Optineurin as an autophagy receptor/adapter
The optineurin protein contains an UBD domain ( 445-502 aa) near its C-terminus (Wagner et al., 2008) and can be ubiquitinated (Liu et al., 2014; Shen et al., 2011). With its ability to bind with ubiquitinated cargos and bring them to the autophagosome-associated protein LC3 via its LIR domain, optineurin fulfils a role as an autophagy receptor or adaptor (Wild et al., 2011). Notably, the optineurin connection with autophagy can also be ubiquitin-independent.
3.1. Ubiquitin-dependent roles
3.1.1. Xenophagy
Xenophagy is the process by which a cell directs autophagy against pathogens. Wild and associates were the first to characterize optineurin as a key component in cellular clearance of Salmonella through an ubiquitin-dependent autophagic pathway (Wild et al., 2011). Cytosolic or vacuole-confined bacteria can be rapidly decorated by ubiquitination (Perrin et al., 2004). Optineurin then links the ubiquitin groups in bacteria and the autophagy protein LC3 (Randow and Youle, 2014). Autophagy adaptors p62 and NDP52 also work along with optineurin, but they all have non-redundant roles. It was further observed that by phosphorylating optineurin at Ser177, TBK1 can significantly enhance the binding affinity of optineurin to LC3 and the autophagic clearance of ubiquitin-coated Salmonella (Wild et al., 2011). Phosphorylation therefore adds another level of optineurin-based autophagy regulation.
3.1.2. Mitophagy
Mitophagy is the selective degradation of defective mitochondria by autophagy. It occurs often to defective mitochondria following damage and stress. This process is regulated by phosphatase and tensin homolog-induced kinase 1 (PINK1) and parkin, a cytosolic E3 ubiquitin ligase. PINK1 is stabilized on the outer membrane of damaged mitochondria, recruiting parkin to the mitochondria (Kane et al., 2014). Once localized at the mitochondria, parkin ubiquitinates the outer membrane proteins (Lazarou, 2015), and in turn recruits optineurin which stably associates with ubiquitinated mitochondria via its UBD domain. Optineurin subsequently induces autophagosome formation to engulf damaged mitochondria via its LIR domain (Wong and Holzbaur, 2014). Disruption of either of these steps leads to inefficient autophagic degradation of mitochondria and accumulation of damaged mitochondria within the cell. Depletion of the endogenous optineurin, for instance, inhibits LC3 recruitment to mitochondria and inhibits mitochondrial degradation. The defects can be rescued by expression of siRNA-resistant wild-type optineurin, but not by ALS-associated E478G optineurin with mutation in the UBD domain, or by optineurin with a mutation in the LIR domain. Another autophagy receptor, p62/SQSTM1, is also recruited, but the recruitment is independent of optineurin in separate domains on damaged mitochondria (Wong and Holzbaur, 2014).
3.1.3. Aggrephagy
Accumulation of misfolded proteins and the associated loss of neurons are considered a hallmark of many neurodegenerative diseases (Aguzzi and O'Connor, 2010). The selective degradation of protein aggregates by autophagy is called aggrephagy, which is a cellular protection mechanism. In a recent study (Shen et al., 2014), optineurin was shown to mediate the degradation of mutant huntingtin (mHtt)-containing inclusion bodies in an ubiquitin-dependent manner. Optineurin colocalized with inclusion bodies formed by mHtt in R6/2 transgenic mice as well as those formed by Htt mutants or the truncated form of TAR DNA-binding protein 43 in Neuro2A cells. This colocalization required the UBD domain of optineurin. Overexpression of wild-type optineurin decreased inclusion bodies through Lys63-linked polyubiquitin-mediated autophagy. In contrast, UBD mutants of optineurin, including E478G and D474N, and UBD deletion mutants (UBD including 411 to 520Δ and 210 to 520Δ) increased inclusion body accumulation while mutants outside of the UBD domain did not (Shen et al., 2014).
