Abstract
Filamin B (FLNB) is a dimeric actin-binding protein that orchestrates the reorganization of the actin cytoskeleton. Congenital mutations of FLNB at the actin-binding domain (ABD) are known to cause abnormalities of skeletal development, such as atelosteogenesis types I and III and Larsen's syndrome, although the underlying mechanisms are poorly understood. Here, using fluorescence microscopy, we characterized the reorganization of the actin cytoskeleton in cells expressing each of six pathological FLNB mutants that have been linked to skeletal abnormalities. The subfractionation assay showed a greater accumulation of the FLNB ABD mutants W148R and E227K than the wild-type protein to the cytoskeleton. Ectopic expression of FLNB-W148R and, to a lesser extent, FLNB-E227K induced prominent F-actin accumulations and the consequent rearrangement of focal adhesions, myosin II, and septin filaments and results in a delayed directional migration of the cells. The W148R protein-induced cytoskeletal rearrangement was partially attenuated by the inhibition of myosin II, p21-activated protein kinase, or Rho-associated protein kinase. The expression of a single-head ABD fragment with the mutations partially mimicked the rearrangement induced by the dimer. The F-actin clustering through the interaction with the mutant FLNB ABD may limit the cytoskeletal reorganization, preventing normal skeletal development.
Keywords: skeletal dysplasia, cell motility, cytoskeleton, F-actin, fluorescence microscopy
dynamic reorganization of actin filaments (F-actin) defines cell shape, regulates force generation, and also mediates mechanotransduction and cell-cell communication, all of which are required for normal organogenesis (1, 16, 35, 36, 55). The structure of the actin cytoskeleton is regulated through interactions with a plethora of actin-binding proteins that define polymerization and assembly of the filaments (8, 27). Filamin (FLN, also named ABP-280) is an actin-binding protein originally isolated as an abundant cytoskeletal protein in smooth muscle and macrophages (18, 53). A large amount of effort has been made to understand the structure-function relationship of FLN (33, 37, 47, 52, 57). The human FLN family consists of three isoforms, FLNA, FLNB, and FLNC, each of which contains an actin-binding domain (ABD) followed by a rod domain with 24 repeats of immunoglobulin (Ig)-like modules (37, 47, 52). The COOH-terminal 24th Ig-like domain connects two FLN monomers, resulting in a FLN dimer with two ABDs (37). The two ABD heads of FLN tether two actin filaments at a 90° angle, defining shapes and stability of cellular F-actin (32). FLN also binds to a subset of membrane proteins, such as glycoproteins and integrins, anchoring the actin cytoskeleton to the plasma membrane (14, 33, 47, 48, 51). Yeast two-hybrid screening and proteomic analyses have revealed ∼100 proteins, including signaling proteins, such as protein kinases, G proteins, and their effectors, interacting with FLN, suggesting that FLN functions as a scaffold connecting signaling pathways and the actin cytoskeleton (3, 33, 37, 47). Accumulating lines of evidence strongly suggest that cellular FLN plays mechanosensing roles in regulating cell motility (5), survival (22), differentiation (56, 58), and mechanoprotection of the cytoskeleton and the plasma membrane (4, 17, 21, 38, 41). Ablation of the FLNA or FLNB gene results in severe malfunctions in cardiovascular or skeletal development (13, 58). Thus cellular FLN is a multipurpose regulator for the actin cytoskeleton and is indispensable to normal organogenesis (57).
Multiple human congenital malfunctions have been linked to missense mutations in FLNA and FLNB (25, 40). FLN ABD consists of two subdomains, calponin-homology (CH) domains 1 and 2 (7, 47). Of particular interest, the majority of the mutations at >30 sites, linked to congenital skeletal dysplasia, such as atelosteogenesis (AO) types I and III (AOI and AOIII), Larsen's syndrome (LS), and boomerang dysplasia (2, 6, 12, 25), lie within the region encoding the CH2 of FLNB. The tandem CH domains have been found in many actin-binding proteins, such as α-actinin, dystrophin, and utrophin. Mutations of α-actinin-4 and dystrophin at ABD CH2 have also been linked to focal segmental glomerulosclerosis (10, 23) and Becker muscular dystrophy/X-linked cardiomyopathy, respectively (10, 19, 23, 39, 43, 44), drawing particular attention to the structural biology of the actin cytoskeleton. In general, CH1 plays a dominant role, whereas CH2 plays a supporting and/or regulatory role, in ABD binding to F-actin (15, 28, 29, 34, 45, 46). The pathological mutations at the CH2 domain elevate the affinity of the ABD to F-actin (10, 15, 19, 42) or induce aggregation of the isolated ABD (44) and cellular FLN (6). Thus, in addition to the gene ablation (58), the gain-of-function mutation at the CH2 domain is most likely a cause of the disruption of skeletal development, although the consequences of the FLNB ABD augmentation are poorly understood at the cellular level.
In the course of the study of platelet functions, we have cloned the full-length (FL) cDNA of human FLNB as an interacting protein of glycoprotein Ib (48). To fully understand the physiological functions of FLNB and the pathological outcomes of the gain of the ABD functions, in the present study, we have assessed cellular impacts of the ectopic expression of six pathological FLNB mutants in AOI, AOIII, and LS. FL and the ABD versions of FLNB with mutations were transiently expressed in fibroblasts, and the cytoskeletal reorganization was monitored using fluorescence microscopy. Our data suggest that expression of the FLNB ABD mutant proteins induces F-actin clustering and cytoskeletal reorganization, which cause a defect in cell migration.
MATERIALS AND METHODS
Materials.
The cDNA insert encoding the human FLNB followed by EGFP (GFP-FLNB) was provided by Dr. Arnoud Sonnenberg (51) and resubcloned between XbaI/KpnI sites of the pcDNA3.1(−) vector. Six mutant FLNB cDNAs (Fig. 1A) were constructed by site-directed mutagenesis using the QuikChange kit (Life Technology). To avoid unwanted mutations in FLNB cDNA and vector regions, the mutagenesis was conducted using a pBluescript vector including a 2.1-kb XbaI/BamHI fragment of FLNB cDNA encoding the ABD region as a template. After confirmation of the site-directed mutation by DNA sequencing, the cDNA segments in pBluescript were reconstructed into the green fluorescent protein (GFP)-tagged FL FLNB in the pcDNA3.1 vector. For hemagglutinin (HA)-tagged-FLNB ABD, the cDNA encoding FLNB residue 1–283 was amplified by PCR using pGFP-FLNB(FL) as template and subcloned into the pRK7-3xHA vector (a gift from Prof. Ian Macara, University of Virginia). pVenus vector encoding the enhanced yellow fluorescent protein mutant was provided by Prof. Atsushi Miyawaki via material transfer agreement (31). HEK-293 and Rat2 cells were purchased from American Type Culture Collection and harvested in DMEM with 10% fetal bovine serum. Transient transfections were conducted for 48 h using X-tremeGENE 9 (Roche) following the manufacturer's protocol.
