Abstract
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that play crucial roles in numerous biological processes. However, the role of miRNAs in antibacterial defence in fish has not been fully determined. Here, we identified that nine miRNAs are differentially expressed in kidney between susceptible and resistant grass carp strains. Analysis of spatial and temporal miRNA expression patterns suggests that cid-miRn-115 and miR-142a-3p are potential regulators of anti-bacterial activity. Overexpressing of cid-miRn-115 and miR-142a-3p results in a visible change in Ctenopharyngodon idella kidney (CIK) cells immune effector activity. Bioinformatics analysis and overexpressing assay shows that cid-miRn-115 and miR-142a-3p directly regulate tlr5 expression. cid-miRn-115 and miR-142a-3p overexpressing leads to a significant decrease in tlr5 expression in CIK, thereby repressing its downstream genes, such as il-1β, il-8 and tnf-α. These findings provide a novel insight into the determination of anti-bacterial compounds in grass carp.
MicroRNAs (miRNAs) are a class of genome-encoded small RNAs that post-transcriptionally regulate the expression of cellular mRNAs exhibiting partial or full complementary miRNA-binding sites1. Several studies have revealed that miRNAs play important roles in the control of many biological processes, such as cell differentiation, proliferation and apoptosis, development, immunity, metabolism and stem cell maintenance2. It is now evident that aberrant miRNA expression in the immune system is sufficient to cause disease, thus indicating that proper regulation of miRNA expression is crucial for disease prevention3. MiRNAs have well-established roles in eukaryotic host responses to viruses, and to extracellular and invasive bacterial pathogens4. The involvement of miRNAs in bacterial infections was first discovered in plants; Arabidopsis miR-393 was shown to contribute to resistance against the extracellular pathogen Pseudomonas syringae, presumably by repressing auxin signalling5. Based on studies of plant immunity and defence mechanisms, it has been suggested that eukaryotes restrict bacterial pathogens by a balanced induction of miRNAs that repress negative defence regulators as well as suppressing miRNAs that repress positive effectors of defence6. Therefore, the study of miRNA-mediated host-bacteria interactions may further elucidate the mechanisms underlying bacterial infection and host counteraction.
The grass carp (Ctenopharyngodon idella) is an economically important cultured fish species in China; its farming results in the largest yield for a single species worldwide7. In recent years, the rapid development of the grass carp aquaculture industry is accompanied with increasingly severe infections, caused by viruses, bacteria and parasites, and resulting in great economic losses8. Investigating mechanisms of host immunity will greatly improve the countrol and prevention of disease. Recently, numerous immune-related genes have been successfully identified in grass carp using various molecular methods. Following the successful completion of furture aquatic animal genome projects, studies of the regulation mechanisms of noncoding RNAs responding to these genes will become increasingly important.
Aeromonas hydrophila, which was first recognized as the causal agent of haemorrhagic septicaemia9, is considered to be the dominant cause of motile aeromonad septivaemia (MAS) in China10, and appears to be distributed globally. A. hydrophila is present in a wide variety of foods (introduced from water, animal faces or food handlers), and, thereby, has the potential to be a significant food-borne pathogen, representing a serious public health concern11. Regardless of the important regulatory role of miRNAs in the host immune system, no studies have been conducted on miRNA transcriptomes and their expression profiles related to immune response to foreign challenge in A. hydrophila. At present, the pathogenic mechanism of A. hydrophila infection is poorly understood and the involvement of miRNAs during A. hydrophila infection has not been reported. We thus aimed to determine the repertoire of miRNAs expressed in the kidney of grass carp and to use this repertoire to study the responses of this teleost to A. hydrophila infection. Solexa sequencing technology was used in this study to sequence and analyse miRNA libraries generated from susceptible grass carp (SGC) and resistant grass carp (RGC) strains.
In this study, we aimed first to characterize the expression of miRNA in the grass carp in relation to MAS, and second to evaluate the diagnostic potential of the investigated miRNAs as biomarker for MAS. Using two kidney microRNA transcriptomes from SGC or RGC infected with a highly pathogenic A. hydrophila, we show that the kidney exhibit different miRNA expression levels. Additionally, we show that these miRNAs are differentially expressed in the immune related tissue and clear time-dependent expression pattern after the bacterial challenge, suggesting their potential use as biomarkers for MAS conditions.
Materials and Methods
Animals used
Grass carp with an average weight of 50 g were cultured individually in Wujiang National Farm of Chinese Four Family Carps, Jiangsu Province, China. Animals were raised at 28 °C in 400 L aerated tanks for one week before the experiment and fed twice daily (in the morning and late in the afternoon) at a ratio of 5% of the total biomass. All experiments were conducted under the guidance of the Care and Use of Laboratory Animals in China. This research was approved by the Committee on the Ethics of Animal Experiments of Shanghai Ocean University, China.
Grass carp were divided into three groups (30 animals per group) for the injection experiments. The conditions were identical among the tanks and the fish were randomly distributed into different tanks. Two groups were maintained in two aquariums and intraperitoneally injected with A. hydrophila AH10 (Aquatic Pathogen Collection Centre of Ministry of Agriculture, China) at a dose of 7.0 × 106 cells suspended in 100 μl PBS per fish. The third group was injected with PBS as control. All fish were observed every 4 h for any mortality and samples were collected until the termination of the experiment at 240 h post-challenge. Grass carp that died in the first 72 h post-challenge were classified as susceptible grass carp (SGC), whereas the animals that survived over 240 h post-challenge were considered resistant grass carp (RGC). The kidney tissues of randomly-selected three fish from both the susceptible and resistant groups were collected and, labeled as SGC and RGC, respectively. Approximately 0.5 g kidney tissue was cut and kept at −80 °C until RNA isolation. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and stored at −80 °C. RNA was quantified using a NanoDrop Spectrophotometer 2000c (NanoDrop Technologies, Wilmington, DE, USA), and its quality was assessed on a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). All samples used had λ260/280 and λ260/230 ratios > 1.8.
