Abstract
At physiological levels, nitric oxide (NO) contributes to the maintenance of normal neuronal activity and survival, thus serving as an important regulatory mechanism in the central nervous system. In contrast, accumulating evidence suggests that exposure to environmental toxins or the normal aging process can trigger excessive production of reactive oxygen/nitrogen species (such as NO), contributing to the etiology of several neurodegenerative diseases. Here we highlight protein S-nitrosylation, resulting from covalent attachment of an NO group to a cysteine thiol of the target protein, as a ubiquitous effector of NO signaling in both health and disease. We review our current understanding of this redox-dependent posttranslational modification under neurodegenerative conditions, and evaluate how targeting dysregulated protein S-nitrosylation can lead to novel therapeutics.
Keywords: Protein S-nitrosylation, transnitrosylation, denitrosylation, NitroMemantine, Alzheimer’s disease, Parkinson’s disease
Neurodegenerative diseases and ROS/RNS
A dominant risk factor for the development of neurodegenerative diseases is advanced age. With extended life expectancy worldwide, patients suffering from these types of neurological conditions will increase dramatically in the coming years. Although symptomatic treatments exist, there are currently no effective drugs to reverse or halt the progression of the diseases. Recent studies, however, have identified several potential molecular targets for neurodegenerative disease therapy, including oxidative/nitrosative stress (mediated by reactive oxygen/nitrogen species [ROS/RNS]), excitotoxicity, protein misfolding, endoplasmic reticulum (ER) stress, and mitochondrial damage [1]. Among these, ROS/RNS has attracted significant attention, as neurons have high oxygen consumption rates, which result in up-regulation of ROS/RNS, while manifesting relatively low antioxidative capacity compared to other cell types [2,3].
Another key risk factor for sporadic forms of neurodegenerative diseases involves aberrant interaction of genetic and environmental factors (G×E). Notably, oxidative/nitrosative stress is also considered to be a central mechanism in this scenario. For instance, environmental toxins, such as agricultural chemicals and heavy metals, stimulate the generation of ROS/RNS typically through impairment of mitochondrial respiration [4]. Thus, an emerging concept in the GxE hypothesis for disease is that genetic predisposition determines the susceptibility of sporadic cases to neurodegenerative disorders by lowering the threshold for oxidative/nitrosative stress engendered by environmental factors [5].
As an example of RNS, nitric oxide (NO)-related species play a critical role in the pathogenesis of a number of neurodegenerative diseases. Although early studies emphasized the fact that NO exerts its biological function via activation of soluble guanylyl cyclase (sGC), which produces cyclic guanosine monophosphate (cGMP), emerging evidence recognizes protein S-nitrosylation as a ubiquitous mediator of NO signaling [6]. Accordingly, in this article, we focus NO-dependent posttranslational modification of cysteine residues (i.e., S-nitrosylation) as an important (patho)-physiological mechanism for NO signaling. Here we provide specific examples of dysregulated S-nitrosylation under neurodegenerative conditions, and illustrate that modulators of protein S-nitrosylation may represent uniquely ‘druggable targets’ for neurodegenerative diseases.
Functional and structural dysregulation of neuronal network activities represents a fundamental cause of neurodegenerative conditions such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease, and amyotrophic lateral sclerosis (ALS). For instance, damage to hippocampal synapses contributes to the cognitive deficits and memory impairment in the most common form of dementia, AD. Additionally, the most common motor disorder, PD, is characterized by the initial loss of dopaminergic (DA) neurons in the substantia nigra pars compacta. Evidence suggests that aberrantly elevated levels of oxidative and nitrosative stress may represent the common molecular denominator leading to these pathological processes. For example, excessively produced ROS/RNS may affect cellular macromolecules, such as proteins and lipids, thereby activating aberrant cellular processes such as accumulation of misfolded/aggregated proteins (e.g., amyloid-β [Aβ], tau, α-synuclein, and TDP-43), dysfunction in proteostasis, mitochondrial injury, and neuroinflammation. These pathological cellular events compromise synaptic and neuronal activity, thus contributing to neurodegenerative conditions. Along these lines, NO can S-nitrosylate specific neuronal proteins in response to neurodegenerative stimuli, leading to protein misfolding, ER stress, and mitochondrial impairment [7–9]. Additionally, reaction of •NO (free radical NO with an electron in the outer pi molecular orbital) and superoxide anion (O2•−) forms peroxynitrite (ONOO−), which can nitrate tyrosine residues of α-synuclein and Aβ to form nitro-tyrosine adducts. This reaction aggravates protein misfolding and aggregation, contributing to neurodegeneration [10]. Of note, this nitration reaction on tyrosine residues is distinct from S-nitrosylation of cysteine thiols, as discussed here.
NO as a second messenger of normal and abnormal neuronal function
Generation of NO in the brain predominantly relies on a family of biosynthetic enzyme, NO synthases (NOSs). Three genetically distinct isoforms of NOS account for NO production in the brain: neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3). These NOSs exist as homodimers, require oxygen, NADPH, and tetrahydrobiopterin as co-factors, and produce NO from L-arginine. The enzymatic activity of nNOS and eNOS is calcium-dependent, whereas iNOS activity is regulated at the transcriptional level [11]. Additionally, as a NOS-independent mechanism, nitrite reductases may participate in NO production in vivo [12].
