Abstract
To discover novel drugs for neuroblastoma treatment, we have previously screened a panel of drugs and identified 30 active agents against neuroblastoma cells. Here we performed microarray gene expression analysis to monitor the impact of these agents on a neuroblastoma cell line and used the connectivity map (cMAP) to explore putative mechanism of action of unknown drugs. We first compared the expression profiles of ten compounds shared in both our dataset and cMAP database and observed the high connectivity scores for 7 of 10 matched drugs regardless of the differences of cell lines utilized. The screen of cMAP for uncharacterized drugs indicated the signature of Epoxy anthraquinone derivative (EAD) matched the profiles of multiple known DNA targeted agents (topoisomerase I/II inhibitors, DNA intercalators, and DNA alkylation agents) as predicted by its structure. Similar result was obtained by querying against our internal NB-cMAP (http://pob.abcc.ncifcrf.gov/cgi-bin/cMAP), a database containing the profiles of 30 active drugs. These results suggest that Epoxy anthraquinone derivative may inhibit neuroblastoma cells by targeting DNA replication inhibition. Experimental data also demonstrate that Epoxy anthraquinone derivative indeed induces DNA double-strand breaks through DNA alkylation and inhibition of topoisomerase activity. Our study indicates that Epoxy anthraquinone derivative may be a novel DNA topoisomerase inhibitor that can be potentially used for treatment of neuroblastoma or other cancer patients.
Keywords: Neuroblastoma, anti-cancer drug, microarray, cMAP, topoisomerase, EAD
1. Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor of childhood. It is highly heterogeneous with diverse clinical behavior [1]. Patients with low-risk disease are essentially curable. However more than half of neuroblastoma patients have high-risk disease. The high risk group comprises patients over one year of age, with stage 4 disease, and MYCN amplification. Current multimodal therapies for high-risk neuroblastoma include surgery, intensive chemotherapy with stem cell rescue, radiation therapy, monoclonal antibody and treatment with a differentiating agent [1,2]. The drugs such as etoposide, doxorubicin, cisplatin, carboplatin, vincristine, topotecan, cyclophosphamide, and 13-cis-retinoic acid are frequently used. However, the disease often returns and only ~30–40% of patients survive despite aggressive therapy. With a better understanding of tumor biology and molecular events governing tumorigenesis, new therapeutics is being developed for neuroblastoma treatment. Recent results of phase III clinical trial show that an antibody-based immunotherapy (anti-GD2 mAb combination therapy) increased event-free survival of patients with high-risk neuroblastoma by 20% [3,4]. Due to a small prevalence of pediatric tumors, novel drug development for pediatric tumors has been very limited especially compared with the number of studies in adult cancers. Therefore there is an urgent need for novel drug development for neuroblastoma treatment.
The connectivity map (cMAP) is a database containing genome-wide transcriptional expression data from cultured human cells treated with bioactive small molecules that enable the discovery of functional connections between drugs, genes, and diseases through the transitory feature of common gene-expression change [5]. Studies have utilized this tool to identify drugs affecting the same molecular pathways and explore putative mechanism of action of unknown drugs [6–8]. The current version of cMAP (build 02) contains more than 7,000 expression profiles representing 1,309 agents (http://www.broadinstitute.org/cmap/).
To identify active agents against neuroblastoma, we have previously screened a panel of drugs with diverse mechanisms of action in neuroblastoma cell lines and identified 30 active agents [9]. In this study we profiled mRNA expression for each of these 30 agents in a neuroblastoma cell line and analyzed the gene expression signatures associated with each drug using the cMAP database and the internal NB-cMAP database (http://pob.abcc.ncifcrf.gov/cgi-bin/cMAP) containing gene expression profiles of the NB cell line for the 30 active drugs. This gene expression profiling-based approach identified an epoxy anthraquinone derivative (EAD) as a potential DNA topoisomerase inhibitor, which was validated by experiments in neuroblastoma cells.
2. Materials and Methods
2.1. Cell culture and microarray experiments
A neuroblastoma cell line SK-N-AS obtained from the American Type Culture Collection (ATCC) was used for microarray analysis. Cells were grown in RPMI 1640 media and other conditions of cell culture were the same as described previously [9]. 600,000 SK-N-AS cells were seeded in each well of 6-well plates. Twenty-four hours after seeding, cells were treated with each of the 30 drugs at two doses as described in Supplemental Table 1. Cells were harvested using Trizole® reagent (Invitrogen, Carlsbad, CA) 6 hours after the drug treatment. Total RNA was purified using Qiagen miRNesay® mini kit on Qiagen Qiacube following manufacture’s protocol (Qiagen, Valencia, CA). For DNA microarray experiment, biotinlyated cRNA was synthesized and hybridized on Affymetrix GeneChip® Human Genome U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA) according to Affymetrix’s protocol.
2.2. Data processing and cMAP analysis
The .CEL files were exported from Affymetrix GCOS software and normalized using the Affymetrix PLIER algorithm with mismatch probe adjustment (Affymetrix Power Tool, Affymetrix, Santa Clara, CA). Values smaller than 4 were replaced with 4. Relative expression values were calculated by dividing with the geometric mean of the corresponding values from the two vehicle control samples with no drug treatment. Each instance was then rank-normalized by sorting the probes and replacing expression values with the rank. For probesets having both ‘treatment’ value and ‘no treatment’ replaced with 4, thus having equal ratios, we determined the order with respect to the ratio of the original values. For cMAP analysis, we used the current version of cMAP (build 02) which contains more than 7,000 expression profiles representing 1,309 drugs. The query signature for each sample was generated using 500 probesets with highest expression and lowest expression, from which the probesets overlapped with Affymetrix HG-U133A were used for querying the cMAP database. In the query result, any mechanism of action represented by two or more instances ranking at the top 20 is shown in the tables.