Small RNA library construction and Illumina sequencing
RNA samples were harvested from kidney of SGC and RGC animals, and immediately frozen in liquid nitrogen. Small RNA libraries were constructed using a TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, Califomia, USA). Approximately 20 μg of small RNA was submitted for sequencing. Briefly, the Solexa sequencing was performed as follows: RNA was purified by polyacrylamide gel electrophoresis (PAGE) to enrich for the molecules in the range of 17–27 nucleotides, and was then ligated with 5′ and 3′ adapters. The resulting samples were used as templates for cDNA synthesis, followed by PCR amplification. The obtained sequencing libraries were subjected to Solexa sequencing-by-synthesis method. After the run, image analysis, sequencing quality evaluation and data production summarization were performed with Illumina/Solexa pipeline.
Basic analysis of sequencing data
The small RNA sequence reads were pre-processed, excluding low-quality reads (ambiguous N and length <18 nt) as well as 3′ adapter, 5′ adapter and poly(A) sequences. The resulting clean reads were aligned against Rfam, allowing a maximum mismatch of 2 nt to remove noncoding RNA, such as rRNA, tRNA, snRNA, and snoRNA. Obtained sequences were then compared with grass carp transcriptome8 to classify mRNA degradation. The remaining sequences were analyzed by BLAST search against Sanger miRBase (version 19.0). Sequences in our libraries that were identical or related (four or fewer nucleotide substitutions) to sequences from grass carp were identified as conserved miRNAs. Reads that did not match any database above were marked as cid-miRn. The secondary structures of the predicted miRNAs12 were confirmed by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Differentially expressed genes between the SGC/RGC libraries
Gene expression levels were calculated using the transcripts per million clean tags (TPM) method13. The calculation of unigene expression levels and the identification of unigenes that were differentially expressed between the libraries were performed by DEGseq14 based on TMM normalized counts. The settings “q.value < 0.0115” and “|log2.Fold change.normalized| > 1” were used as thresholds for judging significant differences in transcript expression.
Real-time RT-PCR analyses of miRNAs
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. RNA integrity was assessed by electrophoresis on 1.0% agarose gel. Three grass carp from each groups were included in qRT-PCR For mRNA quantification, reverse transcription was performed using a High Fidelity primeScript RT-PCR Kit as instructed (Takara, Dalian, China). MiRNA abundance was detected using stem-loop PCR method, and miR-192 expression was detected as internal control16. All reactions were performed in triplicate on the CFX96 Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). The relative gene or miRNA expression was detected using the comparative threshold cycle (CT) method also referred to as the 2−ΔΔCt method. One-way ANOVA tests were performed using SPSS 20 to determine significant differences.
Effect of pEGFP-N1-CiTLR5, cid-miRn-115 and miR-142a-3p on invasion of A. hydrophila in CIK cells
The ORF of Ctenopharyngodon idella tlr5 (Citlr5) was amplified from grass carp cDNA and individually cloned into the pEGFP-N1 vector (Promega) by directional cloning. Invasion and proliferation assays were performed as previously described17. Using the Lipofectamin 2000 (Invitrogen) transfection reagent, 2 × 105 Ctenopharyngodon idella kidney (CIK) cells cellswere transfected with 1 μg pEGFP-N1-CiTLR5 plasmids or 50 nM cid-miRn-115 and miR-142a-3p agomir (GenePharm, China). Twenty-four hours later, the cells were infected with A. hydrophila a the dose of 2.0 × 103 cells for 45 min at 28 °C. The cells were then gently washed by PBS, and fresh M199 containing 100 μg ml−1 gentamicin was supplemented to kill extracellular bacteria for 1 h. After 15 min incubation with 1% Triton X-100, the cell lysates obtainedwere plated in serial dilutions to assess the number of bacteria that successfully entered CIK cells. The experiment was performed in triplicate. The statistical difference was calculated using Student’s t-test and was considered significant at P < 0.05
Induction of pEGFP-N1-CiTLR5, cid-miRn-115 and miR-142a-3p on the expression of three immune factors in vitro
Four micrograms of pEGFP-N1-CiTLR5, cid-miRn-115 and miR-142a-3p was transfected into 1 × 106 CIK cells. After 24 h, total RNA was extracted and reverse transcribed, as described above. Using the sequences from transcriptome8 as templates (Ciil-1β, Ciil-8 and Citnf-α), we designed primers (Table 1) for quantitative reverse transcription PCR (qRT-PCR) to test the gene expressions levels of these immune factors following CiTLR5, cid-miRn-115 and miR-142a-3p overexpression. The relative expression levels of the genes were normalized to the expression of 18s rRNA. Data were calculated by the 2−ΔΔCT method. The assay was performed in triplicate.
