Abstract
Enterovirus D68 (EV-D68) is an emerging pathogen responsible for mild to severe respiratory infections that occur mostly in infants, children and teenagers. EV-D68, one of more than 100 non-polio enteroviruses, is acid-labile and biologically similar to human rhinoviruses (HRV) (originally classified as HRV87). However, there is no approved preventive or therapeutic measure against EV-D68, HRV, or other enteroviruses. In this study, we evaluated the antiviral activity of series of dipeptidyl compounds against EV-D68 and HRV strains, and demonstrated that several peptidyl aldehyde and α-ketoamide peptidyl compounds are potent inhibitors of EV-D68 and HRV strains with high in-vitro therapeutic indices (>1000). One of the α-ketoamide compounds is shown to have favorable pharmacokinetics profiles, including a favorable oral bioavailability in rats. Recent successful development of α-ketoamide protease inhibitors against hepatitis C virus suggests these compounds may have a high potential for further optimization and development against emerging EV-D68, as well as HRV.
Keywords: Enterovirus D68, Human rhinovirus, Aldehyde and α-ketoamide peptidyl compounds, Antivirals
Highlights
-
•
Series of dipeptidyl aldehyde or ketomide compounds were highly effective against enterovirus-D68 and human rhinoviruses.
-
•
The highly effective ketoamide compound is shown to have favorable pharmacokinetics profiles.
-
•
These may have a high potential for further antiviral development against emerging enterovirus-D68 and human rhinoviruses.
1. Introduction
Enteroviruses belong to the largest genera in the Picornaviridae family, one of the most genetically diverse and medically and economically important viruses that are human and animal pathogens (Knowles et al., 2012). The genus Enterovirus is divided into 10 established and 2 proposed species, and among them, at least 7 species infect humans which includes human rhinovirus (HRV) (A–C) and human enteroviruses (A–D) that include enterovirus D68 (EV-D68), and coxsackievirus A and B (CVA and CVB) (Knowles et al., 2012). Viruses in the Enterovirus genus affect millions of people worldwide and cause a wide spectrum of diseases from asymptomatic or mild illnesses to severe illnesses (Tapparel et al., 2012). Although poliovirus (PV) has been nearly eradicated by immunization, non-polio enteroviruses account for more than 85%∼95% of aseptic meningitis cases where a specific viral pathogen is identified (Connolly and Hammer, 1990, Sawyer and Rotbart, 2004). EV-D68, originally classified as human rhinovirus (HRV) 87, was first identified in California in 1962, and is biologically more similar to HRV for its acid-lability and the respiratory track as the target tissue than other enteroviruses (Schieble et al., 1967, Tokarz et al., 2012). EV-D68 can cause mild to severe respiratory illness, which includes fever, runny nose, sneezing, cough, body/muscle aches, wheezing and difficulty breathing at all ages, but infants, children and teenagers are particularly susceptible to severe symptoms. EV-D68 infection causes particularly severe infections in children with asthma or other existing respiratory illnesses. Since its first report in 1962, few outbreaks of EV-D68 had occurred. However, several outbreaks of EV-D68 have been reported in the last 3–4 years in Europe, Asia and North America. Especially, nationwide outbreak of severe respiratory illness due to EV-D68 infection occurred in the US in 2014 (Ayscue et al., 2014, Imamura and Oshitani, 2015, Messacar et al., 2015, Midgley et al., 2014, Tokarz et al., 2012) where EV-D68 has been detected in specimens from 12 patients who died.
HRVs were recently reclassified into the Enterovirus genus. HRV has more than 100 serotypes and one of the most commonly implicated viruses in the common cold, being responsible for 30–50% of the cases (Arruda et al., 1997, Makela et al., 1998, Monto, 1994). HRVs are also important agents for exacerbation of pre-existing respiratory disease such as asthma and chronic obstructive pulmonary disease (Kurai et al., 2013).
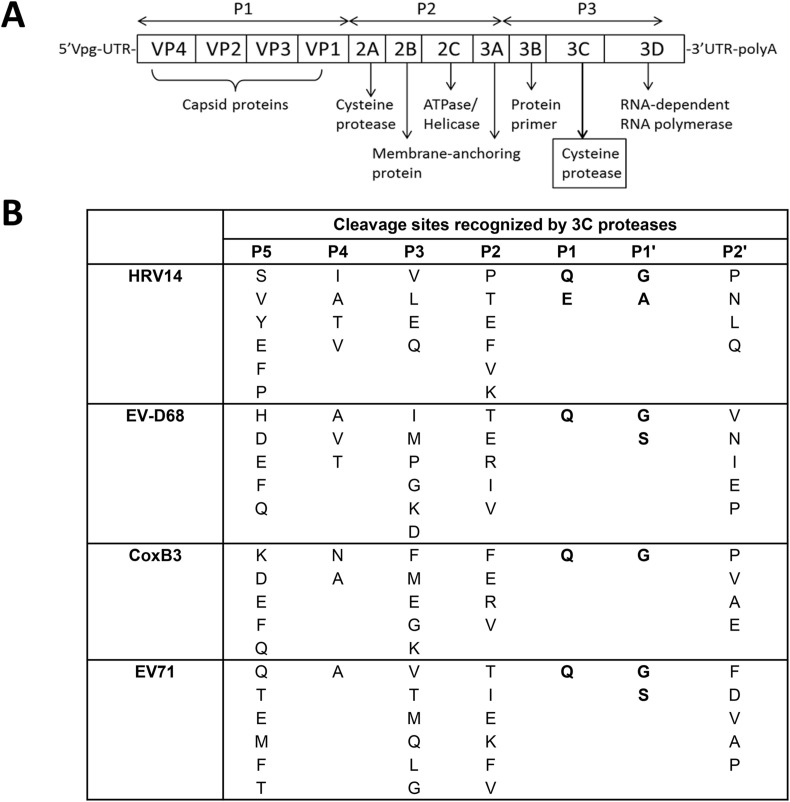
The genome of enteroviruses is a single-stranded, positive sense RNA molecule that consists of one open reading frame (ORF) that encodes a polyprotein. The ORF is divided into three consecutive regions; processing of the P1 region yields the capsid proteins (VP1-4), while processing of the P2 and P3 regions yields the nonstructural replication proteins 2A-2C and 3A-3D, respectively (Fig. 1 ). The polyprotein is first cleaved by a virus-encoded 2A protease at the VP1-2A junction, and other junction sites are cleaved by 3Cpro to generate mature nonstructural and structural proteins (Fig. 1) (Racaniello, 2007). 3Cpro is a chymotrypsin-like cysteine and also a constituent of the replication complex, and helps evade host immunity by interacting with host factors (Chase and Semler, 2012). Because 3Cpro is functionally and structurally highly conserved among enteroviruses, 3Cpro is an attractive target for discovery of anti-enterovirus small molecule therapeutics (Lu et al., 2011).
