Abstract Abstract
The eukaryotic ribosomal DNA cluster consists of multiple copies of three genes, 18S, 5. 8S and 28S rRNAs, separated by multiple copies of two internal transcribed spacers, ITS1 and ITS2. It is an important, frequently used marker in both molecular cytogenetic and molecular phylogenetic studies. Despite this, little is known about intragenomic variations within the copies of eukaryotic ribosomal DNA genes and spacers. Here we present data on intraindividual variations of ITS2 spacer in three species of Agrodiaetus Hübner, 1822 blue butterflies revealed by cloning technique. We demonstrate that a distinctly different intragenomic ITS2 pattern exists for every individual analysed. ITS2 sequences of these species show significant intragenomic variation (up to 3.68% divergence), setting them apart from each other on inferred phylogenetic tree. This variation is enough to obscure phylogenetic relationships at the species level.
Keywords: Agrodiaetus, Lycaenidae, Polyommatus, ITS2, ribosomal DNA, intraindividual variability, cloning
Introduction
The eukaryotic ribosomal DNA (rDNA) cluster consists of three genes, 18S, 5. 8S and 28S rRNAs, separated by two internal transcribed spacers, ITS1 and ITS2. This array forms a transcription unit, which is are typically represented in a genome by several hundred tandemly repeated copies (Long and David 1980, Gerbi 1985). The number of rDNA sequence variants can vary within a wide range both at the species and individual level. For example, different species of Drosophila Linnaeus, 1758 are estimated to have three to 18 variants of rDNA sequences (Stage and Eickbush 2007). The genome of sea sponge Amphimedon queenslandica Hooper & van Soest, 2006 was found to contain approximately 14.5 copies of rDNA sequences per haploid complement (Srivastava et al. 2010). Furthermore, individuals of the same species can have very different numbers of rDNA copies because the clusters display both meiotic rearrangements and somatic mosaicism. It has been shown that in humans for example, the number of rDNA sequences even within a single cluster can vary in an enormous extent, from one repeat unit up to 140 repeats (Stults et al. 2008).
Ribosomal RNA genes have been widely used in taxonomy, biogeographic, phylogenetic analyses, and molecular cytogenetic studies (Hillis and Davis 1986, Mindell and Honeycutt 1990, Wesson et al. 1993, Vogler and DeSalle 1994). In particular, more detailed and precise karyotypes studies became available since fluorescence in situ hybridization (FISH) technology was applied to the chromosomal physical mapping. FISH mapping identifies useful chromosomal markers that can be applied to studies of genome organization and species evolution and can also identify specific chromosomes, homologous chromosomes, chromosome rearrangements and sex chromosomes, among others (Nakajima et al. 2012). Ribosomal RNA genes are among the most mapped sequences in chromosomes in many animal groups including insects (Cabrero and Camacho 2008, Grozeva et al. 2010, 2011, Nguyen et al. 2010, Kuznetsova et al. 2012, Maryańska-Nadachowska et al. 2013, Gokhman et al. 2014, Kuznetsova et al. 2015, Vershinina et al. 2015).
Accordingly, rDNA can be excellent source of cytogenetic markers for comparative genomic studies, evolutionary studies as well as the genetic identification of species (Mantovani et al. 2005, Pedrosa-Harand et al. 2006, Cabral-de-Mello et al. 2011).
At the nucleotide sequence level coding regions and spacers can reveal phylogenetic relationships ranging from the level of major phyla of living organisms to the population level, because they differ widely in their rate of evolution (Hillis and Dixon 1991, Wesson et al. 1992, Kuperus and Chapco 1994, Muccio et al. 2000, Wiegmann et al. 2000). 18S and 28S rDNA genes are reported to be highly informative to reconstruct higher-level phylogenies in plants and animals (see e.g. Soltis et al. 2000, Mukha et al. 2002).
Unlike highly conserved rRNA genes, non-coding fast evolving transcribed spacers have high level of interspecific variability. Therefore, the internal transcribed spacers are considered to be useful phylogenetic markers, specifically for low-level phylogenetic analyses. ITS1 and ITS2 have been used extensively in phylogenetic reconstruction of closely related species and cryptic species complexes (Wilkerson et al. 2004). For instance, ITS have become the standard barcode of choice in most investigations for plants and fungi (Stoeckle 2003, Kress et al. 2005, Sass et al. 2007, Bellemain et al. 2010, Hollingsworth et al. 2011, Schocha et al. 2012, Li et al. 2015).
During PCR all variants of ITS sequences presented in genome are amplified, therefore, direct sequencing could lead to inaccurate or erroneous phylogenetic reconstructions. Accordingly identifying and examination levels of intragenomic and intraspecific variation among ITS sequences are of real importance.
Agrodiaetus is a species-rich subgenus within the Palearctic genus Polyommatus (Talavera et al. 2013). The subgenus includes ca. 130 described species (see Vila et al. 2010, Lukhtanov et al. 2008, 2015a, Vershinina and Lukhtanov 2010, Przybyłowicz et al. 2014, Lukhtanov and Tikhonov 2015). The subgenus was estimated to have originated only about three million years ago (Kandul et al. 2004). Nowadays this rapidly radiated group of butterflies is a model system in studies of speciation (Lukhtanov et al. 2005, Lukhtanov et al. 2015b), and rapid karyotype evolution (Kandul et al. 2007). Several molecular phylogenetic studies have been conducted on Agrodiaetus, also based on ITS2 molecular marker (Wiemers 2003, Wiemers et al. 2009, Wiemers et al. 2010, Lukhtanov et al. 2015a). However, until now rate of ITS2 intragenomic variations in this rapidly evolved group have never been analyzed.
This paper addresses a more detailed analysis of intraindividual variability of ITS2 region in three Polyommatus (Agrodiaetus) species: Polyommatus (Agrodiaetus) peilei Bethune-Baker, 1921, Polyommatus (Agrodiaetus) karindus (Reiley, 1921) and Polyommatus (Agrodiaetus) morgani (Le Cerf, 1909). These three species are closely related to each other (Lukhtanov et al. 2015b), but have clear differences in male wing color and karyotypes (haploid chromosome number are n= 38-39 in Polyommatus (Agrodiaetus) peilei; n=68 and n=73 in different populations of Polyommatus (Agrodiaetus) karindus; and n=25-27 in Polyommatus (Agrodiaetus) morgani) (Lukhtanov et al. 2015b). Direct sequencing of ITS2 give ambiguous results; thus, we sought to clone and sequence ITS2 from these species to quantify the prevalence of intragenomic ITS2 variation and determine its effect on phylogenetic reconstructions.