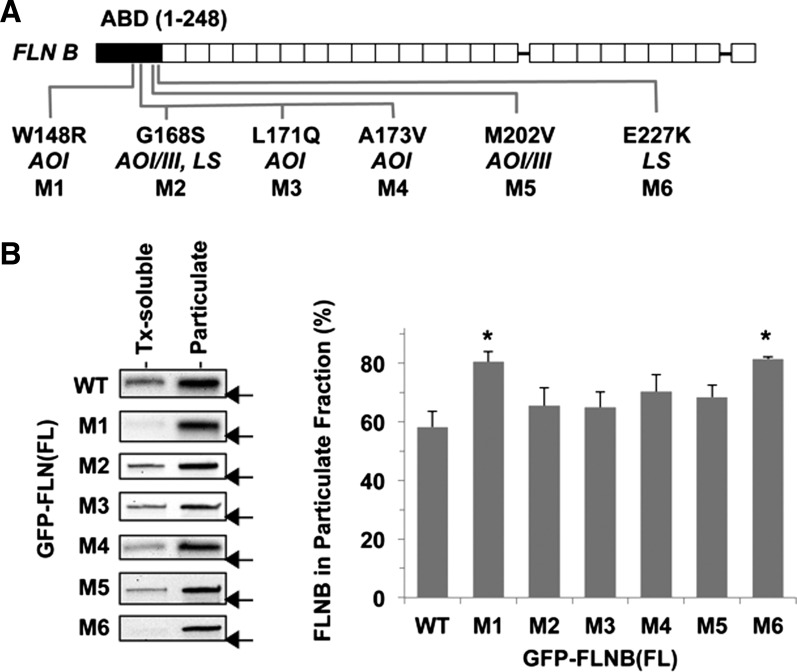
Fig. 1.
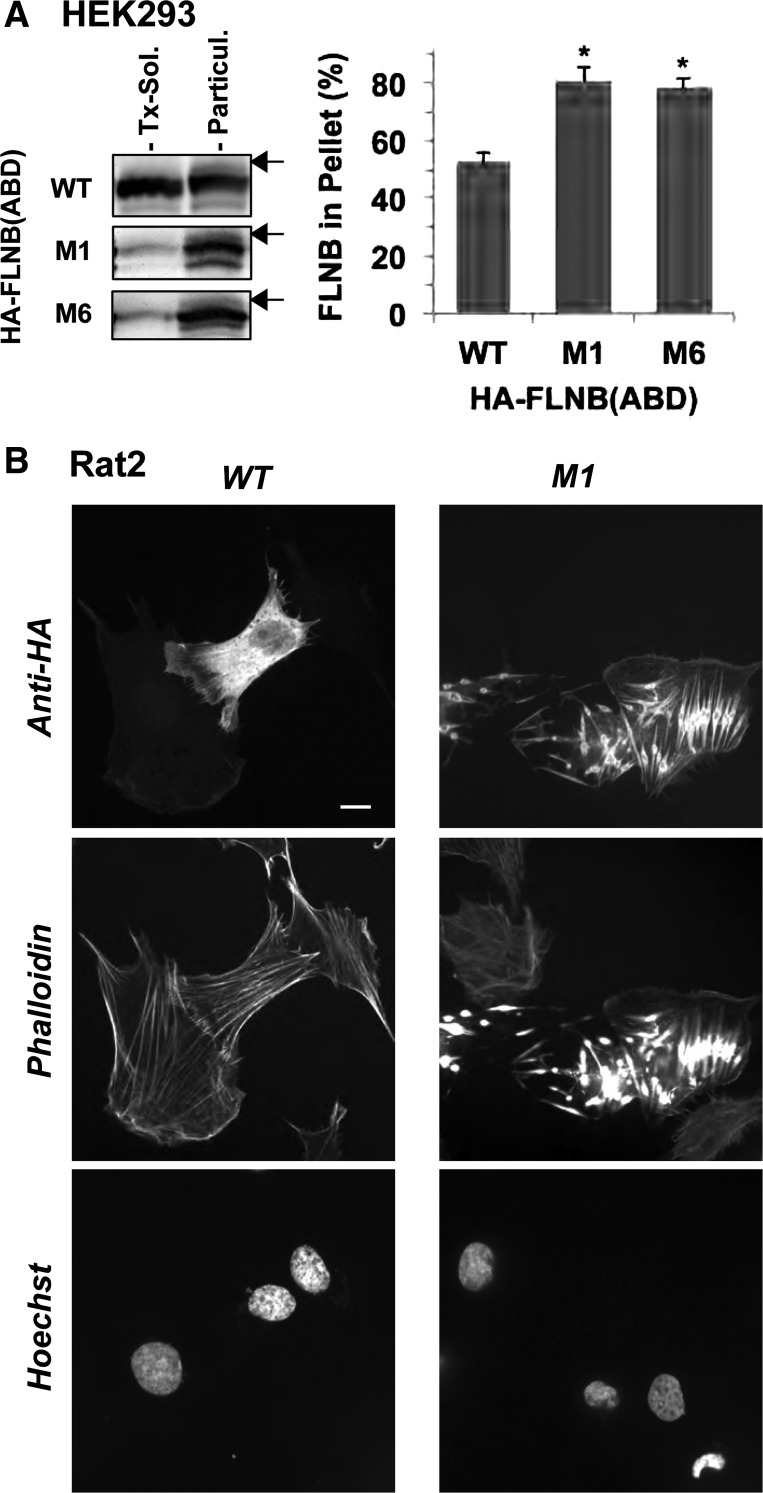
Subcellular fractionation of cells expressing the pathological variants of human filamin B (FLNB). A: human FLNB structure. Amino acid substitutions that have been linked to the genetic disorders atelosteogenesis types I and III (AOI and AOIII) and Larsen's syndrome (LS), M1–M6, are shown below the human FLNB structure. Solid and open boxes indicate actin-binding domain (ABD) and immunoglobulin-like repeats, respectively. B: each of 7 green fluorescent protein (GFP)-tagged full-length (FL) FLNB proteins, wild-type (WT) and M1–M6, was transiently expressed in HEK-293 cells and subjected to subcellular fractionation assay. GFP-FLNB in Triton X-100 (Tx)-soluble and -insoluble (particulate) fractions was detected by immunoblotting using anti-GFP antibody (left), and the percentage of FLNB in the particulate fraction was determined by densitometry (right). Values are means ± SE; n = 3. *P < 0.05 vs. WT (by ANOVA with Dunnett's post hoc test). Arrow indicates the position of a 250-kDa marker on each blot.
Subcellular fractionation.
After removal of the growth medium, in a 6-cm dish, HEK-293 cells expressing GFP- or HA-tagged FLNB proteins were lysed for 30 min on ice with 0.5 ml of lysis buffer [10 mM HEPES (pH 7.4) supplemented with 0.1% Triton X-100, 5 mM MgCl2, 2 mM EGTA, 0.5 mM tris(2-carboxyethyl)phosphine, 10 μg/ml lima bean trypsin inhibitor, 5 μg/ml leupeptin, and 4 mM Pefabloc (Roche)]. Aliquots (0.1 ml) were subjected to immunoblotting as “whole lysates” to verify expression of the proteins (data not shown). Other aliquots (0.25 ml) were processed in an Airfuge for 30 min at 75,000 rpm at room temperature. The pellet was suspended with 0.25 ml of the lysis buffer, and the soluble and particulate fractions were subjected to quantitative immunoblotting using anti-GFP (Aves Labs) or anti-HA (12CA5, University of Virginia), as described previously (49). Relative expression of the ectopic FLNB proteins in the particulate fraction [100 × particulate/(particulate + soluble fraction)] was determined from the densitometric values of each fraction. Values are means ± SE from three independent transfections.
