Summary
Cilia and extracellular vesicles (EVs) are signaling organelles[1]. Cilia act as cellular sensory antennae, with defects resulting in human ciliopathies. Cilia both release and bind to EVs[1]. EVs are submicron-sized particles released by cells and function in both short and long range intercellular communication. In C. elegans and mammals, the Autosomal Dominant Polycystic Kidney Disease (ADPKD) gene products polycystin-1 and polycystin-2 localize to both cilia and EVs, act in the same genetic pathway, and function in a sensory capacity, suggesting ancient conservation[2]. A fundamental understanding of EV biology and the relationship between the polycystins, cilia, and EVs is lacking. To define properties of a ciliated EV-releasing cell, we performed RNAseq on 27 GFP-labeled EV releasing neurons (EVNs) isolated from adult C. elegans. We identified 335 significantly overrepresented genes, of which 61 were validated by GFP reporters. The EVN transcriptional profile uncovered new pathways controlling EV biogenesis and polycystin signaling and also identified EV cargo, which included an antimicrobial peptide and ASIC channel. Tumor necrosis associated factor (TRAF) homologues trf-1 and trf-2 and the p38 mitogen-activated protein kinase (MAPK) pmk-1 acted in polycystin signaling pathways controlling male mating behaviors. pmk-1 was also required for EV biogenesis, independent of the innate immunity MAPK signaling cascade. This first high-resolution transcriptome profile of a subtype of ciliated sensory neurons isolated from adult animals reveals the functional components of an EVN.
The cilium both releases and binds to extracellular vesicles (EVs), suggesting that cilia may be essential in EV-mediated communication as both senders and receivers of information[3-8]. EVs carry specific protein and RNA cargoes that can be transferred between donor and recipient cells without requiring direct contact[9]. EVs mediate a broad range of physiological and pathological processes[10].
The mammalian polycystins localize to cilia as well as urinary EVs released from renal epithelial cells[3, 11-13], and polycystin ciliary trafficking defects may be an underlying cause of ADPKD[14]. In an amazing display of evolutionary conservation, we recently showed that the C. elegans cilium is a source of bioactive polycystin-containing EVs[6]. The mechanisms controlling EV biogenesis, shedding, and release are poorly understood, largely due to technical difficulties in visualizing, isolating, and characterizing these sub-micrometer-sized particles. Here, we defined the unique features of a ciliated extracellular vesicle releasing neuron (EVN).
C. elegans shed and release EVs from 27 ciliated extracellular vesicle releasing neurons (EVNs) including six inner labial type 2 (IL2) neurons and 21 male-specific polycystin-expressing EVNs in the head (four CEM neurons) and tail (16 ray B-type RnB neurons and one hook B-type HOB neuron) (Fig. 1A, B)[6]. In these male-specific EVNs, the polycystins lov-1 and pkd-2 are required for male sex drive, response to mate contact, and vulva location[15-17]. The kinesin-3 protein KLP-6 is exclusively expressed in the 27 EVNs and is required for release of bioactive PKD-2::GFP containing EVs that function in reproductive animal-to-animal communication[6]. Intact cilia but not multivesicular body components are required for EV release, suggesting that PKD-2 containing EVs are not exosomes but rather ectosomes that bud from the plasma membrane at the ciliary base[6]. In addition to klp-6 and the polycystins, only a handful of genes (coexpressed with polycystin genes cwp-1 to -5, alpha-tubulin tba-6, and the EV release regulator cil-7) are exclusively expressed in the EVNs[18-21].
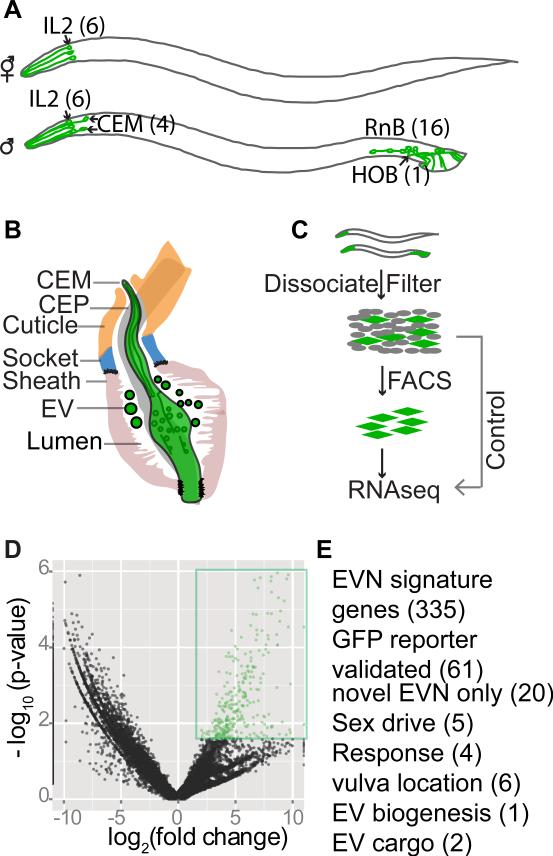
Figure 1. Cell-type RNAseq to define the EVN transcriptome.