Fig. 1.
Genomic organization of picornaviruses (A) and cleavage sites recognized by 3Crpo for human rhinovirus 14 (HRV14), enterovirus D68 (EV-D68), coxsackievirus B3 (CoxB3) and enterovirus 71 (EV71).
We have previously synthesized peptidyl inhibitors based on the conserved key features of 3Cpro or related 3C-like protease (3CLpro) encoded by coronaviruses, caliciviruses or picornaviruses and reported their broad-spectrum antiviral activities against multiple viruses in the enzyme- or cell-based assay systems (Kim et al., 2012). In this study, we evaluated the series of dipeptidyl compounds (Galasiti Kankanamalage et al., 2015, Mandadapu et al., 2013a, Mandadapu et al., 2013b, Mandadapu et al., 2012, Tiew et al., 2011) against the 3Cpro of EV-D68 and HRV. Our results show that dipeptidyl α-ketoamide compounds have potent antiviral activities against EV-D68 as well as multiple strains of HRV with half-maximum concentration (EC50) in the low nanomolar range. These α-ketoamide compounds have excellent in vitro safety indices. One of the α-ketoamide compounds tested in rats was determined to have a favorable oral bioavailability (20%F). Successful development of α-ketoamide protease inhibitors against hepatitis C virus (Telaprevir and Boceprevir) suggests these α-ketoamide compounds may have a high potential for further drug development against emerging EV-D68 as well as HRV.
2. Materials and methods
2.1. Compounds
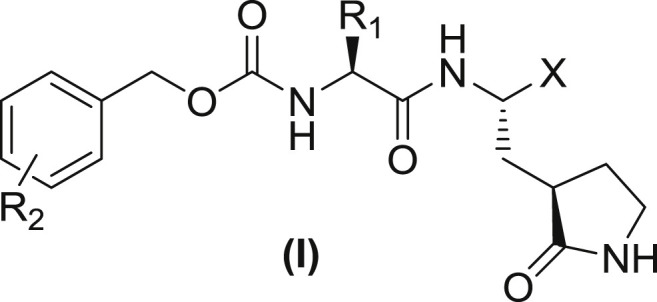
A series of dipeptidyl compounds were synthesized as reported elsewhere (Galasiti Kankanamalage et al., 2015, Mandadapu et al., 2013a, Mandadapu et al., 2013b, Mandadapu et al., 2012, Tiew et al., 2011). In this study, five pairs of aldehyde and ketoamide compounds were focused for their activity against EV-D68 and HRV strains (Table 1 ). Known enterovirus inhibitors, Plecornail, an entry blocker (Pevear et al., 1999), and AG7088, a 3Cpro inhibitor (Patick et al., 1999), were purchased from Sigma-Aldridge (St Louis, MO).
Table 1.
Structures of representative depeptidyl compounds used in this study and their activity against 3Cpro of EV-D68 or HRV14. The IC50 values are indicated as mean and standard variation from two or three separate experiments.
| Compound | R2 | R1 | X | IC50 (μM) |
|
|---|---|---|---|---|---|
| EV-D68 | HRV14 | ||||
| 1 | H | Leu | CHO | 0.17 ± 0.02 | 0.26 ± 0.03 |
| 2 | C(O)C(O)NH(Cyc-Prop) | 0.10 ± 0.01 | 0.18 ± 0.03 | ||
| 3 | Phe | CHO | 0.11 ± 0.02 | 0.09 ± 0.01 | |
| 4 | C(O)C(O)NH(Cyc-Prop) | 0.093 ± 0.01 | 0.085 ± 0.02 | ||
| 5 | Cha | CHO | 0.083 ± 0.01 | 0.073 ± 0.01 | |
| 6 | C(O)C(O)NH(Cyc-Prop) | 0.021 ± 0.01 | 0.02 ± 0.08 | ||
| 7 | m-Cl | Leu | CHO | 0.12 ± 0.02 | 0.2 ± 0.03 |
| 8 | C(O)C(O)NH(Cyc-Prop) | 0.13 ± 0.04 | 0.16 ± 0.01 | ||
| 9 | Cha | CHO | 0.055 ± 0.01 | 0.05 ± 0.02 | |
| 10 | C(O)C(O)NH(Cyc-Prop) | 0.022 ± 0.05 | 0.02 ± 0.01 | ||
| AG7088 | – | – | – | 0.013 ± 0.04 | 0.014 ± 0.05 |
2.2. Cells, viruses, and reagents
WI-38 (human embryonic lung cells) and HeLa cells were maintained in minimal essential medium (MEM) containing 5% fetal bovine serum and antibiotics (chlortetracycline [25 μg/ml], penicillin [250 U/ml], and streptomycin [250 μg/ml]). Both cells were obtained from ATCC (Manassas, VA). Viruses used in the study were EV-D68 and HRV (1B and 51 strains) obtained from ATCC. Hyper immune guinea pig serum raised against EV-D68 was purchased from ATCC.
2.3. Expression and purification of 3Cpro of EV D68
The cDNA encoding full length EV-D68 3Cpro gene was amplified by RT-PCR as previously described (Anand et al., 2002, Schultheiss et al., 1995). The forward primer also contained start codon (underlined) and nucleotide sequences encoding 6 His. Primers are; EVD68Pro-6hisEco-F: 5′- AATTGAATTCAAGGAGATATACCATGCATCATCATCATCATCAT GGACCAGGGTTCGATTTTGCAC and EVD68Pro-Xho-R: 5′-AATTCTCGAGTCATTGTGTATC GGTAAAGTA AGAG-3′. The amplified product was subcloned into pET-28(+) vector using EcoRI and XhoI enzyme sites. The expression and purification of the protease was performed with a standard method described previously by our lab (Takahashi et al., 2011). Recombinant HRV 3Cpro was purchased from EMD chemicals, Inc. (Gibbstown, NJ).
2.4. Fluorescence resonance energy transfer (FRET) protease assays
The FRET substrate (Edans-DFHLQ/GP-Dabcyl) was synthesized by AnaSpec, Inc (Fremont, CA). The FRET protease assay was performed as follows; stock solutions (10 mM) of the substrate and compounds were prepared in DMSO, and diluted in assay buffer. The assay buffer comprised 20 mM HEPES buffer containing NaCl (0–200 mM), 0.4 mM EDTA, glycerol (30–60%), and 6 mM DTT at pH 7. The protease was mixed with serial dilutions of each compound or mock (DMSO) in 25 μl of assay buffer and incubated at 37 °C for 30 min, followed by the addition of 25 μl of substrate. Fluorescence readings were obtained on a fluorescence microplate reader (FLx800, Biotek, Winooski, VT) using an excitation wavelength of 360 nm and an emission wavelength of 460 nm at 1 h following the addition of substrate. The relative fluorescence units (RFU) were determined by subtracting the background values (substrate containing well without protease) from the raw fluorescence values as described previously (Kim et al., 2012, Takahashi et al., 2013). The dose-dependent FRET inhibition curves were fitted with variable slope using the GraphPad Prism software (version 6.04, San Diego, CA) in order to determine the 50% inhibitory concentration (IC50) of the compound.
