Abstract
We analyzed time-series data for fluctuations of intramolecular segments of barcoded E. coli genomic DNA molecules confined in nanochannels with sizes near the persistence length of DNA. These dynamic data allowed us to measure the probability distribution governing the distance between labels on the DNA backbone, which is a key input into the alignment methods used for genome mapping in nanochannels. Importantly, this dynamic method does not require alignment of the barcode to the reference genome, thereby removing a source of potential systematic error in a previous study of this type. The results thus obtained support previous evidence for a left-skewed probability density for the distance between labels, albeit at a lower magnitude of skewness. We further show that the majority of large fluctuations between labels are short-lived events, which sheds further light upon the success of the linearized DNA genome mapping technique. This time-resolved data analysis will improve existing genome map alignment algorithms, and the overall idea of using dynamic data could potentially improve the accuracy of genome mapping, especially for complex heterogeneous samples such as cancer cells.
I. INTRODUCTION
Genomic mapping is an emerging technique for studying large-scale structural variations as a complement to next-generation sequencing (NGS).1 Recently, physical maps from BioNano Genomics and sequences from Pacific Biosciences were combined to generate a high quality assembly that contained no amplification biases that often arise during polymerase chain reaction (PCR).2 Genomic mapping uses DNA molecules orders of magnitude greater in length than in NGS that are stretched close to their contour length. The long, continuous reads of genomic mapping have a resolution of 2–3 kilobasepairs,3 which is sufficient to detect structural variations such as large insertions, deletions, and translocations that can be difficult for NGS to quantify. There are a number of ways to stretch the molecules, including directed surface immobilization,4 extensional flow,5 and nanochannel confinement.6 In the nanochannel method, DNA molecules hundreds of kilobases long are nick-labeled at short sequence motifs with a fluorophore and extended in square nanochannels 45 nm wide,6 which is near the persistence length of DNA in the high ionic strength buffer used for these experiments. This strong confinement linearly orders the labels on the DNA backbone, such that the set of distances between the labels comprises a genomic barcode for each molecule.6–9 To avoid artifacts arising from independent molecules overlapping or touching end-to-end, the backbone is fluorescently stained with a second color to associate the labels with a particular molecule. Since the molecules are in solution in the nanochannels, thermal fluctuations of the distances between labels set an upper bound on the accuracy of the resulting genome map.
The synthesis of the set of DNA barcodes into a consensus genome map requires assigning statistical weights to each label on each barcode. Whether or not an individual molecule adds to the consensus genome map is determined by its p-value, which measures the statistical confidence that one molecule's barcode aligns to the set of barcodes obtained from the other DNA molecules.10 The cutoff for the p-value used to include a molecule in the consensus map depends on a number of factors, such as the genomic length of that molecule and the degree of complexity of the barcode.8 Selecting an appropriate cut-off is especially important when aligning to larger genomes such as human.10 The probability distribution for the distance between labels is a key input to the statistical model used to compute the p-value for each molecule. Thus, an accurate measurement of this probability distribution is critical for creating accurate consensus maps. The goal of this paper is an applied one, namely, to make improved estimates of this probability distribution through the use of time-series data.
In this context, it is worthwhile to review first our previous measurement of the probability density of distances between label pairs of nanoconfined barcoded DNA.11 In the latter work, we used the high-throughput Irys genome mapping system (BioNano Genomics, Inc.) to produce millions of measurements of the distances between fluorescent labels on the DNA backbone from tens of thousands of molecules, and the particular sequence of each molecule was obtained by mapping the barcode pattern to the reference genome—a genomic-based strategy. The resulting large data sets allowed for the calculation of higher moments of the distribution such as the skewness. The distance distribution exhibited non-Gaussian behavior with skew to the left, indicating significantly more bunching of the internal segments of the molecules than would be expected using a typical thermodynamic model based upon Gaussian statistics. Aside from its importance in genome mapping technology, measurements of the distances between labels are important for describing the statics12–16 of polymers confined at a length scale close to their persistence length, especially on the intramolecular scale.
However, the genomic approach used previously involved a potential source of systematic error that could not be controlled in that experimental approach. In order to construct the probability distribution governing the distance between the labels on the DNA backbone, a single snapshot of each barcoded molecule was aligned to the reference genome of the strain of E. coli used in the experiments. Since the alignment method is statistical in nature,17 it is possible that the alignment to the reference genome added a bias to the results. In particular, incomplete dye labeling due to under- or over-nicking, bad fluorophores, or poor nucleotide incorporation could result in a particular DNA molecule aligning to the incorrect region of the genome, and thus leading to an incorrect mapping between the physical distance between labels on the DNA and their genomic distance along the backbone. Similarly, backfolding or knotting of the backbone, while rare and normally preventing alignment of the DNA to the reference in the first place, has been shown to lead to systematic errors in the measured barcodes on the order of 5 kilobases when alignment is successful.18 As a result, this genomic strategy for determining the sequence on a given DNA molecule may have restricted actual large fluctuations from entering the distribution, or, conversely, assigned a large fluctuation to a more relaxed molecule. Since there are possible artifacts that are both larger and smaller than the average value, they may cancel out when computing the mean of the distribution. However, the cancellation effect decreases for higher-order odd moments and is absent for even-order moments. The genomic strategy also struggles to handle labels on the DNA barcode that are not easily resolved by the microscope optics. As we will see shortly, two labels temporarily can become close enough that their intensity peaks are unresolvable, and the image analysis program will record them as a single label at a location offset somewhere between the two labels.
A key limitation of the genomic strategy is that it does not permit any way to control for the aforementioned effects. With only one image of the DNA backbone and its associated labels, the only way to determine the contour length between labels is through a map to the reference genome. Moreover, it is impossible to determine if the thermal fluctuations between two labels led to a loss of optical resolution if there is only a single image.