Material and methods
Butterflies (only males) were collected in NW Iran (Zagros mt., Kordestan provience) in 2007–2014. Bodies were placed in 2 ml plastic vials with 100% ethanol for DNA analysis. Wings were stored in glassine envelopes for morphological study. All samples are stored at Zoological Institute, St Petersburg, Russia.
ITS2 region was amplified using the primer pair: ITS-3 and ITS-4 (White et al. 1990). When ITS-3 and ITS-4 primers failed to amplify a sufficient product, self-designed lepidopteran primers were used:
ILYC2F 5`- GAGAAACATCCAGGACCACT - 3` and
ILYC2RB 5` - CTGATCTGAGGCCA ACG - 3`.
The PCR amplifications were performed either in 50 µl reaction volume containing ca. 10–20 ng genomic DNA and 0.5 mM of each primer, using 26 PCR Master Mix (Fermentas, Lithuania). The temperature profile was as follows: initial denaturation at 94 °C for 1 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 50 °C for 45 s, and extension at 72 °C for 1 min with a final extension at 72 °C for 10 min.
Amplified fragments were purified using GeneJET Gel Extraction Kit (Fermentas, Lithuania). Purification was carried out according to the manufacturer’s protocol. The success of PCR amplification and purification was evaluated by electrophoresis of the products in 1% agarose gel. Purified PCR product was used for direct sequencing or subsequent cloning.
ITS2 PCR products were cloned into blunt-end cloning vector pJET1.2 (Fermentas, Lithuania) according to the manufacturer’s protocol for 10 minutes at room temperature. The pJet1.2 plasmid selects successful ligations through the disruption of an otherwise lethal gene, eco47IR, which enables positive selection of the recombinants. Before ligation, a 3’-A overhang were removed from the PCR products by treating the PCR product with a proofreading DNA polymerase. For transformation 5 µl of the ligation mixture reaction were added to 50 µl of chemo-competent Escherichia coli DH101B cells an incubated for 10 min. on ice. After incubation transformation mixture were pipetted onto pre-warmed LB Anp IPTG agar plate and spread by using inoculation loop. Agar plates with competent Escherichia coli were incubated overnight at 37 °C.
For each cloning, more than 500 clones were obtained. To check if the cloning procedures were successful, PCR with ITS2-speciffic primers were conducted for 20 colonies per cloning reaction. GeneJET Plasmid Miniprep Kit (Fermentas, Lithuania) was used for preparation of plasmid DNA from recombinant Escherichia coli culture. A single colony from a freshly streaked selective plate were picked to inoculate 1–5 mL of LB medium supplemented with ampicillin and incubated for 12–16 hours at 37 °C while shaking at 200–250 rpm. The bacterial culture was harvested by centrifugation at 8000 rpm (6800 × g) in a microcentrifuge for 2 min at room temperature. The supernatant was decanted and all remaining medium was removed. The pelleted cells were resuspended and subjected to SDS/alkaline lysis to liberate the plasmid DNA. The resulting lysate was neutralized to create appropriate conditions for binding of plasmid DNA on the silica membrane in the spin column. Cell debris and SDS precipitate were pelleted by centrifugation, and the plasmid DNA were washed to remove contaminants and eluted.
Sequencing was carried out using 3500xL analyzer (Applied Biosystems). Not less than 300 ng of plasmid DNA template was used for sequencing procedure. Cloned fragments were analyzed edited and aligned in Bioedit Software.
A Bayesian approach for estimating phylogeny was used. Bayesian trees were inferred using partitioned models: GTR for nucleotide substitutions and standard model for indels as implemented in MRBAYES v. 3.2 (Ronquist and Huelsenbeck 2012). Each gap (indel) was treated as a single character regardless of the length of the gap, under the assumption that a given gap is a result from one mutational event (Simmons and Ochoterena 2000).
Results
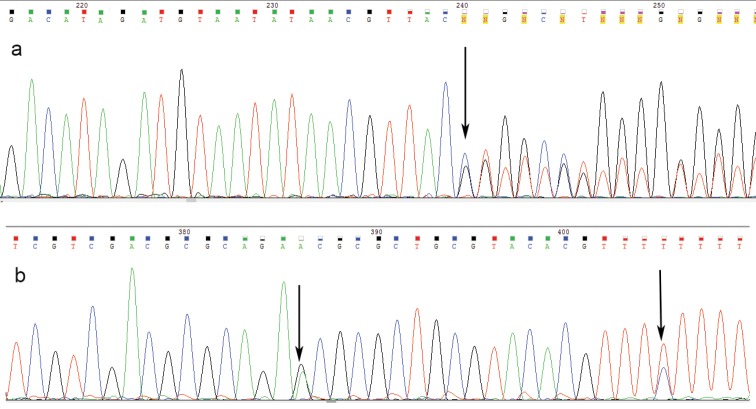
Sequenced region contained 3` end of 5.8S gene, ITS2, and the 5` end of the 28S gene. Direct sequencing of amplicons of 30 individuals (10 individuals per each species) displayed intra-individual heterogeneities in all specimens analyzed. There are two kinds of heterogeneities: single nucleotide substitutions and mono, bi- and multi-nucleotide insertions/deletions. The presence of heterogeneities was indicated by double peaks in substitution positions, and by a series of mixed peaks in case of indel events, both positioned after a sequence of good quality. The examples of heterogeneities revealed by direct sequencing are displayed in Figure 1.
Figure 1.
Examples of results from direct sequencing of ITS2. a Example of polymorphism caused by an indel (black arrow indicate the beginning position of an indel) b Example of single nucleotide substitutions (indicated by black arrows).
To elucidate the visible heterogeneity, the amplicons for 2 specimens of Polyommatus (Agrodiaetus) peilei, 2 specimens of Polyommatus (Agrodiaetus) karindus and one specimen of Polyommatus (Agrodiaetus) morgani were cloned and 10 clones per specimen were sequenced. The summary of the heterogeneities in the ITS region displayed by the clones is depicted in Table 1. Partial sequences of 5,8S and 28S genes were cropped from further analysis. Total length of ITS2 varied from 477 bp up to 512 bp depending on the presence of insertions\deletions. Uncorrected “p” pairwise distances for all clones are given in Table 2.
Table 1.