2.5. Cell-based inhibition assays
WI-38 cell line was used for the cell-based inhibition assay for EV-D68 and HRVs at 37 °C. The inhibitory effects of each compound against HRV strains were also tested in HeLa cells at 33 °C to confirm the results obtained from WI-38 cells. Briefly, confluent or semi-confluent cells were inoculated with each virus at a multiplicity of infection (MOI) of 0.05 for 1 h, and the inoculum was replaced with medium containing mock-DMSO (<0.1%) or each compound (up to 100 μM). The virus infected cells were further incubated for up to 168 h at which time cells show 80–90% cytopathic effects. Viral replication was then determined by the 50% tissue culture infectious dose (TCID50) assay (Reed and Muench, 1938). The EC50 values were determined by using the GraphPad software. Immunofluorescence assay (IFA) was also performed to confirm the anti-EV-D68 activity of one of the α-ketoamide compounds (compound 10) in WI-38 cells. Cells were inoculated with mock-medium or EV-D68 at 0.05 MOI, immediately followed by addition of various concentrations (0.005–0.5 μM) of compound 10 or mock. Cells were then fixed with cold methanol at 72 h post-infection for IFA staining. Primary (hyper immune guinea pig serum raised against EV-D68) and secondary antibody (FITC conjugated goat anti-guinea pig serum) were used for staining.
2.6. Nonspecific cytotoxic effect and enzyme selectivity of compound 10
The 50% cytotoxic concentration (CC50) of each compound was determined in replicating or non-replicating WI-38 cells. Trypsinized cells or confluent cells grown in 96-well plates were treated with various concentrations (1–100 μM) of a compound for 48 or 168 h and cytotoxicity was measured using a CytoTox 96® non-radioactive cytotoxicity assay kit (Promega, Madison, WI) and crystal violet staining. The In vitro therapeutic index was calculated by dividing CC50 by EC50. The enzyme selectivity of compound 10 was evaluated against a panel of representative host proteases including human neutrophil elastase (HNE), chymotrypsin, trypsin, thrombin and carboxypeptidase A (Di Fenza et al., 2007, Groutas et al., 1997, Sidhu et al., 2011, Taggart et al., 2001, Tanaka et al., 1994) as previously reported (Galasiti Kankanamalage et al., 2015). Briefly, various concentrations (2.5–125 μM) of compound 10 was tested against each protease at the ratio of 50 or 250 (10/protease), and the percent inhibition, an index of enzyme selectivity, was determined.
2.7. Pharmacokinetics and oral bioavailability studies in rats
These studies were performed at Washington Biotech Inc using female Sprague-Dawley rats with an IACUC approved protocol (Washington Biotech Inc). Three healthy specific pathogen free (SPF) rats were given orally or intravenously with a single dose of compound 10 at 10 mg/kg (body weight). The drug was dissolved in 10% ethanol and 90% PEG400. Blood was collected in vacutainer tubes containing EDTA at various time points (0.5, 1, 4, 8, 12, 16, and 24 h) and plasma was obtained by centrifugation and stored at – 20 °C until analysis. The concentrations of compound 10 in plasma was measured by liquid-liquid extraction and liquid chromatography/tandem mass spectrometry (LC-MS/MS) method at Washington Biotech Inc. The drug concentration-time graph was generated by GraphPad Prism software. The data were fit to one or two-compartment model with and without a lag time and the best model was selected based on Akaike Information Criteria using PKSolver program (version 2.0) (Zhang et al., 2010). The percent bioavailability (%F) was calculated by dividing the PO plasma AUC0-∞ by the IV plasma AUC0-∞ times 100.
2.8. Comparison of X-ray crystal structures of 3Cpro of EV-D68 and poliovirus
The crystal structure of poliovirus 3Cpro bound with Compound 1 (PDB: 4DCD) that was previously published by our group (Kim et al., 2012) was superimposed with that of EV-D68 (PDB: 3ZV8) (Tan et al., 2013) based on the sequence alignment to minimize the root mean square (RMS) between the 3Cpro of poliovirus and EV-D68 by using PyMol (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC).
3. Results
3.1. Inhibitory effects of the compounds against 3Cpro of EV-D68 or HRV14
The kinetics of EV-D68 3Cpro was comparable to HRV14 3Cpro with the substrate (data not shown). The aldehyde and α-ketomide compounds in Table 1 were highly effective against both 3Cpro of EV-D68 and HRV14 with IC50 values ranging from 0.02 to 0.26 μM in our assay conditions. Each compound showed comparable activity against 3Cpro of EV-D68 or HRV14, suggesting the similar structural requirement for inhibition in these viral proteases (Table 1). These dipeptidyl compounds have the Gln surrogate at a position corresponding to P1 (R1) and leucine (Leu), phenylalanine (Phe) or cyclohexyl alanine (Cha) at P2 position (R2) (Table 1). The compounds with Cha at R2 (compound 5, 6, 9 and 10) were generally more active than those with Leu or Phe at R2 against 3Cpro of EV-D68 and HRV14 (Table 1). Replacing the hydrogen ion at the meta position on the benzene ring (R2) with chlorine did not change the anti-3Cpro-activity. However, an α-ketoamide [C(O)C(O)NH(Cyc-Prop)] moiety at the X position increased the activity of the compounds, compared to the corresponding aldehyde (CHO). The most potent compounds for both EV-D68 and HRV14 3Cpro were compound 6 and 10 and their IC50 values were comparable to those of AG7088 (Table 1).