Confocal imaging.
Rat2 cells were seeded on glass coverslips under the growth conditions described above and subjected to immunofluorescence as described previously (11, 24). Pharmacological pretreatments were conducted for 30 min with DMSO (as control), blebbistatin (20 μM), cytochalasin D (0.5 μM), H1152 (10 μM), and IPA3 (10 μM). Cells were rinsed with PBS, fixed for 15 min with 4% paraformaldehyde, and permeabilized for 5 min with 0.1% Triton X-100 in PBS prior to staining. Cells were stained using anti-vinculin (hVIN, Sigma-Aldrich; 1:200 dilution), anti-nonmuscle myosin heavy chain IIA (myosin heavy chain isoform 9; AbCam; 1:200 dilution), anti-α-actinin (Sigma-Aldrich; 1:100 dilution), anti-septin 2 (Millipore; 1:100 dilution), anti-HA (1:1,000 dilution), phalloidin-Alexa Fluor 647 (Invitrogen; 1:100 dilution), Hoechst 33342 (Sigma-Aldrich; 0.5 μg/ml), and Cy3-conjugated anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch; 1:100 dilution). Quad-color confocal imaging of the specimens was conducted using a fluorescence microscope (Olympus IX70) equipped with a PlanApo ×60 oil objective, a CCD camera (ImagEM, Hamamatsu), and a spinning-disk confocal unit (CARV II). Image acquisition and processing to TIFF format with pseudo color were conducted using IPLab imaging software (BD Biosciences) and ImageJ (http://imagej.nih.gov/ij/), respectively. Images of reproducible phenotypes are shown. The number of pixels occupied by F-actin foci (F-actin accumulation index) was determined using phalloidin-staining images (3–10 fields), which were converted to binary images and measured using the threshold function and the particle analysis tool of ImageJ, respectively.
Scratch wound healing assay.
Rat2 cells were seeded on a glass-bottom 35-mm dish and transiently transfected for 48 h with pGFP-FLNB wild-type (WT), M1, or M6 vector (0.75 μg) plus pVenus vector (0.25 μg) as a tracer under the growth conditions described above. Pipette tips (1 ml) were used to inflict three lines of scratch wounds (∼1 × 5 mm) on the confluent monolayer between pairs of dots marked on the glass bottom of the dish. Cells were harvested for 24 h with fresh growth medium, fixed with 4% paraformaldehyde, and subjected to Hoechst staining. A LUCPlanFL N ×20 objective was used to acquire two images of Venus, Hoechst, and phase contrast for each wound between the marks. Numbers of Venus-positive cells and total cells that migrated into the scratched area and remained in the unscratched area on each image field were counted using IPLab software. Migration index is defined as the ratio of the normalized Venus-positive cell population in the scratched area to that in the unscratched area and expressed as means ± SE from six image fields in each transfection.
Other assays.
Other assays were conducted as described previously (24). Statistical analysis was conducted using Student's t-test or ANOVA with Dunnett's post hoc test. P < 0.05 was considered significant.
RESULTS
W148R and E227K substitutions of FLNB result in augmented binding to the actin cytoskeleton.
Of 31 mutations within the FLNB ABD that have been linked to boomerang dysplasia, AOI, AIIII, and LS (2, 6, 12, 25), 6 were chosen on the basis of the availability of biochemical data (W148R and M202V) (42), the severity of skeletal dysplasia (G168S, L171Q, and A173V), and the frequency of occurrence (E227K) (6) (Fig. 1A). All the residues at the six mutation sites (W148, G168, L171, A173, M202, and E227) are conserved in the ABDs of FLNA, FLNB, and FLNC (42). Each FLNB mutant was transiently expressed in HEK-293 cells as a GFP-fusion protein and subjected to subcellular fractionation (Fig. 1B). In the subcellular fractionation assay, 58.2 ± 9.1% of GFP-FLNB WT was coprecipitated with the cytoskeleton (Fig. 1B). Substitution of W148R (M1) and E227K (M6) increased the amount of FLNB protein precipitated with the particulate fraction to 80%, suggesting increases in FLNB binding to F-actin and/or stability of the actin cytoskeleton (Fig. 1B). The other mutants (M2, M3, M4, and M5) showed a similar tendency, although the differences from WT were not statistically significant. M1 and M6 were subjected to further characterization.
FLNB-W148R and FLNB-E227K proteins cause rearrangement of the cellular F-actin along with focal adhesions, myosin II, and septin 2.
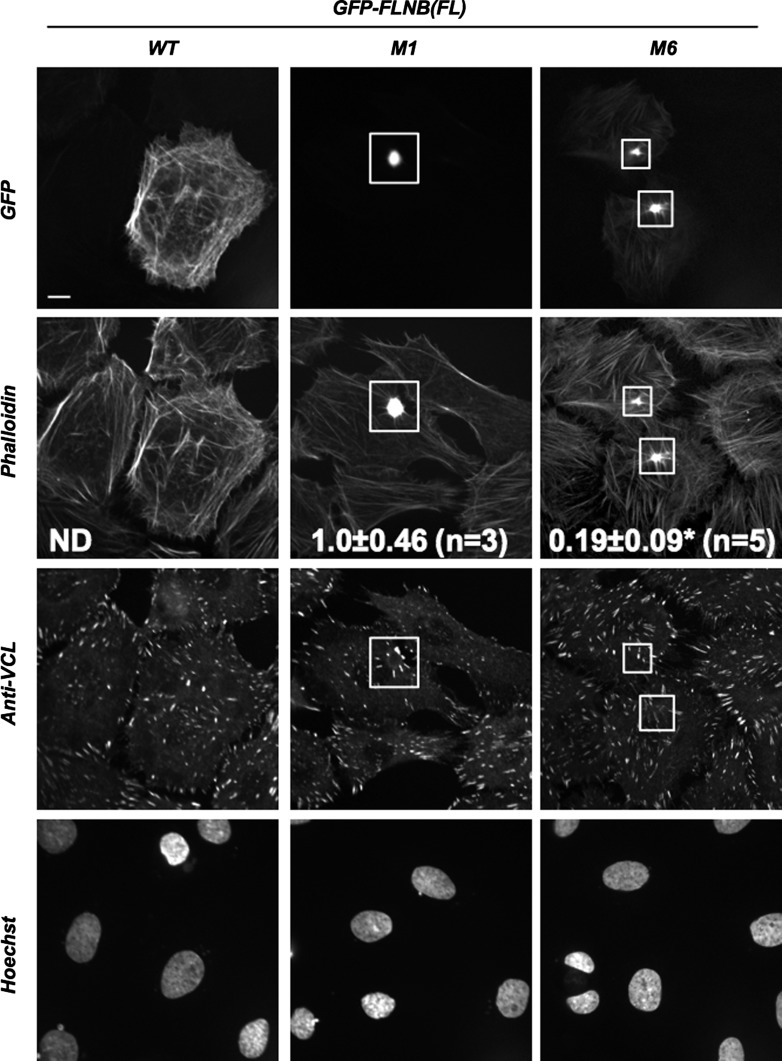
Figures 2 and 3 show immunofluorescence images of Rat2 fibroblasts expressing GFP-FLNB WT, M1, and M6. GFP-FLNB WT was evenly associated with the actin cytoskeleton throughout the cell and caused no noticeable changes in the overall structure of the actin cytoskeleton and focal adhesion, labeled with phalloidin and anti-vinculin, respectively (Fig. 2, left). By sharp contrast, GFP-FLNB M1 was accumulated at foci, which consisted of F-actin. Because of formation of the FLNB-F-actin foci, the structure of the actin cytoskeleton and focal adhesions around the foci was rearranged into a radial shape, without affecting the overall cell shape (Fig. 2, middle). The phenotype induced by expression of GFP-FLNB M6 protein was essentially identical to the M1 protein, although the M6 protein was more visible than the M1 protein on stress fibers (Fig. 2, right). The accumulation of F-actin, defined as the F-actin accumulation index, was significantly greater for the M1 than the M6 protein. The M1 protein appeared more potent in terms of formation of the FLNB-F-actin foci.