Variable positions among sequenced clones.
| Specimen | Clone number | Position | |||||||||||||||||||||||||||||||||||||||||
| 128 | 130 | 131 | 171 | 172 | 173 | 235 | 316 | 326 | 329 | 330 | 331 | 332 | 333 | 334 | 335 | 336 | 337 | 338 | 339 | 340 | 341 | 342 | 343 | 344 | 345 | 346 | 356 | 400 | 414 | 465 | |||||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #01 | T | G | - | C | G | C | A | A | T | T | T | T | T | T | T | T | - | - | C | G | T | T | T | T | T | - | C | G | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #02 | T | G | - | C | G | C | A | A | T | T | T | T | T | T | T | T | - | - | C | G | T | T | T | T | T | - | C | G | G | G | T | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #03 | T | A | A | C | A | C | G | G | C | T | T | T | T | T | T | T | T | T | T | G | T | T | T | T | T | - | C | A | G | A | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #04 | T | G | - | C | G | C | A | A | T | T | T | T | T | T | T | T | - | - | T | G | T | T | T | T | T | - | C | G | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #05 | T | A | A | C | A | C | G | G | C | T | T | T | T | T | T | T | T | T | T | G | T | T | T | T | T | T | - | A | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #06 | T | G | - | C | G | C | A | A | T | T | T | T | T | T | - | - | - | - | T | G | T | T | T | T | T | - | C | G | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #07 | T | G | - | C | G | C | A | A | C | T | T | T | T | T | T | T | T | T | T | G | T | T | T | T | T | T | - | A | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #08 | A | A | - | - | - | - | A | A | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | A | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #09 | T | A | A | C | A | C | G | G | C | T | T | T | T | T | T | T | T | T | T | G | T | T | T | T | T | T | - | A | G | G | C | |||||||||||
| W136 Polyommatus (Agrodiaetus) peilei | #10 | T | G | - | C | G | C | A | A | T | T | T | T | T | T | T | T | - | - | T | G | T | T | T | T | T | - | C | G | G | G | C | |||||||||||
| 14 | 41 | 128 | 131 | 169 | 170 | 171 | 176 | 184 | 185 | 186 | 187 | 188 | 189 | 190 | 191 | 192 | 193 | 194 | 195 | 196 | 197 | 198 | 199 | 200 | 235 | 236 | 326 | 336 | 337 | 338 | 345 | 346 | 356 | ||||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #01 | T | C | T | - | A | C | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | T | T | T | - | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #02 | T | T | T | A | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | C | T | T | T | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #03 | T | T | G | - | - | - | - | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | T | T | T | C | - | C | G | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #04 | T | T | T | - | A | C | C | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | T | T | - | C | - | C | G | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #05 | C | C | T | A | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | C | T | T | - | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #06 | T | T | T | A | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | C | T | T | T | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #07 | T | T | T | A | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | C | T | T | T | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #08 | T | C | T | - | A | C | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | T | - | - | C | - | C | G | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #09 | T | T | T | A | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | G | G | T | T | T | T | T | - | A | ||||||||
| W202 Polyommatus (Agrodiaetus) peilei | #10 | T | C | T | - | A | C | C | T | T | C | G | C | G | T | C | G | G | C | G | A | C | G | T | G | C | A | - | T | T | T | C | - | C | G | ||||||||
| 20 | 150 | 154 | 155 | 166 | 239 | 340 | 341 | 342 | |||||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #01 | A | C | - | - | T | T | T | T | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #02 | A | C | - | - | T | T | T | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #03 | A | T | C | G | T | T | - | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #04 | A | T | C | G | T | T | - | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #05 | A | T | C | G | T | T | T | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #06 | A | C | - | - | T | C | - | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #07 | A | T | C | G | C | T | T | T | T | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #08 | A | C | - | - | T | T | T | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #09 | A | C | - | - | T | T | - | - | - | |||||||||||||||||||||||||||||||||
| V145 Polyommatus (Agrodiaetus) karindus | #10 | G | C | - | - | T | T | - | - | - | |||||||||||||||||||||||||||||||||
| 27 | 84 | 128 | 136 | 169 | 170 | 171 | 331 | 335 | 336 | 337 | 337 | 338 | 339 | 340 | 346 | 351 | 352 | 353 | 354 | ||||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #01 | C | G | T | T | C | C | A | C | T | T | T | T | T | T | T | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #02 | C | G | T | T | C | C | A | T | T | T | T | T | T | T | T | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #03 | C | G | T | T | C | C | A | T | T | T | T | T | T | T | T | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #04 | C | G | T | T | C | C | A | T | T | T | T | T | T | T | T | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #05 | T | G | T | - | C | C | A | T | T | T | T | T | T | T | - | C | A | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #06 | T | G | T | - | C | C | A | T | T | T | T | T | T | T | - | C | A | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #07 | T | A | T | - | C | C | A | T | T | T | T | T | T | T | - | C | A | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #08 | C | G | T | T | C | C | A | T | T | T | T | T | T | T | - | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #09 | C | G | T | T | C | C | A | T | T | T | T | T | T | T | - | C | - | A | A | A | ||||||||||||||||||||||
| Z04 Polyommatus (Agrodiaetus) karindus | #10 | T | G | G | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | ||||||||||||||||||||||
| 7 | 38 | 43 | 79 | 127 | 128 | 148 | 171 | 172 | 173 | 229 | 301 | 318 | 319 | 320 | 321 | 322 | 323 | 324 | 325 | 326 | 327 | 328 | 329 | 330 | 331 | 332 | 333 | 334 | 347 | 348 | 349 | 350 | 352 | 353 | 354 | 355 | 357 | 358 | 391 | 400 | 472 | ||
| W127 Polyommatus (Agrodiaetus) morgani | #01 | A | A | C | - | C | G | T | - | - | - | T | C | A | C | A | C | G | T | T | T | T | T | T | T | T | - | - | - | - | A | A | C | G | - | - | - | A | A | A | A | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #02 | A | A | C | - | C | G | T | - | - | - | C | C | A | C | A | C | G | T | T | T | T | T | T | T | T | T | T | T | T | A | A | C | G | - | - | - | A | A | T | A | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #03 | A | A | C | - | A | T | C | C | G | C | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | A | G | A | G | G | T |
| W127 Polyommatus (Agrodiaetus) morgani | #04 | A | G | C | G | A | T | T | C | G | C | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | A | G | A | G | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #05 | G | G | C | G | C | G | T | - | - | - | T | C | A | C | A | C | G | T | T | C | T | T | T | T | T | T | T | T | - | A | A | C | G | - | - | - | A | A | A | A | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #06 | A | A | C | - | C | G | T | - | - | - | T | C | A | C | A | C | G | T | T | T | T | T | T | T | T | T | T | - | - | A | A | C | G | - | - | - | A | A | A | A | A | G |
| W127 Polyommatus (Agrodiaetus) morgani | #07 | A | A | C | - | C | G | T | C | G | C | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | A | G | A | G | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #08 | A | A | C | G | A | T | T | C | G | C | T | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | - | A | A | A | G | T |
| W127 Polyommatus (Agrodiaetus) morgani | #09 | A | A | T | G | A | T | T | C | G | C | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | A | G | A | G | G | G |
| W127 Polyommatus (Agrodiaetus) morgani | #10 | A | A | C | - | C | G | T | - | - | - | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | A | A | G | A | G | G | G |
Table 2.