3.2. The anti-viral effects of the compounds against EV-D68 or HRV strains in cell culture
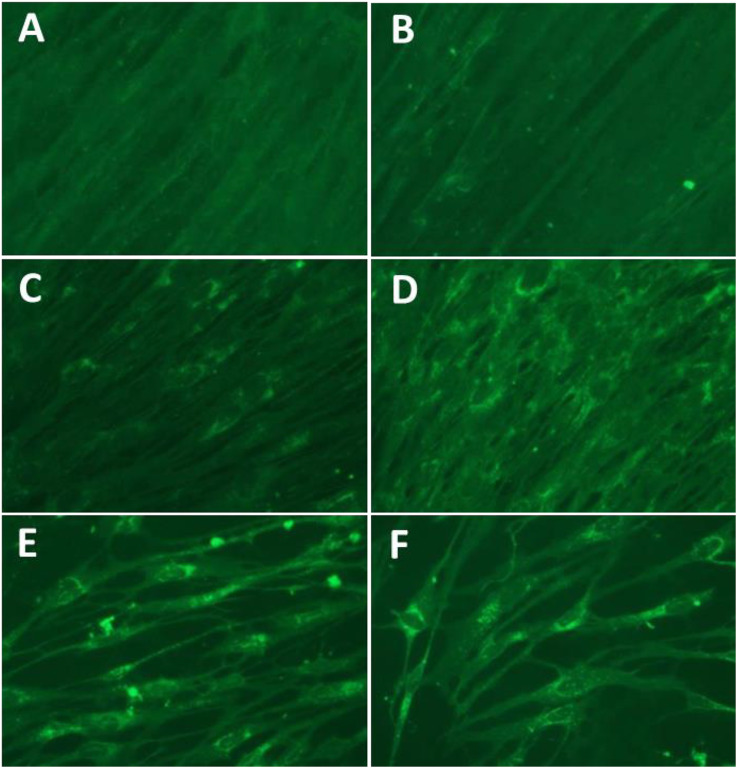
The antiviral effects of the compounds against EV-D68 and HRV strains in cell culture were determined in the human embryonic lung cell line (WI-38 cells) in comparison to AG7088 and Pleconaril (Table 2 ). The antiviral activity of the compounds against HRV strains in HeLa cells at 33 °C was comparable to those in WI-38 cells (data not shown). The EC50 values of the compounds were generally in line with the IC50 values determined by the protease assay (Table 1, Table 2). Among the tested compounds, α-ketoamide compounds, 6 and 10 had the most potent activity against the replication of the viruses (EV-D68, HRV1B and HRV51). Compound 4 also showed similar activity as compound 6 and 10 against the viruses in cell culture. The EC50 values of these two best compounds were comparable to those of AG7088 or Pleconaril (Table 2). IFA staining of EV-D68 in WI-38 cells incubated with various concentrations (0.005–0.5 μM) of compound 10 confirmed the antiviral activity of the compound in the cells (Fig. 2 ). Minimal non-specific staining was observed in the IFA staining (Fig. 2A). In cells infected with EV-D68-without treatment of the compound, cytopathic effects (CPE) were observed and most of the cells still attached to the plate were stained for viral proteins (Fig. 2F). Inhibition of viral replication increased with increasing concentrations of compound 10, demonstrated by the CPE and virus specific staining (Fig. 2B to E).
Table 2.
Effects of the representative dipeptidyl compounds on EV-D68 and HRV strains in cell culture in comparison to those of AG7088 and plecornaril. The EC50 values are indicated as mean and standard variation from two or three separate experiments.
| Compound | EV-D68 | HRV1B | HRV51 |
|---|---|---|---|
| 1 | 0.38 ± 0.25 | 1.90 ± 1.41 | 0.70 ± 0.28 |
| 2 | 0.08 ± 0.02 | 0.25 ± 0.21 | 0.07 ± 0.01 |
| 3 | 0.08 ± 0.02 | 0.95 ± 0.49 | 0.55 ± 0.21 |
| 4 | 0.03 ± 0.01 | 0.19 ± 0.16 | 0.07 ± 0.03 |
| 5 | 0.06 ± 0.01 | 0.65 ± 0.21 | 0.14 ± 0.08 |
| 6 | 0.04 ± 0.01 | 0.15 ± 0.07 | 0.05 ± 0.03 |
| 7 | 0.43 ± 0.04 | 0.75 ± 0.21 | 0.40 ± 0.14 |
| 8 | 0.09 ± 0.01 | 0.16 ± 0.08 | 0.15 ± 0.08 |
| 9 | 0.08 ± 0.02 | 0.70 ± 0.14 | 0.16 ± 0.06 |
| 10 | 0.04 ± 0.02 | 0.08 ± 0.01 | 0.07 ± 0.01 |
| AG7088 | 0.01 ± 0.003 | 0.02 ± 0.01 | 0.05 ± 0.03 |
| Plecornaril | 0.60 ± 0.14 | 0.27 ± 0.18 | 0.07 ± 0.02 |
Fig. 2.
Effects of compound 10 on EV-D68 in cell culture determined by IFA. IFA staining of EV-D68 infected with WI-38 cells with various concentrations (0.005–0.5 μM, B–E) of compound 10. Cells were fixed at 72 h after virus infection for the staining. Controls include no virus infection (A) and virus infection without the compound (F). B-E: treated with compound 10, at 0.5 (B), 0.1 (C), 0.05, or 0.005 μM, respectively.
3.3. Cell toxicity and enzyme selectivity
All listed compounds did not show any cytotoxicity up to 100 μM in proliferating or non-proliferating cells. Since off-target effects for host proteases can be a potential caveat in drug development, the selectivity profile of compound 10 was determined using a representative panel of proteases (Table 3 ). Compound 10 is determined to be a very weak inhibitor of HNE and have no cross-reactivity toward a panel of other host enzymes at the indicated concentrations (Table 3).
Table 3.
Selectivity of compound 10 against important host proteases including human neutrophil elastase (HNE), chymotrypsin, trypsin, thrombin and carboxypeptidase A. Compound 10 was incubated with host proteases at the ration of 50 or 250 and percent inhibition was determined for the enzyme selectivity.
| Enzyme | I/Ea | Compound (% inhibition) |
[I]f (μM) |
|---|---|---|---|
| 10 | |||
| HNE | 50 | 32 | 17.5 |
| Chymotrypsin | 250 | 0 | 2.5 |
| Trypsin | 250 | 0 | 125 |
| Thrombin | 250 | 0 | 2.75 |
| Carboxypeptidase A | 250 | 0 | 43 |
I/E: inhibitor/enzyme; [I]f: final concentrations of inhibitor used in this assay.