3.1.4. Tumor suppression
Optineurin interacts with tumor suppressor HECT domain and ankyrin repeat containing HACE1, an E3 ubiquitin ligase (Liu et al., 2014). It has been shown that HACE1 can ubiquitinate optineurin on Lys193, promoting its interaction with p62/SQSTM1 to form the autophagy receptor complex, activating thereby autophagy and autophagic flux. Coexpression of wild-type HACE1 and optineurin increased the formation of LC3 puncta, removal of oxidatively damaged proteins along with p62, as well as the conversion of LC3-I into lipidated LC3-II (a marker of autophagosome) in human lung cancer cells (Liu et al., 2014). Induction of p62 has been associated with multiple types of human cancer, and the elimination of p62 by autophagy suppresses tumorigenesis (Komatsu and Ichimura, 2010; Mathew et al., 2009). The HACE1-optineurin axis thus promotes tumor suppression in an ubiquitin-dependent fashion (Liu et al., 2014).
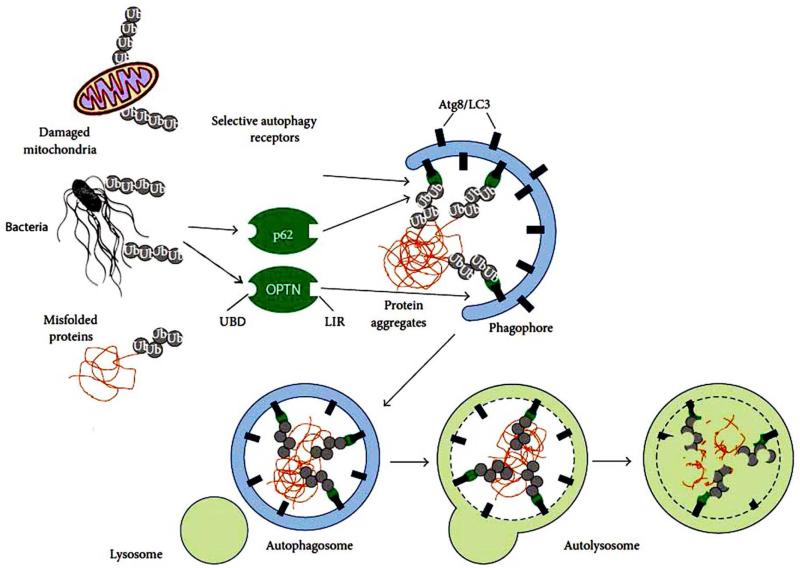
The role of selective autophagy receptors (such as p62 and optineurin) played in the ubiquitin-dependent autophagic clearance of pathogen, removal of misfolded protein or protein aggregate, and elimination of ubiquitinated mitochondria is illustrated in Figure 2.
Figure 2.
The process of ubiquitin-dependent selective autophagy. Ubiquitin coated bacteria, misfolded protein or cellular organelle binds with autophagy receptors such as p62 or optineurin (OPTN) through their ubiquitin binding domain (UBD). The receptors then bind with LC3 and initiate the formation of a double-membrane structure named phagophore. Phagophore closes to form autophagosome and finally, autophagosomes fuse with lysosomes forming autolysosomes where breakdown of the autophagic cargo takes place. Autophagy receptors contain a short LIR sequence responsible for LC3 binding. Recognition of ubiquitinated proteins is mediated by interacting with ubiquitin noncovalently via UBD. Modified from Lippai and Low (2014) with copyright permission.
3.2. Ubiquitin-independent roles
Optineurin can regulate autophagic clearance of protein aggregates also through an ubiquitin-independent pathway (Korac et al., 2013). Short fragment of huntingtin (Htt) protein carrying an extended polyglutamine mutation (Htt ex1 Q103) or superoxide dismutase 1 (SOD1) protein carrying point mutations at position Gly93 (SOD1 G93C or SOD1 G93A), commonly used as models of Huntington disease and ALS, was expressed in HeLa cells. Optineurin was observed to colocalize with both SOD1 G93C and Htt ex1 Q103 inclusions. The ubiquitin binding deficient E478G optineurin mutant was still able to colocalize with GFP-SOD1 G93C protein aggregates, suggesting that optineurin interacts with those aggregates in an ubiquitin-independent manner. Further mapping indicated that the C-terminal coiled-coil domain (454-520 aa) of optineurin recognizes Htt and SOD1 aggregates. Depletion of optineurin increased protein aggregation. Optineurin was in addition shown to actively participate in the degradation of both Htt ex1 Q103 and SOD1 G93C inclusions through the autophagy-lysosome system. This process, interestingly, is also regulated by Ser177 phosphorylation by TBK1 (Korac et al., 2013).