Uncorrected ‘‘p’’ distance matrix of clones.
| Polyommatus (Agrodiaetus) peilei | W136_#01 | W136_#02 | W136_#03 | W136_#04 | W136_#05 | W136_#06 | W136_#07 | W136_#08 | W136_#09 | W136_#10 |
| W136_#01 | - | |||||||||
| W136_#02 | 0.0019 | - | ||||||||
| W136_#03 | 0.0196 | 0.0216 | - | |||||||
| W136_#04 | 0.0019 | 0.0039 | 0.0177 | - | ||||||
| W136_#05 | 0.0216 | 0.0236 | 0.0058 | 0.0196 | - | |||||
| W136_#06 | 0.0058 | 0.0078 | 0.0176 | 0.0039 | 0.0196 | - | ||||
| W136_#07 | 0.0117 | 0.0137 | 0.0157 | 0.0098 | 0.0098 | 0.0098 | - | |||
| W136_#08 | 0.0182 | 0.0202 | 0.0223 | 0.0162 | 0.0202 | 0.0162 | 0.0141 | - | ||
| W136_#09 | 0.0216 | 0.0236 | 0.0058 | 0.0196 | 0 | 0.0196 | 0.0098 | 0.0202 | - | |
| W136_#10 | 0.0019 | 0.0039 | 0.0177 | 0 | 0.0196 | 0.0039 | 0.0098 | 0.0162 | 0.0196 | - |
| Average | 0,0134 | |||||||||
| Polyommatus (Agrodiaetus) peilei | W202_#01 | W202_#02 | W202_#03 | W202_#04 | W202_#05 | W202_#06 | W202_#07 | W202_#08 | W202_#09 | W202_#10 |
| W202_#01 | - | |||||||||
| W202_#02 | 0.0139 | - | ||||||||
| W202_#03 | 0.0200 | 0.0157 | - | |||||||
| W202_#04 | 0.0142 | 0.022 | 0.0140 | - | ||||||
| W202_#05 | 0.0119 | 0.0059 | 0.0119 | 0.0259 | - | |||||
| W202_#06 | 0.0139 | 0 | 0.0157 | 0.0219 | 0.0059 | - | ||||
| W202_#07 | 0.0139 | 0 | 0.0157 | 0.0219 | 0.0059 | 0 | - | |||
| W202_#08 | 0.0122 | 0.0239 | 0.0160 | 0.0061 | 0.0239 | 0.0239 | 0.0239 | - | ||
| W202_#09 | 0.0119 | 0.0019 | 0.0137 | 0.0199 | 0.0078 | 0.0019 | 0.0019 | 0.0219 | - | |
| W202_#10 | 0.0100 | 0.0176 | 0.0177 | 0.0080 | 0.0176 | 0.0176 | 0.0176 | 0.0060 | 0.0157 | - |
| Average | 0,0135 | |||||||||
| Polyommatus (Agrodiaetus) karindus | V145_#01 | V145_#02 | V145_#03 | V145_#04 | V145_#05 | V145_#06 | V145_#07 | V145_#08 | V145_#09 | V145_#10 |
| V145_#01 | - | |||||||||
| V145_#02 | 0.0019 | - | ||||||||
| V145_#03 | 0.0078 | 0.0059 | - | |||||||
| V145_#04 | 0.0078 | 0.0059 | 0 | - | ||||||
| V145_#05 | 0.0059 | 0.0039 | 0.0019 | 0.0019 | - | |||||
| V145_#06 | 0.0059 | 0.0059 | 0.0078 | 0.0059 | 0.0078 | - | ||||
| V145_#07 | 0.0078 | 0.0098 | 0.0078 | 0.0078 | 0.0078 | 0.0137 | - | |||
| V145_#08 | 0.0019 | 0 | 0.0059 | 0.0059 | 0.0039 | 0.0059 | 0.0098 | - | ||
| V145_#09 | 0.0039 | 0.0019 | 0.0039 | 0.0039 | 0.0059 | 0.0019 | 0.0117 | 0.0019 | - | |
| V145_#10 | 0.0059 | 0.0039 | 0.0078 | 0.0059 | 0.0078 | 0.0039 | 0.0137 | 0.0039 | 0.0019 | - |
| Average | 0,0056 | |||||||||
| Polyommatus (Agrodiaetus) karindus | Z704_#01 | Z704_#02 | Z704_#03 | Z704_#04 | Z704_#05 | Z704_#06 | Z704_#07 | Z704_#08 | Z704_#09 | Z704_#10 |
| Z704_#01 | - | |||||||||
| Z704_#02 | 0.0019 | - | ||||||||
| Z704_#03 | 0.0019 | 0 | - | |||||||
| Z704_#04 | 0.0019 | 0 | 0 | - | ||||||
| Z704_#05 | 0.0098 | 0.0078 | 0.0078 | 0.0078 | - | |||||
| Z704_#06 | 0.0098 | 0.0078 | 0.0078 | 0.0078 | 0 | - | ||||
| Z704_#07 | 0.0117 | 0.0098 | 0.0098 | 0.0098 | 0.0019 | 0.0019 | - | |||
| Z704_#08 | 0.0039 | 0.0019 | 0.0019 | 0.0019 | 0.0059 | 0.0059 | 0.0078 | - | ||
| Z704_#09 | 0.0039 | 0.0019 | 0.0019 | 0.0019 | 0.0059 | 0.0059 | 0.0078 | 0 | - | |
| Z704_#10 | 0.0141 | 0.0162 | 0.0162 | 0.0162 | 0.0162 | 0.0162 | 0.0182 | 0.0182 | 0.0182 | - |
| Average | 0,0072 | |||||||||
| Polyommatus (Agrodiaetus) morgani | V127_#01 | V127_#02 | V127_#03 | V127_#04 | V127_#05 | V127_#06 | V127_#07 | V127_#08 | V127_#09 | V127_#10 |
| V127_#01 | - | |||||||||
| V127_#02 | 0.0101 | - | ||||||||
| V127_#03 | 0.0289 | 0.0307 | - | |||||||
| V127_#04 | 0.0267 | 0.0328 | 0.0083 | - | ||||||
| V127_#05 | 0.0122 | 0.0141 | 0.0368 | 0.0246 | - | |||||
| V127_#06 | 0.0041 | 0.0101 | 0.0267 | 0.0205 | 0.0121 | - | ||||
| V127_#07 | 0.0144 | 0.0246 | 0.0083 | 0.0083 | 0.0266 | 0.0185 | - | |||
| V127_#08 | 0.0165 | 0.0266 | 0.0104 | 0.0104 | 0.0246 | 0.0246 | 0.0146 | - | ||
| V127_#09 | 0.0226 | 0.0328 | 0.0083 | 0.0041 | 0.0287 | 0.0267 | 0.0083 | 0.0104 | - | |
| V127_#10 | 0.0124 | 0.0226 | 0.0104 | 0.0104 | 0.0267 | 0.0165 | 0.0021 | 0.0167 | 0.0104 | - |
| Average | 0,0177 | |||||||||
There were 11 single-base substitutions, 3 mono and 4 multi-nucleotide indels, in clones of specimen W136 (Polyommatus (Agrodiaetus) peilei). Interestingly, that clone “W136_#08” differed significantly from all others in having 16-nycleotide polyT deletion at positions “329-344” and 3 base indel at position “171-173”. Clones of second specimen Polyommatus (Agrodiaetus) peilei (W202) had 8 sites with single nucleotide substitutions and 8 positions, where mono multi-nucleotide indels occurred. Three clones had large polymorphic 17-nucleotide indel at positions “184-200”. Variation among clones was significant, with intragenomic differences ranging from 0.0% to 2.39%. The average intragenomic genetic distances for two specimens of Polyommatus (Agrodiaetus) peilei (W136 and W202) were very similar: 1.34% and 1.35% respectively.