3.4. The pharmacokinetics and oral bioavailability of compound 10 in rats
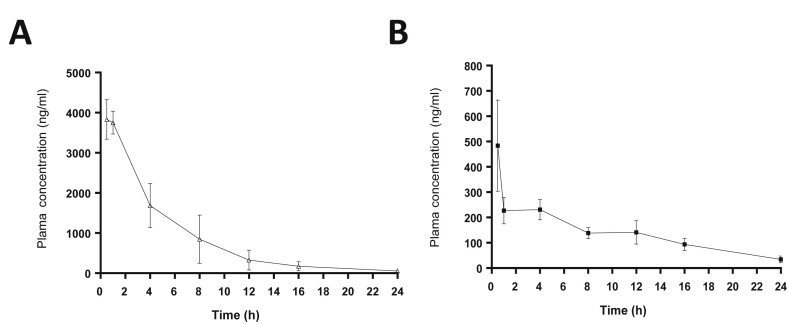
The plasma drug concentration data were fit to a one or two-compartment model with and without a lag time and two–compartment model was selected as a best fit with lowest Akaike Information Criteria using PKSolver program (version 2.0) (Zhang et al., 2010). The plasma concentration-time plot of compound 10 following IV or PO doses is shown in Fig. 3 . The summary of the PK parameters following IV or PO administration of compound 10 is listed in Table 4 . After IV or PO administration, compound 10 declined with a rapid initial distribution phase, followed by a slower elimination phase (Fig. 3). After the IV or PO dose, the concentration of compound 10 maintained above EC90 values against EV-D68 (0.13 μM) for more than 16 h but less than 24 h (Fig. 3). The concentrations of compound 10 also remained above EC90 values against HRV1B (0.45 μM) or HRV51 (0.35 μM) for more than 12 h following the IV dose. However, those after the PO dose rapidly fell below the EC90 values against HRV1B or HRV51 in less than one hr (HRV1B) or 8 h (HRV51) (Fig. 3). The oral bioavailability (%F) of compound 10 was determined to be 20, indicating favorable oral absorption.
Fig. 3.
Plots of the mean plasma concentrations of compound 10 in rats administered with an intravenous (A) or oral (B) route at 10 mg/kg. Values represent mean and standard of error of the means; N = 3 per time point.
Table 4.
A summary of the PK parameters obtained following an intravenous (IV) or oral (PO) dose of compound 10 at 10 mg/kg. SPF rats were orally or intravenously given a single dose of compound 10 and blood was collected at various time points. The plasma drug concentrations were measured and the data were fit to two-compartment model using the PKSolver program. The percent bioavailability (%F) was calculated by dividing the PO plasma AUC0-∞ by the IV plasma AUC0-∞ times 100.
| AUC0-t (ng h/ml)a | AUC0-∞ (ng h/ml) | F (%) | MRT (h) | t1/2 α (h) | t1/2 β (h) | CL [(mg/kg)/(ng/ml)/h] | V [(mg/kg)/ng/ml)] | |
|---|---|---|---|---|---|---|---|---|
| IV | 20465.23 | 21013.35 | NA | 5.55 | 2.81 | 7.52 | 0.00048 | 0.0022 |
| PO | 3369.06 | 4200.66 | 20 | 14.15 | 0.035 | 10.68 | 0.00238 | 0.0019 |
AUC0-t, AUC from time zero to the last measured concentration; AUC0-inf, AUC extrapolated to infinity; F, bioavailability; MRT, mean residence time; t1/2α, distribution half life; t1/2β, elimination half life; CL, clearance; V, volume of distribution.
3.5. Comparison of X-ray crystal structures of 3Cpro of EV-D68 and poliovirus
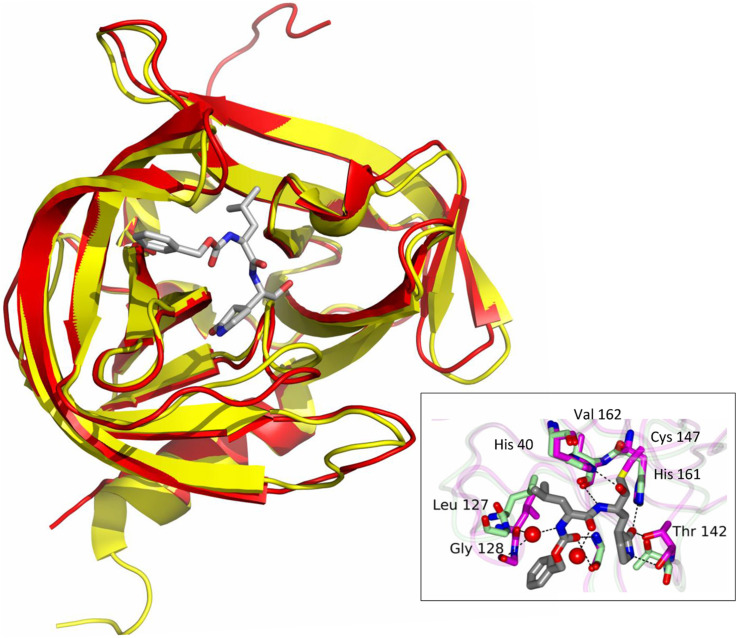
The superimposed crystal structures of EV-D68 and poliovirus 3Cpro show a high degree of similarities in their structures (Fig. 4 ). Poliovirus 3Cpro has an RMS of 0.575 over 945 equivalent residues with EV-D68 3Cpro.
Fig. 4.
Comparison of X-ray crystal structures of poliovirus 3Cpro (yellow) with the bisulfite adduct of compound 1 (gray) (PDB: 4DCD) and EV-D68 3Cpro (red, 3ZV8). Poliovirus 3Cpro was superimposed on EV-D68 3Cpro using PyMol. Image in the insert shows the hydrogen bonds between poliovirus 3Cpro with compound 1. The bisulfite adduct reverts back to aldehyde compound 1, and then interacts with Cys146 to form the enzyme−inhibitor complex (4DCD). The image also shows hydrogen bonds between the compound and enzyme with conformational changes in the enzyme from bound (green) and ligand bound forms (magenta). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
EV-D68 is an emerging medically-important enterovirus and has recently caused several outbreaks associated with severe respiratory illnesses in the US. Many of the affected, lab-confirmed cases were children with underlying airway diseases, such as asthma, suggesting that mildly symptomatic infections with EV-D68 is likely to be much more prevalent than reported. Although respiratory illnesses due to enterovirus are quite common among children and adults, and efforts have been made to develop effective treatments for medically important enteroviruses, including EV-D68, there is no licensed vaccines or drugs available (except for poliovirus vaccines) yet.