2.3. NB-cMAP database
The dataset used for our NB-cMAP (http://pob.abcc.ncifcrf.gov/cgi-bin/cMAP) database is composed of mRNA expression data for 30 active agents at two different concentrations and 2 corresponding vehicle controls applied to human neuroblastoma cell line SK-N-AS for 6 hours. The data processing is described as above and the details of the dataset are available in Supplemental Table 1. The metric based on the Kolmogorov-Smirnov statistics and scoring system were used to build NB-cMAP as described by Lamb et al. [5]. Enrichment of both up- and down-regulated genes from a given signature in the profiles of each treatment instance were estimated with the Kolmogorov-Smirnov statistic and combined to produce an enrichment score as described [5].
2.4. Cell viability and caspase-Glo 3/7 assay
Five neuroblastoma cell lines (SK-N-AS, SH-SY5Y, NBEB, IMR5 and IMR32) were used in cell viability and caspase-Glo 3/7 assay. The cells were seeded in each well on 96-well plates; 24 hours after seeding, cells were treated with drugs which are diluted with the cell culture medium and DMSO to achieve the desired final drug concentration and 0.1% final concentration of DMSO. We evaluated the efficacy of each compound using the CellTiter Blue® cell viability assay (Promega Corporation, Madison, WI) at 24 after drug treatment as prescribed by the manufacturer’s protocol. Apoptosis was measured by Caspase-Glo 3/7® assay (Promega Corporation, Madison, WI) at 24 hours after the drug treatment. The percentage of cell viability or caspase activity was calculated by normalizing the measurement of drug treatment to control (dose of 0 nM). The data shown is a mean representative of the three independent measurements.
2.5. γ-H2AX Western Blotting
SK-N-AS cells were treated with EAD at doses of 0.5, 2, and 5 μM. Positive controls included cells treated with 1 μM camptothecin (CPT) and 10 μM etoposide. Four hours post-treatment, cells were lysed in a buffer containing 1% SDS (sodium dodecyl sulfate), 10 mM Tris-HCl (pH 7.4), 1 mM sodium orthovanadate supplemented with protease inhibitors (Roche Applied Science, Indianapolis, IN) and phosphatase inhibitors (Sigma Chemical Co., St. Louis, MO). Protein concentrations were determined using the BioRad DC Protein Assay. Equal amounts of protein were fractionated on a 4–20% SDS–polyacrylamide gel, transferred to nitrocellulose, and immunoblotted using anti-phospho-H2AX (Upstate, Lake Placid, NY) and anti-GAPDH (Cell Signaling, Beverly, MA) primary antibodies.
2.6. Topoisomerase I/II -Mediated DNA Cleavage Assay
Human recombinant Top1 was purified from baculovirus as previously described [10] and human recombinant Top2α was a generous gift from Dr. Neil Osheroff (Vanderbilt University). DNA cleavage reactions were prepared as previously reported with the exception of the DNA substrate [11]. For topoisomerase I-mediated DNA cleavage assay, a 117-bp DNA oligonucleotide (Integrated DNA Technologies, Coralville, IA) containing a single 5′-cytosine overhang was 3′-end labeled by fill-in reaction with [a-32P]-dGTP in React 2 buffer (50 mM Tris-HCl, pH 8.0, 100 mM MgCl2, 50 mM NaCl) with 0.5 units of DNA polymerase I (Klenow fragment, New England BioLabs, Ipswich, MA). For topoisomerase II-mediated DNA cleavage assay, the recessed strand from the 117-bp DNA oligonucleotide was 5′-labeled using T4 polynucleotide kinase and [γ-32P] ATP. The 5′-labeled single-stranded oligonucleotide were then annealed with its complementary strand to the double-stranded 117-bp oligonucleotide by heating for 5 min at 95°C and slowly cooling to room temperature. Approximately 2 nM of radiolabeled DNA substrate was incubated with or without recombinant Top1 or Top2α in 20 μL of reaction buffer (10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/ml BSA) at 25°C for 20 min in the presence of various concentrations of drugs. The reactions were terminated by adding SDS (0.5% final concentration) followed by the addition of two volumes of loading dye (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol, and 0.1% bromphenol blue).
2.7. DNA Alkylation Assay
A 5′-end labelled 60-bp DNA oligonucleotide (1 nM) was incubated with different doses of EAD for 1 hr at 30 °C in reaction buffer containing 25 mM TEOA (triethanolamine) and 1 mM EDTA. The reaction was quenched by the addition of 5 μg of calf thymus DNA. After ethanol precipitation, the DNA was resuspended in 1 M piperidine and incubated for 15 min at 95 °C. The solution was lyophilized, and the pellet dissolved in formamide loading dye.
2.8. Gel Electrophoresis
Aliquots of each reaction from topoisomerase I-mediated DNA cleavage assay and DNA alkylation assay were subjected to 20% denaturing PAGE. Gels were dried and visualized by using Phosphoimager and ImageQuant software (Molecular Dynamics).