The present study removes the aforementioned problems associated with a genomic strategy for measuring the probability distribution between barcode labels by adopting a physical approach that does not require the sequence of the DNA molecule. Instead of analyzing single snapshots of each molecule, as was done previously (here further denoted as “reference-aligned snapshot” data), videos of the labels fluctuating in time were acquired for each molecule. In this approach, we no longer need to align to the reference genome to determine the contour length between two labels. Rather, we instead measure the physical distance, X, between the labels by fluorescence microscopy. If we make a sufficient number of measurements of this distance, the error in the average value, , becomes small.19 By calibrating the distance measurement with a known standard, we could convert the into a genomic distance. Alternatively, we can simply avoid the genomic distance entirely and consider how the probability distribution for depends on , as the latter quantity should be proportional to the genomic distance. We chose the second approach, as it avoids introducing an additional approximation based on uniform stretching. In the course of our analysis, we found that the removal of frames where nearby dots cannot be resolved, stuck labels and overlapped molecules removed a number of outliers from the distribution that were likely present in the probability distributions obtained by the genomic method.11 While the probability density for still displayed negative skew, the magnitude of the skew was reduced when compared to that obtained from the reference-aligned snapshot data.
The probability density data presented here serve as an improved input for algorithms for genome map assembly, thereby increasing the quality and reliability of the resulting consensus maps. In the course of our analysis, we also developed a number of tools for removing outliers from high-throughput data that may allow massively parallel measurements of polymer dynamics in nanochannels with sizes near the contour length of DNA, going far beyond previous work of this type.12,20–25
II. EXPERIMENTAL METHODS
A. DNA preparation
Barcode-labeled genomic DNA molecules were prepared using methods described in our previous study.11 Explicitly, DNA was extracted from the MG1655 strain of E. coli using the IrysPrep Reagent kit (BioNano Genomics, Inc.). This kit labels the molecules with fluorescent dUTP near the nick site generated by Nt.BspQI, GCTCTTC sequence, using Taq polymerase and subsequent nick repair. Thereafter, there were two key differences with respect to our prior work.11 First, a set of oxygen scavenging compounds were added to the DNA solution, amounting to a final concentration of 1.4 mM Trolox,26 1.4 mM 4-nitrobenzyl alcohol,27 3.6 mg/ml protocatechuic acid,28 and 0.36 μM protocatechuate-3,4-dioxygenase,28 all purchased from Sigma-Aldrich. This strong scavenging system was necessary to minimize photobleaching and quench triplet states during the extended duration of laser excitation required to obtain sufficient video data. This resulting buffer has an overall ionic strength on the order of 100 mM. Second, a fluorophore with emission spectrum farther into the red (similar to cy5) was used to probe the nick sites and was excited by an OBIS 637 nm, 140 mW laser (Coherent).
B. Nanochannel experiments
The DNA molecules were electrophoretically driven into square nanochannels of two sizes, 40 nm and 51 nm, using the version 1 chip from BioNano Genomics. The channel dimensions were characterized by using SEM images of two channels selected at random out of the hundreds in the channel array. Imaging conditions are detailed in Ref. 11 with the following changes. The filter wheel and camera are controlled with Micromanager,29 while the laser selection, CRISP (autofocus), and x-y-stage were controlled manually. Twenty-seven videos, each consisting of 330 frames, were acquired exciting only the barcode labels at 30 frames per second on a Zyla sCMOS 5.5T sensor using a 1040 × 1392 pixel region of the 2560 × 2160 pixel sensor. This standard video frame rate is 5 times faster than the typical exposure time of 150 ms used in genome mapping in nanochannels.18 The duration of the videos is limited by photobleaching. The resolution was 1 pixel = 108.3 nm, which amounted to roughly 350 base pairs per pixel. At the conclusion of the barcode imaging, a single frame was taken while exciting the YOYO dye molecules on the backbone with roughly a few seconds delay after the barcode label video was captured. This backbone image was used to find the locations of the DNA molecules and associate individuals labels to the correct molecule. The raw images were analyzed by the custom image processing program DM-static, which is available from BioNano Genomics.