Table 1. PCR primer sequences used in this study.
| Primer | Sequence (5′–3′) | Application |
|---|---|---|
| Let-7a | tgaggtagtaggttgtatagtt | qRT-PCR |
| Let-7e | tgaggtagtagattgaatagtt | qRT-PCR |
| Let-7i | tgaggtagtagtttgtgctgtt | qRT-PCR |
| miR-142a-3p | tgtagtgtttcctactttatgga | qRT-PCR |
| miR-148 | tcagtgcattacagaactttgt | qRT-PCR |
| miR-21 | tagcttatcagactggtgttggc | qRT-PCR |
| miR-217 | tactgcatcaggaactgattgg | qRT-PCR |
| miR-223 | tgtcagtttgtcaaatacccc | qRT-PCR |
| miR-192 | atgacctatgaattgacagcc | qRT-PCR |
| cid-miRn-115 | tgaaggccgaagtggaga | qRT-PCR |
| cid-miRn-131 | tgcccgcattctccacca | qRT-PCR |
| pEGPF-CiTLR5 | F: CCGGAATTCtgATGGGATTTACATTTATTCTGATCC | Overexpression |
| R: CGCGGATCCgcTACTGATGTGTTTGCATGGACA | vector construction | |
| Citlr5 | F: GAAGATAATCTACTTGGGTGAG | qRT-PCR |
| R: GTCCGAGATGAAGAAGTTGTAG | ||
| Ciil-1β | F: GCCAAGTAGCCGAATCACAGA | qRT-PCR |
| R: AGAAGCCCAAGATATGCAGGA | ||
| Ciil-8 | F: GACGCATTGGTAAACACA | qRT-PCR |
| R: TAACCCAGGGAGCAGTAG | ||
| Citnf-α | TCACGCTCAACAAGTCTCAG | qRT-PCR |
| GAAGTAAATGCCGTCATCAG | ||
| Ci18s rRNA | F: GGACACGGAAAGGATTGACAG | qRT-PCR |
| R: CGGAGTCTCGTTCGTTATCGG |
Results
Overview of the high-throughput sequencing data
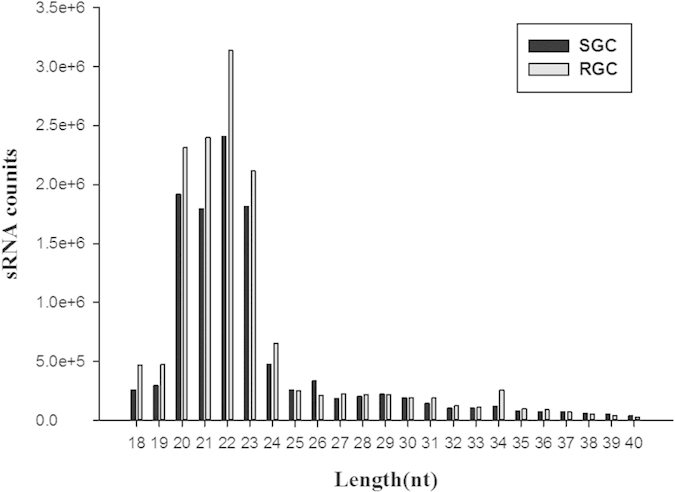
Two sRNA libraries derived from pooled kidney tissues obtained under SGC and RGC, were constructed and sequenced using the Illumina deep-sequencing technology. In total, 13,284,378 and 16,095,116 raw reads were acquired from the SGC and RGC libraries, respectively. After excluding the low-quality tags, adapter sequences, polyA/T/G/C sequences and sequences shorter than 18 nt and longer than 40 nt, 11,218,000 (84.45% of the raw reads) and 13,941,337 (86.62% of the raw reads) clean reads were obtained, respectively (Table 2). The length distribution of the clean reads was analysed (Fig. 1). The majority of the small RNA (sRNA) sequences ranged from 20 to 23 nt in both libraries. The sRNA library under SGC and RGC showed a peak distribution at 22 nt. This supports the notion that mature miRNAs are evolutionary conserved, small, ∼22 nt, noncoding RNAs18.
Table 2. Summary of preliminary analysis of deep sequencing of A. hydrophila-susceptible (SGC) and -resistant grass carp (RGC) small RNA libraries.
| Category | SGC | RGC |
|---|---|---|
| Raw reads | 13,284,378 | 16,095,116 |
| Clean total reads | 11,218,000 | 13,941,337 |
| Total mapped small RNA | 3,301,265 | 4,905,728 |
| Conserved miRNA | 61 | 58 |
| Total conserved miRNA | 61 | |
| Novel miRNA | 99 | 108 |
| Total novel miRNA | 116 |
Figure 1. Length distribution and reads of small RNAs from A. hydrophila- susceptible (SGC) and-resistant grass carp (RGC) libraries.
Discovery of miRNAs in grass carp
After mapping to the grass carp transcriptome, a total of 3,301,265/4,905,728 small RNA were mapped. Among them, 61 miRNAs were known in miRBase 19, and the remaining 116 miRNAs were not found to possess homology to any known metazoan miRNAs, suggesting a possible species-specificity, and were labelled cid-miRn. The miRNA could be distinguished from other sRNAs through a characteristic hairpin structure fold by the flanking sequences. The precursor sequences of the 116 putative candidate miRNAs could form the canonical hairpin structure, and the secondary structure for some candidates (the cid-miRn-100, cid-miRn -101 and cid-miRn-102) were are in Fig. 2.
Figure 2. Predicted hairpin structures of grass carp coding candidate miRNAs.
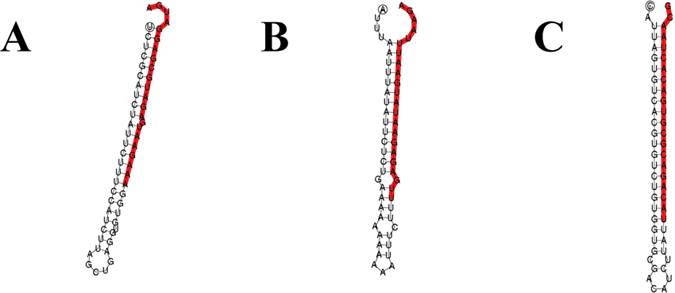
Dominant forms of the mature miRNAs are indicated in red. A: cid-miRn-100, B: cid-miRn-101, C: cid-miRn-102.