3. Results
3.1. Gene expression profiling of active drugs in neuroblastoma cells
We have previously screened a panel of 96 compounds with diverse mechanisms of action in neuroblastoma (NB) cell lines and identified 30 compounds that are active against NB cell lines at concentrations ≤ 10 μM. The details of the screen are described elsewhere [9]. To elucidate the mechanism of action underlying the biological responses of neuroblastoma cells to these active agents, we profiled the gene expression of human neuroblastoma cell line SK-N-AS, a stage 4 MYCN-not-amplified NB cell line, for each of these active compounds using Affymetrix human genome U133plus 2.0 arrays. Our data set includes profiles for 30 different drugs at two different concentrations (a total of 60 gene expression profiles). The details of the dataset are described in Supplemental Table 1. In order to obtain signatures related to direct mechanism of action, we used a relatively early time point (6 hours after compound addition) as described previously [5].
Among 30 active compounds only 10 were also present in the Broad Institute’s connectivity map (cMAP), (http://www.broadinstitute.org/cmap) [5]. Thus we compared expression profiles for these 10 compounds in neuroblastoma cell with those in other cancer cell lines existing in the cMAP database. The query signature for each drug was generated using 500 most up-regulated and 500 most down-regulated probe sets derived from NB cells and searched against the cMAP database. At higher concentration, 7 out of 10 drugs (alsterpaullone, 5-azacytidine, camptothecin, colchicine, doxorubicin, mitoxantrone, and tetrandrine) have high cMAP enrichment scores (p < 0.05), five of them ranking at or near the top despite the differences in cells (NB cells vs. MCF7 and PC3 cells) used to generate the query signature and reference profiles (Table 1). At lower concentration, alsterpaullone, 5- azacytidine, camptothecin, colchicine, and daunorubicin showed high enrichment scores (p < 0.05). There is no profile similarity for the rest of drugs (Table 1). It is plausible that the drugs with dissimilar expression profiles at both low and high concentration could be cell context-dependent.
Table 1.
Comparison of gene expression profiles for common compounds present in NB dataset and cMAP
| Drug | Instances | Higher concentration | Lower concentration | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Rank | Score | p-value | Rank | Score | p-value | ||
| 5-Azacitinide | 3 | 5 | 0.96 | <0.001 | 56 | 0.74 | 0.036 |
| Alsterpaullone | 3 | 1 | 1 | <0.001 | 11 | 0.97 | <0.001 |
| Camptothecin | 3 | 1 | 1 | <0.001 | 2 | 1 | <0.001 |
| Colchicine | 6 | 31 | 0.61 | 0.012 | 47 | 0.56 | 0.027 |
| Daunorubicin | 4 | 389 | 0.38 | 0.508 | 32 | 0.73 | 0.011 |
| Doxorubicin | 3 | 1 | 1 | <0.001 | 952 | -0.46 | 0.430 |
| Etoposide | 4 | 525 | 0.32 | 0.72 | 768 | -0.31 | 0.748 |
| Mitoxantrone | 3 | 1 | 1 | <0.001 | 816 | -0.4 | 0.595 |
| Paclitaxel | 4 | 622 | 0.21 | 0.91 | 444 | -0.27 | 0.860 |
| Tetrandrine | 4 | 39 | 0.68 | 0.023 | 370 | 0.38 | 0.505 |
3.2. NB-cMAP: a database of drug-associated neuroblastoma gene expression profiles
Since 20 active drugs in NB cell lines were not present in the cMAP database and the current Broad cMAP database were mostly generated from carcinoma cell lines (breast cancer and prostate cancer; MCF7 and PC3 respectively), we postulated that the action of some drugs might be cell-context dependent. We therefore developed a web-based database NB-cMAP (http://pob.abcc.ncifcrf.gov/cgi-bin/cMAP) containing 60 neuroblastoma mRNA expression profiles representing 30 active agents as described above with the same structure and algorithm as cMAP [5].
3.3. Identification of Epoxy Anthraquinone Derivative as a potential DNA replication inhibitor
Among 30 active drugs against NB cell lines, most of them have proposed mechanism of action. However, epoxy anthraquinone derivative (EAD), perezone, and helenalin, have not been rigorously evaluated especially in the context of their promising activity in NB treatment. In order to understand the mechanisms of these 3 drugs, we used the connectivity maps to generate hypotheses about the mechanism of action for each of these drugs. We first generated signatures from the gene expression profiles of these drugs in NB cells at higher concentration and queried the cMAP database. For perezone, high connectivity scores were found for multiple instances of three alkylating agents: lomustine, semustine and 15-delta prostaglandin J2 (Supplemental Figure 1A). Comparison of the chemical structures between perezone and these three drugs indicated that they all have a potential alkylating functional group in their structures (Supplemental Figure 1B). The query for helenalin yielded high enrichment scores for multiple topoisomerase I/II inhibitors: camptothecin, daunorubicin, irinotecan, and ellipticine (Supplemental Figure 2), suggesting that helenalin has a mode of action through topoisomerase inhibition.