C. Data filtering
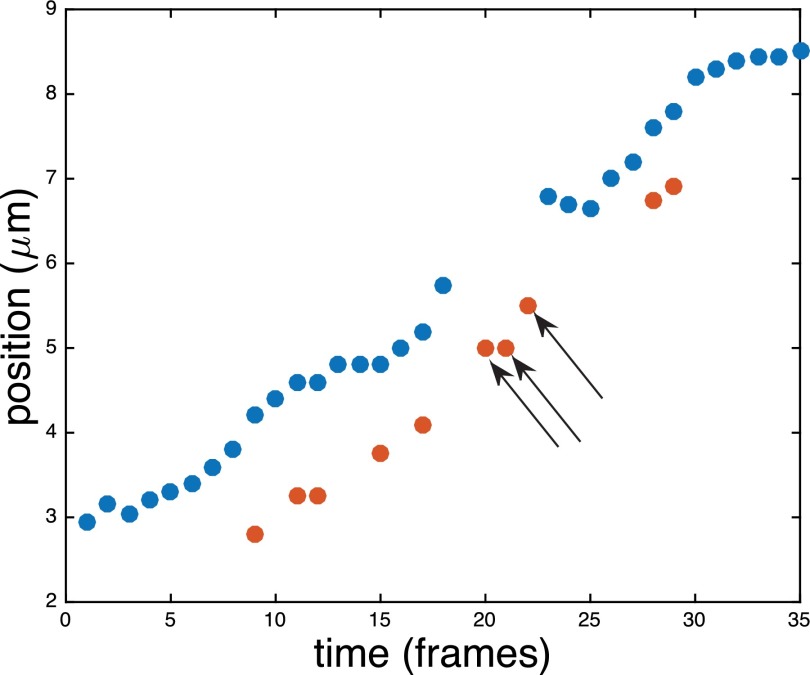
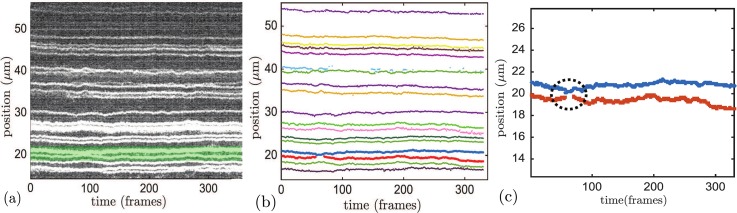
One of the challenges in imaging the barcode labels is photoblinking, a photophysical event which can occur often during long imaging sequences.26–28 Figure 1 is a schematic showing one of the effects blinking has on the measurement of label positions. There was a population of labels that were typically blinked-off more than they were blinked-on, which we call here “intermittent labels.” These occurrences may be the result of transient labeling, excessive localized quenching, or a biological residual from DNA replication. Intermittent labels presented an issue during image processing, especially when they were near a non-intermittent label that happened to blink off, while the intermittent label was blinked-on. This could cause confusion in assigning the labels to the appropriate time trace. This effect poses a significant challenge for image processing, which must be done in an automated way given the very large data sets we have acquired. A second problem occurs when two labels that are in proximity can temporarily become close enough that the image analysis program merges them into one label, which we call here “non-resolved labels.” An example of temporarily non-resolved labels is shown in Fig. 2. There are a number of frames near the 50 frame mark where it appears that the label marked by the red symbols has blinked-off. Concurrently, the position of the label marked by blue symbols has experienced a sudden abnormally large drop, relative to the frames before and after this event. We know empirically that the resolution limit of the image analysis program (2–3 kbp = 0.6–0.9 μm)11 is near the distance between the red and blue label. Thus, we can reasonably attribute this simultaneous blink-off and abrupt change in position within the image analysis software to a merging of the red label into the blue label. In addition to blinking and optically merging, a label can become stuck to the channel wall, whether it is attached to the molecule or is a free, unattached fluorophore that rests alongside the molecule. This is likely a result of non-specific adhesion to the channel walls; this can be quite prevalent in narrow channels with many points of contact between the DNA and the walls. These events also need to be detected and treated appropriately. Finally, as mentioned previously, two independent molecules can become overlapped or touch end-to-end and appear as one molecule, which we here call “overlapping molecules.”
FIG. 1.
Cartoon schematic example of blinking labels' effect on position measurement. The red label is frequently blinked off. The blue label happens to have some blinked off frames at the same frames when the red label has blinked-on frames, denoted by black arrows. In the automated image analysis, these red label frames would be assigned incorrectly to the blue label. Note the overall slope in the position over time. This drift has been added to the schematic to remind the reader that molecules are free to diffuse up and down the channels during a movie, which adds additional complexity to handling blinking events.
FIG. 2.
Example of a label pair with non-resolved frames that would result in an anomalous measurement of position fluctuation, before processing using the filter described in the supplementary material.30 (a) Raw kymograph of the molecule, where each vertical slice is a frame. (b) Time series for all labels on this particular molecule obtained from the standard image processing algorithm prior to any filters, with labels in question (third and fourth from the bottom) denoted with blue and red. (c) Zoom-in on blue and red labels, with other labels removed. Note the region centered in the dotted black circle in the right panel where the lower red label appears to have blinked off for many frames and the upper blue label has exhibited an apparent abrupt large change in position. It is likely that the red label did not blink off and the blue label's actual position is higher than that in the figure.
To avoid all of these artifacts, which are inherent trade-offs as part of the high-throughput data acquisition available in the Irys system, we developed image analysis filters to handle blinking, non-resolved labels, stuck labels, and overlapping molecules. Detailed information about the image analysis protocol, including schematic illustrations of the different filters and examples of experimental data similar to Fig. 2, is provided in the supplementary material.30 Briefly, for each type of filter, empirical thresholds were determined from the distribution of label separation distances and applied to remove individual molecules, labels, and frames.
D. Probability density
Following the methodology in Ref. 11, the filtered data were separated first into bins based upon label pair distance to identify those molecules that should have similar contour length between labels. In the current study, the bin centers were determined by the mean separation distance, , within each video, which differs from our Ref. 11 where the genomic distance was obtained by mapping the DNA barcode back to the reference genome. Within each bin for , the data were further binned into the probability density that a given frame in the movie exhibits a separation distance, X. Since the center of the distribution for X shifts linearly to larger values with , the center of each bin's probability density was shifted to to make data visualization easier. In order to make a clear comparison between the current study's method and the previous reference-aligned snapshot method,11 the bin sizes were kept at 77 nm (∼250 bp) and 25 nm for X and , respectively. Likewise, we only used bins with more than 500 entries to avoid introducing spurious results due to sampling errors.11
In order to explore the effect of the filters, the probability density was also calculated prior to applying the filters for non-resolved, intermittent, and stuck labels, as well as the filter for overlapping molecules. The filter for blinking labels is essential since it is required to rearrange the labels into sensible time series. However, for the purpose of understanding the role of the filters, we relaxed the threshold for blinking filters by approximately 50%.