(A) The ciliated EVNs, six in the hermaphrodite (top) and 27 in male (bottom). (B) The male cephalic sensillum. Glial sheath and socket cells form a continuous lumen surrounding the CEM neuronal cilium, which is exposed to the environment directly through a cuticular opening. The lumen is shared by CEM and CEP neurons. EVs are observed in the lumen. (C) Schematic illustration of the differential RNAseq experiment. EVNs were FACS purified from dissociated and filtered cells isolated from synchronized klp-6p::GFP expressing young adult males and hermaphrodites, followed by mRNA extraction, library construction, and RNAseq. Refer to Supplemental Figure 1 for heat map, principal component analysis, and downsampling in silico. (D) Volcano plot showing 335 genes differentially overrepresented in sorted EVNs (colored and boxed in green) compared to whole worms at a false discovery rate of 10%. (E) 335 EVN gene signature genes were validated by GFP expression pattern and functional analysis (for available mutants). The number of genes for each category is listed in parentheses. Refer to Supplemental Tables S1 and S2 for full list of 335 signature genes and original DEseq data, respectively. See Supplemental Table S3 for GO Analysis of EVN overrepresented genes.
To decipher the biology of a polycystin-expressing EVN in its native environment, we isolated klp-6p::GFP labeled neurons from age-synchronized adult animals by mechanical and proteolytic disruption, filtration, and FACS sorting[22] (Fig. 1C). Animals were alive until the moment they were processed. Therefore samples are as close to the in vivo state as one can get (as opposed to the re-differentiated and fixed cells that other labs have used), which makes profiling results more biologically accurate. RNA from two replicates (8,000 and 11,000 EVNs) was purified and amplified for RNAseq (Methods). RNAseq libraries from the amplification products of EVN cell populations and whole worm lysate control samples were sequenced. Total reads were downsampled in silico to a depth of 4.2–6.7 × 106 reads per library and tested for expression and differential expression (Supplemental Fig. 1C, D). DEseq software identified 9,922 genes expressed in EVNs, 14,455 genes in whole worm (Table S2), and 335 genes significantly overrepresented two to 11-fold in EVNs (Fig. 1D, Table S1). Cluster analysis of the total 14,455 genes expressed between sorted EVNs and whole worm showed high intragroup homogeneity (Supplemental Fig. 1A). Principle Components Analysis showed clear separation between sorted EVNs and whole worm lysates (Supplemental Fig. 1B). Volcano plot of the differential RNAseq transcriptome analysis showed close linearization of log2 fold change and the log 10 p value, indicating most data points were consistently represented in both replicates (Fig. 1D).
To uncover the functions of the 335 overrepresented genes, we used gene ontology (GO) term annotation analysis. Among the GO terms in the 335 overrepresented genes were neuropeptide signaling, ciliogenesis, neuron projection, ion channel activity, and synaptic transmission, which are consistent with the neuronal and ciliated characters of the EVNs (Table S3). Over half of the overrepresented genes including 70 in the top 100 have no assigned name or described RNAi phenotype, likely because global RNAi screens do not examine adult male phenotypes. Thus, many of these EVN signature genes may function specifically in the adult male. 61 of 335 signature genes were expressed in EVNs as demonstrated by published GFP expression analysis or described herein (Table S1). The EVN restricted genes klp-6, lov-1, pkd-2, cil-7 and the five cwp genes[16, 18-20, 23] were up-regulated 5.8-fold or higher. We identified 20 new genes that were exclusively expressed in all EVNs, similar to the EVN release regulators klp-6 and cil-7 (Fig. 2A); in all male-specific EVNs, similar to pkd-2 and lov-1 (Fig. 2B); or in a subset of the EVNs (Fig. 2C, D), including Y70G10A.2 that was expressed in a single EVN (Fig. 2D). Combined, these results indicate that our RNAseq dataset is reliable and highly enriched for EVN genes, and demonstrate the sensitivity of our profiling method.