Polyommatus (Agrodiaetus) karindus had significantly lower rate of intragenomic variability. Specimens V145 and Z704 had 9 and 10 polymorphic positions, respectively. Furthermore, majority number of indels and base substitutions of Z704 specimen is accounted for by one clone (Z704#10). It has one single substitution and 3 multi-nucleotide deletions, which never occurred in other clones. The average intragenomic genetic distances for two specimens of Polyommatus (Agrodiaetus) karindus (V145 and Z704) were: 0.56% and 0.72%, respectively. The highest value was 1.82%.
Clones of Polyommatus (Agrodiaetus) morgani ITS2 showed greater diversity than the other 2 species. For instance, the genetic distance between V127#05 clone and V127#03 was 3.68%. The average intragenomic genetic distance was also significantly higher for this species – 1.77%
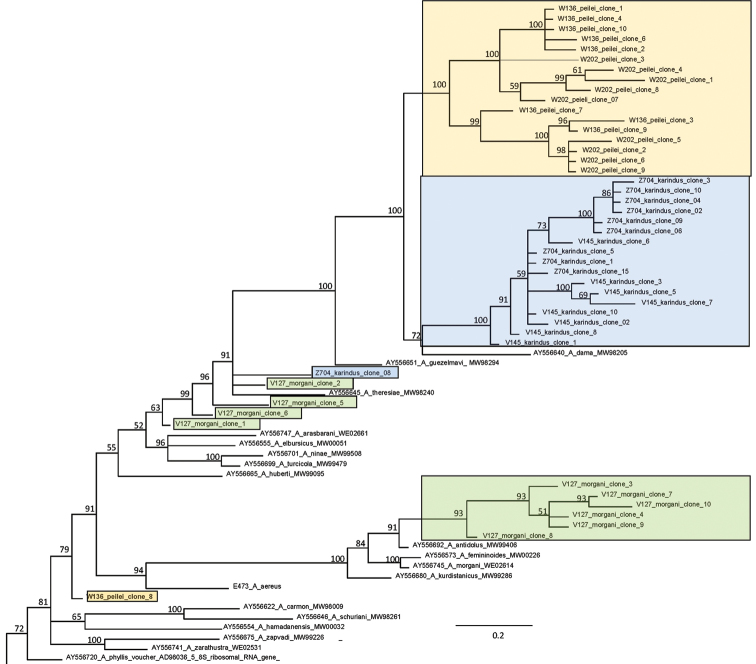
In Bayesian analysis 50 cloned amplicons from Polyommatus (Agrodiaetus) peilei, Polyommatus (Agrodiaetus) karindus, Polyommatus (Agrodiaetus) morgani and ITS2 sequences from all Agrodiaetus species available in the GenBank were included, giving a total of 127 sequences. Since Polyommatus icarus (Rottemburg, 1775) was earlier inferred as sister clade to the subgenus Agrodiaetus (Talavera et al. 2013), we used one specimen (GenBank accession number AY556732) as outgroup to root the phylogeny. Fragment of consensus Bayesian tree, showing clusterization of cloned sequences is given in Figure 2. The complete tree is given online in the Suppl. material 1.
Figure 2.
Fragment of consensus Bayesian tree of the subgenus Agrodiaetus inferred from ITS2 sequences. Posterior probability values >50% are shown. The complete tree is given online in the Suppl. material 1. Cloned sequences of three studied species are highlighted: Polyommatus (Agrodiaetus) peilei – orange colour, Polyommatus (Agrodiaetus) karindus – blue colour, Polyommatus (Agrodiaetus) morgani – green colour.
Discussion
Despite the popularity of the ITS2 nuclear rDNA marker in systematics of different groups of animals and plants, its variability on intraspecific and intraindividual level is still poorly known. The occurrence of multiple ITS2 copies within a single genome should be accounted for before rDNA is used for phylogenetic or population studies. Furthermore, investigation of rates of intra-individual polymorphism can lighten addressing questions regarding speciation, species hybridization end evolutionary history. It is generally considered that multigene families, such as rDNA maintain homogeneity of all copies as a result of concerted evolution (processes of gene conversion and unequal crossing over) (Zimmer et al. 1980, Dover 1982). Mutations rapidly spread to all members of the gene family even if there are arrays located on different chromosomes (Dover 1982, Arnheim1983, Gerbi 1985, Tautz et al. 1988). The efficiency of homogenization of rDNA is usually high (Liao 1999). Concerted evolution of noncoding sequences, such as internal transcribed spacers, can result in fixed interspecific differences and intraspecific homogeneity. Despite this assumption, our results show, that intraindividual variability can be maintained, when mutation rates are higher than rates of homogenization. This can lead to erroneous phylogenetic reconstructions and species misidentification.
Here we contribute with the first insight into the intraspecific ITS2 diversity in the blue butterflies of subgenus Agrodiaetus.
