Abstract
Introduction
Albumin, widely recognized as a highly sensitive and specific marker of hepatocellular carcinoma (HCC) is currently unavailable in the diagnostic laboratory because of the lack of a robust platform. In a prior study we detected albumin mRNA in the majority of intrahepatic cholangiocarcinomas using a novel branched chain RNA in situ hybridization (ISH) platform. We now explore the utility of albumin ISH as a marker of hepatocellular differentiation in hepatocellular carcinomas, and compare its sensitivity with Hep Par 1 and Arginase-1.
Methods
We evaluated 93 HCCs and its mimics including neuroendocrine tumors of the gastrointestinal tract (n= 31), neuroendocrine tumors of the pancreas (n= 163), melanoma (n= 15), and gallbladder carcinoma (n=34). We performed ISH for albumin and immunohistochemistry for Hep Par 1 and Arginase-1. Five previously uncharacterized hepatic neoplasms from our files were also evaluated. Immunohistochemistry for Arginase-1 was performed on 59 intrahepatic cholangiocarcinomas. In addition, 43 HCCs evaluated on the manual platform, were also examined on the automated instrument.
Results
55% of HCCs were moderately differentiated and 39% poorly differentiated. The sensitivity of ISH for Albumin was 99% with 92 of 93 of HCCs staining positive for albumin. In contrast to ISH, the sensitivity of immunohistochemistry for Hep Par1 and Arginase-1 was 84 % and 83 %, respectively. The sensitivity of albumin for poorly differentiated HCCs was 99%, while that for Arginase-1 and Hep Par 1 was 71% and 64%, respectively. 97% of the HCCs showed albumin positivity in >50% of tumor cells using the ISH platform, as compared to 76% and 70% for Hep Par 1 and Arginase-1 immunohistochemistry, respectively. 3 of the 5 previously uncharacterized neoplasms were positive for albumin ISH. Automated albumin ISH platform performed equivalently to the manual format, with albumin reactivity in >50% of tumor cells in all 43 cases that were tested on both platforms. All non- HCCs were negative for albumin. All 59 intrahepatic cholangiocarcinomas were negative for Arginase-1.
Conclusion
Branched chain ISH performed on manual and automated mode is a robust assay for detecting albumin with sensitivity for poorly differentiated HCC superior to Arginase-1 and Hep Par 1. When interpreted in conjunction with Arginase-1, albumin ISH offers a high level of sensitivity as well as specificity.
Keywords: In situ hybridization, albumin, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer related mortality worldwide. (1) The immunohistochemical diagnosis of poorly differentiated and undifferentiated HCC has traditionally been difficult because of a lack of a reliable marker of hepatocellular differentiation.(8, 17) Alpha fetoprotein (AFP) and polyclonal carcinoembryonic antigen (CEA) were the first of many markers of hepatocellular differentiation that have emerged in the last four decades (table 1). However, sensitivity of these markers is low - AFP ranges from 30% to 50%, and polyclonal CEA from 60% to 90%.(9) Background staining can make these markers difficult to interpret, and furthermore, they are more likely to be negative in poorly differentiated and undifferentiated carcinomas. (8, 17) Hep Par 1, a sensitive marker of hepatocellular differentiation represents a significant advance over prior assays. (2, 8, 9, 17) The bright granular cytoplasmic reactivity made interpretation relatively easy, and thus it quickly emerged as the ‘go to’ marker for hepatocellular differentiation. With its widespread adoption two conspicuous failings emerged (5): 1. The specificity, with a significant percentage of gastric, esophageal and lung adenocarcinomas show strong reactivity, a particularly troubling finding since these tumors frequently metastasize to the liver, 2. the sensitivity, especially for poorly differentiated HCCs which was significantly lower than for well-differentiated tumors, thus limiting its diagnostic value.
Table 1. Biomarkers for the detection of hepatocellular differentiation.
| Target | Sensitivity | Other tumors commonly positive | Poorly differentiated HCCs | |
|---|---|---|---|---|
| AFP2,10 | Oncofetal protein | 30-50% | Germ cell tumors | Typically negative |
| Hep Par 15,6 | Carbamoyl phosphatase synthetase (urea cycle) | >90%, staining may be patchy | Lung, colon, esophageal, and gastric carcinoma | <50% |
| CD10 and polyclonal CEA2,8 | Canalicular staining –cross reactivity to biliary glycoprotein | 60-90% | Diffuse cytoplasmic staining often seen in adenocarcinomas | 25% |
| Glypican 38 | Oncofetal antigen | 63-80% | Non seminomatous germ cells tumors, squamous cell carcinoma lung, liposarcoma, Melanoma | 57-83% |
| Arginase-115,23 | Enzyme involved in the hydrolysis of arginine to ornithine and urea | 96% | Rare | 86 |
| ISH Albumin12,13,14 | Major protein in serum | >95% | None | >95% |
More recently, Arginase-1, an enzyme involved in the hydrolysis of arginine to ornithine and urea, was recognized as a sensitive and specific marker for benign and malignant hepatocytes.(23, 24) Subsequent studies have validated its high sensitivity and specificity, as well as its value in evaluating poorly differentiated HCCs.(6, 11, 15, 21, 23, 24) Nevertheless, as evidenced in this study, the sensitivity of Arginase-1 in poorly differentiated HCCs is imperfect, and it is in this category that ancillary support is often required to bolster a diagnosis of HCC.
Albumin, a protein synthesized by hepatocytes, was first proposed as a marker of hepatocellular differentiation in the late 1980s.(10) However, immunohistochemical detection of albumin proved difficult primarily because of its ubiquitous presence, a problem easily overcome by detecting mRNA instead of protein.