MiRNA expression profiles
High-throughput sequencing technology is not only an alternative means to identify small RNAs but also an useful tool to assess their expression profiles, in which the number of reads can serve as an index for the relative abundance of diverse miRNAs19. In the present study, the transcripts per million (TPM) of these miRNA sequences ranged from 0.44 to 155,651.08, indicating great variation in the expression level. The average number of TPMs for novel miRNAs was lower than those of conserved miRNAs (16.04 versus 9,511.31), indicating that the novel miRNAs are usually weakly expressed, whereas conserved miRNAs are highly expressed (see Table S1 and S2 for further details), consistent with the phenomenon observed in other fish species20. Among these identified miRNAs, the most abundant miRNA was miR-101a, with a total TPM value of 308,750.21 in SGC and RGC libraries, followed by miR-146b (TPM value 168,822.24) and miR-126a-3p (TPM value 139,223.22).
To validate the miRNA expression profiles obtained by deep sequencing, the expression levels of eight randomly selected miRNAs were quantified by qPCR. Figure 3 shows significantly increased expression of two miRNAs (miR-21, let-7i) in the SGC library, and of three miRNAs (miR-142a-3p, miR-223, miR-217) in the RGC library. Thus, the qPCR results were consistent with those obtained by deep sequencing.
Figure 3. qPCR of conserved miRNAs in the A. hydrophila-susceptible (SGC) and -resistant grass carp (RGC) libraries.
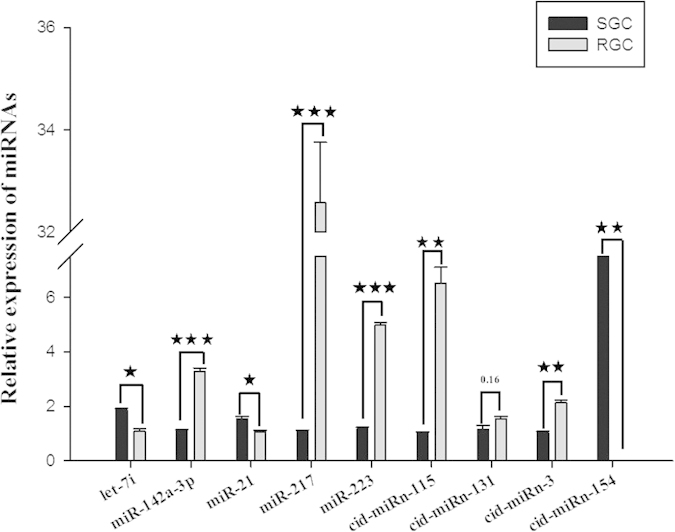
The expression of selected miRNAs in SGC or RGC was validated by qPCR amplification, using a specific primer for each miRNA. The fold change in expression was calculated based on the level of miR-192 expression (used as a control for the normalization). The data were obtained from three independent experiments (mean ± SD). *P < 0.05; **P < 0.01; ***P < 0.001.
TLR5 is a potential target of cid-miRn-115 regulation
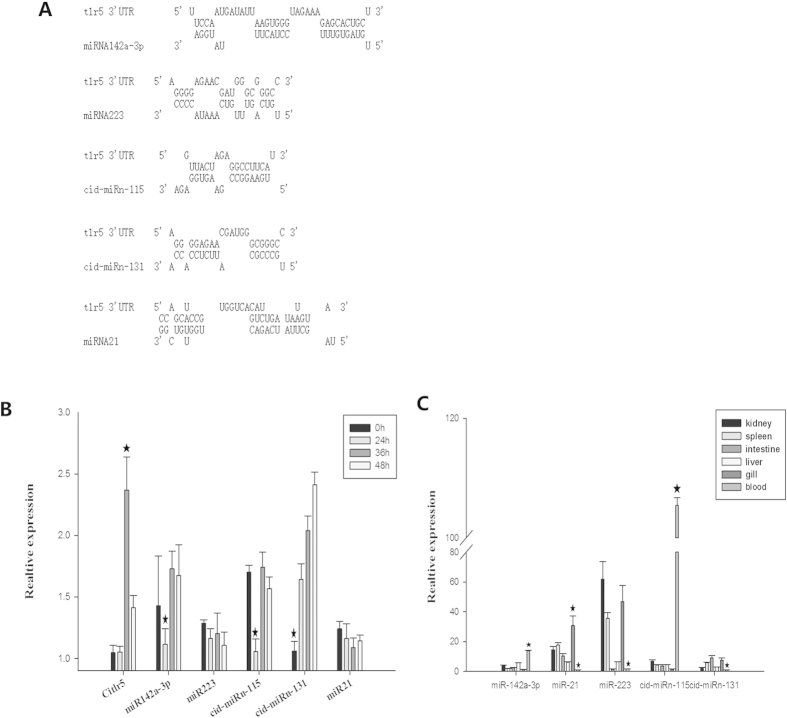
Based on the target scan analysis, we were able to determine miRNA (miR142a-3p, miR-21, miR-223, cid-miRn-115 and cid-miRn-131) binding sites of Citlr5 3′-UTR in grass carp (Fig. 4A). Toll-like receptor 5 (TLR5) binding to bacterial flagellin activates NF-κB signalling and triggers an innate immune response to the invading pathogen21. Thus, we postulated that the expression of Citlr5 would be upregulated and TLR5-inhibitory miRNAs would be downregulated upon flagellin challenge. Our results showed that the Citlr5 expression was time-dependent expression pattern upon flagellin challenge (Fig. 4B), which is consistent with findings from other studies22,23. Meanwhile, we also observed a inverse expression correlation in cid-miRn-115 and miR-142a-3p with tlr5 expression. However, we did not detect any expression change for other miRNAs. The inverse expression correlation between cid-miRn-115, miR-142a-3p and Citlr5 suggests that cid-miRn-115 and miR-142a-3p directly regulate Citlr5 expression in grass carp.