We next studied the effects of EAD using cMAP analysis and found that 7 out of 10 highest scoring instances were topoisomerase I/II inhibitors and DNA intercalators: irinotecan, camptothecin, hycanthone and daunorubicin (Table 2). High connectivity score was also found for ellipticine, another topoisomerase II inhibitor. The analysis of EAD structure along with the structures of these topoisomerase I/II inhibitors and DNA intercalators showed that EAD with its tricyclic fused ring system shares a common pharmacophore displayed with the rest of the DNA intercalators (Supplemental Figure 3). Thus the cMAP analysis suggested the function of EAD as predicted by its structure. In addition, we observed high scores of phenoxybenzamine (alkylating agent) and trichostatin A (HDAC inhibitor) (Table 2). The activity of EAD could be attributed to the presence of an epoxide ring that can easily modify associated enzymes and the ability of the epoxide group to covalently modify its targets; a similar alkylating mode of action can be proposed for phenoxybenzamine (Supplemental Figure 3) containing an active terminal chloro-group. Furthermore, we queried the EAD signature against NB-cMAP to determine whether a similar result could be obtained. As shown in Table 3, within top 10 matches with the highest connectivity scores, EAD at lower concentration is one of them as anticipated, all other 9 matches were known to be DNA replication inhibitors (topoisomerase I/II inhibitors and DNA intercalators) representing 7 drugs: echinomycin, daunorubicin, doxorubicin, mitoxantrone, camptothecin, teniposide, and amonafide. Four drugs (echinomycin, mitoxantrone, teniposide and amonafide) from NB-cMAP query were not present in the cMAP database; their chemical structures suggested their capability of DNA intercalation similar to EAD (Supplemental Figure 4). These results further strengthened our hypothesis that EAD inhibits neuroblastoma cell growth through DNA interaction.
Table 2.
List of compounds showing high connectivity with EAD in cMAP analysis
| Rank | Drug name | Dose | Cell | Score | Instance_id | Mechanism of action |
|---|---|---|---|---|---|---|
| 1 | Phenoxybenzamine | 12 μM | PC3 | 1 | 4652 | Alpha blockade/alkylation |
| 2 | Irinotecan | 100 μM | PC3 | 0.974 | 7535 | Top1 inhibition |
| 3 | Phenoxybenzamine | 12 μM | MCF7 | 0.918 | 5248 | Alpha blockade |
| 4 | Irinotecan | 100 μM | MCF7 | 0.898 | 7530 | Top1 inhibition |
| 5 | Camptothecin | 11 μM | PC3 | 0.86 | 4541 | Top1 inhibition |
| 6 | Camptothecin | 11 μM | MCF7 | 0.819 | 3887 | Top1 inhibition |
| 7 | Phenoxybenzamine | 12 μM | MCF7 | 0.808 | 5613 | Alpha blockade |
| 8 | Hycanthone | 11 μM | MCF7 | 0.74 | 5691 | DNA intercalation |
| 9 | Hycanthone | 11 μM | PC3 | 0.735 | 4630 | DNA intercalation |
| 10 | Daunorubicin | 1 μM | PC3 | 0.695 | 7511 | DNA intercalation/Top2 inhibition |
| 11 | Trichostatin A | 1 μM | PC3 | 0.687 | 5950 | HDAC inhibition |
| 12 | Ellipticine | 16 μM | PC3 | 0.674 | 5779 | DNA intercalation/Top2 inhibition |
| 14 | Trichostatin A | 100 nM | PC3 | 0.649 | 4302 | HDAC inhibition |
| 17 | Irinotecan | 100 μM | MCF7 | 0.624 | 7498 | Top1 inhibition |
| 19 | Trichostatin A | 100 nM | PC3 | 0.612 | 5086 | HDAC inhibition |
Table 3.