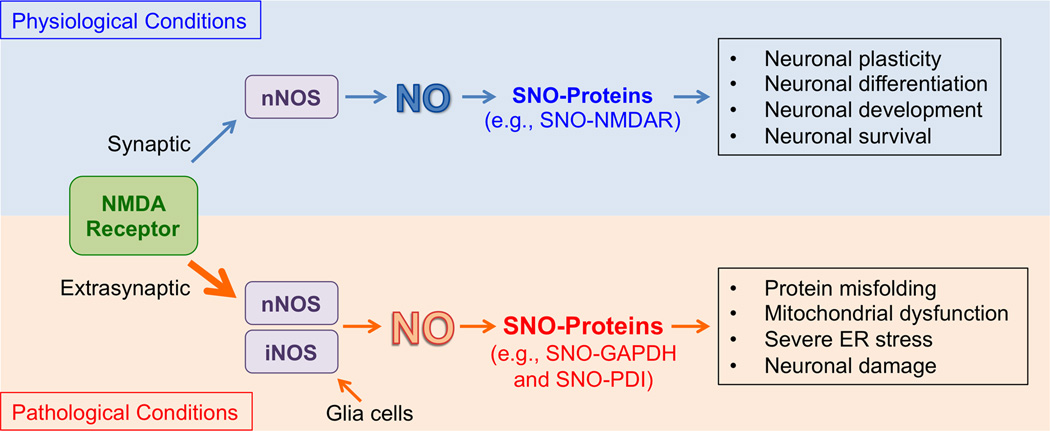
In the nervous system, a well-characterized mechanism for NO production involves activation of N-methyl-d-aspartate (NMDA)-type glutamate receptors. In this cascade, neurotransmitter glutamate stimulates NMDA receptors (NMDARs) to drive Ca2+ influx via receptor-associated ion channels, which in turn activates nNOS to produce NO. Recent studies have shown that the effects of NO on neuronal function in part depend on the cellular location of the NMDARs (Figure 1). Specifically, under physiological conditions, mild activation of “synaptic” NMDARs drives the production of physiological levels of NO, sufficient to maintain normal synaptic plasticity and promote neuronal differentiation or survival. For example, synaptic NO activates the cAMP response element-binding protein (CREB) pathway, modulating long-term potentiation (LTP, an electrical correlate of learning and memory) and mediating neuronal survival [13]. In contrast, accumulating evidence suggests that hyperactivation of “extrasynaptic” NMDARs results in excessive production of NO (and ROS), contributing to the pathophysiology of neurodegenerative diseases. For instance, Aβ oligomers, which can trigger synaptic dysfunction in AD, increase NO concentration to pathological levels primarily via hyperstimulation of extrasynaptic NMDARs [14,15]. In addition, the USA FDA-approved drug, memantine, preferentially blocks extrasynaptic over synaptic NMDARs [16], and has thus shown promise in delaying symptoms not only in AD but also in Lewy body dementia in PD, raising the possibility that the extrasynaptic NMDAR-NO pathway is important in the pathogenesis of dementia in both AD and PD [17,18].
Figure 1.
NO/S-nitrosylation signaling under physiological and pathological conditions in the central nervous system. Physiological activation of NMDA receptors localized at synapses triggers calcium influx through the NMDA receptor-associated ion channel and stimulates nNOS tethered to the NMDA receptor protein complex. Physiological (basal) levels of NO thus produced contribute to normal neuronal functions, e.g., via S-nitrosylation of NMDA receptors (SNO-NMDAR) to prevent their overactivation. Under pathological (neurodegenerative) conditions, excessive activation of extrasynaptic the NMDAR-nNOS pathway (or iNOS expression in glia cells) can lead to overproduction of NO. Under these conditions, excessive generation of ROS can also occur. These pathways cause aberrant SNO-protein formation, such as SNO-GAPDH and SNO-PDI, augmenting pathological processes; in some cases nitrosothiol formation is followed by reaction of the same cysteine residue with ROS to form –SOxH adducts (with × = 1–3).
In addition to NO, other gasoneurotransmitters, such as hydrogen sulfide (H2S) and carbon monoxide (CO), have recently emerged as important regulators of neuronal function [19]. Interestingly, similar to an NO group, H2S can signal at least in part via modification of sulfhydryl groups in target proteins, forming a persulfide bond (R-SSH) in a process termed sulfhydration [20]. Sulfhydration is reported to occur on a specific cysteine residue and affects the activity of target proteins in an analogous manner to S-nitrosylation. Moreover, low levels of H2S can protect neurons, whereas, at high concentration, it produces neurotoxicity [21]. It is anticipated that future investigation will reveal if, like NO, synaptic vs. extrasynaptic signaling accounts in part for the opposing effects of H2S/sulfhydration on neuronal survival.
Molecular mechanisms for production of specific S-nitrosylated proteins
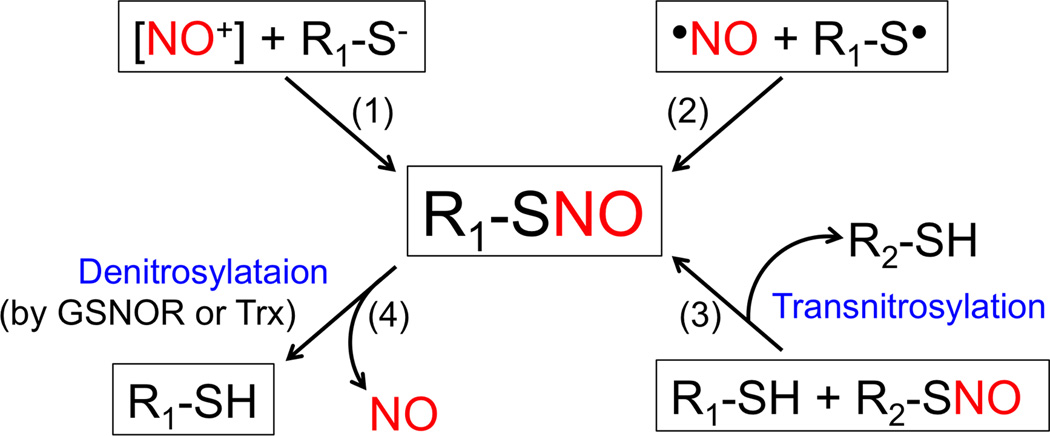
Emerging evidence suggests that NO signals primarily through formation of S-nitrosothiols (SNOs), representing S-nitrosylated proteins (SNO-proteins). Stamler and Lipton’s groups termed this process “S-nitrosylation”, reflecting the biological effects of NO on cellular phenomena, similar to other posttranslational modifications such as phosphorylation [1,6]. The formation of SNO-proteins in vivo entails a reaction between a redox-sensitive thiol (-SH) group (more accurately thiolate anion [-S−]) and nitrosonium cation (NO+) in the presence of transition metals that accept an electron from •NO [1,22,23] (Figure 2). Alternatively, some authorities have argued that radical recombination between •NO and a thiyl radical (RS•) may contribute to the generation of SNO-proteins [1,22–24].
Figure 2.
Biochemical mechanisms of reversible protein S-nitrosylation. (1) Nitrosonium cation [NO+], potentially generated from •NO by metal ion acceptance of the electron, reacts with thiolate anion (R-S−) to generate R-SNO. Note that R-SNO denotes an S-nitrosylated protein (SNO-protein) or S-nitrosothiol (e.g., GSNO and S-nitrosocysteine). (2) Radical recombination of •NO with thiyl radical (RS•) may also produce R-SNO. (3) Transnitrosylation (i.e., transfer of an NO group between two thiol groups). (4) Enzymatic denitrosylation of R-SNO by GSNOR or the Trx system counterbalances R-SNO formation.