Figure 4. TLR5 is a potential target of miRNA regulation.
(A) The alignment between miR142a-3p, miR-21, miR-223, cid-miRn-115, cid-miRn-131 and the 3′-UTR segment of tlr5. (B) CIK was exposed to FLG22 for 0 h, 24 h, 36 h and 48 h. The expression of Citlr5 in CIK was detected using real-time PCR. 18S rRNA expression was detected as internal control for mRNA. MiR-192 expression was detected as the internal control for miRNA. (C) MiRNA samples were extracted from different tissues, including kidney, spleen, intestine, liver, gill and blood. MiRNA expression was detected by qRT-PCR. MiR-192 was used as loading control. The data were obtained from three independent experiments (mean ± SD). *P < 0.05.
In addition, we performed a qRT-PCRs experiment to detect the miRNA expression patterns in grass carp. miR142a-3p, miR-21, miR-223, cid-miRn-115 and cid-miRn-131 were found to be expressed ubiquitously in kidney, spleen, intestine, liver, gill and blood (Fig. 4C). Importantly, cid-miRn-115 and miR-142a-3p expression levels in the blood were higher than that in other tissues. Septicaemia is caused by bacterial infection in the blood (bacteraemia) that often occurs with severe infections. Given that the lowest expression of tlr5 is also found in blood24, we therefore investigated whether a regulatory relationship exists between Citlr5 and cid-miRn-115 and miR-142a-3p.
Altered expression of immune response genes in cid-miRn-115 and miR-142a-3p overexpressing CIK cells
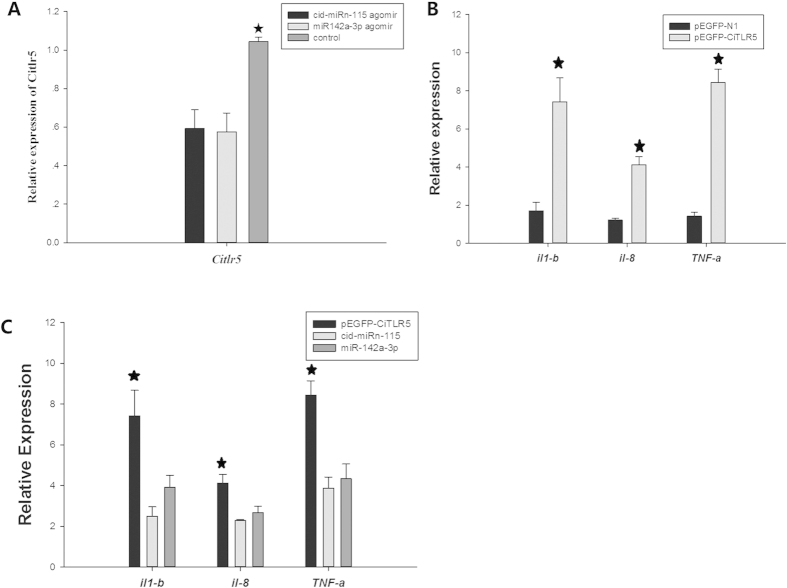
We employed the agomir method to perform a miRNA overexpression of function experiment. We found that in CIK administration of miR-142a-3p and cid-miRn-115 angomir results in a profound up-crease in the endogenous expression of miR-142a-3p and cid-miRn-115 (Fig. S1). Meanwhile, the result showed that the agomir, but not PBS treatment, led to a significant decrease in endogenous Citlr5 expression (Fig. 5A).
Figure 5. cid-miRn-115 and miR-142a-3p overexpression changes the expression of Citlr5 and its downstream genes.
CIK received cid-miRn-115 and miR-142a-3p agomir at a dose of 50 nM for the indicated times. The relative mRNA expression of Citlr5 (A) and of TLR5 downstream genes, including Ciil-1β, Ciil-8 and Citnf-α (B and C) was detected using real-time PCR. 18S rRNA expression was used as internal control for mRNA. MiR-192 was detected as the loading control. The data were obtained from three independent experiments (mean ± SD). *P < 0.05.
The innate immune response is the first line of defence against infections. The principal challenge for the host is to detect the pathogen and mount a rapid defensive response25. TLRs recognise pathogen-associated molecular patterns (PAMPs) and mediate the production of cytokines necessary for the development of effective immunity26. After infection with several strains of enteroinvasive bacteria, cells rapidly up-regulate the expression of a program of host genes, the products of which activate inflammatory and immune responses. This inflammatory program includes the upregulated expression and production of proinflammatory and chemoattractant cytokines, such as IL-1β, IL-8 and TNF-α27.
Cid-miRn-115 and miR-142a-3p overexpression results in downregulation of il-1β, il-8 and tnf-α expression during this process. Indeed, qPCR analysis reveals that cid-miRn-115 and miR-142a-3p overexpression results in a significant decrease in Citlr5 expression (Fig. 5A), thereby downregulating Ciil-1β, Ciil-8 and Citnf-α expression (Fig. 5B,C).