Inhibitors of enterovirus replication are broadly classified into three main groups: inhibitors of attachment, entry, and/or uncoating; replicase inhibitors; and protease inhibitors (Chen and Shih, 2011, Chen et al., 2008, De Palma et al., 2008a, Liu et al., 2015, Norder et al., 2011, Tan et al., 2013, Thibaut et al., 2012). The replication cycle of enterovirus is initiated by the attachment of a virus particle to a specific receptor on the host cell surface. An example of attachment inhibitors include Pleconaril that binds to a hydrophobic pocket within enterovirus VP1, a major capsid protein, and consequently prevents viral attachment and uncoating. Recent report demonstrated that Pleconaril also inhibited the replication of EV-D68 by binding to the hydrophobic pocket of VP1 (Liu et al., 2015). Pleconaril failed to get FDA approval for use in adults for potential adverse effects but there are several other entry inhibitors such as Pocapavir and Vapendavir that are being evaluated for their efficacy for neonatal sepsis associated with enterovirus infection (https://clinicaltrials.gov/ct2/show/NCT00031512). Inhibitors of replicases, such as RNA-dependent RNA-polymerase inhibitors, have been reported, but the viral 2A or 3Cpro has been more actively used as a target for inhibition (De Palma et al., 2008b). The design of enterovirus 3Cpro inhibitors has primarily utilized the known peptidyl recognition element and various warheads, such as aldehyde and α-ketoamide, in the case of transition-state analog inhibitors, which interact with the active site cysteine to yield a tetrahedral adduct (Kim et al., 2012, Ramajayam et al., 2011). The warhead can also be a Michael acceptor, such as an α,β-unsaturated ester or vinyl sulfone which, in contrast to aldehydes and α-ketoamides, reacts with the active site cysteine in a time-dependent manner to form a covalently-bound enzyme-inhibitor adduct (Dragovich et al., 1999, Kuo et al., 2008, Matthews et al., 1999, Patick et al., 1999, Tan et al., 2013). The most extensively studied inhibitors of this class (Michael acceptors) are AG7088 and its variants, which have been shown to be active against various enteroviruses including HRVs (Hung et al., 2011, Matthews et al., 1999, Patick et al., 1999) and EV-D68 (Tan et al., 2013). However, inhibitors of viral proteases haven not yet been licensed for enterovirus infection.
The compounds used in this study belong to a series of dipeptidyl transition state 3Cpro inhibitors and this series was previously reported to have activities against HRVs (Kim et al., 2012). In this study, we have conducted a structure-activity relationship study using the newly generated dipeptidyl derivatives against EV-D68 and HRVs to compare their relative structural requirements for inhibition and determine the potency of the derivatives against these important pathogens. Compound 1, a prototype dipeptidyl aldehyde, was previously reported to have anti-HRV activity (Kim et al., 2012) and was used as a template for structural modification for these new derivatives. Based on the enzyme and cell culture assays, there was no significant difference in the activity of the compounds against 3Cpro of EV-D68 or HRVs, which suggests that the structural requirements for blockage of 3Cpro is similar for EV-D68 or HRVs, although the amino acid homology between 3Cpro of EV-D68 and HRVs is about 50%. This observation may be explained by the overall structural similarity of 3Cpro of EV-D68 and HRVs and the striking similarity in the topography of their active sites determined by X-ray crystallographic studies (Lu et al., 2011). The 3Cpro of EV-D68 and HRVs have similar substrate specificity of a near absolute requirement for Gln residue at P1 and a hydrophobic residue (Leu, Phe) at P2 (Fig. 1). Our structure-activity relationship study also showed that the interchange of Leu and Phe at the P2 position (R1) did not lead to a marked change in activity against 3Cpro of EV-D68 or HRVs. However, the presence of Cha at the P1 position yielded better activity against EV-D68 and HRVs, compared to Leu and Phe. The activity of the compounds did not change upon replacement of hydrogen ion with m-Cl at R2 (cap), suggesting that the presence of the chlorine atom ion at the meta position does not contribute to the binding by interacting with the 3Cpro of EV-D68 or HRVs. In our previous study, the meta-chlorine substituent was found to significantly improve the efficacy as compared to the inhibitor having an unsubstituted phenyl ring against caliciviruses in 3CLpro enzyme and cell based assays (Galasiti Kankanamalage et al., 2015). Interestingly, the α-ketoamide compounds were consistently more potent than their aldehyde counterparts against EV-D68 and HRVs in both enzyme and cell-based assays and the most potent compounds for both EV-D68 and HRV14 were α-ketoamide compounds with Cha at R1 and a benzene cap with or without m-Cl (R2), compound 6 and 10. Previously we have reported that α-ketoamides compounds were less effective than their aldehyde counterparts against 3CLpro of caliciviruses and coronarviruses (Kim et al., 2012, Tiew et al., 2011). The previous and current findings suggest that the choice of warhead that gives a better fit may be different among different viruses encoding 3Cpro or 3CLpro. Further X-ray crystallographic studies may provide a structural basis for our findings. We reported co-crystallization of poliovirus 3Cpro with the bisulfite adduct of compound 1 (Kim et al., 2012, PDB: 4DCD), and Fig. 4 shows the highly conserved active site structure between 4DCD bound with Compound 1 and 3ZV8 (EV-D68 3Cpro) (Fig. 4). The Gln surrogate ring, Leu and carboxybenzyl ring of the compound fit into the S1, S2 and S4 sites, respectively. The cytotoxicity of all tested compounds was minimal at 100 μM in cells, indicating a good safety margin with therapeutic indices >1000. In addition, compound 10 was also shown to have a good enzyme selectivity against basic serine (trypsin, thrombin) and metallo- (carboxypeptidase A) proteases, as well as serine proteases (α-chymotrypsin, human neutrophil elastase) (Table 3).
Compound 10 showed most potent antiviral activity against EV-D68 and HRVs, consequently, its pharmacokinetics and oral bioavailability were evaluated in rats. Unfavorable pharmacokinetics and poor oral bioavailability is one of the major challenges in drug development, exemplified by the unsuccessful development of AG7088 which displays poor aqueous solubility and low oral bioavailability (Dragovich et al., 1999). It was developed as a nasal spray, however, further development was stopped after phase II clinical trials. Another irreversible 3Cpro inhibitor, compound 1 entered clinical trials. Compound 1 was found to be highly effective against HRV strains with EC50 < 100 nM and related enteroviruses and, more importantly, it was orally available (Patick et al., 2005), but it was also dropped during phase 1 clinical trial. Compound 10 is shown to have a favorable oral bioavailability in rats with (%F 20), and the plasma drug concentrations remained higher than in vitro EC90 values against EV-D68 for more than 16 h following oral administration using the simple formulation. The oral bioavailability of the structurally-similar aldehyde compounds was also determined. The %F values for the aldehydes were markedly lower (%F ∼5) with shorter halt-lives in rats, as compared to those of compound 10 (data not shown). Therefore, the α-ketoamide warhead may contribute to improvement of the oral bioavailability and the PK of these dipeptidyl compounds, as well as the smaller size (<600 MW) of these dipeptidyl compounds compared to the bulkier AG7088. The duration of compound 10 in the rat plasma above the EC90 values for HRV1B or HRV51 following an oral dose of compound 10 mg/kg is calculated to be much shorter than that for EV-D68 due to the lower antiviral activity of compound 10 against HRV1B or HRV51 (about 2-fold) compared to EV-D68 in cell culture. HRV1B and HRV51 are the representative HRV strains for a minor (HRV1B) and major receptor group (HRV51) where a minor HRV serotypes use low-density lipoprotein receptor (LDLR) and a major HRV serotypes (about 90%) use ICAM-1 as a receptor. When we tested compound 10 against other major group HRV strains such as HRV18 and HRV68, the EC50 and EC90 were comparable to those against HRV51 (data not shown).