Optineurin moreover mediates the autophagic clearance of extracellular β-amyloid (Aβ) by microglia, a primary immune cell in the brain. Aβ in microglia was revealed by immunoprecipitation to form complexes with LC3-II and optineurin, suggesting that Aβ is targeted to autophagy via LC3/optineurin interactions. The number of LC3-II dots was increased, indicating that autophagy was induced when BV2 microglial cells and primary mouse microglia were treated with Aβ fibrils (Cho et al., 2014). In addition, knocking down LC3 and/or autophagy-related gene 7 homolog by siRNA reduced Aβ degradation, implying that the autophagic process is necessary for the degradation of Aβ fibril. Aβ was not degraded when optineurin was downregulated by siRNA, corroborating further the crucial role of optineurin in the Aβ degradation in microglia (Cho et al., 2014).
4. Optineurin as an autophagy inducer
4.1. Autophagy induction by wild-type and mutated optineurin in vitro
Optineurin not only functions as an autophagy receptor through its ability to interact with ubiquitin and LC3, but also acts as an autophagy inducer. The endogenous optineurin is ubiquitinated and processed through the UPS pathway (Shen et al., 2011). Upon overexpression of wild-type or mutant E50K optineurin in RGC-5 cells, the level of proteasome regulatory β5 subunit (PSMB5, a marker of proteasome activity) was downregulated while that of LC3-II was elevated indicating that the UPS function was compromised and autophagy was induced (Shen et al., 2011). RGC-5 cells transfected with pOPTNWT-GFP or pOPTNE50K-GFP formed fluorescent foci in the perinuclear region. Cells treated with autophagic inhibitor 3-methyladenine showed more foci while treatment with rapamycin reduced the foci formation in optineurin transfectants. Rapamycin is an inhibitor of a Ser/Thr protein kinase named “mammalian target of rapamycin” (mTOR). Inhibition of mTOR mimics cellular starvation that induces autophagy by blocking signals required for cell growth and proliferation. Rapamycin is thus also recognized as an autophagy inducer. The finding that rapamycin reduced optineurin foci formation in transfected RGC-5 cells suggested that the overexpressed or mutated optineurin was cleared, at least in part, via the autophagy pathway (Shen et al., 2011). PSMB5 and LC3 alterations were similarly observed in inducible cell lines when wild-type and E50K optineurin-GFP levels were induced by 10–12 fold (Shen et al., 2011).
Optineurin is also known to be upregulated by proinflammatory cytokines tumor necrosis factors α (Vittitow and Borras, 2002) and interferon γ (Sudhakar et al., 2013). When RGC-5 cells were treated with these cytokines for 24 h, the optineurin level was increased by 2 fold. Foci formation was not apparent, but the PSMB5 level was found reduced by 40-60%, and the LC3-II level was elevated by 1.9-2.5 fold. This indicates that physiologically induced optineurin upregulation can also elicit UPS impairment and autophagy induction (Shen et al., 2011).
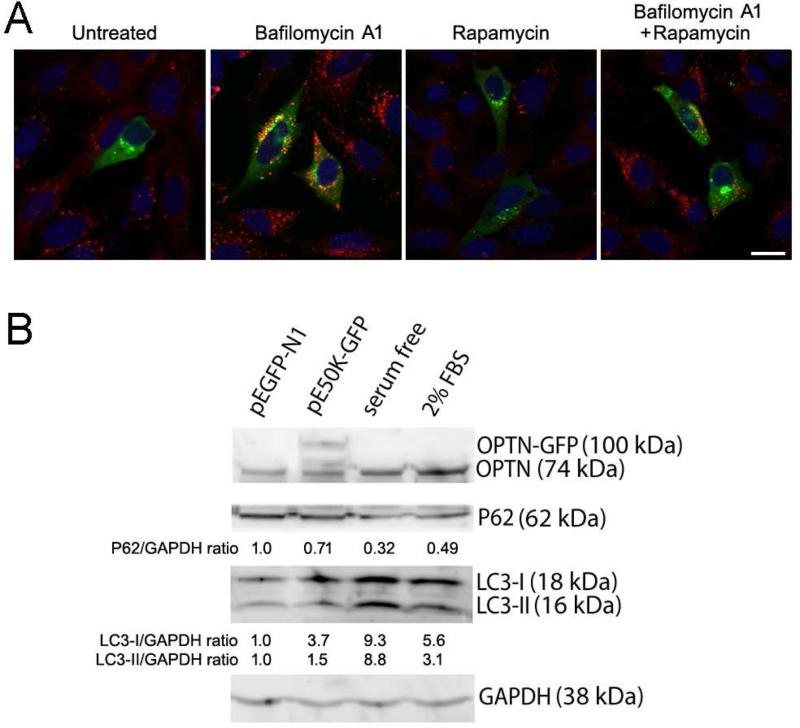