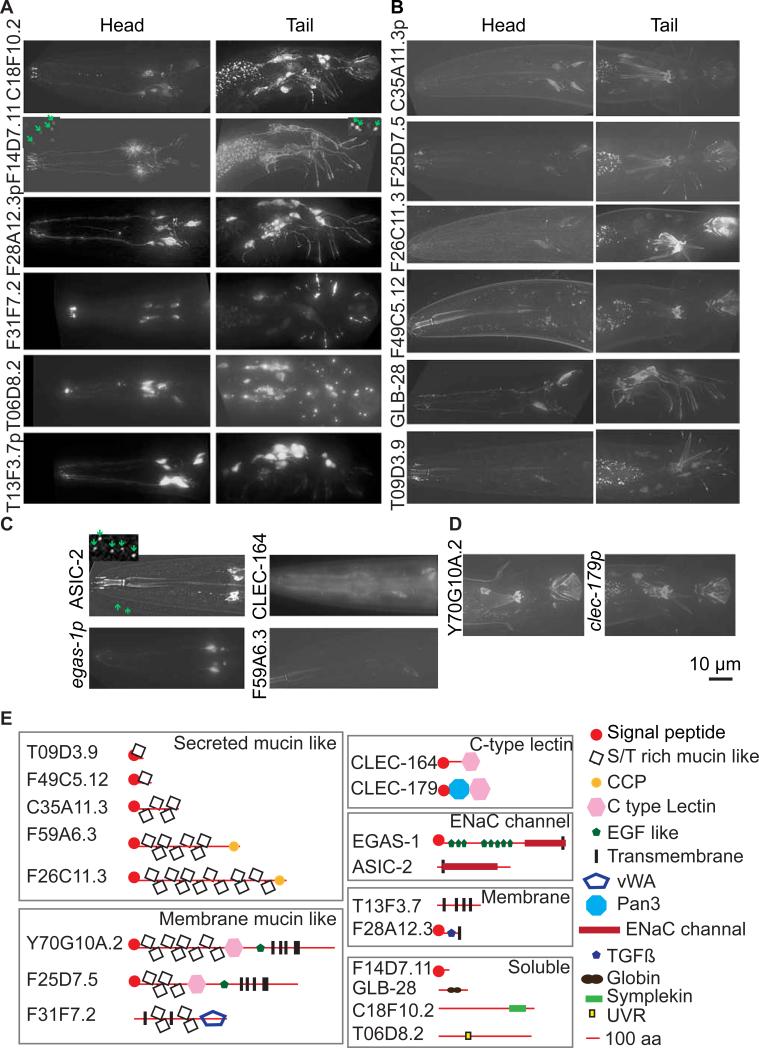
Figure 2. EVN signature genes were expressed in all 27 EVNs (A), the 21 polycystin-expressing EVNs (B), or a subset thereof (C, D).
(A) Head IL2 and CEM (left) and tail HOB and RnB (right) images of GFP reporters that were expressed exclusively in all 27 EVNs. (B) A subset of EVN signature genes showed expression in only the male-specific head CEM (left) and tail HOB and RnB (right) neurons. In a and b, arrowheads point to cell bodies in the head and to ray dendrites in the tail. Scale bar, 10 μm. (C) asic-2 was expressed in the six IL2 neurons, egas-1 in four IL2 quadrant (IL2Q) neurons, clec-164 and F59A6.3 in CEM neurons. (D) Y70G10A.2 was expressed only in the HOB EVN and clec-179 in HOB and a few ray EVNs. In all panels, translational GFP reporters are shown by gene name or open reading frame number, transcriptional (promoter) GFP reporters are shown by gene name or ORF followed by p. For translational reporters, arrows point to ciliary localization. In A and C, dashed line boxes were 4X scaled with enhanced contrast and brightness to visualize EVs (green arrows). F14D7.11::GFP localized in cilia and secreted EVs from all 27 EVNs and ASIC-2::GFP localized in IL2 cilia and secreted EVs. (E) Domain analysis of validated genes. Signal peptide sequences were predicted by SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Mucin-domain containing proteins are predicted to be secreted and extensively O-glycosylated in the Serine (S) or Threonine (T) rich domain. Transmembrane domains were predicted by the TM Pred server (http://www.ch.embnet.org/software/TMPRED_form.html). CCP, C-type lectin (CLEC), Pan3, TGFβ, EGF-like, DEG/ENaC channel, vWA (Von Willebrand factor type A), globin, and UVR domains were predicted by Wormbase. Refer to Supplemental Table S4 listing EVN signature genes with predicted or demonstrated function in stress response or innate immunity.