Effect of cid-miRn-115 and miR-142a-3p on invasion of A. hydrophila in vitro
CIK cells were artificially infected with A. hydrophila for 45 min after transfection with pEGFP-CiTLR5 or cid-miRn-115 and miR-142a-3p agomir. We observed by microscope (20 × ) that the bacteria successfully infected the cells by insertion. After incubation with gentamicin, free bacteria in the culture medium were killed, which was confirmed by the plate count method. The cells were cleared with Triton X-100 and the numbers of invasive pathogens were also calculated using the plate count method. The number of A. hydrophila in cells transfected with cid-miRn-115 and miR-142a-3p was significantly lower than in cells transfected with pEGFP-CiTLR5 (Fig. 6).
Figure 6. Effect of cid-miRn-115, miR-142a-3p and pEGFP-CiTLR5 on A. hydrophila invasion.
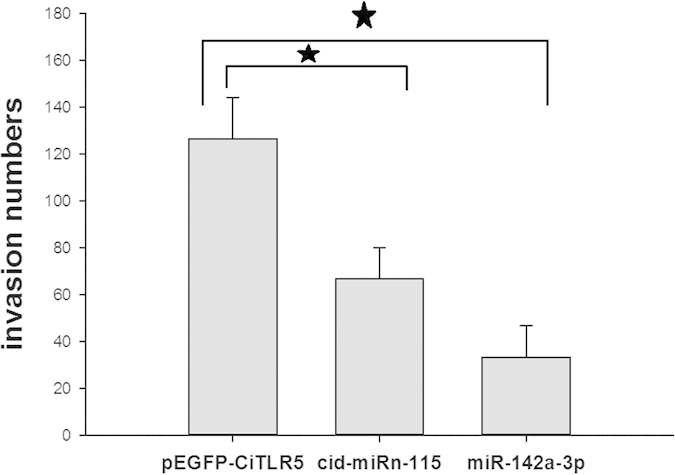
CIK cells were transfected with plasmid pEGFP- pEGFP-CiTLR5 or cid-miRn-115 and miR-142a-3p agomir. After 24 h, the cells were infected with A. hydrophila. The invasion number was counted as the number of entered bacteria. Data are presented as mean ± SE (n = 3). *P < 0.05.
Discussion
miRNAs can affect both the translation and stability of mRNAs28. Consistent the notion that miRNAs play a crucial role in controlling gene expression, misregulation of microRNA expression has been found to correlate with several pathologies29. In addition to these well-established functions in physiological and pathological processes, it is becoming clear that miRNAs also play crucial roles during microbial infections1. To gain further insight into the possible significance of kidney miRNAs in fish, we first identified miRNA expression profiles in the kidney tissue in fish, and then compared miRNAs expression pattern between SGC and RGC kidney samples. We found that nine miRNAs were differentially expressed in the different groups, implying that these differentially expressed miRNAs are involved in bacterial infection. The knowledge of tissue-specific expression pattern of miRNAs can directly inform functional studies30. Here, we found that cid-miRn-115, and miR-142a-3p are significantly highly expressed in immune related tissues. Specifically, cid-miRn-115 and miR-142a-3p display a clear time-dependent expression pattern during FLG22 infection CIK. We thus speculated that cid-miRn-115 and miR-142a-3p might play a key role in regulating the innate immune response.
The innate immune system in fish is considered to be the first line of defence against a broad spectrum of pathogens and is more important in fish than in mammals31. The role of miRNAs in innate immune response has been reported for some species, including Cynoglossus semilaevis, Paralichthys olivaceus, Cyprinus carpio L. and Danio rerio32,33,34,35. In this study, we revealed that cid-miRn-115 and miR-142a-3p are potential regulators of fish innate immune response. Here, we found that cid-miRn-115 and miR-142a-3p are highly expressed in SGC. This study further extends the biological role of cid-miRn-115 and miR-142a-3p in fish.
MiR-142a-3p is a member of the miR-142 family, which has been predicted or experimentally confirmed in a wide range of species. Previous studies have identified miR-142a as a regulator of haematopoiesis, immune system, osteoblast differentiation and fibrosis of the skin by targeting multiple mRNAs36,37,38,39. Sequence alignment suggests that the miR-142 family is highly conserved between invertebrates and vertebrates, which indicates that its function might have been conserved. The findings of our study suggest that role of miR-142 in innate immune response is highly conserved between invertebrates and vertebrates.
MiRNAs control biological processes by regulating the expression of their target genes. Here, we found a binding site of cid-miRn-115 and miR-142a-3p in the 3′-UTR region of Citlr5, and characterized their effects on Citlr5 using overexpression assay. Emerging evidence indicates that the TLR5-flagellin interaction plays a central role in driving the inflammatory response triggered by bacteria, and inhibition of tlr5 normalizes the inflammatory response associated with improved health indicators40. In this study, we found that cid-miRn-115 and miR-142a-3p expression stimulate upregulation by FLG22, which leads to a gradual decrease in the expression of Citlr5.
In that of all known TLRs, only TLR5 can activate proinflammatory gene expression in response to flagellin41. A previous study identified TLR5 signalling is associated with increased susceptibility to Legionnaire’s disease42. These findings indicate the involvement of TLR5 in inducing il-8 and tnf-α, and in response to pathogenic invasion24. Here, we found a similar result, which shows that cid-miRn-115 and miR-142a-3p directly repress Citlr5 expression. TLR5 directly regulates the expression of multiple genes that are necessary for innate immune response, including tnf-α, il-1 and il-8 and so on. We also observed an expression change in the downstream gene Citlr5 when the levels of cid-miRn-115 and miR-142a-3p expression are altered.