(3, 4, 9, 12-14) However, despite the fact that albumin has the potential to be a highly sensitive and specific marker for HCC, it has not found widespread use in the diagnostic laboratory, driven by two unresolved problems: 1) the theoretical possibility that mRNA degrades far more quickly than proteins, and hence a mRNA based biomarker is likely to be less sensitive than immunohistochemistry, and 2) lack of a robust and sensitive automated platform for the detection of RNA that could be adapted for the clinical laboratory.
The introduction of a branched chain in situ hybridization (ISH) assay addresses one of the issues - the assay achieves a high level of sensitivity by amplifying the RNA signal. We have recently utilized this approach for the diagnosis of intrahepatic cholangiocarcinoma (Ferrone et al., submitted), where albumin ISH serves as an indicator of hepatic origin. In this study, we compare albumin to the two leading markers of hepatocellular differentiation - Arginase-1 and Hep Par 1- when used for the diagnosis of HCC in formalin fixed biopsy material. We also assess an automation platform for the detection of albumin mRNA that would potentially allow the adoption of this technology in routine pathology practice.
Materials and Methods
Case selection
The study cases were selected based on a search of the pathology database at the Massachusetts General Hospital from 2003 through 2013. The study was approved by the Institutional Review Board at the Massachusetts General Hospital.
Hepatocellular carcinomas
Tissue microarrays from 94 HCCs were constructed. The arrays were composed of 3 mm cores of paraffin embedded tissue. Moderately and poorly differentiated HCCs were overrepresented in this cohort. Other than 3 tumors that showed clear cells, the other cases represented conventional HCCs. Expression of Arginase-1 and Hep Par 1 was examined on an immunohistochemical platform, while that for albumin on an ISH platform –ViewRNA, Affymetrix, CA (see below for details). The HCCs were graded as well, moderate, and poorly differentiated.(19) The original diagnosis of poorly differentiated HCC was based on the histologic features, specifically the lack of evidence of cholangiocellular differentiation, supplemented in selected cases by the use of immunohistochemistry. The cirrhotic environment within which these tumors arose provided additional support to the diagnosis. 22 examples of benign liver adjacent to the tumor were also evaluated for albumin; 14 cases showed cirrhosis.
Three hepatoid carcinomas, one each from the ovary, stomach, and from the pancreas were also evaluated. Six additional examples of HCC from 2 institutions (UCSF and VA Medical Center, San Francisco, CA and University Hospital Southampton NHS Foundation Trust, Southampton, UK) were also evaluated. Entire sections were evaluated on these cases, as well as the hepatoid carcinomas.
Intrahepatic cholangiocarcinomas
59 resected intrahepatic cholangiocarcinomas were tested for Arginase-1. These tumors have been previously tested by albumin ISH (Ferrone et al, submitted). The intrahepatic cholangiocarcinomas were distinguished from perihilar adenocarcinomas based on the epicenter of the lesion, as determined by imaging, gross and microscopic evaluation. The tumors were placed on a tissue microarrays composed of 3 mm cores of tissue.
Uncharacterized tumors
We examined 5 hepatic neoplasms over a 5-year period from the institutional files of one of the authors (VD) that were classified as undifferentiated carcinomas, and lacked histological and immunohistochemical evidence of hepatocellular differentiation. Entire sections were examined for albumin in this class of tumors.
Non-hepatic tumors
We examined mimics of HCC including neuroendocrine tumors of the gastrointestinal tract (n= 31), neuroendocrine tumors of the pancreas (n= 163), melanoma (n= 15), and gallbladder carcinoma (n=34). The tumors were placed on a tissue microarrays composed of 3 mm cores of tissue.
Immunohistochemistry
Immunohistochemistry for Hep Par 1 (Dako 1:25; Retrieval EDTA pH 9.0 for 20 mins) and Arginase (Cell Marque 1:600) (Retrieval Citrate pH 6.0 for 30 mins) was performed. Details of the procedure are provided in a prior publication from this institution.(18)
In situ hybridization
ISH was performed using both the manual and automated ViewRNA™ platform (Affymetrix, Santa Clara, CA), and results were compared where indicated. This technology utilizes a branched DNA structure for signal amplification to enable detection of RNA in formalin fixed paraffin embedded (FFPE) tissue. Briefly, multiple tiling probes for a gene are hybridized to a specific target RNA. A signal amplification structure is created via a series of hybridization steps where a PreAmplifier (PreAMP) molecule hybridizes to each oligonucleotide probe pair, multiple Amplifier (AMP) molecules hybridize to each pre-amplifier, and multiple alkaline phosphatase -conjugated-label probes (AP-LP) hybridize to each Amplifier (Supplementary material 1,Supplemental Digital Content 1, http://links.lww.com/PAS/A244 ). The branched structure has approximately 400 binding sites for the AP-labeled probe, which can then be visualized with the fast-red substrate as a chromogen to form a red dot or diffuse precipitate that allows the detection of RNA transcripts in situ.
ISH probe (Affymetrix, Santa Clara, CA) was designed against albumin transcripts as identified in the NCBI nucleotide database (Supplementary material 2, Supplemental Digital Content 2, http://links.lww.com/PAS/A245 ). FFPE tissues were sectioned at 5 +/- 1 micron and mounted on Surgipath X-tra glass slides (Leica BioSystems, Buffalo grove, IL), baked for 1 hour at 60°C to ensure tissue attachment to the glass slides, and stored at -80°C until ready for ISH staining. ISH was performed in the manual and automated format as described below.