List of compounds showing high connectivity with EAD in query against internal NB-cMAP database
| Rank | Drug name | Dose | Score | Mechanism of action |
|---|---|---|---|---|
| 1 | Echinomycin | 5 nM | 1 | DNA Intercalation |
| 2 | Daunorubicin | 1 μM | 0.94 | DNA intercalation/Top2 inhibition |
| 3 | Doxorubicin | 1 μM | 0.91 | DNA intercalation/Top2 inhibition |
| 4 | Mitoxantrone | 1 μM | 0.9 | DNA intercalation/Top2 inhibition |
| 5 | Camptothecin | 1 μM | 0.86 | Top1 inhibition |
| 6 | Echinomycin | 0.5 nM | 0.81 | DNA Intercalation |
| 7 | Teniposide | 10 μM | 0.77 | Top2 inhibition |
| 8 | EAD | 0.5 μM | 0.74 | Unknown |
| 9 | Amonafide | 10 μM | 0.71 | DNA intercalation/Top2 inhibition |
| 10 | Daunorubicin | 10 μM | 0.7 | DNA intercalation/Top2 inhibition |
3.4. Epoxy Anthraquinone Derivative inhibits DNA replication and alkylation in neuroblastoma cells
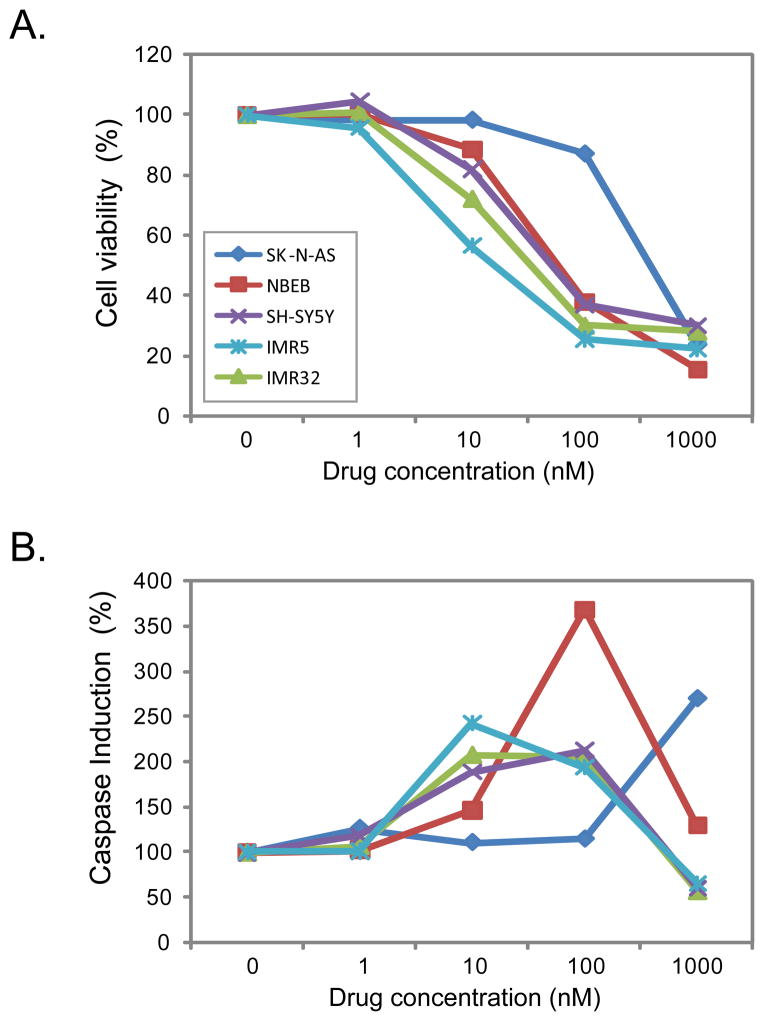
We further examined the cell growth inhibition and pro-apoptotic activity of EAD using multiple doses in a larger panel of NB cell lines including two MYCN-amplified cell lines (IMR32 and IMR5) and three MYCN-not-amplified cell lines (SK-N-AS, SH-SY5Y and NBEB). The inhibition of cell growth was observed for all tested cell lines in a dose-dependent manner (Figure 1A), and the induction of apoptosis started at a dose of 10 nM except SK-N-AS cells which exhibited the induction at 1000 nM (1 μM) (Figure 1B). The induction of apoptosis dropped at a higher concentration after reaching the peak induction, which is probably due to very few viable cells left for the caspase assay.
Figure 1. EAD inhibits cell growth and induces apoptosis in multiple neuroblastoma cell lines.
Five NB cell lines including three MYCN-not-amplified (SK-N-AS, NBEB, and SH-SY5Y) and two MYCN-amplified cell lines (IMR5 and IMR32) were used in this experiment; the cells were treated with EAD at 1, 10, 100 and 1000 nM. The cell viability (A) and caspase-Glo 3/7 activities (B) were measured at 24 hours after the drug treatment. The percentage of cell viability or caspase activity was calculated by normalizing the measurement of drug treatment to no treatment (dose of 0 nM). The data shown is a mean representative of the three independent measurements. SK-N-AS,
 ; NBEB,
; NBEB,
 ; SH-SY5Y,
; SH-SY5Y,
 ; IMR5,
; IMR5,
 ; IMR32,
; IMR32,
 .
.
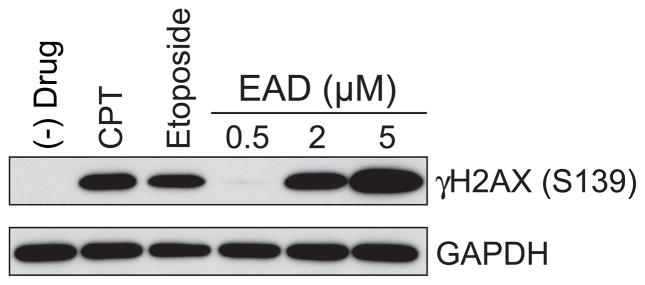
The cMAP analysis on the Broad and NCI database as well as the structure analysis indicated that that EAD functions as DNA replication inhibitor; we therefore next sought to validate this hypothesis experimentally. DNA double-stranded breaks (DSBs) resulting from DNA replication arrest are generally detected by their ability to trigger phosphorylation of histone H2AX (γ-H2AX) [12]. To test whether EAD induces DNA double-stranded breaks, we treated SK-N-AS cells with EAD and measured γ-H2AX levels using Western blot analysis. As predicted, EAD increased γ-H2AX levels in a dose-dependent manner (Figure 2).
Figure 2. EAD increases the level of γH2AX at a dose-dependent manner.
The level of γH2AX protein was measured after SK-N-AS cells were treated with different drugs for 4 hours. Doses of drugs: camptothecin (CPT) at 1 μM; etoposide at 10 μM; EAD at 0.5, 2, and 5 μM.