Importantly, formation of SNO-proteins typically results in alteration in protein conformation, enzymatic activity, protein-protein interactions, or cellular localization [6,25], thus affecting protein function. Compared to other posttranslational modifications such as methylation and acetylation, S-nitrosylation is often a relatively labile modification, depending on temperature and local redox milieu/protein structure, and can be reversed to free thiol in the presence of metal ions and glutathione (GSH). Since NO is chemically a “good leaving group,” it may facilitate subsequent reaction of ROS with the same cysteine residue to the increasingly more stable oxidative products sulfenic (-SOH), sulfinic (-SO2H), and sulfonic acid (-SO3H). Consequently, because of their stability (particularly sulfinic and sulfonic adducts, the latter being irreversible), these oxidations of cysteine thiols can have long-lasting (often pathological) effects on protein function. In contrast, in some cases in both the cardiovascular and nervous systems, S-nitrosylation of a particular cysteine thiol can be relatively stable and thus prevent further irreversible oxidation [26–28]. Hence, it is possible that physiological S-nitrosylation of some targets in the brain can provide neuroprotection in part by shielding reactive cysteine residues from further oxidation.
In general, in cellular context, S-nitrosylation occurs only on specific cysteine residues. Along these lines, recent studies identified at least three different molecular mechanisms that determine the selectivity of cysteine residues for S-nitrosylation. First, proximal localization of the target protein/cysteine(s) to the source of NO production (i.e., NOSs) increases the chance of S-nitrosylation. For instance, in neurons, nNOS is tethered to the NMDAR complex via the adaptor protein, PSD-95, and thus facilitates S-nitrosylation of these proximate proteins [1,22]. Second, the presence of a signature SNO motif (composed of basic and/or acid amino acids) facilitates the electrostatic interaction of the target cysteine residue with acidic/basic side chains, increasing the susceptibility of the thiol to form SNO modification. Third, local hydrophobic compartments near the cysteine residues potentiate the generation of S-nitrosothiols due to the accelerated accumulation of NO and O2 in a hydrophobic phase [6,29].
Moreover, recent studies have revealed new signal transduction pathways, involving transnitrosylation/nitrosylases, for the selective S-nitrosylation of particular proteins. Protein-to-protein transnitrosylation, whereby an NO group is transferred from a donor protein (serving as a nitrosylase) to a specific acceptor protein (being S-nitrosylated and, in this case, acting as a denitrosylase), may be the principal mechanism to produce SNO-proteins in vivo [30,31]. In this scheme, the transnitrosylation reaction occurs when the two proteins are present in the same protein complex, and thereby only a specific subset of proteins is S-nitrosylated. For instance, in several neurodegenerative diseases, SNO-caspase-3 and SNO-GAPDH can transnitrosylate XIAP and nuclear proteins (such as SIRT1 and DNA-PK), respectively, augmenting cell death-signaling pathways [32,33].
In addition, at least two major classes of denitrosylases, namely S-nitrosoglutathione (GSNO) reductase (GSNOR) and the thioredoxin (Trx) family of proteins, control the degree of protein S-nitrosylation via thiol denitrosylation [34]. With NADH as a coenzyme, GSNOR reduces GSNO to the intermediate S-hydroxylaminoglutathione (GSNHOH), which then forms glutathione sulfinamide (GSONH2) via spontaneous rearrangement, or in the presence of GSH, yields GSSG (oxidized glutathione) [35,36]. Because GSNO (or S-nitrosocysteine) functions as a physiological NO donor and as an intracellular bioavailable NO pool, GSNOR-dependent degradation of GSNO contributes to decreased levels of SNO-proteins, such as SNO-PPARγ [37,38]. In addition, GSNOR, also known as formaldehyde dehydrogenase (or class III alcohol dehydrogenase), efficiently detoxifies both endogenous and exogenous formaldehyde. Although GSNOR is expressed in every tissue examined [39], the effects of its GSNO catalytic activity on brain function remain unclear.
The Trx system, comprised of Trx, Trx reductase (TrxR), and NADPH, represents a major cytosolic oxidoreductase for protein disulfides. In this scheme, reduced Trx employs its conserved Cys-Gly-Pro-Cys active site to donate a proton to an oxidized substrate (such as peroxiredoxins (Prxs), catalyzing the reduction of disulfides; during this process, Trx itself becomes oxidized. To regenerate Trx activity, TrxR receives electrons from NADPH and reduces oxidized Trx [40]. Notably, the Trx system also exhibits denitrosylating activity. For instance, Trx denitrosylates caspase-3, caspase-9, eNOS, and nNOS [41–43]. In addition, other Trx-like proteins, including PDI and TRP14, can serve as denitrosylases [44,45]. These Trx-related denitrosylating enzymes bear evolutionally conserved domains and are highly expressed in the brain, suggesting their physiologically relevant role in neurons. In the future, further work on transnitrosylation/S-nitrosylation/denitrosylation will provide important insights into their functions in the central nervous system, may lead to the discovery of additional enzymes catalyzing these reactions, and may represent promising therapeutic targets for neurodegenerative diseases.
Protein S-nitrosylation as an important mediator of neuronal survival and damage
S-Nitrosylation-dependent NO signaling affects a variety of cellular processes both under physiological and pathological conditions. Below, we briefly review specific examples of SNO-proteins, and then discuss how they regulate neuronal survival or damage. For a more detailed description of the SNO-proteins that are formed in specific disease conditions, the reader is referred to the following review articles [1,6].
When produced at low (physiological) levels, NO generally fosters normal neuronal function/activity and thus facilitates neuronal differentiation, development, and survival. For instance, S-nitrosylation of caspases and HDAC2 function in this manner [46,47]. As an example, the well-known executioner of apoptosis, caspase-3, is constitutively S-nitrosylated in some tissues at its active site under basal conditions, inhibiting its protease activity and thus offering cytoprotection. However, in apoptotically-stimulated neurons, the Trx system denitrosylates caspase-3 to amplify cell death signaling [47,48].
Additionally, when production of NO is moderately increased after exposure to mild neurotoxic stimuli, NO can still mediate neuroprotective effects. In part, this effect is mediated by S-nitrosylation of overly excited NMDARs or the redox-sensitive chaperone and peptidase protein DJ-1 [22,49,50]. These nitrosylation reactions serve as a form of negative feedback on degenerative processes. For instance, S-nitrosylation of NMDARs downregulates their excessive activity and can thereby provide neuroprotection in experimental models of neurodegenerative conditions [22,50–53]. Consistent with this idea, hypo-S-nitrosylation of NMDARs under mild stress can aggravate pathological processes, due to a lack of the neuroprotective effects of NO [54].