Manual ISH Assay
For the manual (non-automated) format for ISH, baked tissue sections were subjected to xylene deparaffinization followed by ethanol dehydration. To unmask the RNA targets, dewaxed sections were incubated in 500 ml pretreatment buffer at 90-95°C for 10 minutes and digested with 1:100 dilution protease at 40°C for 10 minutes, followed by fixation with 10% neutral buffered formlin at room temperature for 5 minutes. Unmasked tissue sections were subsequently hybridized with 1:50 dilution of the albumin probe set for 2 hours at 40°C, followed by series of post-hybridization washes. Signal amplification was achieved via a series of sequential hybridizations and washes as described in the user manual (see below). The specific conditions were as follow: pre-amplifier: 25 min at 40°C; amplifier: 15 min at 40°C; hybridization with labeled probe: 1:1000 dilution for 15 min at 40°C; signal detection with Fast Red substrate: 30 min at 40°C. Slides were post-fixed in 10% neutral buffered formalin, counterstained with Gill's hematoxylin, mounted using Dako Ultramount (Dako, Carpinteria, CA), and visualized using a standard bright-field microscope. Additional details of the assay are provided at -https://www.panomics.com/downloads/15891_RevB%2013%2008%2030_ViewRNA%20eZ%20Assay%20Manual_low%20res3.pdf
Automated albumin ISH assay
43 cases evaluated on the manual assay were repeated on the automated platform. In 20 cases, entire sections were evaluated, while 3 mm microarray samples were evaluated in the other 23 cases.
The albumin probe used in the automated assay was identical to that used in the manual assay. Automated ISH assays for albumin mRNA were performed using ViewRNA eZ Detection Kit (Affymetrix, Santa Clara, CA) on the Bond RX immunohistochemistry and ISH Staining System with BDZ 6.0 software (Leica Biosystems, Buffalo grove, IL). Paraffin embedded tissue sections on slides were processed automatically from deparaffinization, through ISH staining to hematoxylin counterstaining; sections were coverslipped off-instrument. Briefly, 5 micron sections of formalin fixed tissue were baked for 1 hour at 60°C and placed on the Bond RX for processing. The Bond RX user-selectable settings were as follows: ViewRNA 1 protocol; ViewRNA Dewax1; ViewRNA HIER 10 min, ER1 (95); ViewRNA Enzyme1 (20); ViewRNA Probe Hybridization. With these settings, the RNA unmasking conditions for the liver tissue consisted of a 10 min incubation at 95°C in Bond Epitope Retrieval Solution 1 (Leica Biosystems, Buffalo grove, IL) followed by 20 min incubation with Proteinase K from the Bond Enzyme Pretreatment Kit at 1:1000 dilution (Leica Biosystems, Buffalo Grove, IL). Human albumin (Cat#ZVA1-16742) and a house keeping mRNA-targeting Probe Sets composed of a cocktail of GAPDH, PPIB and ACTB were diluted 1:20 in ViewRNA Probe Diluent (Affymetrix, Santa Clara, CA); house keeping gene set were evaluated in selected cases. Post-run, slides were rinsed with water; air dried for 30 min at room temperature, dipped in xylene and mounted using HistoMount solution (Life Technologies, Grand Island, NY). Additional details on the automation are provided at - https://www.panomics.com/downloads/UM18849_RevB_QGViewRNA_ISH_Tissue_2-plex_ThermoBrite_121014.pdf
Quantification
A semi-quantitative method of scoring was devised and used to score both immunohistochemistry and in situ hybridization. The quantitative analysis was performed only on the tumor tissue samples on the microarray. Tumors demonstrating no staining at all were given a score of 0; < 5% of cells staining – score 1+, 5% to 50% - score 2+ and more than 50% - score 3.
Results
Albumin (in situ hybridization)
Normal liver
Similar to a prior study (Ferrone et. al. submitted) the 22 examples of benign liver (14 with cirrhosis)– tissue adjacent to the hepatocellular carcinomas were diffusely positive for albumin. Periportal hepatocytes stained more intensely than zone 3 hepatocytes. While the interlobular bile ducts were occasionally positive, bile ductules were uniformly positive for albumin.
Hepatocellular carcinoma
In order to determine the utility of albumin ISH for the diagnosis of hepatocellular carcinoma we tested a cohort of 94 additional HCC cases selected for a wide range of hepatocellular differentiation. The mean age of the cohort of patients with HCC was 64 years (range 44 -81) with 65 males and 28 females. The majority of the HCCs were moderately differentiated (55%) or poorly differentiated (39%). The background liver showed cirrhosis in 83 cases.
92 of 94 HCCs were positive for albumin (Figures 1, 2 and 3) (table 2). One of the tumors negative for albumin failed GAPDH testing and was not included in the final analysis. The suboptimal mRNA preservation was likely related to radiofrequency ablation. Among the positive cases, 89 cases showed reactivity in >50% of the tumor, and all 92 cases showed characteristic dot like reactivity in > 5% of the tumor cells. All 3 examples of HCC with clear cells were positive for albumin with >50% of the tumor positive in all 3 cases. We also evaluated 6 additional HCCs from 2 other institutions, all of which were diffusely positive for albumin. 5 of these tumors were poorly differentiated and one moderately differentiated.