Autophagy activation and autophagic flux were further investigated by p62 immunostaining and Western blotting in RGC-5 cells. It is shown that activation of autophagy and autophagic flux correlates with a decreased, and autophagic suppression correlates with an increased, p62 level. RGC-5 cells transfected overnight with pOPTNE50K-GFP or pEGFP-N1 control were untreated or treated with bafilomycin A1 (an autophagic inhibitor that inhibits the fusion between autophagosomes and lysosomes and prevents thereby maturation of autophagic vacuoles), rapamycin, or both for 24 h. Immunostaining demonstrated that the p62 level was lower in pOPTNE50K-GFP transfected cells than the surrounding non-transfected non-green (Figure 3A) as well as GFP control (not shown) cells, confirming an activated autophagy and enhanced autophagic flux by E50K transfection. Bafilomycin A1 treatment which blocks the fusion of autophagosomes with lysosomes dramatically increased the p62 staining in all cells. Autophagic inducer rapamycin reduced p62 staining (Figure 3A), but the staining was heightened back to the bafilomycin A1 level when cells were treated with both bafilomycin A1 and rapamycin (Figure 3A). Quantification by Western blotting verified that the LC3-II level was increased in pOPTNE50K-GFP transfected cells, consistent with an autophagy activation (Figure 3B). Concomitantly, the p62 level was reduced, indicating an augmented autophagic flux in pOPTNE50K-GFP cells (Figure 3B).
Figure 3.
Levels of p62. A. Immunostaining of p62 in RGC-5 cells. RGC-5 cells transfected overnight with pOPTNE50K-EGFP were untreated or treated with 100 nM of bafilomycin A1, 2 μM of rapamycin or both for 24 h. Cells were fixed and immunostained with anti-p62 polyclonal antibody (MBL International) and rhodamine conjugated goat anti rabbit secondary antibody. pOPTNE50K-EGFP transfected cells (green) that displayed green fluorescence foci showed weaker p62 staining (red) than non-transfected non-green cells; indicating an induced autophagy and enhanced autophagic flux by E50K transfection. Autophagy flux inhibitor bafilomycin A1 dramatically elevated p62 staining in both non-transfected and transfected cells. Autophagy inducer rapamycin decreased p62 staining in all cells, while double treatment of bafilomycin A1 and rapamycin reverted the staining to the elevated level. The E50K optineurin foci formation, similar to p62, was also increased by bafilomycin A1 but reduced by rapamycin. Scale bar, 10 μm. B. Western blotting of RGC-5 cells transfected with pEGFP-N1 (control), or pOPTNE50K-EGFP. Cells serum starved (serum free) overnight or cultured in medium containing 2% fetal bovine serum (FBS) were used as autophagy induction positive controls. Membranes were blotted either simultaneously with rabbit anti-optineurin (OPTN, Cayman) and anti-p62, or singly with anti-LC3 (MBL International) or anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Trevigen). The relative ratios to pEGFP-N1 control (normalized against GAPDH) were presented. LC3-II was increased by approximately 50% in pOPTNE50K-EGFP transfected cells. It was also highly elevated in autophagy induction serum free and 2% FBS samples as expected. The level of p62 was decreased by about 30% in pOPTNE50K-EGFP cells, indicating an enhanced autophagy flux.
In a recent study by Chalasani and associates (2014), the LC3-II level was also demonstrated to be increased upon overexpression of E50K in RGC-5 cells. However, Western blotting showed an increase of p62 in adenovirus infected E50K-expressing cells. The disparity seen in the p62 data could be related to differences in E50K expression levels (high versus moderate) and/or experimental conditions.