Fig. 2.
Effects of ectopic expression of GFP-FLNB on the actin cytoskeleton in Rat2 fibroblasts. Rat2 fibroblasts expressing FL GFP-FLNB WT, M1, and M6 were visualized by confocal immunofluorescence microscopy. Top to bottom: localization of GFP-FLNB, F-actin (phalloidin-Alexa Fluor 647), focal adhesions (anti-vinculin/anti-mouse IgG-Cy3), and nuclei (Hoechst dye). Boxes show location of accumulated GFP-FLNB M1 and M6. Scale bar, 10 μm. Values (means ± SE) represent F-actin accumulation index. ND, not detected. *P < 0.05 vs. M1 (by Student's t-test).
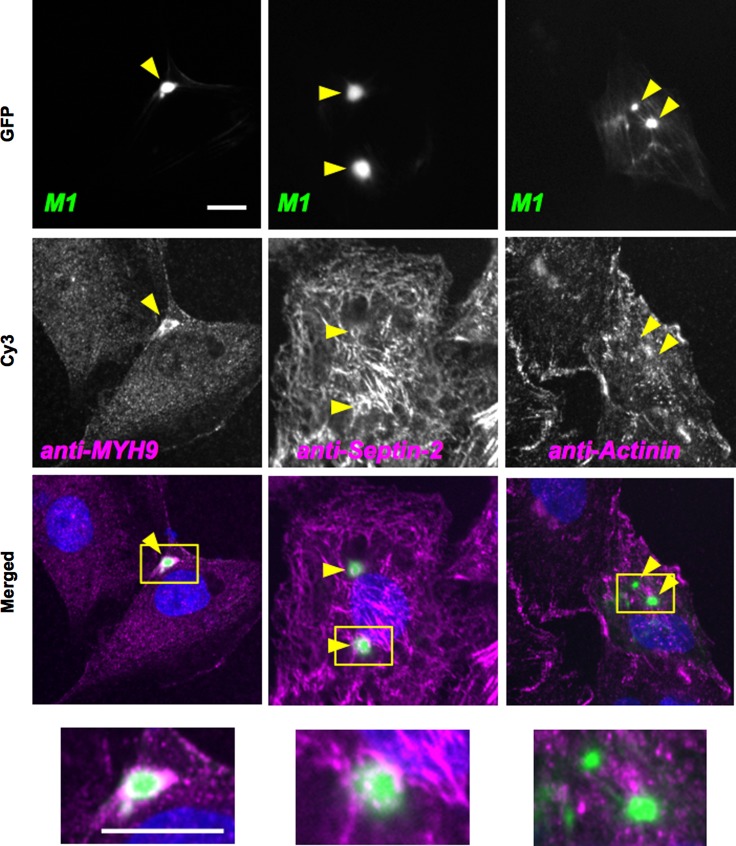
Fig. 3.
Localization of myosin, septin 2, and α-actinin in cells expressing FL GFP-FLNB M1. Confocal immunofluorescence microscopy of Rat2 cells expressing FL GFP-FLNB M1 were conducted using anti-myosin heavy chain [anti-myosin heavy chain isoform 9 (MYH9)], anti-septin 2, and anti-α-actinin antibodies with Cy3-labeled secondary antibodies. Green and magenta in merged images show GFP and Cy3, respectively. Arrowheads and boxes in merged images show location of FLNB foci and position of zoomed image at bottom, respectively. Scale bar, 10 μm.
Myosin II and septin filaments bind to F-actin and contribute to regulation of cell motility. The endogenous nonmuscle myosin II and septin 2 were labeled with anti-myosin heavy chain isoform 9 (Fig. 3, left) and anti-septin 2 (Fig. 3, middle) antibodies. Myosin II and septin filaments were colocalized at the FLNB-F-actin foci induced by the M1 protein (Fig. 3). It should be noted that myosin II and septin 2 filaments surrounded the FLNB-F-actin foci, unlike F-actin, which was incorporated into the core of the foci (Fig. 3, bottom). Next, we tested whether FLNB ABD mutation affects F-actin binding of α-actinin, which also possesses the ABD with the tandem CH domains. Immunofluorescence images show that α-actinin was not incorporated into the FLNB-F-actin complexes, suggesting that their binding to F-actin is mutually exclusive (Fig. 3, right). Thus the pathological mutations at FLNB ABD trigger the formation of the FLNB-F-actin foci, leading to a selective rearrangement and displacement of actin-binding proteins.
Reorganization of the actin cytoskeleton induced by FLNB-W148R is associated with delayed directional migration of the cell.