TNF receptor associated factors (TRAFs) act as signaling adaptors for the TNFR superfamily, Toll like receptor and many other receptors[24]. We found that the C. elegans TRAF homologues trf-1 and trf-2 were overrepresented 8.6 fold in EVNs (Table S1) and exclusively expressed in the male-specific EVNs (Fig. 3A-D). TRF-1 is a canonical TRAF, with a RING domain, a TRAF zinc finger domain, and a MATH (meprin-associated TRAF homology) domain, while trf-2 (Y110A7.2) encodes a MATH domain only protein (Fig. 3A, C). GFP-tagged TRF-1 and TRF-2 localized through out male-specific EVNs including cilia, dendrites, cell bodies, and axons, but were excluded from nuclei (Fig. 3B, D). We did not observe TRF-1::GFP or TRF-2::GFP released in the extracellular environment, indicating that, unlike the polycystins LOV-1 and PKD-2, TRF-1 and TRF-2 are not EV-cargo.
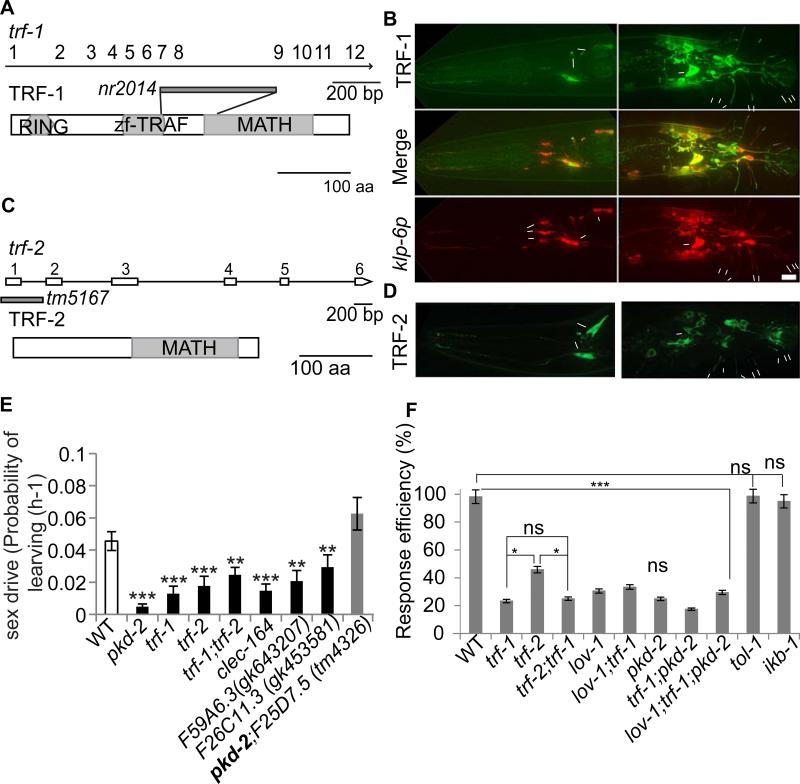
Figure 3. trf-1 and trf-2 coexpressed in male-specific EVNs and acted in the polycystin-signaling pathways.
(A, C) Schematic of trf-1 and trf-2 genomic and protein structure, with molecular lesions indicated in genomic structure. (A) The trf-1 locus encodes all common domains shared by mammalian TRAFs: a RING finger domain that may act as an E3 ubiquitin-ligase; a TRAF zinc finger and MATH domain that may mediate protein-protein interaction or oligomerization. The trf-1 (nr2041) mutation deletes part of the zf-TRAF domain and part of the MATH domain and may affect mRNA splicing. (C) TRF-2 has only a MATH domain. The trf-2(tm5167) deletion removes upstream of first exon to the end of the first intron. (B) trf-1 was co-expressed with klp-6 in male-specific, but not IL2, EVNs. (D) TRF-2::GFP was expressed and localized in the same pattern as TRF-1::GFP. (E) TRAF, lectins, and adhesion molecules were required for male sex drive. The tm4326 deletion allele of F25D7.5 suppresses the sex drive defect of pkd-2 mutant males (in bold). The probability of the male leaving the food lawn per hour PL(h-1) is indicated on the Y-axis, the male's genotype on the X-axis. Data was analyzed by Kruskal-Wallis test. * p<0.05, ** p<0.01, *** p<0.001. (F) Response Efficiency (RE) of lov-1, pkd-2, trf-1, trf-2 and Toll pathway genes tol-1 and ikb-1. Data was analyzed with Fisher's exact test, Bonferroni-Holm corrected. * p<0.05, *** p<0.001. ns not significant. The minimum number (n) of individual males analyzed per genotype was 37 for sex drive and 59 for response behavior.
trf-1 and trf-2 were required for all polycystin-mediated male behaviors: sex drive, response and vulva location (Fig. 3E-F, 4G). Neither trf-1 nor trf-2 was required for PKD-2::GFP localization to cilia or EVs, or for ciliogenesis in EVNs. The trf-2; trf-1 double mutant was not more response or location of vulva (Lov) defective than the trf-1 single mutant, indicating that trf-1 and trf-2 act non-redundantly. The lov-1; pkd-2; trf-1 triple mutant and lov-1; trf-1 and trf-1; pkd-2 doubles were indistinguishable from each other, indicating that polycystins and TRAFs act in the same genetic pathway (Fig. 3F). tol-1 (the sole Toll-like receptor in C. elegans) and ikb-1 (inhibitor of kappa beta homologue) were not overrepresented in EVNs and were not required for response or vulva location (Fig. 3E). These data indicate that the C. elegans TRAFs but not other components of the Toll pathway act in polycystin-mediated sensory signaling.