In contrast, excessive production of NO can result in aberrant protein S-nitrosylation, even reacting with cysteines lacking the full nitrosylation motif, and thus contributing to the progression of neurodegeneration. Persistent hyperactivation of extrasynaptic NMDARs or increased iNOS activity in glial cells can typically result in overproduction of NO. Aberrantly formed SNO-proteins occurring under pathological nitrosative stress include, to name but a few, SNO-MEF2, SNO-parkin, SNO-PDI, SNO-GAPDH, SNO-XIAP, SNO-Cdk5, and SNO-Drp1. Our group and others have demonstrated that these SNO-proteins indeed accumulate in human brains from patients with neurodegenerative diseases (but not in controls), contributing to an array of degenerative processes such as protein misfolding (e.g., mediated by SNO-parkin and SNO-PDI), mitochondrial dysfunction (SNO-MEF2, SNO-Drp1, and SNO-parkin), synaptic damage (SNO-Drp1, SNO-Cdk5, and SNO-MEF2), and neuronal cell death (SNO-MEF2, SNO-PDI, SNO-GAPDH, SNO-XIAP, and SNO-Drp1) [5,7,8,32,55–57]. Additionally, NO inhibits cGMP production via S-nitrosylation of sGC, possibly representing a negative-feedback loop to suppress the sGC/cGMP pathway [58].
Taken together, these findings imply that the biological effects of NO on neurological function are largely dependent on the level, timing, duration, and/or location of its production. Moreover, SNO-proteins can act as either neuroprotective or neurodestructive effectors, depending on the target proteins and the site of modification; therefore, careful strategies have to be designed to develop therapeutics that regulate protein S-nitrosylation in neurodegenerative diseases. Below, we delineate possible pharmacological approaches to maintain physiological SNO-protein activity, while preventing aberrant S-nitrosylation, in order to ameliorate neurodegenerative diseases.
Possible approaches to pharmacologically modulate formation of SNO-proteins
NOS inhibitors or antioxidants
One way to develop effective therapies for neurodegenerative diseases may involve decreasing the detrimental effects of SNO-proteins derived from nitrosative/oxidative stress. Based on this hypothesis, emphasis has been placed on pharmacological inhibition of NOSs or application of antioxidants (reacting with and scavenging ROS/RNS), as potential therapeutic options. However, the majority of NOS inhibitors available have limited specificity towards each of the NOS isoforms. Moreover, NOS inhibitors or antioxidants can block physiological production of NO (as well as ROS), which are associated not only with the formation of neuroprotective SNO-proteins in the brain but also with the health of other tissues and organs such as the cardiovascular system. Along these lines, NOS inhibition therapies and antioxidant treatments have displayed limited success in human clinical trials. For instance, NOS inhibition by NG-monomethyl-l-arginine (l-NMMA; a competitive inhibitor of all NOS isoforms) in healthy volunteers resulted in an increase in blood pressure without showing significant beneficial effects to brain function [59,60]. In addition, antioxidant clinical trials have had mixed outcomes; in some studies, antioxidants (such as vitamin E [α-tocopherol]) reportedly slowed the progression of moderate-to-severe AD [61] or early PD [62], while other studies did not find any influence of antioxidants on cognitive impairment or cerebrospinal fluid (CSF) biomarkers related to Aβ or tau pathology [63,64]. Thus, clinical trials to date have not supported these approaches for neurodegenerative diseases.
Targeting denitrosylases
Recent drug discovery efforts, employing high-throughput screening plus structure-based lead optimization, have identified small molecule inhibitors of GSNOR [65]. Among these inhibitors, N6022 fits into the enzyme’s substrate-binding pocket, thus acting as a potent and selective inhibitor of GSNOR [66,67]. Through inhibition of GSNOR, N6022 enhances GSNO levels and the intracellular SNO-protein pool, and thus potentiates physiological (basal) NO activity; however, the drug also inhibits GSNOR-mediated aldehyde reduction. Importantly, N6022 inhibition of GSNOR has shown safety and efficacy in animal models of asthma [68], suggesting that this drug may be tested in human clinical trials for CNS disease if it or a congener is permeable to the blood-brain-barrier (BBB). Moreover, for neurodegenerative diseases, this drug would have to be used as a preventive measure at a pre-symptomatic stage to augment the neuroprotective action of NO, as toxic levels of NO are already present under disease conditions.
As an alternative approach to regulate protein denitrosylation, recent studies have identified Trx-mimetic peptides (TXMs) that catalyze the reduction of SNO, protecting cells from nitrosative stress [69,70]. For instance, TXMs reversed the inhibitory effects of (S)NO on the Prx-Trx system, suggesting that TXMs may hold potential to improve pathological conditions related to SNO stress. However, TXMs also imitate the oxidoreductase activity of Trx, regulating oxidation (i.e., disulfide formation) of Trx substrate proteins [71,72]. Moreover, similar to NOS inhibitors and antioxidant therapies, TXMs can affect basal SNO (and oxidation) levels. Accordingly, significant caution should be exercised when engaging these regulators of denitrosylases in both animal and possibly clinical settings, since alterations of basal SNO levels may cause undesirable effects. Additionally, whether TXMs cross the BBB remains unknown, although dysfunction of the BBB in these diseases [73] may allow access to affect neurodegenerative diseases. As delineated below, in order to overcome some of these possible issues, we propose that alteration of specific SNO-protein levels may be more beneficial because it may minimize effects on physiological S-nitrosylation.
Targeting the formation of specific SNO-protein(s)
To date, proteomics-based identification of SNO-proteins has revealed hundreds, if not thousands, of such proteins both under physiological and pathological conditions [74–78]. Although these studies characterized a large number of targets for S-nitrosylation, alteration of S-nitrosylation of a single key protein appeared to be sufficient, at least in certain animal models, to influence neuronal function and activity. Here we provide two examples of pharmacological techniques that can preferentially control SNO-NMDAR or SNO-GAPDH formation, thus preventing neuronal injury or damage.
Hyperactivation of the NMDARs (especially extrasynaptic NMDARs) is thought to play a critical role in a wide range of neurodegenerative diseases via pathological activation of downstream enzymes and generation of ROS/RNS. An important mechanism to downregulate this toxic pathway is S-nitrosylation of NMDARs. S-Nitrosylation can occur on various cysteine residues on different NMDAR subunits, including Cys744 and Cys798 on the GluN1 (NR1) subunit, and Cys87, Cys320, and Cys399 on the GluN2A (NR2A) subunit [22,50–53]. Ambient oxygen levels are known to facilitate disulfide formation between Cys744 and Cys798 on the GluN1, blocking S-nitrosylation of these cysteine residues. Under physiological (or even more hypoxic) conditions, however, the relatively reducing state in the brain favors free thiol groups on these GluN1 subunit cysteines over disulfide. The presence of free thiol enhances S-nitrosylation of Cys744 and Cys798, leading to increased NO sensitivity at the other NMDAR sites (i.e., Cys87, Cys320, and Cys399 on the GluN2A); this series of polynitrosylation reactions inhibits the receptor’s activity, providing a negative feedback system since NMDAR stimulation results in the generation of NO from nNOS. Interestingly, S-nitrosylation per se of Cys744 and Cys798 on GluN1 has little direct influence on the ion channel’s activity. However, Cys744 and Cys798 on GluN1comprise a “redox sensor,” exerting an allosteric affect on the other SNO sites on GluN2A (or homologous cysteine residues on GluN2B) [22,50–53].