Disease outbreaks, some of them caused by pathogenic bacteria, are considered to be one of the largest constraints to development of the aquaculture sector43. Similar to terrestrial animal production, antibiotics are also used in aquaculture in an attempt to control bacterial disease44. An alternative to killing pathogenic bacteria with antibiotics is to prevent them from attacking the host, without the need to kill them45. Quorum sensing pathogens, like the aquatic pathogen A. hydrophila, probably increase their chances to infect their host successfully by delaying virulence factor production until the population density is high enough to overwhelm the host immune system46. Recognition of microbial pathogens is an essential element for the initiation of innate immune responses. TLR5-deficient mice show increased survival, resulting from decreased migration of bacteria from the intestinal tract to the mesenteric lymph nodes47. Thus, TLR5 can be either beneficial or detrimental to the host, depending on the bacterial dose and route of infection48. Our results show that A. hydrophila in grass carp leads to the activation of tlr5. This is similar to a previously reported study in other fish. Cid-miRn-115 and miR-142a-3p overexpression affects the level of tlr5 expression. We here propose a model for the avoidance of bacterial injury in fish: Once a fish is exposed to a bacterial infection, cid-miRn-115 and miR-142a-3p expression is rapidly upregulated, earlier than other innate immune genes. Cid-miRn-115 and miR-142a-3p upregulation could release tlr5 inhibition, thus inhibiting its downstream pathway and inflammation reaction. MiRNA-mediated gene regulation operates earlier than most transcriptional responses. The fast regulation of miRNAs after bacterial infection indicates that miRNA-mediated gene silencing acts earlier than most gene transcriptional responses after bacterial damage. Fish have evolved numerous strategies for effectively escaping bacterial injury through distinct signalling pathways. MiRNAs are implicated in buffering developmental processes against the effects of environmental fluctuations.
In summary, we here revealed a novel regulatory mechanism for bacterial infections in fish from miRNA viewpoint. We found that cid-miRn-115 is differentially expressed between SGC and RGC, and its overexpressing using agomir leads to a significant change in A. hydrophila invasion and proliferation rates. The post-transcriptional regulation of tlr5 by cid-miRn-115 and miR-142a-3p could affect the expression of Citlr5 and its downstream genes, including il-1β, il-8 and tnf-α, which in turn affect the immune functions in grass carp. However, samples number and more accurate experimental program should be taken into account in future studies as revealed by miRNA-mediated regulation.
Additional Information
How to cite this article: Xu, X.-Y. et al. MicroRNA-induced negative regulation of TLR-5 in grass carp, Ctenopharyngodon idella. Sci. Rep. 6, 18595; doi: 10.1038/srep18595 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from China’s Agricultural Research System (CARS-46-04), the National Key Technology R&D Program of China (2012BAD26B02) and Shanghai Universities First-class Disciplines Project of Fisheries.
Footnotes
Author Contributions X.X., L.L. and J.L. study concept and design, analyses and interpretation of data; X.X. drafting of manuscript and obtaining funding; Y.S., J.F., H.Y. and W.H. provided experiment materials; X.X. and W.H. performed the experiments; all authors approved the final version of the manuscript.
References
- Eulalio A., Schulte L. & Vogel J.. The mammalian microRNA response to bacterial infections. Rna Biol 9, 742–750 (2012). [DOI] [PubMed] [Google Scholar]
- Bueno M. J., Perez de Castro I. & Malumbres M.. Control of cell proliferation pathways by microRNAs. Cell Cycle 7, 3143–3148 (2008). [DOI] [PubMed] [Google Scholar]
- O’Connell R. M., Rao D. S., Chaudhuri A. A. & Baltimore D.. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122 (2010). [DOI] [PubMed] [Google Scholar]
- Schulte L. N., Eulalio A., Mollenkopf H. J., Reinhardt R. & Vogel J.. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30, 1977–1989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006). [DOI] [PubMed] [Google Scholar]
- Ruiz-Ferrer V. & Voinnet O.. Roles of plant small RNAs in biotic stress responses. Annual review of plant biology 60, 485–510 (2009). [DOI] [PubMed] [Google Scholar]
- FAO, The State of World Fisheries and Aquaculture. Opportunities and challenges (Rome, 2014).