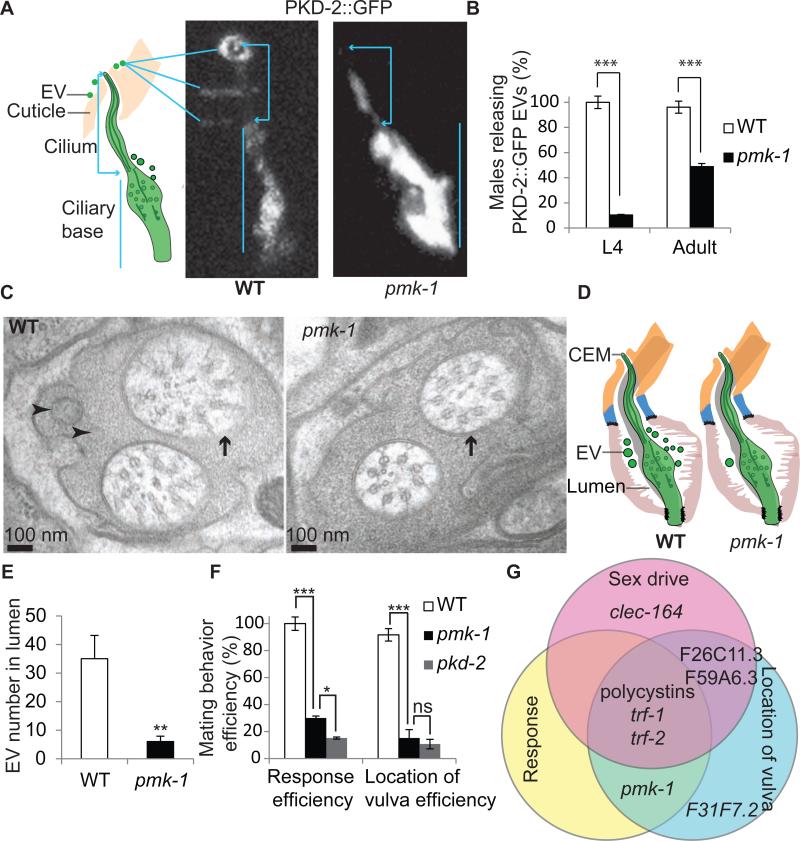
Figure 4. The p38 MAPK pmk-1 was required for EV biogenesis and polycystin-mediated response and vulva location behaviors.
(A) PKD-2::GFP CEM ciiary localization in wild-type and pmk-1 males. Cartoon shows the CEM cilium protruding from the cuticular pore and EVs surrounding the cuticle. In wild type, PKD-2::GFP localized to the ciliary base, cilium, and EVs released outsjde the male's nose. Note in wild type, EVs accumulated and surrounded the ciliary pore and cuticle. In pmk-1 males PKD-2::GFP localized normally to the ciliary base and cilium, but was absent from EVs. To show the full length of PKD-2::GFP labeled cilia and the EVs, images of wild type and pmk-1 males were taken with different exposure times, and adjusted with different brightness and contrast. The original images were enlarged six times by the Photoshop CS5 image transformation tool. Scale bar = 1 μm (B) pmk-1 mutant males were defective in PKD-2::GFP labeled EV release in both L4 (n=20 for wild-type, n=29 for pmk-1) and adult stage (n=52 for wild-type and n=43 for pmk-1). Data was analyzed with Fisher's exact test, Bonferroni-Holm corrected; *** p<0.001. (C, D) TEM and cartoon of wild-type and pmk-1 cephalic sensillum at the level of the ciliary transition zone. In wild type, EVs were present in the extracellular lumen. In pmk-1 males, EVs numbers are significantly reduced in the cephalic lumen. In pmk-1 males, the CEM axoneme contained eight rather then nine doublet microtubules of wild-type males. Black arrowheads = EVs, white arrows = CEM transition zone, black arrows = CEP transition zone. Scale bar = 100nm. (E) Bar graph of number of EVs (mean + SEM) in wild-type and pmk-1. Mann-Whitney test was used for statistical analysis; n=7 cephalic sensillae for wild type from two worms, n=4 cephalic sensillae for pmk-1 from one worm. **p=0.01. (F) pmk-1 mutant males are response and Lov defective. Statistical analysis was done by Fisher's exact test, Bonferroni-Holm corrected for response efficiency, RE (n=184 for wild type, 60 for pmk-1 and 264 for pkd-2), and one-way ANOVA for location of vulva efficiency, LE (n=20 for wild type, 20 for pmk-1 and 18 for pkd-2). * p<0.05, *** p<0.001. ns = not significant. (F) The C. elegans polycystins function in a multi-layered signaling network. Venn diagram showing polycystin-mediated mating behaviors are genetically separable. lov-1, pkd-2, and the TRAFs played a central role in sex drive, response, and vulva location while other genes act in specific behavioral pathways. Refer to Supplemental Table S5 for RE and LE of mutants corresponding to new EVN signature genes and pathways.