It is in addition important to stress that RGC-5 cells from an immortalized rat cell line created originally by transforming postnatal day 1 rat retinal cells with E1A adenovirus (Agarwal, 2013; Van Bergen et al., 2009) have been re-characterized and shown to be in fact mouse SV-40 T antigen transformed photoreceptor 661W cells (Krishnamoorthy et al., 2013). They cannot and are not considered to be retinal ganglion cells (RGCs) or RGC models. Further of note is that besides RGC-5 cells, the findings reported by Shen et al. (2011) and those described in Figure 3 have been reproduced in rat neuronal pheochromocytoma PC12 (Shen et al., 2011) and/or mouse brain neuroblastoma Neuro2A (data not shown) cell lines. Optineurin foci have also been observed in other cell types such as human trabecular meshwork cells (Park et al., 2006).
4.2. Autophagy induction by wild-type and mutated optineurin in vivo
The level of PSMB5 and LC3 was examined on the retinal slides of E50K transgenic mice in which E50K optineurin was overexpressed under the CMV early enhancer/chicken β-actin promoter (Chi et al., 2010). These mice developed phenotypes that mimic the clinical features of NTG patients, including neuropathy of the optic disc and degeneration of the RGCs without an increased intraocular pressure (Chi et al., 2010), and are thought to be a NTG mouse model.
Tissue sections from 12-month-old E50K transgenic mice displayed a fainter staining of PMSB5 but a stronger staining of LC3 in RGCs compared with those from control littermate mice. Staining with anti-optineurin also yielded a higher intensity in the transgenic tissues as expected. The staining results were confirmed by Western blotting of retinal extracts. By electron microscopy, autophagosome-like structures were demonstrated in RGCs of eyes of E50K transgenic mice, indicating autophagic induction (Shen et al., 2011).
To establish another animal model, recombinant adeno-associated type 2 viral (AAV2) vector that carries GFP, OPTNWT-GFP or OPTNE50K-GFP was used to introduce intravitreally optineurin (wild-type and E50K mutant) gene into RGCs of Norway Brown rats (Ying et al., 2015). Moderate to strong GFP expression in RGCs was observed in rat eyes 5 weeks after a single injection of AAV2 viral vectors. In wild-type and E50K optineurin-vector injected eyes, the intraocular pressure was normal as expected from the clinical perspective. However, their retina was noted to be thinner; the RGC density was lower; the apoptosis level was higher; the axons were degenerated; and the axon counts were much reduced; indicating that upregulated and mutated optineurin induced toxic effects such as apoptosis and the toxic effects were more dramatically seen with the E50K mutation (Koga et al., 2010; Meng et al., 2012; Park et al., 2006; Turturro et al., 2014). Furthermore, the level of PSMB5 was found declined and that of LC3 was induced, confirming that in vivo (Ying et al., 2015), as in vitro (Shen et al., 2011), the UPS was downregulated and autophagy was induced upon overexpression and/or mutation of optineurin.
5. Implication of autophagy in optineurin associated diseases
Autophagy contributes to basic cellular homeostasis such as the turnover of mitochondria (through mitophagy) and other organelles (for example endoplasmic reticulum and peroxisomes) (Johansen and Lamark, 2011; Wang and Klionsky, 2011); the clearance of protein aggregates accumulated during stress, aging, and disease owing to perturbations in protein structure or folding (Lamark and Johansen, 2012); and the regulation of lipid metabolism (lipophagy) (Singh et al., 2009). With an autophagy connection, optineurin, when mutated or when its expression level is altered, may lead to glaucoma, ALS, and other neurodegenerative diseases, in which mitochondria dysfunction and protein aggregation are implicated; as well as cancer (Liu et al., 2014). Paget's disease of the bone, a chronic disorder characterized by increased bone turnover, is associated with p62 mutations and optineurin. Such an association suggests that Paget's disease may likewise be developed via an autophagy related mechanism. Of special significance to the ocular fields, optineurin may also play an autophagy receptor role in age-related macular degeneration, a disease resulting from accumulation of environmental stress induced protein misfolding and aggregation in the retinal pigment epithelium (Wang et al., 2014).
6. Concluding remarks
Optineurin is a classical autophagy receptor characterized by its UBD and LIR motifs. It can also act as a non-classical receptor undergoing ubiquitin-independent autophagy. Similar to p62, optineurin has an inherent ability to polymerize and can also itself undergo autophagic degradation. These findings highlight the multiple roles of optineurin as a cargo carrier, regulator, and a substrate for autophagy. Mediated by its interaction with myosin VI, optineurin in addition links myosin VI to autophagosome maturation and subsequent fusion with the lysosome (Tumbarello et al., 2012).