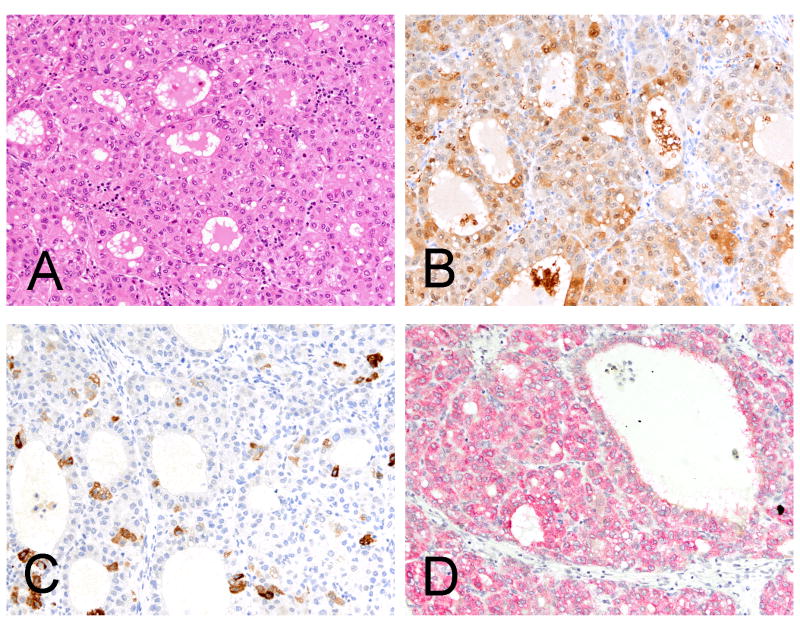
Figure 1.
A moderately differentiated hepatocellular carcinoma (A, H&E) with diffuse reactivity for Arginase-1 and focal reactivity for Hep Par 1 (B, immunohistochemistry Arginase-1 and C, immunohistochemistry Hep Par 1). The in situ hybridization stain for albumin is diffusely and strongly positive (D, in situ hybridization albumin).
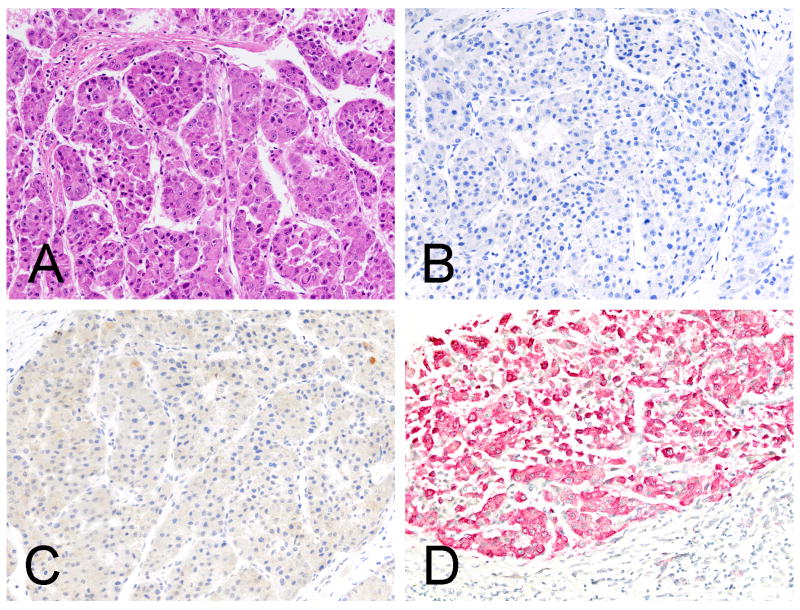
Figure 2.
A moderately differentiated hepatocellular carcinoma (A, H&E) negative for Arginase-1 and Hep Par 1, respectively (B, immunohistochemistry Arginase-1 and C, immunohistochemistry Hep Par 1). The in situ hybridization stain for albumin is diffusely and strongly positive (D, in situ hybridization albumin).
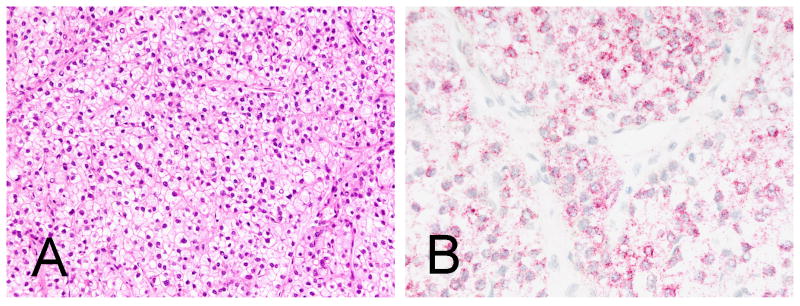
Figure 3.
Hepatocellular carcinoma with clear cells (A, H&E) is diffusely and strongly positive for albumin (B, in situ hybridization for albumin). Note the dot like pattern of reactivity.
Table 2. Comparison of bISH for albumin, Hep Par1 and Arginase for the diagnosis of hepatocellular carcinoma.
| ALBUMIN + | HEP PAR1+ | ARGINASE+ | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N=93 | N=89 | N=87 | |||||||||||||
| %+ | Score0 | Score1 | Score2 | Score3 | %+ | Score0 | Score1 | Score2 | Score3 | %+ | Score0 | Score1 | Score2 | Score3 | |
| All cases | 99% | 1 | 0 | 3 | 89 | 84% | 14 | 7 | 8 | 60 | 83% | 15 | 11 | 18 | 43 |
| Grade 1 | 100% | 0 | 0 | 0 | 6 | 100% | 0 | 0 | 0 | 4 | 100% | 0 | 1 | 2 | 1 |
| Grade 2 | 100% | 0 | 0 | 1 | 50 | 98% | 1 | 5 | 4 | 39 | 90% | 5 | 5 | 11 | 27 |
| Grade 3 | 99% | 1 | 0 | 2 | 33 | 64% | 13 | 2 | 4 | 17 | 71% | 10 | 5 | 5 | 15 |
Score 1=< 5% of tumor staining positive
Score 2=5% to 50% of tumor staining positive
Score 3> 50% of tumor staining positive
Albumin was evaluated using in situ hybridization
Hep Par1 and Arginase-1 was evaluated on an immunohistochemistry platform
Grade = grade of differentiation
In radiofrequency ablated tumors, the necrotic regions of tumor, both foci of coagulated and liquifactive necrosis, were negative for albumin. The adjacent foci of viable tumor were strongly positive for albumin. In addition, tissues demonstrating visible electrothermy artifact showed considerable decrease in the intensity of reactivity for albumin.