In addition to the TRAFs, a significant number of EVN signature genes were implicated in cellular stress or innate immune responses (Table S4). The stress-activated p38 MAPK homologue pmk-1 was overrepresented in EVNs. pmk-1 mutant males were defective in EV biogenesis and release (Fig. 4A-E). 100% of wild-type late L4 larval and adult males released PKD-2::GFP-containing EVs from the ciliated sensory neurons in the head (CEMs) and tail (RnBs and HOB) (Fig. 4A-B). In contrast, less than 10% of L4 pmk-1 and 50% of adult pmk-1 mutant males released PKD-2::GFP labeled EVs (Fig. 4B). Ciliary localization of PKD-2::GFP appeared similar between wild-type and pmk-1 males (Fig. 4A). We conclude that pmk-1 regulates release of PKD-2::GFP-containing EVs but not ciliary localization of PKD-2::GFP.
To examine pmk-1 ciliary and EV ultrastructure, we used serial thin cut transmission electron microscopy. In the wild-type cephalic sensillum, EVs are found in the lumen formed by glial-like support cells and surrounding the CEM and CEP cilia[6] (Fig. 4C-D). In the pmk-1 cephalic sensillum, EVs were largely absent from the lumen and CEM cilia had fewer doublets than the expected nine (Fig. 4C). The quantity of lumen EVs in pmk-1 mutant males was significantly reduced, indicating an EV biogenesis defect (Fig. 4D-E). pmk-1 mutant males were also response and Lov defective but displayed normal sex drive (Fig. 4F). Similarly, the EV release regulators klp-6 and cil-7 are required for response and vulva location behaviors but not sex drive[20]. For the EVN-specific genes regulating mating behaviors, response and vulva location depended on EV biogenesis or cilia-EV interactions, while sex drive was independent. These results also suggest that polycystin-mediated behaviors involve different signaling networks (Fig 4G).
In the innate immune response pmk-1 is the most downstream component in a MAP kinase cascade pathway[25]. However, expression of only pmk-1 but not the upstream kinase cascade genes tir-1, nsy-1 and sek-1 was overrepresented in EVNs. We examined tir-1, nsy-1 and sek-1 mutant males for mating behaviors and release of PKD-2::GFP containing-EVs and observed no defects (Table S4), indicating that pmk-1 acts via an unknown EV biogenesis pathway. In cultured mammalian cells, p38 MAPK is implicated in the production of EVs from macrophages after exposure to tobacco smoke, from aortic endothelial cells induced by TNF-α, and from glial cells [26-28]. We conclude that pmk-1 is a conserved regulator of EV biogenesis that acts independent of the innate immune MAPK cascade. These data also reveal the potential of EVN signature gene dataset to uncover new pathways in EV biology.
Our EVN-profile identified many transmembrane (TM) proteins (Table S1 with annotation of signal peptide + TM domains containing proteins) and several channels (Fig. 2E, Table S1), including two acid sensing/amiloride sensitive ion channels (ASICs). asic-2 and egas-1, are expressed in the shared IL2 neurons (Fig.2C). Neither asic-2 nor egas-1 was required for remodeling of IL2 neurons in the reproductively-arrested dauer state[29] or for male mating behaviors (Table S5). ASIC-2::GFP localized to IL2 cilia and EVs secreted from IL2 neurons in reproductively growing animals. Hence, our EVN-profile also identified EV cargo, including the known (LOV-1, PKD-2, CWP-1, and CIL-7)[6, 20] and new cargoes.