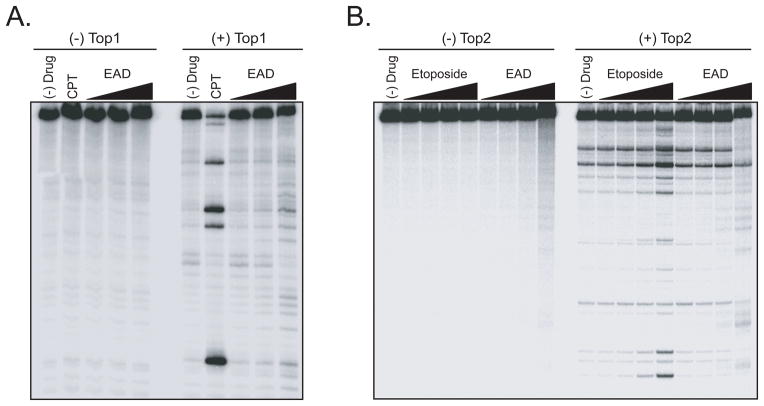
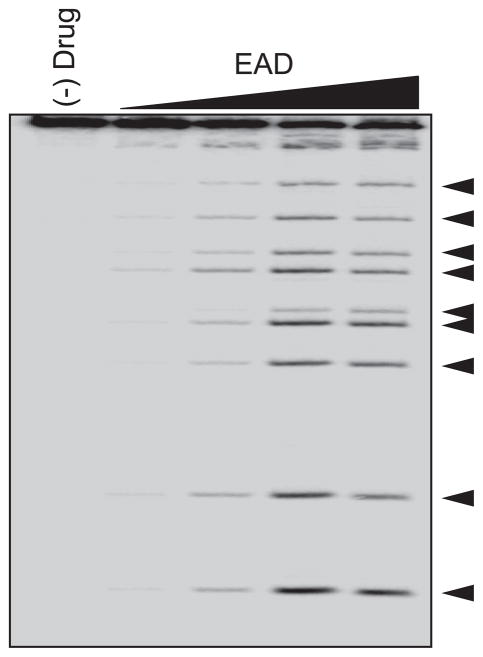
We then tested whether EAD inhibit Topoisomerase I/II activities using a topoisomerase-mediated DNA cleavage assays [13]. These assays allow comparison of the effectiveness of the topoisomerase inhibitors in stabilizing topoisomerase-DNA cleavage complex. The formation of topoisomerase I- DNA complexes by EAD is only observed by the weakly increased intensity of the DNA cleavage bands at a high dose of 100 μM; the stabilizing effect of EAD is much less effective than camptothecin (CPT), a known topoisomerase I inhibitor (Figure 3A). However, we observed the induction of topoisomerase II-DNA complexes by EAD at higher concentrations, although weaker than etoposide, a topoisomerase II inhibitor (Figure 3B). Interestingly, EAD retards the mobility of the DNA substrate either in the presence or absence of topoisomerase I/II enzymes, indicating that EAD may bind covalently to the DNA probably as a result of the reactive epoxide group. In addition, we observed that EAD has a strong connectivity with phenoxybenzamine, further evidence of covalent modification mechanism of action common for EAD and phenoxybenzamine. Therefore we conducted piperidine cleavage assay to confirm the alkylating ability of EAD a, in which the specific breaks at sites of N7-guanine alkylation are created by piperidine [14]. As shown in Figure 4, the resulting cleavages by EAD were in a dose-dependent manner, proving the N7-guanine alkylation by EAD. Therefore, these experiments proofed that EAD is a strong DNA alkylating agent, and a weak topoisomerase inhibitor.
Figure 3. EAD induces Topoisomerase I/II-mediated DNA cleaving activity.
A. The induction of topoisomerase I (Top 1)DNA complexes by EAD was measured using DNA cleavage assays as described in Materials and Methods. Doses of drugs: camptothecin (CPT) at 1 μM; EAD at 1, 10, and 100 μM. B. The induction of topoisomerase II (Top2)-DNA complexes by EAD was measured using DNA cleavage assays as described in Materials and Methods. Doses of drugs: etoposide at 0.1, 1, 10, and 100 μM; EAD at 0.1, 1, 10, and 100 μM.
Figure 4. EAD induces DNA alkylation.
The guanine-alkylating ability of EAD was examined using a piperidine cleavage assay at doses of 0.1, 1, 10, and 100 μM as described in Materials and Methods.