Comparison of ISH albumin with immunohistochemistry for Hep Par 1 and Arginase-1
In addition to the presence or absence of reactivity, a semi-quantitative analysis was performed to document the percentage of tumor cells staining on each preparation. The semi-quantitative analysis was performed only on the tissue microarray samples of HCC.
Hep Par 1 (immunohistochemistry)
75 of the 89 HCCs were positive for Hep Par1. The sensitivity of the assay was 84%. 7 of the 75 (9%) positive cases showed focal (1+) reactivity. Among the 14 tumors negative for Hep Par 1, 1 was moderately differentiated and 13 poorly differentiated.
Arginase-1 (immunohistochemistry)
72 of the 87 HCCs were positive for Arginase-1 - sensitivity of the assay was 83%. 11 (15%) tumors that were positive for Arginase-1 were only focally (1+) positive. Among the 15 tumors negative for Arginase-1, 5 were moderately differentiated and 10 poorly differentiated.
Albumin (in situ hybridization) vs. Hep Par 1 and Arginase-1 (immunohistochemistry)
The only HCC (poorly differentiated) that was negative for albumin was positive for both Hep Par 1 and Arginase-1. The 9 tumors that were negative for both Hep Par1 and Arginase-1, were positive for albumin - 8 of these tumors were grade 3.
Hepatoid carcinoma
Although not the primary focus of this paper, we evaluated 3 cases of hepatoid carcinomas, one each from the ovary, pancreas, and the stomach, all of which showed diffuse positivity using Albumin ISH. In comparison, 2 of the 3 tumors were positive for Hep Par 1 while all of the 3 tumors were negative for Arginase-1. The one tumor (pancreas) that was negative for Hep Par1 showed diffuse strong positive staining using AFP immunohistochemistry (Figure 4).
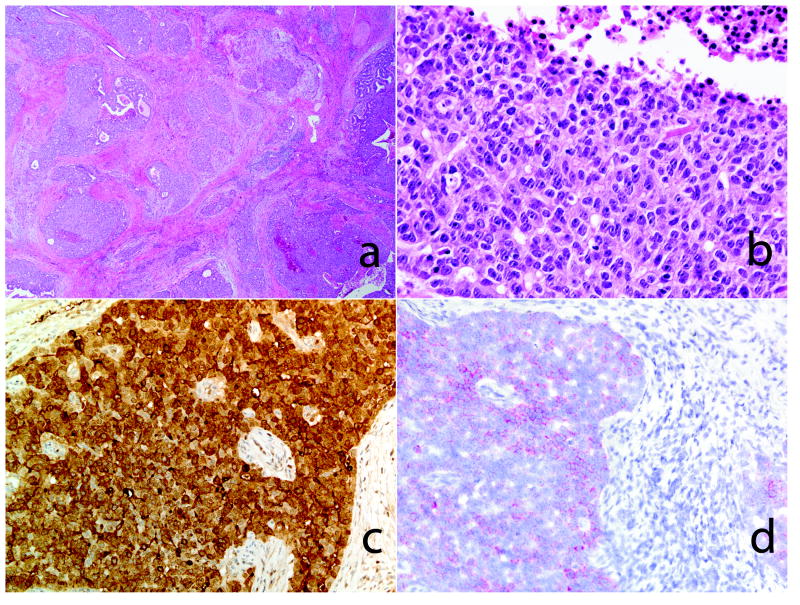
Figure 4.
Hepatoid carcinoma of the pancreas. Low and high power view of the pancreatic neoplasm (A&B, H&E stain). Overt morphological features of hepatocellular differentiation are not seen. An immunohistochemical stain for alpha feto-protein (AFP) was strongly positive (C, AFP, immunohistochemistry). The tumor cells were positive for albumin (D, in situ hybridization).
Intrahepatic cholangiocarcinomas
The mean age of this cohort was 64 years (range 37-77); M:F =1:1. The median tumor size was 5.5 cm. We previously found these tumors to be positive for albumin ISH (Ferrone et al, submitted). The 59 tumors were evaluated on an immunohistochemical platform for Arginase-1 and 1 was found to be positive for this marker.
Automated platform
In order to assess the utility of albumin ISH for routine pathology practice, 43 HCCs (26 moderate and 17 poorly differentiated) that stained positive for Albumin using the manual ISH platform were re-tested on the automated Leica Bond RX platform. Using the manual staining platform, all cases had shown albumin ISH staining in >50% of the tumor component. Albumin ISH using the automated platform showed similar staining pattern to what was seen using the manual platform for all tested cases, and the concordance between the 2 platforms was 100%.
Specificity of albumin as a marker of hepatocellular differentiation
We further evaluated cases that could potentially mimic HCC, including neuroendocrine tumors of the gastrointestinal tract, neuroendocrine tumors of the pancreas, melanoma, and gallbladder carcinoma (n=143). These tumors were uniformly negative for albumin.