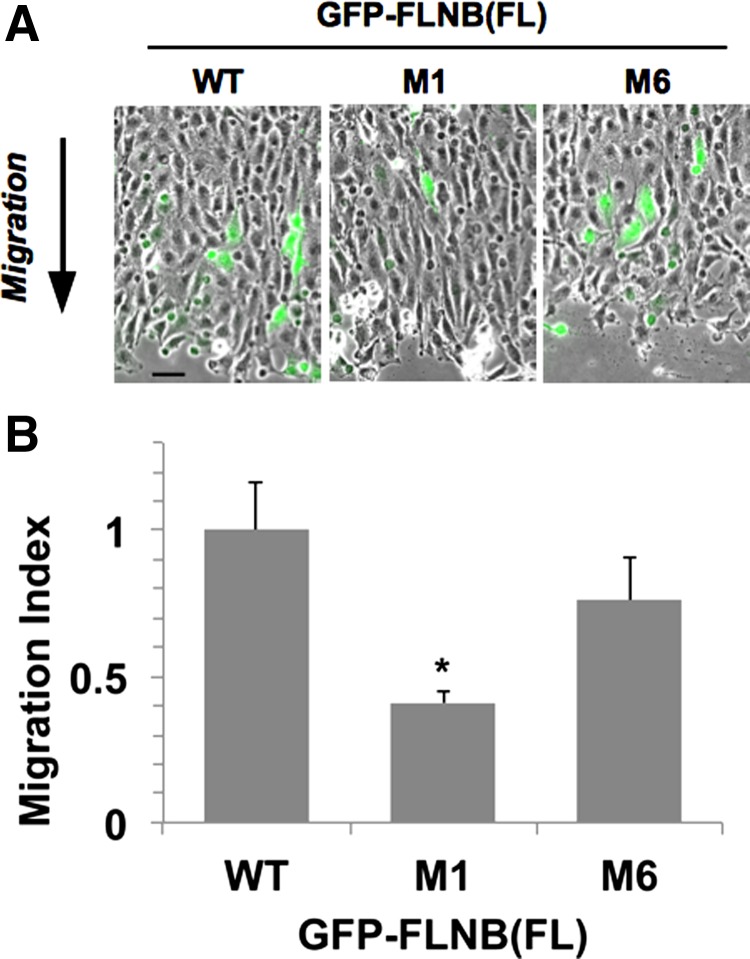
Using the scratch wound healing assay, we sought to determine whether the highly localized disturbance of the F-actin network induced by the FLNB M1 protein affects the global reorganization of the cytoskeleton during cell migration. The scratch wound healing assay is an established experimental model for evaluating directional cell migration that governs normal organ development. GFP-FLNB WT, M1, or M6 was coexpressed with Venus yellow fluorescent protein as a tracer, and the directional migration of the cell expressing Venus was determined by fluorescence microscopy. In response to formation of the scratch wounds on the confluent monolayer, Rat2 cells directionally migrated into the wound area (Fig. 4A). Cells expressing GFP-FLNB WT migrated into the wound area, whereas fewer cells expressing the M1 protein were found in the area (Fig. 4A, green). Figure 4B shows the migration index, the relative number of migrated cells expressing GFP-FLNB, which is normalized against the transfection efficiency of each experiment. Fewer M1 protein- than WT protein-expressing cells migrated into the wound area (Fig. 4B). These data show a biological link between the pathological M1 mutation of FLNB ABD and the dysfunction of the cells. The M6 protein showed a similar tendency, although the reduction in migration was not statistically significant.
Fig. 4.
Scratch wound healing assay of Rat2 cells expressing FL GFP-FLNB WT, M1, and M6 with Venus. At 24 h after scratching, cells were fixed and stained with Hoechst dye. A: epifluorescence images of Venus signals (green) at the wound site superimposed on the corresponding phase-contrast images. Arrow indicates direction of cell migration. Scale bar, 50 μm. B: ratio of the relative population of Venus-positive cells at the wound area to that at the unscratched area (migration index). Number of Venus-positive cells in each image field was normalized against number of total cells, which was counted on Hoechst fluorescence images. Values (means ± SE from 6 image fields of 3 independent wounds) were normalized against WT. *P < 0.05 (by ANOVA with Dunnett's post hoc test).
Actin polymerization, PAK, and ROCK are involved in formation of F-actin foci induced by FLNB-W148R.
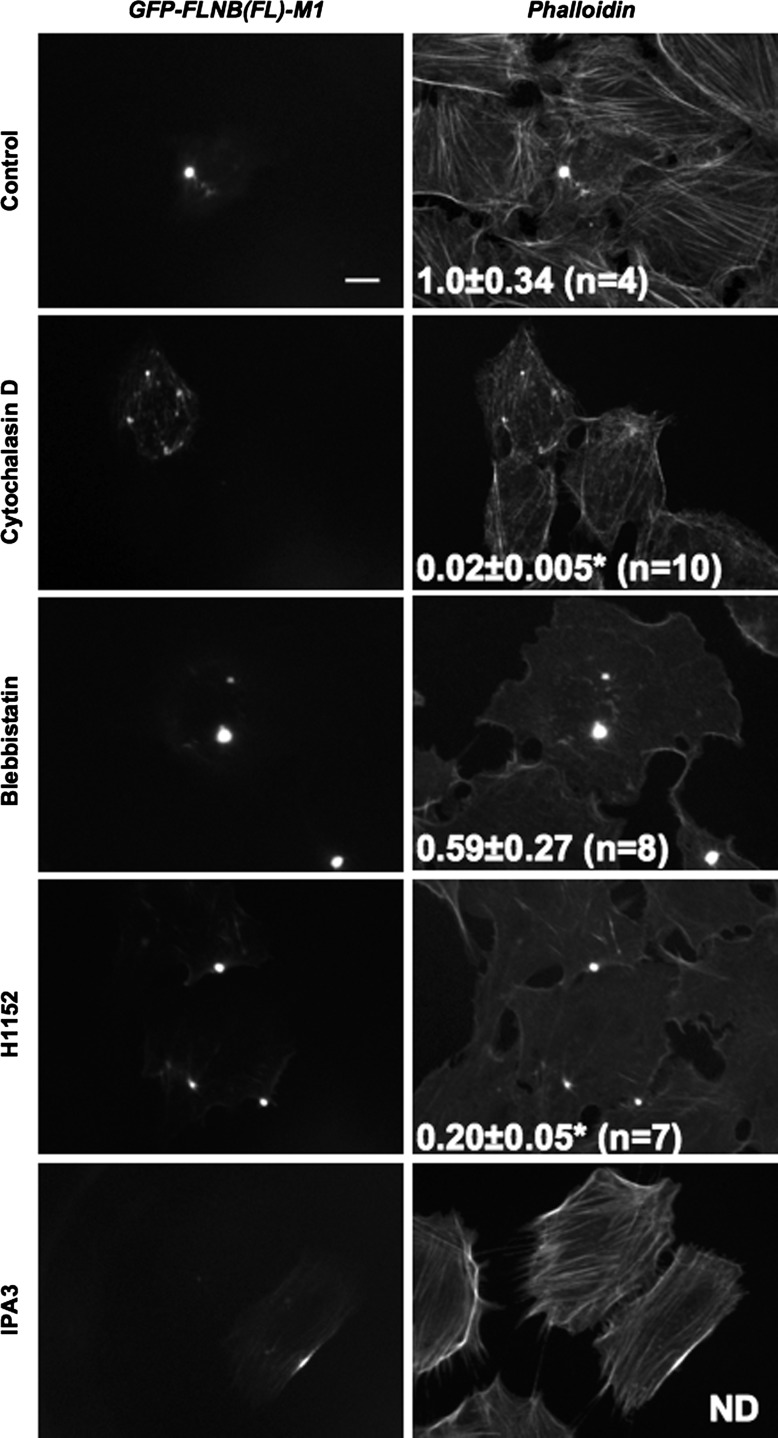
To further characterize the pathological FLNB-F-actin foci induced by the M1 mutation, we tested the stability of the complex against treatments with antagonists for the actin cytoskeleton. Treatment with cytochalasin D (an actin polymerization inhibitor) disturbed the formation of FLNB-F-actin foci, resulting in punctate accumulation of GFP-FLNB on F-actin, with a significant reduction in the F-actin accumulation index to 0.02 ± 0.005 (Fig. 5). On the other hand, treatment with blebbistatin, which inhibits myosin II ATPase, had little effect on formation of F-actin foci, although the actin cytoskeleton network was disrupted. A higher dose of blebbistatin (100 μM) caused detachment of the cells (data not shown). Inhibition of ROCK by use of H1152 disrupted the actin cytoskeleton in parallel with a significant reduction in F-actin accumulation. By contrast, inhibition of PAK with IPA3 blocked formation of FLNB-F-actin foci, although GFP-FLNB M1 was accumulated on augmented F-actin bundles. Thus, actin polymerization, PAK, and ROCK are, at least partially, involved in formation of the pathological FLNB-F-actin foci. It should be also noted that the formation of the foci is reversible and clearly distinguishable from the aggresome consisting of denatured proteins (26).