In summary, new derivative compounds of a dipeptidyl compound series that have anti-HRV activity (Kim et al., 2012) were synthesized to further improve their inhibitory activity against EV-D68 and HRVs. The antiviral potency and the favorable pharmacokinetic properties of compound 10 suggest that this compound maybe used as a viable lead candidate for further development for EV-D68 and/or HRV infections as a single agent or for a combination treatment with an entry inhibitor.
Acknowledgment
This work was generously supported by an NIH grant (R01AI109039). We thank David for technical assistance.
References
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda E., Pitkaranta A., Witek T.J., Doyle C.A., Hayden F.G. Frequency and natural history of rhinovirus infections in adults during autumn. J. Clin. Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscue P., Van Haren K., Sheriff H., Waubant E., Waldron P., Yagi S., Yen C., Clayton A., Padilla T., Pan C., Reichel J., Harriman K., Watt J., Sejvar J., Nix W.A., Feikin D., Glaser C., Centers for Disease, C., Prevention Acute flaccid paralysis with anterior myelitis - California, June 2012–June 2014. MMWR. Morb. Mortal. Wkly. Rep. 2014;63:903–906. [PMC free article] [PubMed] [Google Scholar]
- Chase A.J., Semler B.L. Viral subversion of host functions for picornavirus translation and RNA replication. Future virol. 2012;7:179–191. doi: 10.2217/fvl.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.C., Shih S.R. Drug discovery in enteroviral infections. Infect. Disord. Drug Targ. 2011;11:337–345. doi: 10.2174/187152611795768060. [DOI] [PubMed] [Google Scholar]
- Chen T.C., Weng K.F., Chang S.C., Lin J.Y., Huang P.N., Shih S.R. Development of antiviral agents for enteroviruses. J. Antimicrob. Chemother. 2008;62:1169–1173. doi: 10.1093/jac/dkn424. [DOI] [PubMed] [Google Scholar]
- Connolly K.J., Hammer S.M. The acute aseptic meningitis syndrome. Infect. Dis. Clin. North Am. 1990;4:599–622. [PubMed] [Google Scholar]
- De Palma A.M., Heggermont W., Lanke K., Coutard B., Bergmann M., Monforte A.M., Canard B., De Clercq E., Chimirri A., Purstinger G., Rohayem J., van Kuppeveld F., Neyts J. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 2008;82:4720–4730. doi: 10.1128/JVI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma A.M., Vliegen I., De Clercq E., Neyts J. Selective inhibitors of picornavirus replication. Med. Res. Rev. 2008;28:823–884. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- Di Fenza A., Heine A., Koert U., Klebe G. Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions. ChemMedChem. 2007;2:297–308. doi: 10.1002/cmdc.200600185. [DOI] [PubMed] [Google Scholar]
- Dragovich P.S., Prins T.J., Zhou R., Webber S.E., Marakovits J.T., Fuhrman S.A., Patick A.K., Matthews D.A., Lee C.A., Ford C.E., Burke B.J., Rejto P.A., Hendrickson T.F., Tuntland T., Brown E.L., Meador J.W., 3rd, Ferre R.A., Harr J.E., Kosa M.B., Worland S.T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J. Med. Chem. 1999;42:1213–1224. doi: 10.1021/jm9805384. [DOI] [PubMed] [Google Scholar]
- Galasiti Kankanamalage A.C., Kim Y., Weerawarna P.M., Uy R.A., Damalanka V.C., Mandadapu S.R., Alliston K.R., Mehzabeen N., Battaile K.P., Lovell S., Chang K.O., Groutas W.C. Structure-guided design and optimization of dipeptidyl inhibitors of norovirus 3CL protease. Structure-activity relationships and biochemical, X-ray crystallographic, cell-based, and in Vivo studies. J. Med. Chem. 2015;58:3144–3155. doi: 10.1021/jm5019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groutas W.C., Kuang R., Venkataraman R., Epp J.B., Ruan S., Prakash O. Structure-based design of a general class of mechanism-based inhibitors of the serine proteinases employing a novel amino acid-derived heterocyclic scaffold. Biochemistry. 1997;36:4739–4750. doi: 10.1021/bi9628937. [DOI] [PubMed] [Google Scholar]
- Hung H.C., Wang H.C., Shih S.R., Teng I.F., Tseng C.P., Hsu J.T. Synergistic inhibition of enterovirus 71 replication by interferon and rupintrivir. J. Infect. Dis. 2011;203:1784–1790. doi: 10.1093/infdis/jir174. [DOI] [PubMed] [Google Scholar]
- Imamura T., Oshitani H. Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev. Med. Virol. 2015;25:102–114. doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-Like proteases of picornaviruses, noroviruses, and coronaviruses. J. virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles N.J., Hovi T., Hyypiä T., King A.M.Q., Lindberg A.M., Pallansch M.A., Palmenberg A.C., Simmonds P., Skern T., authors, o . Picornaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. , Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. tenth Ed. Elsevier; San Diego, CA: 2012. pp. 855–880. [Google Scholar]
- Kuo C.J., Shie J.J., Fang J.M., Yen G.R., Hsu J.T., Liu H.G., Tseng S.N., Chang S.C., Lee C.Y., Shih S.R., Liang P.H. Design, synthesis, and evaluation of 3C protease inhibitors as anti-enterovirus 71 agents. Bioorg. Med. Chem. 2008;16:7388–7398. doi: 10.1016/j.bmc.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurai D., Saraya T., Ishii H., Takizawa H. Virus-induced exacerbations in asthma and COPD. Front. Microbiol. 2013;4:293. doi: 10.3389/fmicb.2013.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sheng J., Fokine A., Meng G., Shin W.H., Long F., Kuhn R.J., Kihara D., Rossmann M.G. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science. 2015;347:71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Qi J., Chen Z., Xu X., Gao F., Lin D., Qian W., Liu H., Jiang H., Yan J., Gao G.F. Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J. virol. 2011;85:10319–10331. doi: 10.1128/JVI.00787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimaki M., Blomqvist S., Hyypia T., Arstila P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadapu S.R., Weerawarna P.M., Gunnam M.R., Alliston K.R., Lushington G.H., Kim Y., Chang K.O., Groutas W.C. Potent inhibition of norovirus 3CL protease by peptidyl alpha-ketoamides and alpha-ketoheterocycles. Bioorg. Med. Chem. Lett. 2012;22:4820–4826. doi: 10.1016/j.bmcl.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadapu S.R., Gunnam M.R., Galasiti Kankanamalage A.C., Uy R.A., Alliston K.R., Lushington G.H., Kim Y., Chang K.O., Groutas W.C. Potent inhibition of norovirus by dipeptidyl alpha-hydroxyphosphonate transition state mimics. Bioorg. Med. Chem. Lett. 2013;23:5941–5944. doi: 10.1016/j.bmcl.2013.