III. RESULTS AND DISCUSSION
A. Filter statistics
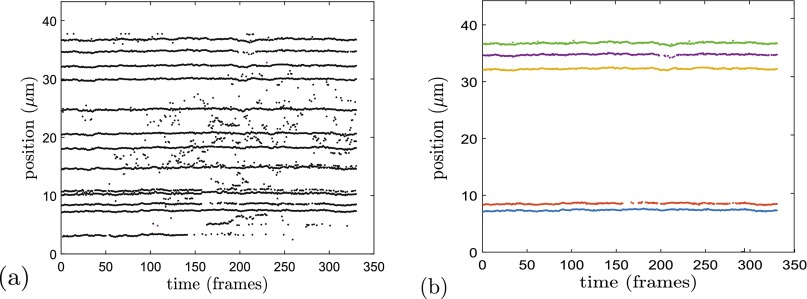
The numbers of molecules, labels, and frames before and after the various filters are presented in Table I for both nanochannels. The distribution of molecule lengths is presented in the supplementary material;30 the molecules lengths ranged from 19 to 124 μm. In terms of the total raw data, the filters removed 229 of the 3122 molecules, 8380 of the 28 357 labels, and 6 432 966 of the 10 021 788 individual label images. This reflects the conservative thresholds set for each filter, which when combined as described in the supplementary material30 leads to data attrition. The largest contributor to the reduction in data is the filter that dealt with blinking labels. There were a substantial number of intermittent labels that appeared to be free labels unattached to the DNA molecule, either non-conjugated or conjugated to small, separate DNA fragments. These labels were free to move among many of the attached labels, which would lead to erroneous measurements of the attached labels' positions. An example of this effect is shown in Fig. 3; the substantial data attrition caused by stray labels is apparent from this figure. Due to the high labeling density of the nicking enzyme used and the prevalence of free labels, many labels were prone to erroneous position measurements. However, the high-throughput nature of this experiment afforded a large amount of data with which to filter; beyond the values in the bottom row of Table I, each label image equates to multiple measurements of label pair separation. We thus chose a conservative approach, removing both the intermittent labels and nearby non-intermittent labels from the data set. Even so, there are still over 1 × 106 individual label images for each channel size. This leads to over 7 × 106 measurements of label pair separation, which is several times that used in Ref. 11.
TABLE I.
Filtered data statistics. The raw numbers are those before the filters. The raw number of labels is a lower bound based upon the number of labels after the first blinking-labels filter that associated the raw label positions with individual time series; this was necessary for counting. The number of label images is the number of labels multiplied by the number of blinked-on frames in each label's time series.
| 40 nm channels | 51 nm channels | |||
|---|---|---|---|---|
| Raw data | Post filters | Raw data | Post filters | |
| Number of molecules | 1783 | 1773 | 1339 | 1120 |
| Number of labels | 16 008 | 11 577 | 12 349 | 8400 |
| Number of label images | 5 714 472 | 2 111 088 | 4 307 316 | 1 473 734 |
FIG. 3.
Example of the effect of the filters for a barcoded DNA molecule with a large number of “intermittent labels.” (a) Raw label positions before they are associated with individual time series. Note the intermittent labels, those that are rarely blinked-on, many of which appear to be stray labels that are likely unattached to the molecule. (b) Remaining label positions that have passed through the filters and are associated with individual time series, denoted by different colors. The filters have removed the labels with a threshold number of nearby intermittent labels. This has substantially reduced the total quantity of data but ensured that we do not have abnormal fluctuations associated with misplaced intermittent labels.
B. Probability density
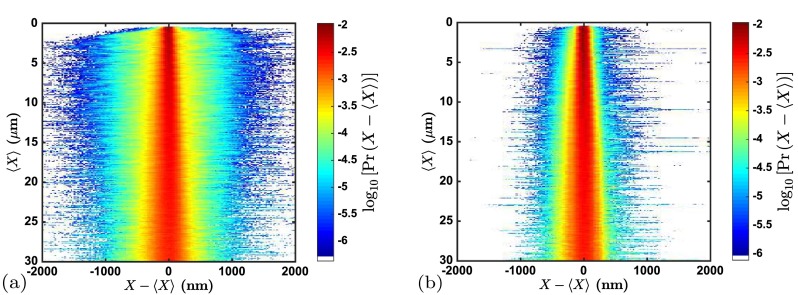
The probability densities for the instantaneous distance X between two labels relative to their average distance for the 40 nm nanochannels are plotted in Fig. 4 after (a) the blinking filter and (b) after the remaining filters. As expected, the filters have substantially reduced the spread in the distribution by removing various artifacts in the automated image processing pipeline, most notably large errors in the location of labels caused by free labels such as in Fig. 3. In comparison with the distributions obtained from reference alignment,11 the pre-filtered distribution in Fig. 4(a) is also much wider. This indicates that the conservative threshold p-values used in genome alignment8 naturally filter out many of the artifacts. Indeed, removing artifacts is critical to genome mapping in nanochannels.
FIG. 4.
One-dimensional probability densities for the separation distance X between labels relative to their average distance in the 40 nm nanochannels (a) before and (b) after applying the data filters for non-resolved, intermittent, and stuck labels, as well as the filter for overlapping molecules. Horizontal slices across this figure correspond to the probability density for a given average separation distance between labels. Bin size for is 77 nm. Bin size for is 25 nm. Only bins for which there were 500 counts are displayed.
The key result in Fig. 4 is the post-filtered probability density in panel b. The most obvious difference between this intensity map and those of our previous study is the lack of extreme left-lying outliers at around = −1700 nm, which appear in the reference-aligned snapshot data11 but not in the data filtered from the videos. In the previous work,11 these outliers resulted from only a handful of individual molecules out of thousands, which would presumably be removed by the filters as experimental artifacts if video data had been available in the previous study.