Fig. 5.
Effects of antagonists for the actin cytoskeleton on accumulation of the FLNB-F-actin complex. Confocal immunofluorescence images of GFP and phalloidin of Rat2 cells expressing FL GFP-FLNB M1 treated with DMSO (control), cytochalasin D (0.5 µM), blebbistatin (20 µM), H1152 (10 µM), and IPA3 (10 µM) are shown. Scale bar, 10 μm. Values (means ± SE) represent F-actin accumulation index. *P < 0.05 vs. control (by ANOVA with Dunnett's post hoc test).
The single-head ABD fragment of FLNB-W148R partially represents the action of the FL FLNB dimers.
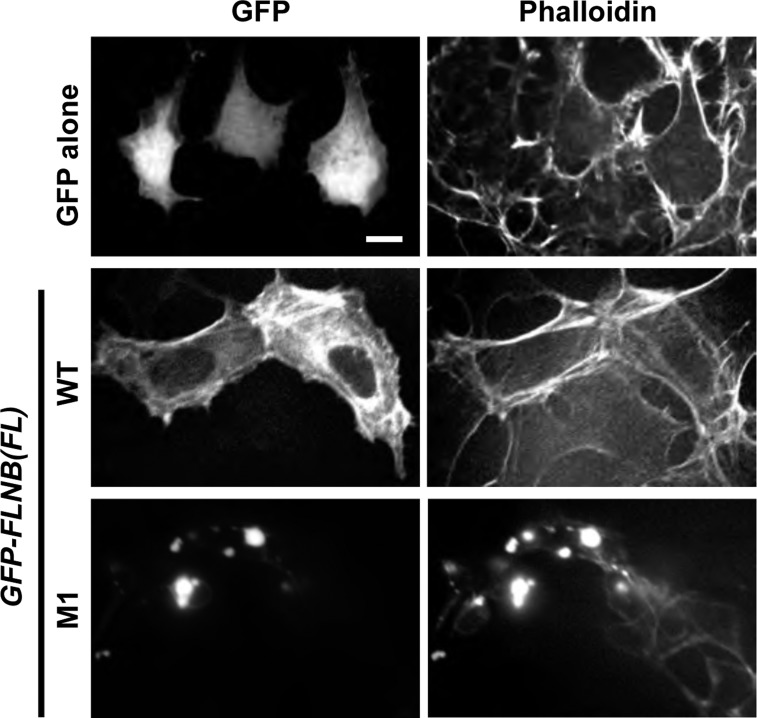
Cellular FLNB forms a dimer with two ABDs that cross-link actin filaments and define the structure of the cytoskeleton. Recombinant M1 ABD protein has been reported to bind to F-actin with higher affinity than WT ABD (42). To test whether the augmented tethering activity by two ABD heads is a cause of the formation of the pathological FLNB-F-actin foci, we characterized the single-head ABD protein, including residue 1–283, expressed in HEK-293 and Rat2 cells. In the subfractionation assay, HA-tagged M1 and M6 ABD proteins coprecipitated with the cytoskeletal fraction to a greater extent than the WT ABD protein (Fig. 6A), essentially in agreement with data from the in vitro F-actin cosedimentation assay (42). Immunofluorescence images showed that the WT ABD protein was distributed mostly in the cytoplasm, with subtle accumulation on the actin filaments (Fig. 6B, left). There was no noticeable change in the cytoskeletal structure. On the other hand, the M1 ABD protein clearly altered the structure of the actin cytoskeleton (Fig. 6B, right). The M1 ABD protein formed multiple spindle structures with F-actin in the cells, causing prominent rearrangement of the cytoskeleton. Each spindle structure consists of filamentous extensions from a thickened center, which is reminiscent of the FLNB-F-actin accumulation induced by the FL M1 protein. Thus the interaction of the M1 ABD is capable of triggering F-actin clustering, partially mimicking the FL protein, although the cross-linking activity of two-headed FLNB is needed for stable formation of the FLNB-F-actin foci. As shown in Fig. 7, the formation of FLNB-F-actin induced by the M1 protein was reproducible at least in HEK-293 cells.
Fig. 6.
F-actin clustering induced by actin-binding domain (ABD) fragments of the pathological FLNB mutants. HEK-293 and Rat2 cells expressing hemagglutinin (HA)-tagged FLNB residue 1–248, including ABD, were subjected to subcellular fractionation (A), and images were visualized by confocal immunofluorescence (B). HA-FLNB ABD was detected with anti-HA antibody. Extent of HA-FLNB ABD on immunoblot was determined by densitometry. Values are means ± SE from 3 independent transfections. *P < 0.05 (by Student's t-test). Arrow indicates position of a 37-kDa marker on each blot. Scale bar, 10 μm.
Fig. 7.
Confocal microscopy images of HEK-293 cells expressing FL FLNB WT and M1. HEK-293 cells were transiently transfected with plasmids encoding GFP alone, GFP-FLNB WT, and the M1 protein and stained with rhodamine-labeled phalloidin (1:50 dilution). Representative confocal fluorescence images of GFP and F-actin are shown. Scale bar, 10 μm.
DISCUSSION
In this study we demonstrated a rearrangement of the actin cytoskeleton and consequent dysmotility of cells expressing the FLNB protein with a mutation at ABD (W148R) that has been linked to skeletal dysplasia in AOI. Dynamic reorganization of the actin cytoskeleton is fundamental to virtually every aspect of cellular function required for normal development, such as proliferation, differentiation, and migration. The abnormal formation of the FLNB-F-actin complex induced by the W148R (M1) mutation of FLNB may underlie dysfunctions in cells responsible for skeletal development, such as osteoblasts and chondrocytes, in which FLNB is expressed (20, 54).
It is clear that none of M1–M6 proteins within the ABD CH2 domain of FLNB is a loss-of-function mutation. The CH2 subdomain in the ABD is a hot spot of mutations of FLNA, FLNB, α-actinin-4, and dystrophin, which have been linked to human congenital disorders (23, 25, 39, 40). Sawyer et al. showed that the M1 mutation causes an elevated affinity of the isolated ABD to F-actin and a decreased thermal stability of the domain, without affecting the overall three-dimensional structure (42), likely through changes in the regulatory role of CH2 (15). Therefore, the increased extent of the M1 and M6 proteins co-sedimented with the cytoskeletal fraction is most likely attributed to an augmented interaction with F-actin and/or a consequent stabilization of the actin cytoskeleton in the cells.