4. Discussion
High-risk neuroblastoma remains largely incurable despite intensive multimodality therapies [1,2], which include cytostatic and cytotoxic agents. Many of them are alkylating drugs targeting at a broad range of DNA and proteins, which belong to the earliest class of anticancer therapies that still play an important role in the treatment of pediatric malignancies [15]. Unfortunately there is also a significant toxicity associated with these drugs due to non-selective effects in healthy tissue. Therefore there is a pressing need to identify effective drugs with the least side effects. We have previously screened a panel of agents and identified 30 active drugs against neuroblastoma cells (Supplemental Table 1); most of them have well-characterized mechanisms of action. In order to elucidate the mechanism of action of uncharacterized drugs, we used a gene expression-based chemical genomic approach that has been proven to be effective [5–8]. Among the drugs tested, EAD has been identified to induce significant cell death in neuroblastoma cell lines in our previous study [9], it has also been reported that EAD inhibits angiogenesis [16]. Both cMAP database from Broad Institute and the internal NB-cMAP database as well as its structure analysis demonstrated that EAD may inhibit neuroblastoma cells through DNA alkylation, DNA intercalation, and topoisomerase I/II inhibition. We validated these results experimentally to show that EAD induced DNA double-strand breaks, DNA alkylation and inhibited topoisomerase II activity (Figure 2–4); however the inhibition of toposomerase I is only weakly observed at a high dose of EAD. Topoisomerase I and II (Top1 and Top2) are well established molecular targets of anticancer drugs and have been the focus for the treatment of various cancers [17–19], and inhibition of these critical enzymes will result in DNA strand breaks which cause arrest in the cell cycle [17–19]. Among the well established Top1 inhibitors, camptothecin and its water-soluble derivative irinotecan were shown to have similar expression profiles with EAD. Irinotecan is approved by the FDA for colorectal tumors and is under clinical trial in treating children with refractory solid tumors including neuroblastoma. Multiple Top2 inhibitors including daunorubicin, doxorubicin, mitoxantrone, ellipticine, amonafide and teniposide were found to have a strong connectivity with EAD. Teniposide is a non-intercalating Top2 inhibitor and all other 5 drugs are intercalating Top2 inhibitors. All these drugs have been shown to have anticancer activity [18]. Daunorubicin, doxorubicin, mitoxantrone and teniposide are FDA-approved, but amonafide and ellipticine are not currently in clinical use [18]. In addition, EAD was shown to have a strong connectivity with phenoxybenzamine and trichostatin A. Phenoxybenzamine is a α2 adrenoceptor agonist and its molecular mode of action is proposed to be through alkylation of the receptor. Trichostatin A, a histone deacetylase inhibitor, is proposed to block the catalytic reaction by chelating a zinc ion in the active-site pocket through its hydroxamic acid group [20]. It has been reported to induce cell growth arrest, apoptosis and differentiation in tumor cells [20]. It is important to note that EAD apart from its alkylating characteristics, can also act by chelating Zn2+ ions through the planar 1,3-carbonyl groups and probably share similar mode of action with trichostatin A. These important findings suggest that EAD can act through multiple modes of action which may increase the potency of the anti-cancer effect; therefore it may be developed into an effective drug against neuroblastoma and other cancers. Although this study was done in neuroblastoma but it is likely that this novel drug may act on all dividing cells given their nature of action, further study is warranted to investigate the effect of EAD in other tumor cells.
In this study we also revealed the drug action mechanism for another two uncharacterized drugs, perezone and helenalin, which have been identified as cytotoxic agents against neuroblastoma cells based on our previous study [9]. Perezone is a sesquiterpenoic benzoquinone isolated from the roots of Perezia adnata [21], and its anti-cancer mechanism is unknown. cMAP analysis has shown that three high scored matches for perezone expression profile are the alkylation agents: lomustine, semustine and 15-delta prostaglandin J2, suggesting that perezone might act as an alkylating agent. Perezone is also a redox active compound that can possibly involved in cellular redox processes. Helenalin is a naturally occurring sesquiterpene lactone extracted from Arnica montana and Arnica chamissonis ssp. foliosa. It has been reported that Helenalin and its analogs exert their anti-cancer and anti-inflammatory effects via inhibiting NF-κB and telomerase activity, and impairing protein and DNA synthesis [22–24]. However, our gene expression-based chemical genomic analysis demonstrated similar expression profile changes of helenalin as those of camptothecin, irinotecan, daunorubicin and ellipticine, which are topoisomerase I/II inhibitors. Four topoisomerase I/II inhibitors contain fused aromatic rings capable of DNA intercalation via π-stacking (Supplemental 2B), however helenalin with its active α,β–unsaturated terpenoid ring system can exert its topoisomerase inhibition by alkylating any of the DNA repairing enzymes or damaged DNA itself. Helenalin also has a strong connectivity with GW-8510, a relatively selective CDK2 inhibitor that shows antitumor activity by inhibition of cell growth and induction of apoptosis [25]. Even though a direct chemical structural comparison seems not evident, the fact that the GW-8510 possesses extended aromatic ring systems as well as the exocyclic lactam ring may possibly contribute to its biological activity (Supplemental 2B). Common pharmacophores that were identified for these drugs and the similarity in their gene expression profiles warrants further experimental validation and this information could be useful in the rational design of the next generation of specific and selective topoisomerase inhibitors. Validation of these two compounds will be the subject of future studies.
The connectivity Map (cMAP) developed by the Broad Institute is a robust approach for compound activity prediction that uses gene expression signature enrichment analysis to identify compounds which act with similar mode of action. We used this tool to identify the putative mechanisms for three uncharacterized drugs and validated the mechanism of EAD action by our internal NB-cMAP, the structure analysis and experimental data. NB-cMAP, created with same structure and algorithm as cMAP, uses the dataset of neuroblastoma gene expression data for 60 instances representing 30 active agents. There are 10 common drugs existing in both cMAP and NB-cMAP. Thus we used the gene expression signatures of these 10 drugs in the NB-cMAP dataset to query cMAP and test if all 10 drugs could be retrieved from cMAP with high connectivity scores. Seven drugs at higher concentration and five at lower concentration returned with high connectivity scores for the same drugs. For those drugs with no connectivity, they may induce a set of context specific genes in neuroblastoma cells different from leukemia, breast, and prostate cancer cell lines used in the cMAP dataset. This context dependency was also observed in the previous study [5]. Therefore NB-cMAP dataset, generated in neuroblastoma cell line and included 20 drugs not existing in cMAP, might provide useful information in addition to cMAP. Despite the cell context dependency for some agents, the majority of the agents appear to be cell line independent as shown in this study and the previous study [5].
In conclusion, we have used a gene expression-based chemical genomic approach combined with structure analysis to identify the putative mechanism of action for three uncharacterized drugs that are cytotoxic against neuroblastoma cells. Our study demonstrated that EAD inhibits DNA replication in neuroblastoma cells through DNA alkylation, DNA intercalation, and topoisomerase inhibition. EAD may prove to be a good candidate for the potential treatment of high-risk neuroblastoma as well as other cancer patients, but further studies are warranted to evaluate its in vivo therapeutic potential and possible side effects.