The ITS2 of all specimens of three Agrodiaetus species - (Polyommatus (Agrodiaetus) peilei, Polyommatus (Agrodiaetus) karindus and Polyommatus (Agrodiaetus) morgani) were intragenomically variable. There were a number of indels and base substitutions accounting for both the length and sequence variabilities. Numerous indels lead to length variation (477-512 bp) of studied sequences. Bayesian phylogenetic reconstruction revealed that cloned sequences of certain individuals did not form a monophyletic unanimity, but the majority of clones clustered together within species borders. In particular, clones of Polyommatus (Agrodiaetus) peilei and Polyommatus (Agrodiaetus) karindus individuals are recovered as two distinct separated clusters, both with a Bayesian posterior probability of 1.00. The position of 6 clones of Polyommatus (Agrodiaetus) morgani specimen on the ITS2 tree support the conclusion that abovementioned species belong to “antidolus” species-group which comprise 5 allopatric in distribution, closely related taxa: Polyommatus (Agrodiaetus) femininoides (Eckweiler, 1987), Polyommatus (Agrodiaetus) antidolus (Rebel, 1901), Polyommatus (Agrodiaetus) aereus (Eckweiler, 1998), Polyommatus (Agrodiaetus) kurdistanicus (Forster, 1961) and Polyommatus (Agrodiaetus) morgani. “Antidolus” clade revealed with a high level of posterior probability. However, when considering all cloning data, in some cases differences between cloned sequences of the same individual were greater than that between species. For instance, the remainder of Polyommatus (Agrodiaetus) morgani clones are placed as the basal taxa to clade, consist of Polyommatus (Agrodiaetus) guezelmavi (Olivier, Puplesiene, van der Poorten, De Prins & Wiemers, 1999), Polyommatus (Agrodiaetus) dama (Staudinger, 1892) and majority of Polyommatus (Agrodiaetus) peilei and Polyommatus (Agrodiaetus) karindus clones. One clone of Polyommatus (Agrodiaetus) karindus (Z704_#08) also was recovered as sister taxa to abovementioned clade. Finally, W136_#08 clone of Polyommatus (Agrodiaetus) peilei is found to be more genetically distant from other clones of this individual than the great number of other species of the subgenus Agrodiaetus (Figure 2).
Recent works showed that tandem arrays of rRNA genes in most Lepidoteran species form one or two so-called rDNA clusters, although some exceptions in cluster number exist (Nguen et al. 2010). Data on the number and distribution of rDNA clusters in genomes of lycaenid butterflies are very scarce. Previous investigation by Vershinina et. al. (2015) examined ribosomal clusters in seven blue butterflies of the genus Polyommatus and showed the presence of two different variants of the location of major rDNA clusters in Polyommatus species: with one or two rDNA-carrying chromosomes in haploid karyotype (Vershinina et al. 2015). Polyommatus (Agrodiaetus) peilei, Polyommatus (Agrodiaetus) karindus and Polyommatus (Agrodiaetus) morgani were among studied species, which bear a single rDNA cluster. Thus, all intragenomic ITS2 patterns for every individual analysed, belong to a single rDNA cluster, which means that examined level of intragenomic variability not caused by sequencing ITS2 copies located on different chromosomes.
To conclude, our study demonstrates that the results of direct sequencing may not describe the actual and entire set of sequence variants. Level of divergence between clones of one individual can be comparable to interspecific genetic differences variations or even exceed them. Hence, cloning and subsequent intraindividual haplotypes handling are required for reliable phylogenetic reconstructions.
Acknowledgements
The complete financial support for this study was provided by the grant from the Russian Science Foundation no. 14-14-00541 to the Zoological Institute of Russian Academy of Sciences. We thank Boris Anokhin and Alsu Saifitdinova for help in DNA cloning. The work was partially performed using equipment of the ‘Chromas’ Core Facility and Centre for Molecular and Cell Technologies of St. Petersburg State University. Postdoctoral fellowship was provided to Nazar Shapoval from St. Petersburg State University.
Citation
Shapoval NA, Lukhtanov VA (2015) Intragenomic variations of multicopy ITS2 marker in Agrodiaetus blue butterflies (Lepidoptera, Lycaenidae). Comparative Cytogenetics 9(4): 483–497. doi: 10.3897/CompCytogen.v9i4.5429
Supplementary materials
Consensus Bayesian tree of the subgenus Polyommatus (Agrodiaetus) inferred from ITS2 sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Nazar A. Shapoval, Vladimir A. Lukhtanov
Data type: TIFF image
Explanation note: Consensus Bayesian tree of the subgenus Polyommatus (Agrodiaetus) inferred from ITS2 sequences. Posterior probability values >50% are shown.
References
- Arnheim N. (1983) Concerted evolution of multigene families. In: Nei M, Koehn RK. (Eds) Evolution of genes and proteins. Sinauer, Sunderland (MA), 38–61. [Google Scholar]
- Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiology 10: . doi: 10.1186/1471-2180-10-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral-de-Mello DC, Cabrero J, Lopez-Leon MD, Camacho JP. (2011) Evolutionary dynamics of 5S rDNA location in acridid grasshoppers and its relationship with H3 histone gene and 45S rDNA location. Genetica 139: 921–931. doi: 10.1007/s10709-011-9596-7 [DOI] [PubMed] [Google Scholar]
- Cabrero J, Camacho JPM. (2008) Location and expression of ribosomal RNA genes in grasshoppers: abundance of silent and cryptic loci. Chromosome Research 16: 595–607. doi: 10.1007/s10577-008-1214-x [DOI] [PubMed] [Google Scholar]
- Dover GA. (1982) Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. doi: 10.1038/299111a0 [DOI] [PubMed] [Google Scholar]
- Gerbi SA. (1985) Evolution of ribosomal DNA. In: MacIntyre RJ. (Ed.) Molecular evolutionary genetics. Plenum Press, London, New York, 419–517. doi: 10.1007/978-1-4684-4988-4_7 [Google Scholar]
- Gokhman VE, Anokhin BA, Kuznetsova VG. (2014) Distribution of 18S rDNA sites and absence of the canonical TTAGG insect telomeric repeat in parasitoid Hymenoptera. Genetica 142: 317–322. doi: 10.1007/s10709-014-9776-3 [DOI] [PubMed] [Google Scholar]
- Grozeva S, Kuznetsova V, Anokhin B. (2010) Bed bug cytogenetics: karyotype, sex chromosome system, FISH mapping of 18S rDNA, and male meiosis in Cimex lectularius Linnaeus, 1758 (Heteroptera: Cimicidae). Comparative Cytogenetics 4: 151–160. doi: 10.3897/compcytogen.v4i2.36 [Google Scholar]