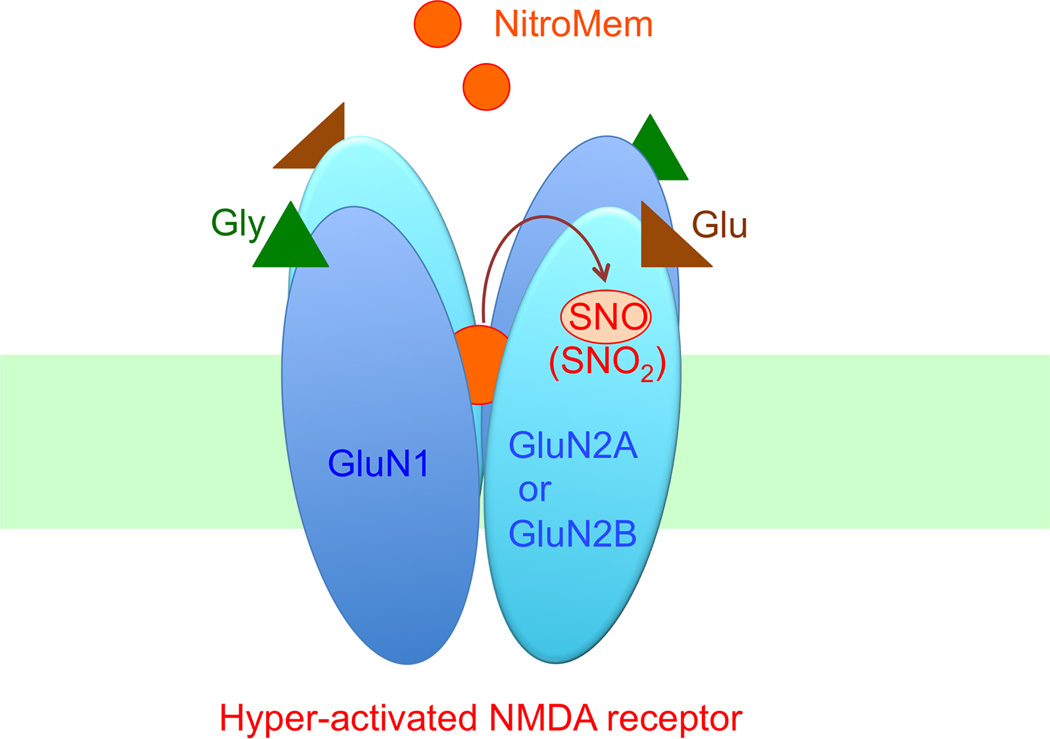
Previously, we and our colleagues had discovered that the drug memantine preferentially blocks extrasynaptic NMDAR-associated ion channels because they are excessively/tonically open in a number of neurodegenerative conditions [14,15]. Memantine affords neuroprotection by blocking tonically-activated NMDAR-operated channels. Memantine has the unique propensity to do this because we discovered that it is an “open-channel blocker,” i.e., it blocks ion channels only when they are open, and, statistically, more channels are open during tonic activation of the NMDAR. Additionally, memantine manifests a relatively fast off-rate from the channels at physiological resting potential, meaning that the drug does not accumulate in the channels and thus block their normal function. Accordingly, unlike other NMDAR antagonists that persistently block channel activity and cause severe side effects, memantine spares synaptic function under physiological conditions [79,80]. Recently, we took advantage of the fact that this drug prefers to bind in excessively open, extrasynaptic channels in order to target an NO group specifically to the redox-sensitive thiol sites of these overstimulated NMDARs. This new type of drug, termed NitroMemantines, contains a nitro group that has been tethered to the memantine moiety, thus serving not only as an open-channel blocker but also as an NO donor. In this manner, a NitroMemantine drug can provide increased blockade of hyperactivated NMDARs through S-nitrosylation in addition to channel block, thus affording more neuroprotection that that offered by memantine [15,81] (Figure 3). Consistent with this notion, NitroMemantine protected neurons more efficiently than memantine in cellular and animal models of AD and cerebrovascular disease [15,82]. The lead NitroMemantine candidate developed to date, designated YQW-036/NMI-6979, has shown a favorable pharmacokinetic profile, excellent CNS penetration, and good safety index in early preclinical studies [81,82]. The NitroMemantines are currently being evaluated for clinical trials for AD and other neurological disorders.
Figure 3.
NitroMemantine-mediated inhibition of hyperactivated NMDA receptors. NitroMemantine was synthesized by addition of a nitro group (-NO2) to the memantine moiety. This allows NitroMemantine to antagonize excessively activated NMDA receptors via two sites of action: 1) the ion channel where memantine itself binds, and 2) an extracellular redox-sensitive cysteine thiol groups of the receptor where the nitro group reacts to inhibit NMDAR activity (forming –SNO or –SNO2). Thus, in this scenario, NitroMemantine serves as an NO (or, more precisely, a nitro group) donor specifically targeting NMDA receptors. This figure also shows that the NMDA receptor is a heterodimer, composed of two GluN1 and two GluN2 subunits. Excessive concentrations of glycine (Gly) and glutamate (Glu), co-agonists of the receptor, can trigger pathological activation of the NMDA receptor.