In our previous study,11 we found that the probability distribution data were reasonably well fit by a normal-inverse Gaussian function. For the present study, in order to remove any bias from the fit, we calculated the skew directly from the data in Fig. 4(b). This was due to the poor fit of the reference-aligned snapshot data to the normal-inverse Gaussian distribution (see supplementary material30). Given this shortcoming in the fitting of this ad hoc choice of probability distribution to our video data, we decided to calculate the skew directly from the non-fitted video data via
| (1) |
Extreme values of can drastically increase the skewness due to the fact that skewness involves the cube of . However, these occurrences are few in number, and this method of calculating the skewness is not impaired by the goodness (or lack thereof) of any fit.
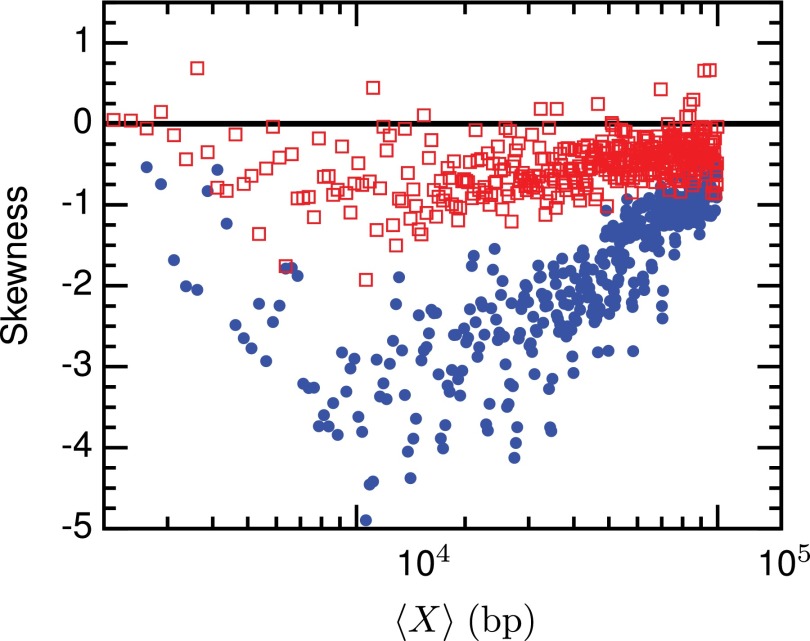
Fig. 5 displays the skewness from the video data computed directly from Fig. 4(b) as well as that of the reference-aligned snapshot data;11 the skewness data for the 51 nm channels are provided in the supplementary material.30 As was the case with the reference-aligned snapshot data,11 we observed negative skewness for most bins. In general, the video data skew's magnitude is reduced compared to the values of the reference-aligned snapshot data.11
FIG. 5.
Skewness for 40 nm nanochannels. Blue dots are the values from the reference-aligned snapshot data,11 and red squares are the skew in the video data calculated directly from Fig. 4(b). The horizontal black line indicates zero skew.
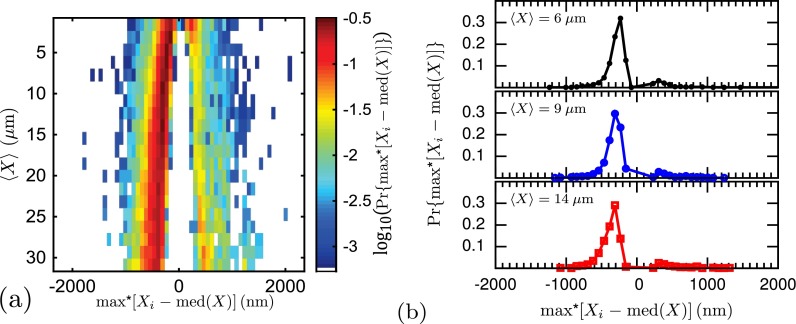
To provide an alternate metric for quantifying the skewness of the data, we also decided to estimate the distribution's asymmetry by the maximum fluctuation within each time series. This parameter also avoids the use of any distribution or fitting method. It contains important technical information for mapping that advances the applied aims of this manuscript, since it represents the maximum spread in position a label will exhibit, and thus sets the upper bound on the error in a given snapshot. Figure 6 plots the probability density of the maximum deviation from the median, positive or negative, within a time-series, as well as three individual slices in . Even though this method reduces the data set for a given molecule down to one frame from each time-series, thereby reducing the data by more than two orders of magnitude, the results are stark. The weight of the distribution for the maximum fluctuation from the mean is almost completely skewed towards extensions less than the median. This strongly supports the key result of our initial study,11 namely, a skew-left distribution. Furthermore, we find that for the majority of molecules (70% or more), the maximum fluctuation during a 10 s video for labels separated by 10–15 kbp (3090–4635 nm) is in the range 500–1000 bp (155–309 nm) less than the median. For labels separated by 100–105 kbp (30 900–32 445 nm), this range increases to 1250–1750 bp (386–540 nm) less than the median. Figure 6 is the first of two key results of this manuscript, providing a bound on the error in the distance between labels within a single DNA barcode.
FIG. 6.
(a) One-dimensional probability density for the maximum fluctuation within a time series for 40 nm nanochannels. (b) Individual slices in . The fluctuation is measured from the median of the label separation within a time series, where max* denotes the signed maximum deviation from the median. The bin sizes are 1545 nm and 77 nm for and max*[Xi -med(X)], respectively. For reference, in our system we estimate a conversion of 3.3 bp/nm for the stretched DNA.