- Grozeva S, Kuznetsova V, Anokhin B. (2011) Karyotypes, male meiosis and comparative FISH mapping of 18S ribosomal DNA and telomeric (TTAGG)n repeat in eight species of true bugs (Hemiptera: Heteroptera). Comparative Cytogenetics 5(4): 355–374. doi: 10.3897/compcytogen.v5i4.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva S, Anokhin B, Kuznetsova VG. (2014) Bed bugs (Hemiptera). In: Sharachov I. (Ed.) Protocols for Cytogenetic Mapping of Arthropod Genomes. Taylor & Francis, CRC press, Boca Raton, 285–326. [Google Scholar]
- Hillis DM, Davis SK. (1986) Evolution of ribosomal DNA: fifty million years of recorded history in the frog genus Rana. Evolution 40: 1275–1288. doi: 10.2307/2408953 [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology 66: 411–453. doi: 10.1086/417338 [DOI] [PubMed] [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. (2011) Choosing and using a plant DNA barcode. PLoS ONE 6: . doi: 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul NP, Lukhtanov VA, Dantchenko AV, Coleman JWS, Sekercioglu CH, Haig D, Pierce NE. (2004) Phylogeny of Agrodiaetus Hübner 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences of COI and COII and nuclear sequences of EF1-α: Karyotype diversification and species radiation. Systematic Biology 53(2): 278–298. doi: 10.1080/10635150490423692 [DOI] [PubMed] [Google Scholar]
- Kandul NP, Lukhtanov VA, Pierce NE. (2007) Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 61(3): 546–559. doi: 10.1111/j.1558-5646.2007.00046.x [DOI] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. (2005) Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America 102(23): 836–8374. doi: 10.1073/pnas.0503123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperus WR, Chapco W. (1994) Usefulness of internal transcribed spacer regions of ribosomal DNA in Melanopline (Orthoptera. Acrididae) systematics. Annals of the Entomological Society of America 87: 751–754. doi: 10.1093/aesa/87.6.751 [Google Scholar]
- Kuznetsova VG, Grozeva SM, Anokhin BA. (2012) The first finding of (TTAGG)n telomeric repeat in chromosomes of true bugs (Heteroptera, Belostomatidae). Comparative Cytogenetics 6(4): 341–346. doi: 10.3897/compcytogen.v6i4.4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova VG, Maryańska-Nadachowska A, Anokhin B, Aguin-Pombo D. (2015) Evidence for TTAGG telomere repeats and rRNA gene clusters in leafhoppers of the genus Alebra (Hemiptera: Auchenorrhyncha: Cicadellidae). European Journal of Entomology 112(2): 207–214. doi: 10.14411/eje.2015.045. [Google Scholar]
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. (2015) Plant DNA barcoding: from gene to genome. Biological Reviews 90: 157–166. doi: 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- Liao A. (1999) Molecular evolution ’99 Concerted evolution: molecular mechanism and biological implications. The American Journal of Human Genetics 64: 24–30. doi: 10.1086/302221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, David ID. (1980) Repeated genes in eukaryotes. Annual Review of Biochemistry 49: 727–764. doi: 10.1146/annurev.bi.49.070180.003455 [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Kandul NP, Plotkin JB, Dantchenko AV, Haig D, Pierce NE. (2005) Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature 436(7049): 385–389. doi: 10.1038/nature03704 [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Shapoval NA, Dantchenko AV. (2008) Agrodiaetus shahkuhensis sp. n. (Lepidoptera, Lycaenidae), a cryptic species from Iran discovered by using molecular and chromosomal markers. Comparative Cytogenetics 2(2): 99–114. [Google Scholar]
- Lukhtanov VA, Shapoval NA, Dantchenko AV. (2014) Taxonomic position of several enigmatic Polyommatus (Agrodiaetus) species (Lepidoptera, Lycaenidae) from Central and Eastern Iran: insights from molecular and chromosomal data. Comparative Cytogenetics 8(4): 313–322. doi: 10.3897/CompCytogen.v8i4.8939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Dantchenko AV, Vishnevskaya MS, Saifitdinova AF. (2015a) Detecting cryptic species in sympatry and allopatry: analysis of hidden diversity in Polyommatus (Agrodiaetus) butterflies (Lepidoptera: Lycaenidae). Biological Journal of the Linnean Society. doi: 10.1111/bij.12596
- Lukhtanov VA, Shapoval NA, Anokhin BA, Saifitdinova AF, Kuznetsova VG. (2015b) Homoploid hybrid speciation and genome evolution via chromosome sorting. Proceedings of the Royal Society B 282: . doi: 10.1098/rspb.2015.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Tikhonov AV. (2015) Chromosomal and molecular evidence for presence of Polyommatus (Agrodiaetus) poseidon (Lepidoptera, Lycaenidae) in Caucasus region. Comparative Cytogenetics 9(2): 249–255. doi: 10.3897/CompCytogen.v9i2.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani M, Dos L, Abel S, Moreira-Filho O. (2005) Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123: 211–216. doi: 10.1007/s10709-004-2281-3 [DOI] [PubMed] [Google Scholar]
- Maryańska-Nadachowska A, Kuznetsova VG, Karamysheva TV. (2013) Chromosomal location of rDNA clusters and TTAGG telomeric repeats in eight species of the spittlebug genus Philaenus (Hemiptera: Auchenorrhyncha: Aphrophoridae). European Journal of Entomology 110(3): 411–418. doi: 10.14411/eje.2013.055 [Google Scholar]
- Mindell DP, Honeycutt RL. (1990) Ribosomal RNA in vertebrates: evolution and phylogenetic applications. Annual Review of Ecology and Systematics 21: 541–566. doi: 10.1146/annurev.es.21.110190.002545 [Google Scholar]
- Muccio T, Marinucci M, Fruster L, Maroli M, Pesson B, Gramiccia M. (2000) Phylogenetic analysis of Phlebotomus species belonging to the subgenus Larrousius (Diptera: Psychodidae) by ITS2 rDNA sequences. Insect Biochemistry and Molecular Biology 30: 387–393. doi: 10.1016/S0965-1748(00)00012-6 [DOI] [PubMed] [Google Scholar]
- Mukha D, Wiegmann BM, Schal C. (2002) Evolution and phylogenetic information content of the ribosomal DNA repeat unit in the Blattodea (Insecta). Insect Biochemistry and Molecular Biology 32: 951–960. doi: 10.1016/S0965-1748(01)00164-3 [DOI] [PubMed] [Google Scholar]
- Nakajima RT, Cabral-de-Mello DC, Valente GT, Venere PC, Martins C. (2012) Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evolutionary Biology 12: . doi: 10.1186/1471-2148-12-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Sahara K, Yoshido A, Marec F. (2010) Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica 138(3): 343–354. doi: 10.1007/s10709-009-9424-5 [DOI] [PubMed] [Google Scholar]