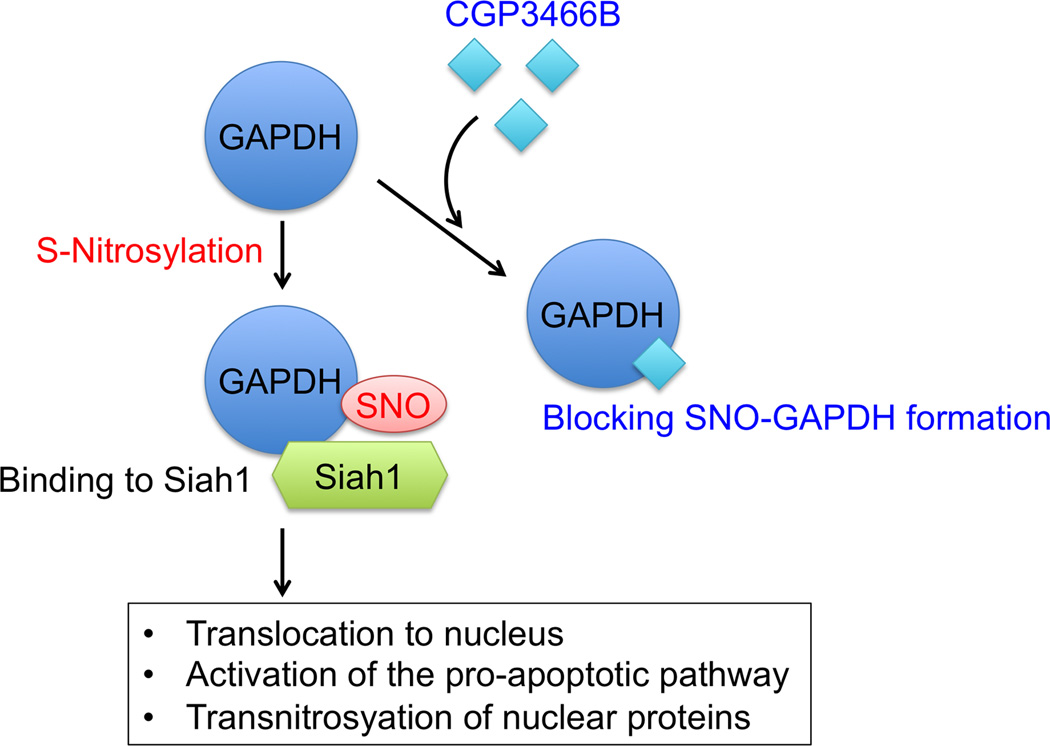
As a second example, S-nitrosylation of glyceraldehyde-3-phophate dehydrogenase (GAPDH) enhances its ability to bind to the ubiquitin E3 ligase Siah [56]. Because Siah possesses a nuclear localization signal, the SNO-GAPDH/Siah complex translocates to the nucleus, activating the cascade of apoptotic cell death via degradation of nuclear proteins [56,83]. GAPDH is also regarded as a putative direct target of the drug, deprenyl (Selegiline®) [84]. Deprenyl is used clinically for its reported ability to delay the progression of symptoms in early PD [62,85]. Possible neuroprotection by deprenyl was initially thought to derive from its inhibitory activity of monoamine oxidase-B (MAO-B); however, subsequent studies have suggested that deprenyl may exert a protective action through its antioxidant effects or prevention of GAPDH nuclear translocation, rather than inhibition of MAO-B [86,87]. Consistent with this notion, the deprenyl derivative CGP3466B (TCH346 or Omigapil), which lacks MAO-B inhibitory activity, has displayed potent neuroprotective activity in PD models [84,88]. Solomon Snyder’s group further showed that direct binding of deprenyl or CGP3466B to GAPDH prevents the formation of SNO-GAPDH, thus inhibiting the interaction of GAPDH and Siah, and hindering nuclear localization of GAPDH [89,90] (Figure 4). Unfortunately, despite these encouraging preclinical findings, CGP3466B failed to show neuroprotective effects in human clinical trials for PD [91]. Hence, although the neuroprotective action of CGP3466B (or related compounds) may in part arise from its ability to block the SNO-GAPDH-Siah pathway, the drug has not yet proven useful in human clinical trials for PD.
Figure 4.
CGP3466B exerts its neuroprotective effect via inhibition of SNO-GAPDH formation. S-Nitrosylation of GAPDH enhances its interaction with Siah1 (bearing a nuclear localization signal) and promotes its translocation to the nucleus, whereas SNO-GAPDH triggers cellular events leading to neurodegeneration. CGP3466B (TCH346 or Omigapil) potently and selectively blocks GAPDH S-nitrosylation and thereby Siah binding.
Concluding Remarks
Recent advancements in the understanding of protein S-nitrosylation suggest that this regulatory posttranslational modification might represent a potentially viable therapeutic target for a wide range of neurodegenerative diseases. Supporting this view, NOS inhibitors or antioxidants that indirectly modulate the formation of SNO-proteins have shown some promise in preclinical studies. Moreover, direct and specific alterations of SNO-NMDAR or SNO-GAPDH formation with NitroMemantine or CGP3466B, respectively, show some efficacy in modulating disease-related processes. Nonetheless, additional optimization of these drugs and further clinical trials will be needed to prove their potential safety and efficacy in humans.
Ideally, the goal of effective S-nitrosylation (or NO)-based therapy is to i) only block pathological (typically aberrantly-produced) formation of SNO-proteins, while sparing normal and physiological functions of SNO-proteins, and ii) only enhance NO’s neuroprotective function without potentiating its neurodestructive actions. Future research in the field is expected to address challenging questions for the development of small-molecule drugs that meet these requirements (see Outstanding Questions Box). For instance, NOS inhibitors or antioxidant treatments will have to be employed only under conditions where NO exerts a neurotoxic activity. To this end, elucidation of optimal timing in the administration of these medications may significantly improve the safety and efficacy of these treatments. Moreover, identification of previously unknown SNO-protein targets using new sensitive probes [77,78,92,93] may facilitate the development of new SNO-targeting small molecules. Importantly, implementation of interdisciplinary approaches, ranging from protein biochemistry to medicinal chemistry, should further enhance the discovery of such new drugs. Finally, since S-nitrosylation appears to regulate multiple signaling pathways leading to neurodegeneration, future studies should assess whether a combinatorial therapeutic approach, employing multiple SNO-targeting drugs, might potentiate their clinical utility for neurodegenerative diseases.
Outstanding Questions Box.
What is the optimal timing with relation to insult for the administration of NOS inhibitors or antioxidants to significantly improve the safety and efficacy of these treatments? These medications will have to be employed only under conditions where NO exerts a neurotoxic activity, as physiological formation of SNO-proteins is important for normal neuronal function and should not be inhibited under basal conditions.
What are the critical SNO-proteins for the pathophysiology of neurodegenerative disorders that have not yet been identified and characterized? Due to technical limitations, proteomic detection of endogenous SNO-proteins is currently challenging; however, adoption of newly developed SNO probes [69,70,83,84] may facilitate the discovery of additional SNO-proteins that may serve as critical targets for future therapeutic intervention. Moreover, towards this end, implementation of interdisciplinary approaches, ranging from protein biochemistry to medicinal chemistry, will facilitate development of small molecules that target specific SNO-proteins.
Trends Box.
Protein S-nitrosylation represents an important component of NO signaling.