GFP-tagged F14D7.11 was also EV cargo, found in cilia and EVs released from all 27 EVNs (Fig. 2A), similar to the EV biogenesis regulator CIL-7 [20]. F14D7.11 resembles an antimicrobial peptide (AMP) and contains a CYSTM (cysteine rich transmembrane) module proposed to play a role in stress tolerance[30]. Our EVN-profile identified 13 AMPs, with F14D7.11 being the most highly overrepresented (Tables S1 and S4). Intriguingly, human urinary exosomes contain AMPs that have bactericidal activity[31]. An intriguing possibility is that EVs possess antimicrobial activity and play protective roles during the mating process. Alternatively, C. elegans could release EVs that have deleterious effects on other organisms. Consistent with this idea, unidentified C. elegans male secretions reduce hermaphrodite lifespan[32] and parasitic nematodes use EVs to transfer small RNAs to mammalian cells and modulate host innate immunity[33].
Several EVN signature genes encode lectins or adhesion molecules (Fig. 2E, Table S4). In our dataset, 13 clec genes were significantly overrepresented and do not overlap with those upregulated upon infection or expressed at higher levels in the bodies of adult males compared to hermaphrodites (Tables S1, S4). To determine whether these lectins or adhesion molecules play a role in polycystin-mediated behaviors, we examined their expression patterns (Fig. 2A-D) and functions in sex drive, response, and vulva location for available mutants (Fig. 3C, 4G). clec-164 was specifically expressed in the four CEMs and required for male sex drive (Fig. 2C, 3C). The single transmembrane (TM) protein F28A12.3 resembles an adhesion receptor with a cysteine-rich extracellular domain and was expressed in all 27 EVNs (Fig. 2A, 2E), yet mutant males displayed normal mating behaviors (Table S5). F25D7.5 and its paralog Y70G10A.2 are predicted to encode a long extracellular region containing a C-type lectin fold and EGF domain, followed by five TM spanning domains (Fig. 2E). F25D7.5 was expressed in 21 male-specific EVNs (Fig. 2B) and may negatively regulate sex drive, as its mutation suppressed pkd-2 sex drive but not response or Lov defects (Fig. 3E, Table S5). Y70G10A.2 was expressed exclusively in the HOB EVN (Fig. 2D) but not required for HOB-mediated vulva location behavior (Table S5). F26C11.3 and F59A6.3 encode secreted mucin-like serine-threonine rich proteins that contain a complement control protein (CCP) domain at their C-termini. F26C11.3 was expressed in 21 male-specific EVNs (Fig. 2B) while F59A6.3 was only expressed in the CEMs (Fig. 2C). F59A6.3 and F26C11.3 single mutants displayed decreased sex drive (Fig. 3E); the latter were also Lov (Table S5). F31F7.2, a predicted two TM protein with a von Willebrand factor type A domain, was expressed in all 27 EVNs (Fig. 2A) but required only for vulva location and not sex drive or response behavior (Table S5).
C. elegans EVNs, EVs, and EVN-expressed genes serve male reproductive signaling functions (Fig. 4G). The polycystin-expressing, male specific EVNs are required for sex drive, response, and vulva location. Some EVN-expressed genes (the TRAFs trf-1 and trf-2) were required for all polycystin-mediated behaviors, while other played more specific behavioral functions (Fig. 4G). For example, CEM-expressed clec-164 was required only for sex drive and pan EVN-expressed F31F7.2 only for vulva location. The p38 MAPK pmk-1 was required for response and vulva location behavior, but not sex drive. Intriguingly, the known regulators of EV biogenesis: pmk-1, cil-7, and klp-6 display this property[20], suggesting that luminal EVs may be required for the integrity of the male sensory organs mediating these behavioral functions. EVs play diabolical roles in the spread of toxic cargo in cancer, infectious diseases, and neurodegenerative disorders[10, 34]. We speculate that EVs may play a similar role ADPKD and other ciliopathies, We conclude that C. elegans polycystin-mediated signaling pathways are genetically separable, and that our EVN-expressed gene list identified new components and pathways regulating mating behavior as well as EV biogenesis, EV cargo sorting, and EV signaling.
Supplementary Material
Acknowledgements
We thank WormBase; the National Bioresource Project for strains; Christina DeCoste for assistance with FACS; Ken Nguyen, Leslie Gunther and Geoff Perumal for help in HPF-FS, embedding and serial section protocols performed at Einstein, and William Rice and Ed Eng at the New York Structural Biology Center (NYSBC), for help in electron tomography; Lillian Hunter for assistance with strain construction; members of the Barr and Murphy labs and Rutgers C. elegans community for discussion and insight, more than we ever learned in school; Dr. Emily Troemel for critical reading of the manuscript; Rutgers Genetics Department for sabbatical time and critical bridge funding. Use of the NYSBC facilities was supported by the Albert Einstein College of Medicine. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was funded by NIH DK059418 and DK074746 (to M.M.B.), NIH OD 010943 (to D.H.H.), and the Medical Research Council (to M.G.-N. and J.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes supplemental document (legends, experimental procedure, strain list, and references), one supplemental figure, five supplemental tables (two of which are data sets).