- Pedrosa-Harand A, de Almeida CC, Mosiolek M, Blair MW, Schweizer D, Guerra M. (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. TAG Theoretical and Applied Genetics 112: 924–933. doi: 10.1007/s00122-005-0196-8 [DOI] [PubMed] [Google Scholar]
- Przybyłowicz Ł, Lukhtanov V, Lachowska-Cierlik D. (2014) Towards the understanding of the origin of the Polish remote population of Polyommatus (Agrodiaetus) ripartii (Lepidoptera: Lycaenidae) based on karyology and molecular phylogeny. Journal of Zoological Systematics and Evolutionary Research 52(1): 44–51. doi: 10.1111/jzs.12040 [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass C, Little DP, Stevenson DW, Specht CD. (2007) DNA barcoding in the cycadales: testing the potential of proposed barcoding markers for species identification of cycads. PLoS ONE 2: . doi: 10.1371/journal.pone.0001154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schocha CL, Seifert KA, Huhndorf S, Robert V, Spougea JL, Levesque CA, Chen W. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109(16): 6241–6246. doi: 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49(2): 369–381. doi: 10.1093/sysbio/49.2.369 [PubMed] [Google Scholar]
- Soltis DE, Slotis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, Axtell M, Swensen SM, Princ LM, Kress WJ, Nixon KC, Farris JS. (2000) Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Botanical Journal of the Linnean Society 133(4): 381–461. doi: 10.1006/bojl.2000.0380 [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726. doi: 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage DE, Eickbush TH. (2007) Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome research 17(12): 1888–1897. doi: 10.1101/gr.6376807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle M. (2003) Taxonomy, DNA, and the bar code of life. BioScience 53: 796–797. [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ. (2008) Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Research 18: 13–18. doi: 10.1101/gr.6858507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Lukhtanov VA, Pierce NE, Vila R. (2013) Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics 29: 166–192. doi: 10.1111/j.1096-0031.2012.00421.x [DOI] [PubMed] [Google Scholar]
- Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA. (1988) Complete sequences of the ribosomal RNA genes of Drosophila melanogaster. Molecular Biology and Evolution 5: 366–376. [DOI] [PubMed] [Google Scholar]
- Vershinina AO, Lukhtanov VA. (2010) Geographical distribution of the cryptic species Agrodiaetus alcestis alcestis, A. alcestis karacetinae and A. demavendi (Lepidoptera, Lycaenidae) revealed by cytogenetic analysis. Comparative Cytogenetics 4(1): 1–11. doi: 10.3897/compcytogen.v4i1.21 [Google Scholar]
- Vershinina AO, Anokhin BA, Lukhtanov VA. (2015) Ribosomal DNA clusters and telomeric (TTAGG)n repeats in blue butterflies (Lepidoptera, Lycaenidae) with low and high chromosome numbers. Comparative Cytogenetics 9(2): 161–171. doi: 10.3897/CompCytogen.v9i2.4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila R, Lukhtanov VA, Talavera G, Gil-T F, Pierce NE. (2010) How common are dot-like distribution ranges? Taxonomical oversplitting in Western European Agrodiaetus (Lepidoptera, Lycaenidae) revealed by chromosomal and molecular markers. Biological Journal of the Linnean Society 101: 130–154. doi: 10.1111/j.1095-8312.2010.01481.x [Google Scholar]
- Vogler AP, DeSalle R. (1994) Evolution and phylogenetic information content of the ITS-1 region in the tiger beetle Cicindela dorsalis. Molecular Biology and Evolution 11: 393–405. [DOI] [PubMed] [Google Scholar]
- Wesson DM, Porter CH, Collins FH. (1992) Sequence and secondary structure comparisons of ITS rDNA in mosquitoes (Diptera: Culicidae). Molecular Phylogenetics and Evolution 1: 253–269. doi: 10.1016/1055-7903(92)90001-W [DOI] [PubMed] [Google Scholar]
- Wesson DM, McLain DK, Oliver JH, Piesman J, Collins FH. (1993) Investigation of the validity of species status of Ixodes dammini (Acadri: Ixodidae) using rDNA. Proceedings of the National Academy of Sciences USA 90: 10221–10225. doi: 10.1073/pnas.90.21.10221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee, Taylor JS. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols, a Guide to Methods and Applications. Academic Press, New York, 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1 [Google Scholar]
- Wiegmann BM, Mitter C, Regier JC, Friedlander TP, Wagner DM, Nielsen ES. (2000) Nuclear genes resolve Mesozoic-aged divergences in the insect order Lepidoptera. Molecular Phylogenetics and Evolution 15: 242–259. doi: 10.1006/mpev.1999.0746 [DOI] [PubMed] [Google Scholar]
- Wiemers M. (2003) Chromosome differentiation and the radiation of the butterfly subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus) a molecular phylogenetic approach. Ph.D. Dissertation, University of Bonn, Bonn, Germany, 203 pp http://hss.ulb.uni-bonn.de/2003/0278/0278.htm [Google Scholar]
- Wiemers M, Keller A, Wolf M. (2009) ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evolutionary Biology 9: . doi: 10.1186/1471-2148-9-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Stradomsky BV, Vodolazhsky DI. (2010) A molecular phylogeny of Polyommatus s. str. and Plebicula based on mitochondrial COI and nuclear ITS2 sequences (Lepidoptera: Lycaenidae). European Journal of Entomology 107: 325–336. doi: 10.14411/eje.2010.041 [Google Scholar]
- Wilkerson RC, Reinert JF, Li C. (2004) Ribosomal DNA ITS2 Sequences Differentiate Six Species in the Anopheles crucians Complex (Diptera: Culicidae). Journal of Medical Entomology 41(3): 392–401. doi: 10.1603/0022-2585-41.3.392 [DOI] [PubMed] [Google Scholar]
- Zimmer EA, Martin SL, Beverly SM, Kan YW, Feder JL. (1980) Rapid duplication and loss of genes coding for the chains of hemoglobin. Proceedings of the National Academy of Sciences of the United States of America 77: 2158–2162. doi: 10.1073/pnas.77.4.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consensus Bayesian tree of the subgenus Polyommatus (Agrodiaetus) inferred from ITS2 sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Nazar A. Shapoval, Vladimir A. Lukhtanov
Data type: TIFF image
Explanation note: Consensus Bayesian tree of the subgenus Polyommatus (Agrodiaetus) inferred from ITS2 sequences. Posterior probability values >50% are shown.