Optineurin has been reported to be a key player in xenophagy, mitophagy, aggrephagy, as well as tumor suppression. It is unclear whether optineurin is also involved in other autophagic process such as removal of ribosomes (ribophagy), peroxisomes (pexophagy), and surplus endoplasmic reticulum (reticulophagy). The spatial-temporal relationship between optineurin and other autophagic receptors and how posttranslational modifications including phosphorylation, acetylation, and ubiquitination coordinate selective autophagy are likewise open questions.
Gene therapy in humans is still a long term goal in treating neurodegenerative diseases by inducing autophagy (Frake et al., 2015). There have, nevertheless, been efforts in inducing autophagy by drugs. Rapamycin, a compound approved by the U.S. Food and Drug administration (Vafai and Mootha, 2013) and its analogs have been used previously to reduce Htt levels and attenuated the mutant Htt toxicity in the cell, fly and mouse models of disease (Ravikumar et al., 2004). The protective effect was attributed to enhanced clearance of the mutant protein via autophagy. A similar strategy was applied to inject rapamycin intraperitoneally to the rats that had received injection of AAV2-E50K-GFP viral vector. The optineurin level, RGC and axonal counts, and apoptosis in AAV2-E50K-GFP-injected rat eyes were averted to closer to normal limits after treatment with rapamycin. It appears that the accumulated mutant optineurin could be cleared, at least partially, by the rescuing strategy involving rapamycin (Ying et al., 2015). While intriguing, the use of rapamycin is expected to bring unwanted side effects. Imbalance in the autophagic flux is also problematic. Further insights into the roles of optineurin in autophagy and the pathophysiological mechanism of optineurin-related diseases may lead to safer and more efficient therapeutic approaches.
Optineurin is a gene linked to normal tension glaucoma and amyotrophic lateral sclerosis.
Autophagy is an intracellular degradation system that delivers cytoplasmic components to the lysosomes for degradation.
Containing ubiquitin binding and LC3 interacting domains, the optineurin protein is a selective autophagy receptor.
Optineurin is involved in xenophagy, mitophagy, aggrephagy, and tumor suppression in an ubiquitin-dependent manner. It can also mediate the removal of protein aggregates through an ubiquitin-independent mechanism.
Optineurin is an autophagy inducer, inducing autophagic process upon overexpression and mutation.
Acknowledgments
This work has been supported by grants (EY018828 and EY001792) from the National Eye Institute, Bethesda, MD, an unrestricted departmental grant from Research to Prevent Blindness, New York, NY; and a grant award G2013110 from BrightFocus Foundation, Clarksburg, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal N. RGC-5 cells. Invest Ophthalmol Vis Sci. 2013;54:7884. doi: 10.1167/iovs.13-13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A, O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Aung T, Rezaie T, Okada K, Viswanathan AC, Child AH, Brice G, Bhattacharya SS, Lehmann OJ, Sarfarazi M, Hitchings RA. Clinical features and course of patients with glaucoma with the E50K mutation in the optineurin gene. Invest Ophthalmol Vis Sci. 2005;46:2816–2822. doi: 10.1167/iovs.04-1133. [DOI] [PubMed] [Google Scholar]
- Chalasani ML, Kumari A, Radha V, Swarup G. E50K-OPTN-induced retinal cell death involves the Rab GTPase-activating protein, TBC1D17 mediated block in autophagy. PLoS One. 2014;9:e95758. doi: 10.1371/journal.pone.0095758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi ZL, Akahori M, Obazawa M, Minami M, Noda T, Nakaya N, Tomarev S, Kawase K, Yamamoto T, Noda S, Sasaoka M, Shimazaki A, Takada Y, Iwata T. Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum Mol Genet. 