References
- 1.Wood CR, Rosenbaum JL. Ciliary ectosomes: transmissions from the cell's antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Hagan R, Wang J, Barr MM. Mating behavior, male sensory cilia, and polycystins in Caenorhabditis elegans. Semin Cell Dev Biol. 2014;33:25–33. doi: 10.1016/j.semcdb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, Charlesworth MC, Torres VE, LaRusso NF, Harris PC, Ward CJ. Characterization of PKD protein- positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans Ciliated Sensory Neurons Release Extracellular Vesicles that Function in Animal Communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakeberg JL, Tammachote R, Woollard JR, Hogan MC, Tuan HF, Li M, van Deursen JM, Wu Y, Huang BQ, Torres VE, Harris PC, Ward CJ. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol. 2011;22:2266–2277. doi: 10.1681/ASN.2010111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 9.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 11.Yoder BK, Hou X, Guay-Woodford LM. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- 12.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol. 2002;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 13.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Fedeles SV, Dong K, Anyatonwu G, Onoe T, Mitobe M, Gao JD, Okuhara D, Tian X, Gallagher AR, Tang Z, Xie X, Lalioti MD, Lee AH, Ehrlich BE, Somlo S. Altered trafficking and stability of polycystins underlie polycystic kidney disease. J Clin Invest. 2014;124:5129–44. doi: 10.1172/JCI67273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- 16.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 17.Barrios A, Nurrish S, Emmons SW. Sensory Regulation of C. elegans Male Mate-Searching Behavior. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portman DS, Emmons SW. Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev Biol. 2004;270:499–512. doi: 10.1016/j.ydbio.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Miller RM, Portman DS. A latent capacity of the C. elegans polycystins to disrupt sensory transduction is repressed by the single-pass ciliary membrane protein CWP-5. Dis Model Mech. 2010;3:441–450. doi: 10.1242/dmm.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguire JE, Silva M, Nguyen KC, Hellen E, Kern AD, Hall DH, Barr MM. Myristoylated CIL-7 Regulates Ciliary Extracellular Vesicle Biogenesis. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurd DD, Miller RM, Nunez L, Portman DS. Specific alpha- and beta- tubulin isotypes optimize the functions of sensory Cilia in Caenorhabditis elegans. Genetics. 2010;185:883–896. doi: 10.1534/genetics.110.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaletsky R, Williams AB, Arey R, Lakhina V, Landis JL, Murphy CT. Transcriptional profiling of isolated adult C. elegans neurons identifies the neuronal IIS/FOXO transcriptome and a new regulator of axon regeneration. in review. [Google Scholar]
- 23.Peden EM, Barr MM. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr Biol. 2005;15:394–404. doi: 10.1016/j.cub.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 24.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewbank JJ. Signaling in the immune response. 2006:1–12. doi: 10.1895/wormbook.1.83.1. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CJ, Liu Y, Chen Y, Yu D, Williams KJ, Liu ML. Novel proteolytic microvesicles released from human macrophages after exposure to tobacco smoke. Am J Pathol. 2013;182:1552–1562. doi: 10.1016/j.ajpath.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis AM, Wilkinson PF, Gui M, Gales TL, Hu E, Edelberg JM. p38 mitogen-activated protein kinase targets the production of proinflammatory endothelial microparticles. J Thromb Haemost. 2009;7:701–709. doi: 10.1111/j.1538-7836.2009.03304.x. [DOI] [PubMed] [Google Scholar]
- 28.Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, Matteoli M, Verderio C. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009;28:1043–1054. doi: 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder NE, Androwski RJ, Rashid A, Lee H, Lee J, Barr MM. Dauer-Specific Dendrite Arborization in C. elegans Is Regulated by KPC-1/Furin. Curr Biol. 2013;23:1527–1535. doi: 10.1016/j.cub.2013.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venancio TM, Aravind L. CYSTM, a novel cysteine-rich transmembrane module with a role in stress tolerance across eukaryotes. Bioinformatics. 2010;26:149–152. doi: 10.1093/bioinformatics/btp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiemstra TF, Charles PD, Gracia T, Hester SS, Gatto L, Al-Lamki R, Floto RA, Su Y, Skepper JN, Lilley KS, Karet Frankl FE. Human Urinary Exosomes as Innate Immune Effectors. J Am Soc Nephrol. 2014;25:2017–27. doi: 10.1681/ASN.2013101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, Ceroni A, Babayan SA, Blaxter M, Ivens A, Maizels RM. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vader P, Breakefield XO, Wood MJ. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med. 2014;20:385–93. doi: 10.1016/j.molmed.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.