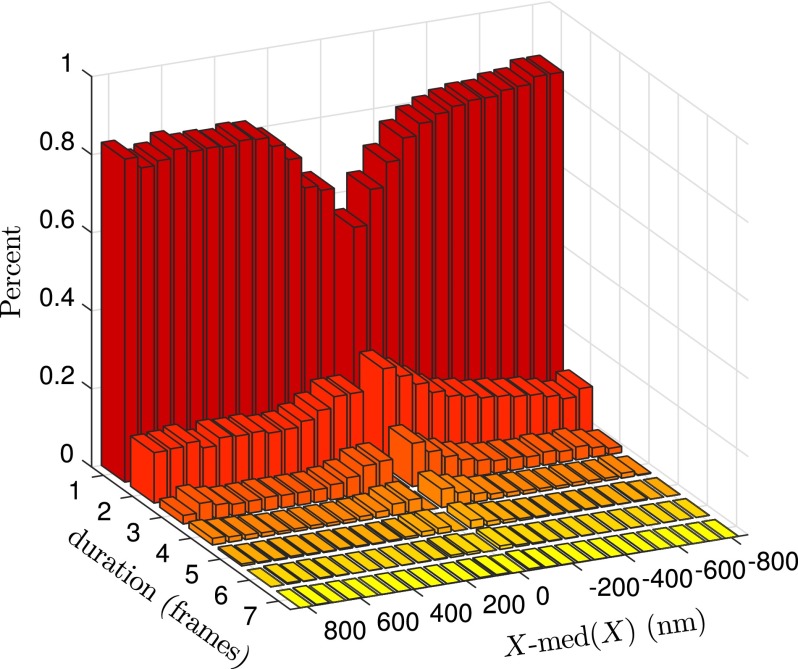
In addition to measuring the distribution of distances between barcodes, the video data allowed us to investigate the label position dynamics. Eventually, these data could advance our understanding of confined polymer dynamics12,20–25 in nanochannels with sizes near the contour length of DNA. For our present purposes, which are aimed towards providing improved probability distributions for genome alignment algorithms, the dynamic data allow us to understand better the role of exposure time on the measured probability distribution and thereby produce the second of our two key results. To this end, we calculated the duration of large label separation fluctuations. The distribution of these events for the 40 nm nanochannels is plotted in Fig. 7 where the fluctuation has been measured relative to the median position within its time series. For all sizes of fluctuations in both sizes of channel, 70% or more last only one frame, which is approximately 30 ms. This indicates that molecule segments are mainly residing in the same general conformation and not spending significant time in bunched or overextended states, which is consistent with the degree of linearization the nanochannels produce. The small proportion of long-lived large fluctuations further indicates that there is little backfolding and corroborates recent simulations on the likelihood of backfolding in channels of this size.14 The ionic strength in our experiments is well below 1 M, meaning that a channel size of 40 nm resides in the non-backfolded Odijk regime (cf. Fig. 11 of Ref. 14). There is also some variation in the X - med(X) direction.
FIG. 7.
Distribution of duration of label separation fluctuations for the 40 nm nanochannels. The fluctuation is measured from the median of the label separation within a time series. Bin size in X - med(X) is 62 nm. Data acquired at 30 frames per second. For reference, in our system we estimate a conversion of 3.3 bp/nm for the stretched DNA.
The dominant effect in Fig. 7 is that of an oscillator, with label pair distances near the median value tending to have a longer occupancy than those that are over-stretched or compressed. This leads to the dip in the distribution for durations of one frame near the median value, and the tendency for these states to persist for several frames. If the distance between label pairs is close to the median, there is no entropic restoring force and their distance only changes due to thermal fluctuations. There is also an asymmetry for longer-lived states, with those in compression tending to last longer than those in extension. The asymmetry could be the result of friction, since compressed states experience monomer-monomer friction in addition to monomer-wall friction while extended states tend to experience the latter friction.
The temporal distribution in Fig. 7 sheds further light on how nanochannels near the persistence length of DNA lead to the success of the mapping algorithm in the presence of thermal fluctuations of label pair separations. Most large fluctuations have been suppressed, and even when they occur they are quite transient. Moreover, by obtaining a sufficient number of replicates of each segment, the rare events fail to add to the consensus map. Inspection of Fig. 4(b) suggests that large fluctuations comprise roughly 1% of the data. Since the current nanochannel technology algorithm uses an image duration of 150 ms,18 the majority of the remaining large fluctuations are averaged out by the longer image exposure.
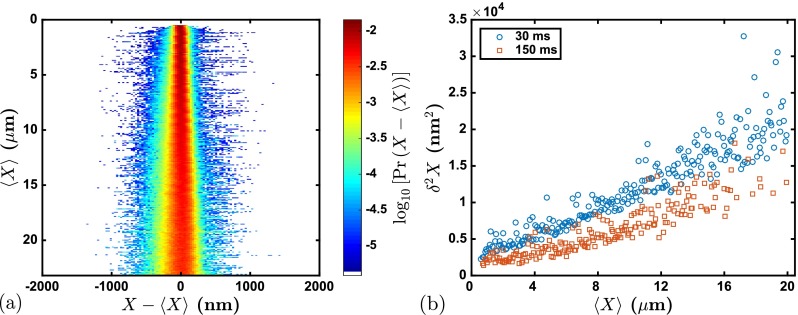
To make this point clear, Fig. 8 displays the probability density and variance for the data in the 40 nm nanochannels when a moving window average of five frames (i.e., 150 ms) is used to downsample the data originally acquired at 30 ms intervals. The downsampled data approximate what one would obtain during conventional genome mapping in nanochannels, which uses a 150 ms exposure time. Note that there are still some differences between the downsampled data and that obtained in genome mapping, since the filters used to produce Fig. 4(b) from video data are different than the image processing used to link labels to DNA molecules during genome mapping. Nevertheless, the overall objectives of both image processing methods are the same, namely, to remove artifacts due to free or missing labels. The distribution in Fig. 8(a) is narrower than that in Fig. 4(b) due to the increased (effective) exposure time in the downsampled data, since the large but brief fluctuations in Fig. 7 are averaged out by the more frequent and longer-lived states with distances closer to the median distance. This effect is easily understood from Fig. 8(b), which shows the reduction in the variance by increasing the exposure time from 30 ms to an effective value of 150 ms. Figure 8(a) represents the second key result of our analysis, a probability distribution that reflects the role of the longer exposure time used for genome mapping in nanochannels.