2010;19:2606–2615. doi: 10.1093/hmg/ddq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY. Autophagy in microglia degrades extracellular beta-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy. 2014;10:1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic Control of Autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Ohtsubo M, Hotta Y, Minoshima S. Oligomerization of optineurin and its oxidative stressor E50K mutation-driven covalent cross-linking: possible relationship with glaucoma pathology. PLoS One. 2014;9:e101206. doi: 10.1371/journal.pone.0101206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Hauser MA, Sena DF, Flor J, Walter J, Auguste J, Larocque-Abramson K, Graham F, Delbono E, Haines JL, Pericak-Vance MA, Rand Allingham R, Wiggs JL. Distribution of optineurin sequence variations in an ethnically diverse population of low-tension glaucoma patients from the United States. J Glaucoma. 2006;15:358–363. doi: 10.1097/01.ijg.0000212255.17950.42. [DOI] [PubMed] [Google Scholar]
- Iida A, Hosono N, Sano M, Kamei T, Oshima S, Tokuda T, Nakajima M, Kubo M, Nakamura Y, Ikegawa S. Novel deletion mutations of OPTN in amyotrophic lateral sclerosis in Japanese. Neurobiol Aging. 2012;33:1843, e1819–1824. doi: 10.1016/j.neurobiolaging.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachaner D, Filipe J, Laplantine E, Bauch A, Bennett KL, Superti-Furga G, Israel A, Weil R. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Mol Cell. 2012a;45:553–566. doi: 10.1016/j.molcel.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Kachaner D, Genin P, Laplantine E, Weil R. Toward an integrative view of Optineurin functions. Cell Cycle. 2012b;11:2808–2818. doi: 10.4161/cc.20946. [DOI] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Shen X, Park JS, Qiu Y, Park BC, Shyam R, Yue BYJT. Differential effects of myocilin and optineurin, two glaucoma genes, on neurite outgrowth. Am J Pathol. 2010;176:343–352. doi: 10.2353/ajpath.2010.090194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. Journal of cell science. 2013;126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy RR, Clark AF, Daudt D, Vishwanatha JK, Yorio T. A forensic path to RGC-5 cell line identification: lessons learned. Invest Ophthalmol Vis Sci. 2013;54:5712–5719. doi: 10.1167/iovs.13-12085. [DOI] [PubMed] [Google Scholar]
- Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M. Keeping the immune system in check: a role for mitophagy. Immunol Cell Biol. 2015;93:3–10. doi: 10.1038/icb.2014.75. [DOI] [PubMed] [Google Scholar]
- Leung YF, Fan BJ, Lam DS, Lee WS, Tam PO, Chua JK, Tham CC, Lai JS, Fan DS, Pang CP. Different optineurin mutation pattern in primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2003;44:3880–3884. doi: 10.1167/iovs.02-0693. [DOI] [PubMed] [Google Scholar]
- Li Y, Kang J, Horwitz MS. Interaction of an adenovirus E3 14.7-kilodalton protein with a novel tumor necrosis factor alpha-inducible cellular protein containing leucine zipper domains. Mol Cell Biol. 1998;18:1601–1610. doi: 10.1128/mcb.18.3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippai M, Low P. The Role of the Selective Adaptor p62 and Ubiquitin-Like Proteins in Autophagy. Biomed Res Int. 2014;2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, Fan L, Chen S, Zhou J, Sun Y, Ruan K, Cheng S, Komatsu M, White E, Li L, Ji H, Finley D, Hu R. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen GH, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, DiPaola RS, Karantza-Wadsworth V, White E. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Lv J, Ge H, Zhang L, Xue F, Zhu Y, Liu P. Overexpressed mutant optineurin(E50K) induces retinal ganglion cells apoptosis via the mitochondrial pathway. Mol Biol Rep. 2012;39:5867–5873. doi: 10.1007/s11033-011-1397-7. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Nagabhushana A, Bansal M, Swarup G. Optineurin is required for CYLD-dependent inhibition of TNF-α-induced NF-κB activation. PLoS One. 2011;6:e17477. doi: 10.1371/journal.pone.0017477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, Okamoto K. Optineurin in neurodegenerative diseases. Neuropathology. 2011;31:569–574. doi: 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- Park B, Ying H, Shen X, Park JS, Qiu Y, Shyam R, Yue BYJT. Impairment of protein trafficking upon overexpression and mutation of optineurin. PLoS One. 2010;5:e11547. doi: 10.1371/journal.pone.0011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BC, Shen X, Samaraweera M, Yue BYJT. Studies of optineurin, a glaucoma gene: Golgi fragmentation and cell death from overexpression of wild-type and mutant optineurin in two ocular cell types. Am J Pathol. 