- Xu X., Shen Y., Fu J., Lu L. & Li J.. De novo assembly of the grass carp Ctenopharyngodon idella transcriptome to identify miRNA targets associated with motile aeromonad septicemia. PloS one 9, e112722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B. & Austin D. A.. In Disease of Farmed and Wild Fish. (Springer Netherlands, 2012). [Google Scholar]
- Nielsen M. et al. Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis Aquat Organ 46, 23–29 (2001). [DOI] [PubMed] [Google Scholar]
- Edberg S. C., Browne F. A. & Allen M. J.. Issues for microbial regulation: Aeromonas as a model. Critical reviews in microbiology 33, 89–100 (2007). [DOI] [PubMed] [Google Scholar]
- Ryu S. et al. Discovery of novel human breast cancer microRNAs from deep sequencing data by analysis of pri-microRNA secondary structures. PloS one 6, e16403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PloS one 5, e15224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W.. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D. & Tibshirani R.. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. Y., Shen Y. B., Fu J. J., Lu L. Q. & Li J. L.. Determination of reference microRNAs for relative quantification in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 36, 374–382 (2014). [DOI] [PubMed] [Google Scholar]
- Yu H. et al. Molecular cloning and functional characterization of the NFIL3/E4BP4 transcription factor of grass carp, Ctenopharyngodon idella. Dev Comp Immunol 47, 215–222 (2014). [DOI] [PubMed] [Google Scholar]
- Bartel D. P.. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Qi P., Guo B., Zhu A., Wu C. & Liu C.. Identification and comparative analysis of the Pseudosciaena crocea microRNA transcriptome response to poly(I:C) infection using a deep sequencing approach. Fish Shellfish Immunol. 39, 483–491 (2014). [DOI] [PubMed] [Google Scholar]
- Xu Z. Q. et al. Identification and Characterization of MicroRNAs in Channel Catfish (Ictalurus punctatus) by Using Solexa Sequencing Technology. PloS one 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. I. et al. Structural basis of TLR5-flagellin recognition and signaling. Science 335, 859–864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. D. et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247–1253 (2003). [DOI] [PubMed] [Google Scholar]
- Andersen-Nissen E., Smith K. D., Bonneau R., Strong R. K. & Aderem A.. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. The Journal of experimental medicine 204, 393–403 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M., Swain B., Maiti N. K., Routray P. & Samanta M.. Inductive expression of toll-like receptor 5 (TLR5) and associated downstream signaling molecules following ligand exposure and bacterial infection in the Indian major carp, mrigal (Cirrhinus mrigala). Fish Shellfish Immunol. 32, 121–131 (2012). [DOI] [PubMed] [Google Scholar]
- Aderem A. & Ulevitch R. J.. Toll-like receptors in the induction of the innate immune response. Nature 406, 782–787 (2000). [DOI] [PubMed] [Google Scholar]
- Hayashi F. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001). [DOI] [PubMed] [Google Scholar]
- Elewaut D. et al. NF-kB Is a Central Regulator of the Intestinal Epithelial Cell Innate Immune Response Induced by Infection with Enteroinvasive Bacteria. The Journal of Immunology 163, 1457–1466 (1999). [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N.. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. Genetics 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- Amoretti M. et al. Production and detection of cold antihydrogen atoms. Nature 419, 456–459 (2002). [DOI] [PubMed] [Google Scholar]
- Yan B. et al. microRNA regulation of skin pigmentation in fish. J. Cell Sci. 126, 3401–3408 (2013). [DOI] [PubMed] [Google Scholar]
- Saurabh S. & Sahoo P. K.. Lysozyme: an important defence molecule of fish innate immune system. Aquac Res 39, 223–239 (2008). [Google Scholar]
- Sha Z. et al. Identification and characterization of Cynoglossus semilaevis microRNA response to Vibrio anguillarum infection through high-throughput sequencing. Dev Comp Immunol 44, 59–69 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang B. C., Zhang J. & Sun L.. In-depth profiling and analysis of host and viral microRNAs in Japanese flounder (Paralichthys olivaceus) infected with megalocytivirus reveal involvement of microRNAs in host-virus interaction in teleost fish. BMC genomics 15, 878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. et al. Identification and characterization of microRNAs in the spleen of common carp immune organ. J. Cell. Biochem. 115, 1768–1778 (2014). [DOI] [PubMed] [Google Scholar]
- Ordas A. et al. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC genomics 14, 696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T. et al. miR-142-3p is essential for hematopoiesis and affects cardiac cell fate in zebrafish. Biochem. Biophys. Res. Commun. 425, 755–761 (2012). [DOI] [PubMed] [Google Scholar]
- Hu W. et al. miR1423p promotes osteoblast differentiation by modulating Wnt signaling. Mol Med Rep 7, 689–693 (2013). [DOI] [PubMed] [Google Scholar]
- Makino K. et al. Circulating miR-142-3p levels in patients with systemic sclerosis. Clin Exp Dermatol 37, 34–39 (2012). [DOI] [PubMed] [Google Scholar]
- Huang B. et al. miR-142-3p restricts cAMP production in CD4 + CD25- T cells and CD4 + CD25 + TREG cells by targeting AC9 mRNA. EMBO Rep 10, 180–185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohmke C. J. et al. TLR5 as an anti-inflammatory target and modifier gene in cystic fibrosis. J Immunol 185, 7731–7738 (2010). [DOI] [PubMed] [Google Scholar]
- Gewirtz A. T., Navas T. A., Lyons S., Godowski P. J. & Madara J. L.. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. The Journal of Immunology 167, 1882–1885 (2001). [DOI] [PubMed] [Google Scholar]
- Metcalfe H. J., Best A., Kanellos T., La Ragione R. M. & Werling D.. Flagellin expression enhances Salmonella accumulation in TLR5-positive macrophages. Dev Comp Immunol 34, 797–804 (2010). [DOI] [PubMed] [Google Scholar]
- Defoirdt T., Boon N., Bossier P. & Verstraete W.. Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture 240, 69–88 (2004). [Google Scholar]
- Defoirdt T.. Virulence mechanisms of bacterial aquaculture pathogens and antivirulence therapy for aquaculture. Reviews in Aquaculture 6, 100–114 (2014). [Google Scholar]
- Clatworthy A. E., Pierson E. & Hung D. T.. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 (2007). [DOI] [PubMed] [Google Scholar]
- Donabedian H.. Quorum Sensing and its Relevance to Infectious Diseases. Journal of Infection 46, 207–214 (2003). [DOI] [PubMed] [Google Scholar]
- Uematsu S. et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c + lamina propria cells. Nat. Immunol. 7, 868–874 (2006). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S.. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.