Supplementary Material
Acknowledgments
We thank Sophic Systems Alliance (East Falmouth, MA) for helping to develop NB-cMAP database. We thank Drs. Adam Cheuk and Patricia Tsang for their technical assistance in cell viability assay. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. This work was supported by the NIH Intramural Program, Center for Cancer Research, National Cancer Institute.
Abbreviation List
- NB
neuroblastoma
- cMAP
connectivity map
- EAD
Epoxy anthraquinone derivative
Footnotes
5. Conflict of interest
The authors declare no conflict of interest for this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, Mosseri V, Simon T, Garaventa A, Castel V, Matthay KK. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman S, Chen HX, Matthay KK, Cohn SL, Maris JM, Sondel P. A phase III randomized trial of the chimeric anti-GD2 antibody ch14.18 with GM-CSF and IL2 as immunotherapy following dose intensive chemotherapy for high-risk neuroblastoma: Children’s Oncology Group (COG) study ANBL0032; ASCO Annual Meeting; 2009. p. 15. [Google Scholar]
- 4.Trial watch: Immunotherapy shows promise in phase III neuroblastoma trial. Nature reviews drug discovery. 2009;8:604. doi: 10.1038/nrd2951. [DOI] [PubMed] [Google Scholar]
- 5.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 6.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 8.De Preter K, De Brouwer S, Van Maerken T, Pattyn F, Schramm A, Eggert A, Vandesompele J, Speleman F. Meta-mining of neuroblastoma and neuroblast gene expression profiles reveals candidate therapeutic compounds. Clin Cancer Res. 2009;15:3690–3696. doi: 10.1158/1078-0432.CCR-08-2699. [DOI] [PubMed] [Google Scholar]
- 9.Gheeya JS, Chen QR, Benjamin CD, Cheuk AT, Tsang P, Chung JY, Metaferia BB, Badgett TC, Johansson P, Wei JS, Hewitt SM, Khan J. Screening a panel of drugs with diverse mechanisms of action yields potential therapeutic agents against neuroblastoma. Cancer Biol Ther. 2009;8 doi: 10.4161/cbt.8.24.10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pourquier P, Jensen AD, Gong SS, Pommier Y, Rogler CE. Human DNA topoisomerase I-mediated cleavage and recombination of duck hepatitis B virus DNA in vitro. Nucleic Acids Res. 1999;27:1919–1925. doi: 10.1093/nar/27.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dexheimer TS, Kozekova A, Rizzo CJ, Stone MP, Pommier Y. The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 2008;36:4128–4136. doi: 10.1093/nar/gkn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dexheimer TS, Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat Protoc. 2008;3:1736–1750. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattes WB, Hartley JA, Kohn KW. DNA sequence selectivity of guanine-N7 alkylation by nitrogen mustards. Nucleic Acids Res. 1986;14:2971–2987. doi: 10.1093/nar/14.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izbicka E, Izbicki T. Therapeutic strategies for the treatment of neuroblastoma. Curr Opin Investig Drugs. 2005;6:1200–1214. [PubMed] [Google Scholar]
- 16.Takano S, Gately S, Jiang JB, Brem S. A diaminoantraquinone inhibitor of angiogenesis. J Pharmacol Exp Ther. 1994;271:1027–1033. [PubMed] [Google Scholar]
- 17.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 18.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba T, Yokosuka O, Fukai K, Kojima H, Tada M, Arai M, Imazeki F, Saisho H. Cell growth inhibition and gene expression induced by the histone deacetylase inhibitor, trichostatin A, on human hepatoma cells. Oncology. 2004;66:481–491. doi: 10.1159/000079503. [DOI] [PubMed] [Google Scholar]
- 21.Tellez JF, Carvajal K, Cruz D, Carabez A, Chavez E. Effect of perezone on arrhythmias and markers of cell injury during reperfusion in the anesthetized rat. Life Sci. 1999;65:1615–1623. doi: 10.1016/s0024-3205(99)00411-7. [DOI] [PubMed] [Google Scholar]
- 22.Farhana L, Dawson MI, Fontana JA. Apoptosis induction by a novel retinoid-related molecule requires nuclear factor-kappaB activation. Cancer Res. 2005;65:4909–4917. doi: 10.1158/0008-5472.CAN-04-4124. [DOI] [PubMed] [Google Scholar]
- 23.Lyss G, Knorre A, Schmidt TJ, Pahl HL, Merfort I. The anti-inflammatory sesquiterpene lactone helenalin inhibits the transcription factor NF-kappaB by directly targeting p65. J Biol Chem. 1998;273:33508–33516. doi: 10.1074/jbc.273.50.33508. [DOI] [PubMed] [Google Scholar]
- 24.Huang PR, Yeh YM, Wang TC. Potent inhibition of human telomerase by helenalin. Cancer Lett. 2005;227:169–174. doi: 10.1016/j.canlet.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 25.Dai Y, Grant S. CDK inhibitor targets: a hit or miss proposition?: cyclin-dependent kinase inhibitors kill tumor cells by downregulation of anti-apoptotic proteins. Cancer Biol Ther. 2006;5:171–173. doi: 10.4161/cbt.5.2.2568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.