2006;169:1976–1989. doi: 10.2353/ajpath.2006.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:404–412. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Heon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Waitzman DM, Seeman JL, Kaufman PL, Sarfarazi M. Molecular cloning and expression profiling of optineurin in the rhesus monkey. Invest Ophthalmol Vis Sci. 2005;46:2404–2410. doi: 10.1167/iovs.04-1243. [DOI] [PubMed] [Google Scholar]
- Rogov VV, Suzuki H, Fiskin E, Wild P, Kniss A, Rozenknop A, Kato R, Kawasaki M, McEwan DG, Lohr F, Guntert P, Dikic I, Wakatsuki S, Dotsch V. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem J. 2013;454:459–466. doi: 10.1042/BJ20121907. [DOI] [PubMed] [Google Scholar]
- Shen WC, Li HY, Chen GC, Chern Y, Tu PH. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2014:e36098. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ying H, Qiu Y, Park JS, Shyam R, Chi ZL, Iwata T, Yue BYJT. Processing of optineurin in neuronal cells. J Biol Chem. 2011;286:3618–3629. doi: 10.1074/jbc.M110.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara MT, Girardin SE. Emerging themes in bacterial autophagy. Curr Opin Microbiol. 2014;23C:163–170. doi: 10.1016/j.mib.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Sudhakar C, Vaibhava V, Swarup G. IRF-1-binding site in the first intron mediates interferon-γ-induced optineurin promoter activation. Biochem Biophys Res Commun. 2013;437:179–184. doi: 10.1016/j.bbrc.2013.06.065. [DOI] [PubMed] [Google Scholar]
- Turturro S, Shen X, Shyam R, Yue BYJT, Ying H. Effects of mutations and deletions in the human optineurin gene. Springerplus. 2014;3:99. doi: 10.1186/2193-1801-3-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Medicine. A common pathway for a rare disease? Science. 2013;342:1453–1454. doi: 10.1126/science.1248449. [DOI] [PubMed] [Google Scholar]
- Vittitow J, Borras T. Expression of optineurin, a glaucoma-linked gene, is influenced by elevated intraocular pressure. Biochem Biophys Res Commun. 2002;298:67–74. doi: 10.1016/s0006-291x(02)02395-1. [DOI] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Elazar Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci Signal. 2011;4:pe39. doi: 10.1126/scisignal.2002355. [DOI] [PubMed] [Google Scholar]
- Weishaupt JH, Waibel S, Birve A, Volk AE, Mayer B, Meyer T, Ludolph AC, Andersen PM. A novel optineurin truncating mutation and three glaucoma-associated missense variants in patients with familial amyotrophic lateral sclerosis in Germany. Neurobiol Aging. 2013;34:1516, e1519–1515. doi: 10.1016/j.neurobiolaging.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde IB, Brack M, Winget JM, Mayor T. Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J Proteome Res. 2011;10:1062–1072. doi: 10.1021/pr1008543. [DOI] [PubMed] [Google Scholar]
- Willoughby CE, Chan LL, Herd S, Billingsley G, Noordeh N, Levin AV, Buys Y, Trope G, Sarfarazi M, Heon E. Defining the pathogenicity of optineurin in juvenile open-angle glaucoma. Invest Ophthalmol Vis Sci. 2004;45:3122–3130. doi: 10.1167/iovs.04-0107. [DOI] [PubMed] [Google Scholar]
- Wojcik S. Crosstalk between autophagy and proteasome protein degradation systems: possible implications for cancer therapy. Folia Histochem Cyto. 2013;51:249–264. doi: 10.5603/FHC.2013.0036. [DOI] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Turturro S, Nguyen T, Shen X, Zelkha R, Johnson EC, Morrison JC, Yue BYJT. Induction of autophagy in rats upon overexpression of wild-type and mutant optineurin gene. BMC Cell Biol. 2015;16:14. doi: 10.1186/s12860-015-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Yue BYJT. Cellular and molecular biology of optineurin. Int Rev Cell Mol Biol. 2012;294:223–258. doi: 10.1016/B978-0-12-394305-7.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Geetha T, Babu JR. Failure of ubiquitin proteasome system: risk for neurodegenerative diseases. Neuro-degenerative diseases. 2014;14:161–175. doi: 10.1159/000367694. [DOI] [PubMed] [Google Scholar]