S-Nitrosylation mediates both physiological and pathological brain signaling.
NitroMemantine protects neurons via S-nitrosylation of NMDA receptors.
CGP3466B (or TCH346) is neuroprotective by inhibiting formation of SNO-GAPDH.
Acknowledgments
This work was supported in part by grants from the National Institute of Health, Brain & Behavior Research Foundation, and the Michael J. Fox Foundation.
Conflicts of Interest
S.A.L. is the inventor on worldwide patents for the use of memantine and NitroMemantine for neurodegenerative disorders. Per guidelines of Harvard University, where the work was initially performed, S.A.L. participates in a royalty-sharing agreement with his former institution Boston Children’s Hospital/Harvard Medical School, which licensed the drug memantine (Namenda®) to Forest Laboratories, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakamura T, et al. Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004;10(Suppl):S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 3.Barnham KJ, et al. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 4.Migliore L, Coppede F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Mutat. Res. 2009;674:73–84. doi: 10.1016/j.mrgentox.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Ryan SD, et al. Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hess DT, et al. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 7.Cho DH, et al. S-Nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uehara T, et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, et al. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349:500–506. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ischiropoulos H, et al. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 11.Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu. Rev. Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg JO, et al. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 13.Lu YF, et al. Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J Neurosci. 1999;19:10250–10261. doi: 10.1523/JNEUROSCI.19-23-10250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molokanova E, et al. Differential effects of synaptic and extrasynaptic NMDA receptors on Aβ-induced nitric oxide production in cerebrocortical neurons. J. Neurosci. 2014;34:5023–5028. doi: 10.1523/JNEUROSCI.2907-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talantova M, et al. Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proc. Natl. Acad. Sci. USA. 2013;110:E2518–E2527. doi: 10.1073/pnas.1306832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia P, et al. Memantine preferentially blocks extrasynaptic over synaptic NMDA receptor currents in hippocampal autapses. J. Neurosci. 2010;30:11246–11250. doi: 10.1523/JNEUROSCI.2488-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisberg B, et al. Memantine in moderate-to-severe Alzheimer's disease. N. Engl. J. Med. 2003;348:1333–1341. doi: 10.1056/NEJMoa013128. [DOI] [PubMed] [Google Scholar]
- 18.Aarsland D, et al. Memantine in patients with Parkinson's disease dementia or dementia with Lewy bodies: a double-blind, placebo-controlled, multicentre trial. Lancet Neurol. 2009;8:613–618. doi: 10.1016/S1474-4422(09)70146-2. [DOI] [PubMed] [Google Scholar]
- 19.Mustafa AK, et al. Signaling by gasotransmitters. Sci. Signal. 2009;2:re2. doi: 10.1126/scisignal.268re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul BD, Snyder SH. H(2)S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 21.Chen MJ, et al. Gene profiling reveals hydrogen sulphide recruits death signaling via the N-methyl-D-aspartate receptor identifying commonalities with excitotoxicity. J. Cell. Physiol. 2011;226:1308–1322. doi: 10.1002/jcp.22459. [DOI] [PubMed] [Google Scholar]
- 22.Lipton SA, et al. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Ruiz A, et al. Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic. Biol. Med. 2011;51:17–29. doi: 10.1016/j.freeradbiomed.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr. Opin. Chem. Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamler JS, et al. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 26.Chen YY, et al. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, et al. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid. Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J. Clin. Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc. Natl. Acad. Sci. USA. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Lipton SA. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid. Redox Signal. 2013;18:239–249. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia J, et al. Target-selective protein S-nitrosylation by sequence motif recognition. Cell. 2014;159:623–634. doi: 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura T, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornberg MD, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benhar M, et al. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 35.Staab CA, et al. The Janus face of alcohol dehydrogenase 3. Chem Biol Interact. 2009;178:29–35. doi: 10.1016/j.cbi.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 36.Hedberg JJ, et al. Reduction of S-nitrosoglutathione by human alcohol dehydrogenase 3 is an irreversible reaction as analysed by electrospray mass spectrometry. Eur J Biochem. 2003;270:1249–1256. doi: 10.1046/j.1432-1033.2003.03486.x. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, et al. S-Nitrosoglutathione reductase-dependent PPARγ denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J. Clin. Invest. 2015;125:1679–1691. doi: 10.1172/JCI73780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 39.Estonius M, et al. Alcohol dehydrogenase in human tissues: localisation of transcripts coding for five classes of the enzyme. FEBS Lett. 1996;397:338–342. doi: 10.1016/s0014-5793(96)01204-5. [DOI] [PubMed] [Google Scholar]
- 40.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem. Biophys. Res. Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 41.Benhar M, et al. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erwin PA, et al. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 43.Qu ZW, et al. N-methyl-D-aspartate receptor-dependent denitrosylation of neuronal nitric oxide synthase increase the enzyme activity. PLoS One. 2012;7:e52788. doi: 10.1371/journal.pone.0052788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zai A, et al. Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J. Clin. Invest. 1999;103:393–399. doi: 10.1172/JCI4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pader I, et al. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc. Natl. Acad. Sci. USA. 2014;111:6964–6969. doi: 10.1073/pnas.1317320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nott A, et al. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 47.Mannick JB, et al. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 48.Tenneti L, et al. Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci. Lett. 1997;236:139–142. doi: 10.1016/s0304-3940(97)00780-5. [DOI] [PubMed] [Google Scholar]