FIG. 8.
(a) Probability density and (b) variance in the label distances obtained by applying a 150 ms moving-window average to the data appearing in Fig. 4(b). The variance data for the 150 ms moving-window average are lower than the variance data for the original 30 ms imaging time.
IV. CONCLUSIONS
In this study, we have obtained time-series data of barcoded DNA in nanochannels using high-throughput imaging to provide further evidence for a non-Gaussian distribution of internal fluctuations. Using videos allowed us to detect and remove many experimental artifacts including non-resolved label pairs, stuck labels, and overlapping molecules. After removing these artifacts, we were able to construct a probability density for the distance between barcode labels using over a thousand molecules and over 7 × 106 measurements of label pair separation distances. The distribution displayed left-skewness, corroborating previous work11 using single snapshots of molecules aligned to a reference genome. However, the magnitude of the skewness measured from dynamic data without alignment to the genome was in general reduced when compared to values obtained from the reference-aligned snapshot data.
Accurate measurements of the probability distributions are critical for genome alignment algorithms, which use these probability distributions as part of the algorithm for determining whether a particular DNA molecule aligns to the consensus genome map created by all of the other molecules in the ensemble. In this respect, the key results of our paper are (i) Fig. 6, which provides a bound on the expected error (the maximum fluctuation) when a single snapshot is used to estimate the distance between two barcode labels and (ii) Fig. 8(a), which provides the expected probability density for barcode labels using the 150 ms exposure time from the commercial technology. We anticipate that both of these results will prove useful in the further maturation of alignment algorithms for genome mapping in nanochannels.
While our results are useful for the current technology, which uses snapshots of the DNA to construct the genome map, videos might provide a method to obtain greater accuracy for certain subsets of molecules. In a potential future application where genome mapping is used as a diagnostic in cancer detection/identification, taking movies of molecules from a targeted region may increase the accuracy of the genomic map, even if the rate of data acquisition and analysis might be somewhat slower. This method might be advantageous for heterogeneity in chromosome populations.31–33 For cases where significant heterogeneity in the population is suspected, for example, tumor cells, using videos to remove these experimental artifacts would illuminate minute differences only present in a small number of individuals or cells.
The development of the filters we used here to remove artifacts from the ultrahigh-throughput data produced by the Irys system is an important step towards future work focused on intramolecular dynamics. Removing the non-resolved labels, stuck labels, and overlapping molecules will allow us to calculate parameters such the relaxation time and diffusion of internal segments of DNA defined by label pairs. Altering our methodology could allow for analysis of dynamics at longer timescales. Stroboscopic imaging would increase the lifetime of the labels by spreading the total excitation dose across a longer duration. This would allow us to capture the longer modes of oscillation, across many multiples of the longest relaxation time, that are inaccessible within the current 10 s window.
ACKNOWLEDGMENTS
This work was supported by the NIH (Grant No. R01-HG006851) and the NSF (Grant No. CBET-1262286) and was carried out in part using computing resources at the University of Minnesota Supercomputing Institute. Jeffrey G. Reifenberger and Han Cao are employees of BioNano Genomics, which is commercializing nanochannel genome mapping.
References
- 1. Levy-Sakin M. and Ebenstein Y., Curr. Opin. Biotechnol. 24, 690 (2013). 10.1016/j.copbio.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 2. Pendleton M., Sebra R., Pang A. W. C., Ummat A., Franzen O., Rausch T., Stütz A. M., Stedman W., Anantharaman T., Hastie A., Dai H., Fritz M. H.-Y., Cao H., Cohain A., Deikus G., Durrett R. E., Blanchard S. C., Altman R., Chin C.-S., Guo Y., Paximos E. E., Korbel J. O., Darnell R. B., McCombie W. R., Kwok P.-Y., Mason C. E., Schadt E. E., and Bashir A., Nat. Methods 12, 780 (2015). 10.1038/nmeth.3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neely R. K., Dedecker P., Hotta J.-I., Urbanaviciute G., Klimasauskas S., and Hofkens J., Chem. Sci. 1, 453 (2010). 10.1039/c0sc00277a [DOI] [Google Scholar]
- 4. Teague B., Waterman M. S., Goldstein S., Potamousis K., Zhou S., Reslewic S., Sarkar D., Valouev A., Churas C., Kidd J. M., Kohn S., Runnheim R., Lamers C., Forrest D., Newton M. A., Eichler E. E., Kent-First M., Surti U., Livny M., and Schwartz D. C., Proc. Natl. Acad. Sci. 107, 10848 (2010). 10.1073/pnas.0914638107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meltzer R. H., Krogmeier J. R., Kwok L. W., Allen R., Crane B., Griffis J. W., Knaian L., Kojanian N., Malkin G., Nahas M. K., Papkov V., Shaikh S., Vyavahare K., Zhong Q., Zhou Y., Larson J. W., and Gilmanshin R., Lab Chip 11, 863 (2011). 10.1039/c0lc00477d [DOI] [PubMed] [Google Scholar]