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadapu S.R., Gunnam M.R., Tiew K.C., Uy R.A., Prior A.M., Alliston K.R., Hua D.H., Kim Y., Chang K.O., Groutas W.C. Inhibition of norovirus 3CL protease by bisulfite adducts of transition state inhibitors. Bioorg. Med. Chem. Lett. 2013;23:62–65. doi: 10.1016/j.bmcl.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D.A., Dragovich P.S., Webber S.E., Fuhrman S.A., Patick A.K., Zalman L.S., Hendrickson T.F., Love R.A., Prins T.J., Marakovits J.T., Zhou R., Tikhe J., Ford C.E., Meador J.W., Ferre R.A., Brown E.L., Binford S.L., Brothers M.A., DeLisle D.M., Worland S.T. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messacar K., Schreiner T.L., Maloney J.A., Wallace A., Ludke J., Oberste M.S., Nix W.A., Robinson C.C., Glode M.P., Abzug M.J., Dominguez S.R. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;63:798–799. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- Midgley C.M., Jackson M.A., Selvarangan R., Turabelidze G., Obringer E., Johnson D., Giles B.L., Patel A., Echols F., Oberste M.S., Nix W.A., Watson J.T., Gerber S.I. Severe respiratory illness associated with enterovirus D68-Missouri and Illinois, 2014. MMWR. Morb. Mortal. Wkly. Rep. 2014;63:798–799. [PMC free article] [PubMed] [Google Scholar]
- Monto A.S. Studies of the community and family: acute respiratory illness and infection. Epidemiol. Rev. 1994;16:351–373. doi: 10.1093/oxfordjournals.epirev.a036158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H., De Palma A.M., Selisko B., Costenaro L., Papageorgiou N., Arnan C., Coutard B., Lantez V., De Lamballerie X., Baronti C., Sola M., Tan J., Neyts J., Canard B., Coll M., Gorbalenya A.E., Hilgenfeld R. Picornavirus non-structural proteins as targets for new anti-virals with broad activity. Antivir. Res. 2011;89:204–218. doi: 10.1016/j.antiviral.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Patick A.K., Binford S.L., Brothers M.A., Jackson R.L., Ford C.E., Diem M.D., Maldonado F., Dragovich P.S., Zhou R., Prins T.J., Fuhrman S.A., Meador J.W., Zalman L.S., Matthews D.A., Worland S.T. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999;43:2444–2450. doi: 10.1128/aac.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patick A.K., Brothers M.A., Maldonado F., Binford S., Maldonado O., Fuhrman S., Petersen A., Smith G.J., 3rd, Zalman L.S., Burns-Naas L.A., Tran J.Q. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2005;49:2267–2275. doi: 10.1128/AAC.49.6.2267-2275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevear D.C., Tull T.M., Seipel M.E., Groarke J.M. Activity of pleconaril against enteroviruses. Antimicrob. agents Chemother. 1999;43:2109–2115. doi: 10.1128/aac.43.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2015, https://clinicaltrials.gov/ct2/show/NCT00031512. Pleconaril Enteroviral Sepsis Syndrome.
- Racaniello V.R. Picornaviridae: the viruses and their replication. In: Fields B.N., Knipe D.M., Howley P.M., editors. Fields Virology. fifth ed. Lippincott Wlliam & Wilkins; Philadelphia, PA: 2007. pp. 796–801. [Google Scholar]
- Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 2011;39:1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Sawyer M., Rotbart H. third ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2004. Viral Meningitis and the Aseptic Meningitis Syndrome; pp. 75–93. [Google Scholar]
- Schieble J.H., Fox V.L., Lennette E.H. A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol. 1967;85:297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- Schultheiss T., Sommergruber W., Kusov Y., Gauss-Muller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J. Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu P.S., Liang A., Mehta A.Y., Abdel Aziz M.H., Zhou Q., Desai U.R. Rational design of potent, small, synthetic allosteric inhibitors of thrombin. J. Med. Chem. 2011;54:5522–5531. doi: 10.1021/jm2005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart C.C., Lowe G.J., Greene C.M., Mulgrew A.T., O'Neill S.J., Levine R.L., McElvaney N.G. Cathepsin B, L, and S cleave and inactivate secretory leucoprotease inhibitor. J. Biol. Chem. 2001;276:33345–33352. doi: 10.1074/jbc.M103220200. [DOI] [PubMed] [Google Scholar]
- Takahashi D., Kim Y., Chang K.O., Anbanandam A., Prakash O. Backbone and side-chain (1)H, (15)N, and (13)C resonance assignments of Norwalk virus protease. Biomol. NMR Assign. 2011;6:19–21. doi: 10.1007/s12104-011-9316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D., Kim Y., Lovell S., Prakash O., Groutas W.C., Chang K.O. Structural and inhibitor studies of norovirus 3C-like proteases. Virus Res. 2013;178:437–444. doi: 10.1016/j.virusres.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., George S., Kusov Y., Perbandt M., Anemuller S., Mesters J.R., Norder H., Coutard B., Lacroix C., Leyssen P., Neyts J., Hilgenfeld R. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013;87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y.I.G., Dakoji S., Cho Y., Mobashery S. Conscripting the active-site zinc ion in carboxypeptidase a in inactivation chemistry by a new type of irreversible enzyme inactivator. J. Am. Chem. Soc. 1994;116:7475–7480. [Google Scholar]
- Tapparel C., Siegrist F., Petty T.J., Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2012;14C:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Thibaut H.J., De Palma A.M., Neyts J. Combating enterovirus replication: state-of-the-art on antiviral research. Biochem. Pharmacol. 2012;83:185–192. doi: 10.1016/j.bcp.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Tiew K.C., He G., Aravapalli S., Mandadapu S.R., Gunnam M.R., Alliston K.R., Lushington G.H., Kim Y., Chang K.O., Groutas W.C. Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorg. Med. Chem. Lett. 2011;21:5315–5319. doi: 10.1016/j.bmcl.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R., Firth C., Madhi S.A., Howie S.R., Wu W., Sall A.A., Haq S., Briese T., Lipkin W.I. Worldwide emergence of multiple clades of enterovirus 68. J. General Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huo M., Zhou J., Xie S. PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 2010;99:306–314. doi: 10.1016/j.cmpb.2010.01.007. [DOI] [PubMed] [Google Scholar]