- 49.Choi MS, et al. Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models. J. Neurosci. 2014;34:15123–15131. doi: 10.1523/JNEUROSCI.4751-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi YB, et al. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 51.Kim WK, et al. Attenuation of NMDA receptor activity and neurotoxicity by nitroxyl anion, NO. Neuron. 1999;24:461–469. doi: 10.1016/s0896-6273(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi H, et al. Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei SZ, et al. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8:1087–1099. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 54.Gasperini L, et al. Prion protein and copper cooperatively protect neurons by modulating NMDA receptor through S-nitrosylation. Antioxid. Redox Signal. 2015;22:772–784. doi: 10.1089/ars.2014.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao D, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hara MR, et al. S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 57.Qu J, et al. S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc Natl Acad Sci USA. 2011:14330–14335. doi: 10.1073/pnas.1105172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayed N, et al. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc. Natl. Acad. Sci. USA. 2007;104:12312–12317. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamler JS, et al. Nitric oxide regulates basal systemic and pulmonary vascular resistance in healthy humans. Circulation. 1994;89:2035–2040. doi: 10.1161/01.cir.89.5.2035. [DOI] [PubMed] [Google Scholar]
- 60.Haynes WG, et al. Inhibition of nitric oxide synthesis increases blood pressure in healthy humans. J. Hypertens. 1993;11:1375–1380. doi: 10.1097/00004872-199312000-00009. [DOI] [PubMed] [Google Scholar]
- 61.Sano M, et al. A controlled trial of selegiline, alpha-tocopherol, or both as treatment for Alzheimer's disease. The Alzheimer's Disease Cooperative Study. N. Engl. J. Med. 1997;336:1216–1222. doi: 10.1056/NEJM199704243361704. [DOI] [PubMed] [Google Scholar]
- 62.Shoulson I. DATATOP: a decade of neuroprotective inquiry. Parkinson Study Group. Deprenyl And Tocopherol Antioxidative Therapy Of Parkinsonism. Ann. Neurol. 1998;44:S160–S166. [PubMed] [Google Scholar]
- 63.Galasko DR, et al. Antioxidants for Alzheimer disease: a randomized clinical trial with cerebrospinal fluid biomarker measures. Arch. Neurol. 2012;69:836–841. doi: 10.1001/archneurol.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen RC, et al. Vitamin E and donepezil for the treatment of mild cognitive impairment. N. Engl. J. Med. 2005;352:2379–2388. doi: 10.1056/NEJMoa050151. [DOI] [PubMed] [Google Scholar]
- 65.Sun X, et al. Discovery of s-nitrosoglutathione reductase inhibitors: potential agents for the treatment of asthma and other inflammatory diseases. ACS Med. Chem. Lett. 2011;2:402–406. doi: 10.1021/ml200045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Green LS, et al. Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry. 2012;51:2157–2168. doi: 10.1021/bi201785u. [DOI] [PubMed] [Google Scholar]
- 67.Colagiovanni DB, et al. A nonclinical safety and pharmacokinetic evaluation of N6022: a first-in-class S-nitrosoglutathione reductase inhibitor for the treatment of asthma. Regul. Toxicol. Pharmacol. 2012;62:115–124. doi: 10.1016/j.yrtph.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Blonder JP, et al. Pharmacologic inhibition of S-nitrosoglutathione reductase protects against experimental asthma in BALB/c mice through attenuation of both bronchoconstriction and inflammation. BMC Pulm. Med. 2014;14:3. doi: 10.1186/1471-2466-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kronenfeld G, et al. Thioredoxin-mimetic peptides as catalysts of S-denitrosylation and anti-nitrosative stress agents. Free Radic. Biol. Med. 2015;79:138–146. doi: 10.1016/j.freeradbiomed.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 70.Cohen-Kutner M, et al. Thioredoxin-mimetic peptides (TXM) reverse auranofin induced apoptosis and restore insulin secretion in insulinoma cells. Biochem. Pharmacol. 2013;85:977–990. doi: 10.1016/j.bcp.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Cohen-Kutner M, et al. Thioredoxin-mimetic peptide CB3 lowers MAPKinase activity in the Zucker rat brain. Redox Biol. 2014;2:447–456. doi: 10.1016/j.redox.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bachnoff N, et al. Alleviation of oxidative stress by potent and selective thioredoxin-mimetic peptides. Free Radic. Biol. Med. 2011;50:1355–1367. doi: 10.1016/j.freeradbiomed.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 73.Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 74.Paige JS, et al. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao G, et al. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zahid S, et al. Differential S-nitrosylation of proteins in Alzheimer's disease. Neuroscience. 2014;256:126–136. doi: 10.1016/j.neuroscience.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 77.Raju K, et al. Regulation of brain glutamate metabolism by nitric oxide and S-nitrosylation. Sci. Signal. 2015;8:ra68. doi: 10.1126/scisignal.aaa4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doulias PT, et al. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci. Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lipton SA. Pathologically activated therapeutics for neuroprotection. Nat. Rev. Neurosci. 2007;8:803–808. doi: 10.1038/nrn2229. [DOI] [PubMed] [Google Scholar]
- 80.Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat. Rev. Drug. Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, et al. The pharmacology of aminoadamantane nitrates. Curr. Alzheimer Res. 2006;3:201–204. doi: 10.2174/156720506777632808. [DOI] [PubMed] [Google Scholar]
- 82.Takahashi T, et al. Pharmacologically targeted NMDA receptor antagonism by NitroMemantine for cerebrovascular disease. Sci. Rep. doi: 10.1038/srep14781. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawa A, et al. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kragten E, et al. Glyceraldehyde-3-phosphate dehydrogenase, the putative target of the antiapoptotic compounds CGP 3466 and R-(−)-deprenyl. J. Biol. Chem. 1998;273:5821–5828. doi: 10.1074/jbc.273.10.5821. [DOI] [PubMed] [Google Scholar]
- 85.Birkmayer W, et al. Increased life expectancy resulting from addition of L-deprenyl to Madopar treatment in Parkinson's disease: a longterm study. J. Neural. Transm. 1985;64:113–127. doi: 10.1007/BF01245973. [DOI] [PubMed] [Google Scholar]
- 86.Tatton W, et al. Neuroprotection by deprenyl and other propargylamines: glyceraldehyde-3-phosphate dehydrogenase rather than monoamine oxidase B. J. Neural. Transm. 2003;110:509–515. doi: 10.1007/s00702-002-0827-z. [DOI] [PubMed] [Google Scholar]
- 87.Tatton WG, Chalmers-Redman RM. Modulation of gene expression rather than monoamine oxidase inhibition: (−)-deprenyl-related compounds in controlling neurodegeneration. Neurology. 1996;47:S171–S183. doi: 10.1212/wnl.47.6_suppl_3.171s. [DOI] [PubMed] [Google Scholar]
- 88.Waldmeier PC, et al. Neurorescuing effects of the GAPDH ligand CGP 3466B. J. Neural. Transm. Suppl. 2000:197–214. doi: 10.1007/978-3-7091-6301-6_13. [DOI] [PubMed] [Google Scholar]
- 89.Xu R, et al. Behavioral effects of cocaine mediated by nitric oxide-GAPDH transcriptional signaling. Neuron. 2013;78:623–630. doi: 10.1016/j.neuron.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hara MR, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc. Natl. Acad. Sci. USA. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olanow CW, et al. TCH346 as a neuroprotective drug in Parkinson's disease: a double-blind, randomised, controlled trial. Lancet Neurol. 2006;5:1013–1020. doi: 10.1016/S1474-4422(06)70602-0. [DOI] [PubMed] [Google Scholar]
- 92.Seneviratne U, et al. Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs. J. Am. Chem. Soc. 2013;135:7693–7704. doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J, et al. An unexpected Bis-ligation of S-nitrosothiols. J. Am. Chem. Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]