Difficult to Characterize Hepatic Neoplasms
Of the 5 undifferentiated tumors, 3 were positive for albumin (Figure 5) (table 3). One of the cases negative for ISH was eventually diagnosed as gallbladder carcinoma. The biopsy from case 3 was largely necrotic tissue; the rare viable cells were negative for albumin. In retrospect, the presence of Arginase-1 (in one case) and an elevated AFP level in all 3 cases support the diagnosis of HCC.
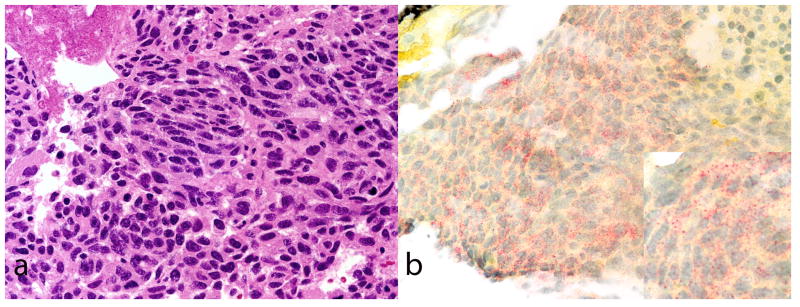
Figure 5.
A poorly differentiated hepatocellular carcinoma (A, H&E) that was negative for Arginase-1 and Hep Par-1 (not shown) is positive for albumin (B, in situ hybridization for albumin). Note the dot like pattern of reactivity.
Table 3. bISH for albumin in selected challenging hepatic neoplasms.
| Age | Sex | 0riginal diagnosis | Albumin | 0ther IHCs | Heppar1 | Serum AFP (ng/ml) | Cirrhosis | Final Diagnosis | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 52 | M | Poorly differentiated carcinoma | Positive | Positive: AE1/3, Cam5.2, CDX2, Arginase-1Negative: CD10,CEA, AFP | Negative | 2782 | Absent | Hepatocellular Carcinoma |
| 2 | 82 | M | Poorly differentiated carcinoma (gallbladder fossa) | Negative | None | 1.2 | Absent | Gallbladder carcinoma | |
| 3 | 52 | M | Poorly differentiated carcinoma (largely necrotic) | Negative | Positive: keratin 19Negative: AFP, CK7, Arginase-1 | Negative | 1572 | Present | Hepatocellular Carcinoma |
| 4 | 52 | M | Poorly differentiated carcinoma | Positive | Positive: CK19Negative: Hep par1, HMB45, LCA, S100,, ArginaseVimentin | Negative | 2397.3 | Absent | Hepatocellular Carcinoma |
| 5 | 75 | M | Undifferentiated carcinoma | Positive | Positive: AFP, AE1/3, Cam5.2Negative: Glypican3, B catenin, Polyclonal CEA, CK7, CK20, Arginase-1Ki67-90% | Negative | n/a | Absent | Hepatocellular Carcinoma |
Discussion
The two common challenges encountered by the pathologist evaluating a hepatic neoplasm are: 1) the distinction between a benign and malignant hepatic neoplasm, and 2) at the poorly differentiated/undifferentiated end of the spectrum, distinguishing HCCs from metastatic adenocarcinoma. There are a wide variety of markers that assist in unraveling the latter diagnostic problem including AFP, polyclonal CEA, CD10, Hep Par-1, and Arginase-1 (table 1).
Amongst these, Arginase-1 has emerged as the marker of choice - its high sensitivity for hepatocellular differentiation combined with the virtual absence of reactivity of non-hepatic neoplasms makes it an almost ideal marker.(24) Importantly, Arginase-1 also offers a high sensitivity for poorly differentiated HCC - 86% in a recent analysis.(24) In that study, Arginase-1 outperformed Hep Par 1 with an overall sensitivity of 96% and specificity of 84%, respectively.(24) Among poorly differentiated HCCs, however, the two stains may prove complimentary, since a significant percentage of HCCs negative for Arginase-1 were positive for Hep Par 1.
Our study suggests that albumin ISH offers sensitivity for poorly differentiated HCC superior to Arginase-1. A higher percentage of tumor cells were positive for albumin. Additionally, almost a fifth of the tumors that were positive for Arginase-1 showed only faint reactivity for this protein. In our experience this low intensity reactivity is often best regarded as equivocal evidence of hepatocellular differentiation. The red dot-like staining pattern makes the ISH assay easier to interpret than immunohistochemistry. The interpretation was further simplified by the virtual absence of non-specific signal. It should be noted that this analysis was performed on routinely processed paraffin embedded tissue, and there were no specific efforts to ensure preservation of RNA. Poorly differentiated hepatocellular neoplasms were overrepresented in this cohort, constituting 39% of cases. This was deliberately done to test the assay on a cohort of cases most likely requiring the use of ancillary testing. Within the class of poorly differentiated HCCs, all but 1 poorly differentiated HCC stained positively for albumin. In comparison 71% and 64 % of poorly differentiated HCCs stained for Arginase-1 and Hep Par 1, respectively.