Importantly, these CH2 mutations of FLNB are capable of reorganizing the actin cytoskeleton. The deformation of the actin cytoskeleton is mimicked by the single-head ABD M1. Therefore, the M1 mutation appears to alter F-actin characteristics without F-actin cross-linking. Timely docking of actin-binding proteins at specific loci defines the stability, structure, and function of F-actin (8). The CH2 mutants of FLNB may dominate on F-actin and limit the accessibility of other actin-regulating proteins, forcing rearrangement of the cytoskeleton. This idea is supported by the fact that the endogenous α-actinin is excluded from the accumulated FLNB-F-actin complex. It will be interesting to see whether the CH2 mutations of other proteins also affect interactions with other actin-binding proteins and result in a rearrangement of cellular F-actin.
The M1-induced pathological accumulation of the FLNB-F-actin complex is associated with dysmotility of cells. The FLNB-F-actin complex is wrapped with myosin II and septin, and the formation is partially suppressed by the inhibition of actin polymerization, PAK, and ROCK. Lazaro-Dieguez et al. (26) showed that treatment of cells with jasplakinolide, which stabilizes F-actin, induces formation of the aggresome of actin filaments. The FLNB-F-actin foci induced by the M1 mutation are clearly distinguished from the actin aggresome because of the reversibility in response to the antagonists. PAK and ROCK are effectors of Rac1 and RhoA, respectively, regulating the cytoskeletal reorganization. Both kinases are known to phosphorylate and activate LIM kinase (9, 30). LIM kinase then phosphorylates cofilin, promoting actin polymerization (1). Thus, inhibition of PAK and ROCK results in dephosphorylation/disinhibition of cofilin, leading to actin depolymerization, similar to the action of cytochalasin D. Therefore, actin polymerization appears to be necessary for formation of the FLNB-F-actin assembly induced by the M1 mutation.
It is noteworthy that rearrangement of the actin cytoskeleton occurs at limited areas around only one or two FLNB-F-actin foci formed in each cell. We presume that accumulation of the FLNB-F-actin complex with myosin II and septin likely produces irregular tension in the cytoskeletal system that disrupts orchestrated force generation and interferes the directional migration. Our data show the importance of PAK in the deformation of the actin cytoskeleton-induced M1 mutant. This may be due to the phosphorylation of FLNA at Ser2152 (50) in addition to the activation of LIM kinase and actin polymerization. The PAK-FLNB axis deserves further investigation for a full understanding of the pathogenesis.
In this study we mainly characterized the phenotype induced by expression of M1. The ectopic expression of M2, M3, M4, and M5 also causes formation of FLNB-F-actin foci in HEK-293 cells (data not shown). This is consistent with the report by Daniel et al. (6) that accumulation of FLNB, which is similar to the FLNB-F-actin foci with the M1 mutation (Fig. 7), occurs in HEK-293 cells expressing FLNB with the mutation of M5, as well as L171R, S235P, and G751R. Thus the phenotype induced by the M1 protein appears to represent the pathological dysfunctions of cells expressing FLNB CH2 mutants. To more precisely predict the phenotype of the congenital disorders, we must comprehensively characterize interaction of each FLNB CH2 mutant with F-actin and its cellular impacts on the rearrangement of the actin cytoskeleton. Furthermore, we must establish links between the disturbance of the actin cytoskeleton and the skeletal dysplasia. Zheng et al. (56) showed the pathological link between FLNB and Runx2, a transcription factor that plays a dominant role in osteogenesis and chondrogenesis. Accumulating lines of evidence suggest that the actin cytoskeleton mediates mechanotransduction during organogenesis (35, 55, 57). The FLNB mutation may affect gene regulation, such as Runx2 signaling, through the mechanical stresses due to the localized rearrangement of F-actin.
GRANTS
This work is supported by National Heart, Lung, and Blood Institute Grant R01 HL-072439, the Brandywine Valley Hemophilia Foundation Research and Education Fund, a Pennsylvania C.U.R.E. Grant, and the Sidney Kimmel Cancer Center Genomics Facility, funded in part by National Cancer Institute Grant P30 CA-056036.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Y.Z., S.S.S., and M.E. developed the concept and designed the research; Y.Z. and M.E. performed the experiments; Y.Z., S.S.S., and M.E. analyzed the data; Y.Z., S.S.S., and M.E. interpreted the results of the experiments; Y.Z. and M.E. prepared the figures; Y.Z. and M.E. drafted the manuscript; Y.Z. and M.E. edited and revised the manuscript; Y.Z., S.S.S., and M.E. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Toshiro Takafuta (National Hospital Organization Kure Medical Center, Japan) for valuable discussion.
REFERENCES
- 1.Arber S, Barbayannis FA, Hanser H, Schnelder C, Stanyon CA, Bernards O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805–809, 1998. [DOI] [PubMed] [Google Scholar]
- 2.Bicknell LS, Morgan T, Bonafe L, Wessels MW, Bialer MG, Willems PJ, Cohn DH, Krakow D, Robertson SP. Mutations in FLNB cause boomerang dysplasia. J Med Genet 42: e43, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. The BioGRID interaction database: 2015 update. Nucleic Acids Res 43: D470–D478, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cranmer SL, Ashworth KJ, Yao Y, Berndt MC, Ruggeri ZM, Andrews RK, Jackson SP. High shear-dependent loss of membrane integrity and defective platelet adhesion following disruption of the GPIbα-filamin interaction. Blood 117: 2718–2727, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255: 325–327, 1992. [DOI] [PubMed] [Google Scholar]
- 6.Daniel PB, Morgan T, Alanay Y, Bijlsma E, Cho TJ, Cole T, Collins F, David A, Devriendt K, Faivre L, Ikegawa S, Jacquemont S, Jesic M, Krakow D, Liebrecht D, Maitz S, Marlin S, Morin G, Nishikubo T, Nishimura G, Prescott T, Scarano G, Shafeghati Y, Skovby F, Tsutsumi S, Whiteford M, Zenker M, Robertson SP. Disease-associated mutations in the actin-binding domain of filamin B cause cytoplasmic focal accumulations correlating with disease severity. Hum Mutat 33: 665–673, 2012. [DOI] [PubMed] [Google Scholar]
- 7.Djinovic Carugo K, Banuelos S, Saraste M. Crystal structure of a calponin homology domain. Nat Struct Biol 4: 175–179, 1997. [DOI] [PubMed] [Google Scholar]
- 8.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 83: 433–473, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol 1: 253–259, 1999. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlicher AJ, Krishnan R, Guo M, Bidan CM, Weitz DA, Pollak MR. α-Actinin binding kinetics modulate cellular dynamics and force generation. Proc Natl Acad Sci USA 112: 6619–6624, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eto M, Kirkbride JA, Chugh R, Karikari NK, Kim JI. Nuclear localization of CPI-17, a protein phosphatase-1 inhibitor protein, affects histone H3 phosphorylation and corresponds to proliferation of cancer and smooth muscle cells. Biochem Biophys Res Commun 434: 137–142, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrington-Rock C, Firestein MH, Bicknell LS, Superti-Furga A, Bacino CA, Cormier-Daire V, Le Merrer M, Baumann C, Roume J, Rump P, Verheij JB, Sweeney E, Rimoin DL, Lachman RS, Robertson SP, Cohn DH, Krakow D. Mutations in two regions of FLNB result in atelosteogenesis I and III. Hum Mutat 27: 705–710, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA 103: 19836–19841, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox JE. Identification of actin-binding protein as the protein linking the membrane skeleton to glycoproteins on platelet plasma membranes. J Biol Chem 260: 11970–11977, 1985. [PubMed] [Google Scholar]