- 6. Lam E. T., Hastie A., Lin C., Ehrlich D., Das S. K., Austin M. D., Deshpande P., Cao H., Nagarajan N., Xiao M., and Kwok P.-Y., Nat. Biotechnol. 30, 771 (2012). 10.1038/nbt.2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hastie A. R., Dong L., Smith A., Finklestein J., Lam E. T., Huo N., Cao H., Kwok P.-Y., Deal K. R., Dvorak J., Luo M.-C., Gu Y., and Xiao M., PLoS One 8, e55864 (2013). 10.1371/journal.pone.0055864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao H., Hastie A. R., Cao D., Lam E. T., Sun Y., Huang H., Liu X., Lin L., Andrews W., Chan S., Huan S., Tong X., Requa M., Anantharaman T., Krogh A., Yang H., Cao H., and Xu X., GigaScience 3, 34 (2014). 10.1186/2047-217X-3-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Majesta O., Searles V. B., Dickens C. M., Astling D., Albracht D., Mak A. C., Lai Y. Y., Lin C., Chu C., Graves T., Kwok P.-Y., Wilson R. K., and Sikela J. M., BMC Genomics 15, 387 (2014). 10.1186/1471-2164-15-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valouev A., Schwartz D. C., Zhou S., and Waterman M. S., Proc. Natl. Acad. Sci. 103, 15770 (2006). 10.1073/pnas.0604040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reinhart W. F., Reifenberger J. G., Gupta D., Muralidhar A., Sheats J., Cao H., and Dorfman K. D., J. Chem. Phys. 142, 064902 (2015). 10.1063/1.4907552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reisner W., Morton K. J., Riehn R., Wang Y. M., Yu Z., Rosen M., Sturm J. C., Chou S. Y., Frey E., and Austin R. H., Phys. Rev. Lett. 94, 196101 (2005). 10.1103/PhysRevLett.94.196101 [DOI] [PubMed] [Google Scholar]
- 13. Odijk T., Phys. Rev. E 77, 060901 (2008). 10.1103/PhysRevE.77.060901 [DOI] [PubMed] [Google Scholar]
- 14. Muralidhar A., Tree D. R., and Dorfman K. D., Macromolecules 47, 8446 (2014). 10.1021/ma501687k [DOI] [Google Scholar]
- 15. Gupta D., Sheats J., Muralidhar A., Miller J. J., Huang D. E., Mahshid S., Dorfman K. D., and Reisner W., J. Chem. Phys. 140, 214901 (2014). 10.1063/1.4879515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dai L., Renner C. B., and Doyle P. S., Macromolecules 48, 2812 (2015). 10.1021/acs.macromol.5b00280 [DOI] [Google Scholar]
- 17. Valouev A., Shotgun Optical Mapping: A Comprehensive Statistical and Computational Analysis ( University of Southern California, 2006). [Google Scholar]
- 18. Reifenberger J. G., Dorfman K. D., and Cao H., Analyst 140, 4887 (2015). 10.1039/C5AN00343A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riehn R., Lu M., Wang Y.-M., Lim S. F., Cox E. C., and Austin R. H., Proc. Natl. Acad. Sci. U. S. A. 102, 10012 (2005). 10.1073/pnas.0503809102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tree D. R., Wang Y., and Dorfman K. D., Biomicrofluidics 7, 054118 (2013). 10.1063/1.4826156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muralidhar A. and Dorfman K. D., Macromolecules 48, 2829 (2015). 10.1021/acs.macromol.5b00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y.-L., Lin Y.-H., Chang J.-F., and Lin P.-K., Macromolecules 47, 1199 (2014). 10.1021/ma401923t [DOI] [Google Scholar]
- 23. Carpenter J. H., Karpusenko A., Pan J., Lim S. F., and Riehn R., Appl. Phys. Lett. 98, 253704 (2011). 10.1063/1.3602922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karpusenko A., Carpenter J. H., Zhou C., Lim S. F., Pan J., and Riehn R., J. Appl. Phys. 111, 024701 (2012). 10.1063/1.3675207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su T., Das S. K., Xiao M., and Purohit P. K., PLoS One 6, e16890 (2011). 10.1371/journal.pone.0016890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasnik I., McKinney S. A., and Ha T., Nat. Methods 3, 891 (2006). 10.1038/nmeth934 [DOI] [PubMed] [Google Scholar]
- 27. Dave R., Terry D. S., Munro J. B., and Blanchard S. C., Biophys. J. 96, 2371 (2009). 10.1016/j.bpj.2008.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aitken C. E., Marshall R. A., and Puglisi J. D., Biophys. J. 94, 1826 (2008). 10.1529/biophysj.107.117689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N., Computer Control of Microscopes Using μManager ( John Wiley & Sons, Inc., Hoboken, NJ, USA, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.See supplementary material at http://dx.doi.org/10.1063/1.4938732E-BIOMGB-9-025506 for a detailed description of the filters, as well as additional data for the 40 nm nanochannels and all the data for the 51 nm nanochannels.
- 31. Ye C. J., Stevens J. B., Liu G., Bremer S. W., Jaiswal A. S., Ye K. J., Lin M.-F., Lawrenson L., Lancaster W. D., Kurkinen M., Liao J. D., Gairola C. G., Shekhar M. P., Naryan S., Miller F. R., and Heng H. H., J. Cell Physiol. 219, 288 (2009). 10.1002/jcp.21663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrison C. D., Liu P., Woloszynska-Read A., Zhang J., Luo W., Qin M., Bshara W., Conroy J. M., Sabatini L., Vedell P., Xiong D., Liu S., Wang J., Shen H., Li Y., Omilian A. R., Hill A., Head K., Guru K., Kunnev D., Leach R., Eng K. H., Darial C., Hoeflich C., Veeranki S., Glenn S., You M., Pruitt S. C., Johnson C. S., and Trump D. L., Proc. Natl. Acad. Sci. 111, E672 (2014). 10.1073/pnas.1313580111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Friedrich A., Jung P., Reisser C., Fischer G., and Schacherer J., Mol. Biol. Evol. 32, 184 (2015). 10.1093/molbev/msu295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4938732E-BIOMGB-9-025506 for a detailed description of the filters, as well as additional data for the 40 nm nanochannels and all the data for the 51 nm nanochannels.