Graham and coworkers using a platform that is likely similar to that presented in this study, found similar results with majority of the HCCs being positive for albumin.(7) However, the results of this study, published only in an abstract form, found that the sensitivity of Arginase-1 and albumin to be comparable. The reason underlying these differences is uncertain, particularly since technical details are unavailable.(7)
In conjunction with a recent study on intrahepatic cholangiocarcinomas by our group (Ferrone et al, submitted), we confirm that the increased sensitivity of the ISH assay does not lead to a loss of specificity, and a diverse group of carcinomas including those arising from the lung, breast, esophagus, stomach, colon, ovary, renal cell carcinoma and urinary bladder lacked reactivity for albumin (16). Other potential mimics of HCC tested in the present study, including melanoma and neuroendocrine tumors, were also negative for albumin. However, albumin reactivity does not distinguish between HCC and cholangiocarcinoma: based on our data the majority of intrahepatic cholangiocarcinomas are also positive for albumin, albeit at a lower intensity (16). Arginase-1 appears to represent a more specific marker for distinguishing HCC from cholangiocarcinoma, however, occasional cholangiocarcinomas were positive in our study (1 of 59 cases) as well as in other studies (2 of 18 cases). (15,24)
Another potential application of the assay is the diagnosis of hepatoid carcinomas. The current ancillary tests for the diagnosis of hepatoid carcinoma is far from satisfactory - Arginase-1 is generally negative, while Hep Par 1 lacks the required specificity to distinguish between adenocarcinomas and hepatoid carcinomas.(15, 21) Nevertheless, a larger cohort of hepatoid carcinomas must be analyzed before employing albumin as a definitive marker of hepatoid carcinoma. Although, we did not stain examples of hepatocellular dysplasia and hepatic adenomas, we anticipate that these neoplasms, like all neoplasms with evidence of hepatocellular differentiation would stain positively for albumin. Thus, this assay cannot distinguish HCC from adenoma and hepatic dysplasia.
Our results are similar to prior efforts at detecting albumin in hepatic neoplasms.(3, 4, 9, 12-14, 22) The current in situ hybridization platform, however, differs considerably from the albumin ISH assay introduced in the 1990s. In this prior assay, oligonucleotide probes were labeled with digoxigenin while the current assay amplifies the mRNA signal using branched DNA amplification, theoretically allowing for the detection of a single copy of RNA and considerably increasing the sensitivity of the assay. While high sensitivity for HCC can be achieved with traditional ISH assays (93-95%)(9, 12, 14), difficulties of implementing and automating this technique have prevented its wide acceptance. As demonstrated in this study a branched chain ISH approach can be readily automated and detects albumin in poorly differentiated cases. Signal amplification provides a means of uncovering albumin expression at low levels and accounts for the almost perfect sensitivity of the current assay.(12-14, 20)
We validate our findings by evaluating a cohort of poorly differentiated/undifferentiated carcinomas of the liver. The five cases that lacked unequivocal histological or immunohistochemical evidence of hepatocellular differentiation were identified in the files of one of the authors (VD). Three of the five cases were strongly (>50% tumor cells stained) positive for albumin (although in retrospect Arginase-1 was positive in 1 case). Case number 2 represented a gallbladder carcinoma, and was negative for albumin. However, the possibility that some of these tumors represent an undifferentiated hepatic tumor that do not display hepatic differentiation (such as a cholangiocarcinoma) cannot be excluded, although this is unlikely given the markedly elevated alpha-fetoprotein level.
The ISH platform used here offers pathologists a novel means of detecting mRNA, including targets that are expressed at low levels. The availability of this technology is particularly valuable for detecting secreted proteins such as albumin, as well as targets against which an immunohistochemical approach has been unsuccessful. The bright red dots generated by this technology make the assessment of these preparations straightforward. It should be emphasized that the presence of even rare dots (> 3 per cell) is strong evidence of a positive result since non-specific signal (in our experience < 1 dot per 10 cells) is minimal.
To conclude, branched chain albumin ISH offers a robust means of detecting a tumor of liver origin, and is particularly valuable when confronted with a poorly differentiated HCC. However, the assay cannot distinguish HCCs from intrahepatic cholangiocarcinomas. The combined use of albumin and Arginase-1 could represent a powerful means of identifying tumors with hepatocellular differentiation- with tumors that are positive for albumin but lack Arginase-1, a careful evaluation of the morphological features is necessary to exclude an intrahepatic cholangiocarcinoma. The ability to automate the assay opens up the prospect of its use in the diagnostic clinical laboratory.
Supplementary Material
Acknowledgments
This work was supported by Howard Hughes Medical Institute (M.N.R.), the Burroughs Wellcome Trust (D.T.T., M.N.R.), K12CA087723-11A1 (D.T.T.), and Affymetrix, Inc. (D.T.T., M.N.R., V.D.).