- 15.Galkin VE, Orlova A, Salmazo A, Djinovic-Carugo K, Egelman EH. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat Struct Mol Biol 17: 614–616, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol 26: 315–333, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem 273: 1689–1698, 1998. [DOI] [PubMed] [Google Scholar]
- 18.Hartwig JH, Stossel TP. Isolation and properties of actin, myosin, and a new actin-binding protein in rabbit alveolar macrophages. J Biol Chem 250: 5696–5705, 1975. [PubMed] [Google Scholar]
- 19.Henderson DM, Lee A, Ervasti JM. Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. Proc Natl Acad Sci USA 107: 9632–9637, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Lu J, Lian G, Zhang J, Hecht JL, Sheen VL. Filamin B regulates chondrocyte proliferation and differentiation through Cdk1 signaling. PLos One 9: e89352, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson WM, Jaasma MJ, Tang RY, Keaveny TM. Mechanical loading by fluid shear is sufficient to alter the cytoskeletal composition of osteoblastic cells. Am J Physiol Cell Physiol 295: C1007–C1015, 2008. [DOI] [PubMed] [Google Scholar]
- 22.Kainulainen T, Pender A, D'Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin A in connective tissues after challenge by applied tensile forces. J Biol Chem 277: 21998–22009, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR. Mutations in ACTN4, encoding α-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 24: 251–256, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Khasnis M, Nakatomi A, Gumpper K, Eto M. Reconstituted human myosin light chain phosphatase reveals distinct roles of two inhibitory phosphorylation sites of the regulatory subunit, MYPT1. Biochemistry 53: 2701–2709, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, Wachsmann-Hogiu S, Acuna D, Shapiro SS, Takafuta T, Aftimos S, Kim CA, Firth H, Steiner CE, Cormier-Daire V, Superti-Furga A, Bonafe L, Graham JM Jr, Grix A, Bacino CA, Allanson J, Bialer MG, Lachman RS, Rimoin DL, Cohn DH. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet 36: 405–410, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Lazaro-Dieguez F, Aguado C, Mato E, Sanchez-Ruiz Y, Esteban I, Alberch J, Knecht E, Egea G. Dynamics of an F-actin aggresome generated by the actin-stabilizing toxin jasplakinolide. J Cell Sci 121: 1415–1425, 2008. [DOI] [PubMed] [Google Scholar]
- 27.Lee SH, Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol Cells 29: 311–325, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehman W, Craig R, Kendrick-Jones J, Sutherland-Smith AJ. An open or closed case for the conformation of calponin homology domains on F-actin? J Muscle Res Cell Motil 25: 351–358, 2004. [DOI] [PubMed] [Google Scholar]
- 29.Lin AY, Prochniewicz E, James ZM, Svensson B, Thomas DD. Large-scale opening of utrophin's tandem calponin homology (CH) domains upon actin binding by an induced-fit mechanism. Proc Natl Acad Sci USA 108: 12729–12733, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285: 895–898, 1999. [DOI] [PubMed] [Google Scholar]
- 31.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90, 2002. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol 179: 1011–1025, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr 5: 160–169, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norwood FL, Sutherland-Smith AJ, Keep NH, Kendrick-Jones J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure 8: 481–491, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11: 353–365, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 11: 633–643, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci 31: 411–419, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Razinia Z, Makela T, Ylanne J, Calderwood DA. Filamins in mechanosensing and signaling. Annu Rev Biophys 41: 227–246, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts RG, Gardner RJ, Bobrow M. Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations. Hum Mutat 4: 1–11, 1994. [DOI] [PubMed] [Google Scholar]
- 40.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet 33: 487–491, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Rognoni L, Most T, Zoldak G, Rief M. Force-dependent isomerization kinetics of a highly conserved proline switch modulates the mechanosensing region of filamin. Proc Natl Acad Sci USA 111: 5568–5573, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: insights from the crystal structures of filamin B actin binding domains. J Mol Biol 390: 1030–1047, 2009. [DOI] [PubMed] [Google Scholar]
- 43.Sawyer GM, Sutherland-Smith AJ. Crystal structure of the filamin N-terminal region reveals a hinge between the actin binding and first repeat domains. J Mol Biol 424: 240–247, 2012. [DOI] [PubMed] [Google Scholar]
- 44.Singh SM, Kongari N, Cabello-Villegas J, Mallela KM. Missense mutations in dystrophin that trigger muscular dystrophy decrease protein stability and lead to cross-β-aggregates. Proc Natl Acad Sci USA 107: 15069–15074, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh Surinder M, Mallela Krishna MG. The N-terminal actin-binding tandem calponin-homology (CH) domain of dystrophin is in a closed conformation in solution and when bound to F-actin. Biophys J 103: 1970–1978, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjöblom B, Ylänne J, Djinović-Carugo K. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr Opin Struct Biol 18: 702–708, 2008. [DOI] [PubMed] [Google Scholar]
- 47.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2: 138–145, 2001. [DOI] [PubMed] [Google Scholar]
- 48.Takafuta T, Wu G, Murphy GF, Shapiro SS. Human β-filamin is a new protein that interacts with the cytoplasmic tail of glycoprotein Ibα. J Biol Chem 273: 17531–17538, 1998. [DOI] [PubMed] [Google Scholar]
- 49.Terai M, Eto M, Young GD, Berd D, Mastrangelo MJ, Tamura Y, Harigaya K, Sato T. Interleukin 6 mediates production of interleukin 10 in metastatic melanoma. Cancer Immunol Immunother 61: 145–155, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, Kumar R. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol 4: 681–690, 2002. [DOI] [PubMed] [Google Scholar]
- 51.van der Flier A, Kuikman I, Kramer D, Geerts D, Kreft M, Takafuta T, Shapiro SS, Sonnenberg A. Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin β subunits. J Cell Biol 156: 361–376, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta 1538: 99–117, 2001. [DOI] [PubMed] [Google Scholar]
- 53.Wang K, Ash JF, Singer SJ. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc Natl Acad Sci USA 72: 4483–4486, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson SG, Jones MR, Mullin BH, Dick IM, Richards JB, Pastinen TM, Grundberg E, Ljunggren O, Surdulescu GL, Dudbridge F, Elliott KS, Cervino AC, Spector TD, Prince RL. Common sequence variation in FLNB regulates bone structure in women in the general population and FLNB mRNA expression in osteoblasts in vitro. J Bone Miner Res 24: 1989–1997, 2009. [DOI] [PubMed] [Google Scholar]
- 55.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 27: 355–371, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng L, Baek HJ, Karsenty G, Justice MJ. Filamin B represses chondrocyte hypertrophy in a Runx2/Smad3-dependent manner. J Cell Biol 178: 121–128, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou AX, Hartwig JH, Akyürek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol 20: 113–123, 2010. [DOI] [PubMed] [Google Scholar]
- 58.Zhou X, Tian F, Sandzen J, Cao R, Flaberg E, Szekely L, Cao Y, Ohlsson C, Bergo MO, Boren J, Akyurek LM. Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci USA 104: 3919–3924, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]