References
- 1.Bosch FX, Ribes J, Diaz M, et al. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Chu PG, Ishizawa S, Wu E, et al. Hepatocyte antigen as a marker of hepatocellular carcinoma: an immunohistochemical comparison to carcinoembryonic antigen, CD10, and alpha-fetoprotein. Am J Surg Pathol. 2002;26:978–988. doi: 10.1097/00000478-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 3.D'Errico A, Baccarini P, Fiorentino M, et al. Histogenesis of primary liver carcinomas: strengths and weaknesses of cytokeratin profile and albumin mRNA detection. Hum Pathol. 1996;27:599–604. doi: 10.1016/s0046-8177(96)90169-0. [DOI] [PubMed] [Google Scholar]
- 4.D'Errico A, Deleonardi G, Fiorentino M, et al. Diagnostic implications of albumin messenger RNA detection and cytokeratin pattern in benign hepatic lesions and biliary cystadenocarcinoma. Diagn Mol Pathol. 1998;7:289–294. doi: 10.1097/00019606-199812000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Fan Z, van de Rijn M, Montgomery K, et al. Hep par 1 antibody stain for the differential diagnosis of hepatocellular carcinoma: 676 tumors tested using tissue microarrays and conventional tissue sections. Mod Pathol. 2003;16:137–144. doi: 10.1097/01.MP.0000052103.13730.20. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara M, Kwok S, Yano H, et al. Arginase-1 is a more sensitive marker of hepatic differentiation than HepPar-1 and glypican-3 in fine-needle aspiration biopsies. Cancer Cytopathol. 2012;120:230–237. doi: 10.1002/cncy.21190. [DOI] [PubMed] [Google Scholar]
- 7.Graham RP, Vrana ME, Law KL, et al. Comparison of novel in situ hybridization RNA scope method to other markers of hepatocellular phenotype. Modern Pathology. 2014;27(Supplement 2):421A. [Google Scholar]
- 8.Kakar S, Gown AM, Goodman ZD, et al. Best practices in diagnostic immunohistochemistry: hepatocellular carcinoma versus metastatic neoplasms. Arch Pathol Lab Med. 2007;131:1648–1654. doi: 10.5858/2007-131-1648-BPIDIH. [DOI] [PubMed] [Google Scholar]
- 9.Kakar S, Muir T, Murphy LM, et al. Immunoreactivity of Hep Par 1 in hepatic and extrahepatic tumors and its correlation with albumin in situ hybridization in hepatocellular carcinoma. Am J Clin Pathol. 2003;119:361–366. doi: 10.1309/8l872rphejrkf5jj. [DOI] [PubMed] [Google Scholar]
- 10.Kojiro M, Kawano Y, Isomura T, et al. Distribution of albumin- and/or alpha-fetoprotein positive cells in hepatocellular carcinoma. Laboratory investigation. 1981;44:221–226. [PubMed] [Google Scholar]
- 11.Krings G, Ramachandran R, Jain D, et al. Immunohistochemical pitfalls and the importance of glypican 3 and arginase in the diagnosis of scirrhous hepatocellular carcinoma. Mod Pathol. 2013;26:782–791. doi: 10.1038/modpathol.2012.243. [DOI] [PubMed] [Google Scholar]
- 12.Krishna M, Lloyd RV, Batts KP. Detection of albumin messenger RNA in hepatic and extrahepatic neoplasms. A marker of hepatocellular differentiation. Am J Surg Pathol. 1997;21:147–152. doi: 10.1097/00000478-199702000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Murray GI, Paterson PJ, Ewen SW, et al. In situ hybridisation of albumin mRNA in normal liver and hepatocellular carcinoma with a digoxigenin labelled oligonucleotide probe. J Clin Pathol. 1992;45:21–24. doi: 10.1136/jcp.45.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira AM, Erickson LA, Burgart LJ, et al. Differentiation of primary and metastatic clear cell tumors in the liver by in situ hybridization for albumin messenger RNA. Am J Surg Pathol. 2000;24:177–182. doi: 10.1097/00000478-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Radwan NA, Ahmed NS. The diagnostic value of arginase-1 immunostaining in differentiating hepatocellular carcinoma from metastatic carcinoma and cholangiocarcinoma as compared to HepPar-1. Diagn Pathol. 2012;7:149. doi: 10.1186/1746-1596-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice-Stitt T, Mubeen A, Shahid M, et al. Chromogenic In Situ Hybridization for Albumin Distinguishes Intrahepatic Cholangiocarcinoma from Non-Hepatic Neoplasms. Laboratory Investigation. 2014;94:199A. [Google Scholar]
- 17.Roskams T, Katoonizadeh A, Komuta M. Hepatic progenitor cells: an update. Clin Liver Dis. 2010;14:705–718. doi: 10.1016/j.cld.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Routhier CA, Mochel MC, Lynch K, et al. Comparison of 2 monoclonal antibodies for immunohistochemical detection of BRAF V600E mutation in malignant melanoma, pulmonary carcinoma, gastrointestinal carcinoma, thyroid carcinoma, and gliomas. Hum Pathol. 2013;44:2563–2570. doi: 10.1016/j.humpath.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Theise ND, Curado MP, Franceschi S, et al. Hepatocellular carcinoma. In: Bosman FT, Carneiro F, Hruban RH, et al., editors. WHO Classification of Tumours of the Diagestive System. Lyon, France: International Agency for Reseach on Cancer; 2010. pp. 205–216. [Google Scholar]
- 20.Tickoo SK, Zee SY, Obiekwe S, et al. Combined hepatocellular-cholangiocarcinoma: a histopathologic, immunohistochemical, and in situ hybridization study. Am J Surg Pathol. 2002;26:989–997. doi: 10.1097/00000478-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Timek DT, Shi J, Liu H, et al. Arginase-1, HepPar-1, and Glypican-3 are the most effective panel of markers in distinguishing hepatocellular carcinoma from metastatic tumor on fine-needle aspiration specimens. Am J Clin Pathol. 2012;138:203–210. doi: 10.1309/AJCPK1ZC9WNHCCMU. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi K, Nalesnik MA, Carr BI. In situ hybridization of albumin mRNA in normal liver and liver tumors: identification of hepatocellular origin. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:361–365. doi: 10.1007/BF02915135. [DOI] [PubMed] [Google Scholar]
- 23.Yan BC, Gong C, Song J, et al. Arginase-1: a new immunohistochemical marker of hepatocytes and hepatocellular neoplasms. Am J Surg Pathol. 2010;34:1147–1154. doi: 10.1097/PAS.0b013e3181e5dffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan BC, Hart JA. Recent developments in liver pathology. Arch Pathol Lab Med. 2009;133:1078–1086. doi: 10.5858/133.7.1078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.