Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that have unique functions at post-transcriptional level (epigenetics). MiRNAs play a pivotal role in controlling gene expression at various levels including differentiation, cell-cycle regulation, apoptosis and many others in mammals as well as in many organisms. Recently, there has been greater understanding of the contribution of dysregulation of miRNA into disease status in particular carcinogenesis. In this review, we will discuss miRNA discovery, nomenclature, function, contribution of their dysregulation into disease status in particular carcinogenesis and their potential role as biomarkers.
Key words: miRNA, breast cancer, neo-adjuvant chemotherapy
Introduction
Oncogenic mutations are known to contribute to the process of carcinogenesis. More recently the epigenetic alterations in gene expression caused by mechanisms other than changes in DNA sequence were found to be central to the process of tumorigenesis. These epigenetic changes include DNA methylation, genomic imprinting, histone modifications, and altered MicroRNAs (miRNAs) expression. The epigenetics effects of miRNAs expression alteration were shown to be central to the process of carcinogenesis in various cancers.1,2 The miRNAs are found to exhibit a significant pathological consequence dually, either by enhancing oncogene expression facilitating tumorigenesis or repressing the tumor suppressor genes expression resulting in oncogene overexpression, hence tumor progression. These miRNA are thought to originate from either debris of apoptosis or necrosis or from miRNA-containing microvesicles that are produced by cancer cells.2 MicroRNAs (miRNAs) are a class of small non-coding RNA of ≈25 nt that have unique functions at post-transcriptional level.3,4 Recently it has been discovered that miRNAs play a pivotal role in controlling gene expression at various levels including differentiation, cell-cycle regulation, apoptosis and many others in mammals as well as in many organisms.5 It is highly conserved within distantly related organisms and it is speculated that miRNAs govern nearly 30% of the human protein-coding genes suggesting their central role in cell function regulation.6,7 The latest version of the miRBase 21 (www.miRbase.org), where the reporting of annotated mature miRNA genes are constantly updated, shows 2588 unique mature human miRNAs, an increase of 10 miRNAs in comparison to version 20 (www.miRbase.org).8 Of note, this number is constantly increasing from 218 annotated miRNAs at the establishment of miRBase 2002. Since most of these miRNA have been annotated based of high-throughput sequencing, Kim et al., have suggested that most of these miRNAs require further experimental and functional validation. Moreover, it is estimated that the number of miRNA deposited in miRBase is inflated since many of the misannotated ones have been removed.8
Moreover an exponential increase in miRNAs related publications took place from 2001 up to date. Initially, only 2 miRNA related papers before 2000 were published,9 however the number has jumped from 87 in 2000 up to 19,388 in 2013 and the number is increasing where by the end of 2014 it was 24,490.
miRNA genomic distribution and gene structure
miRNA genes are dispersed throughout the chromosomes in humans except for the Y chromosome. About 50% of known miRNAs are found in clusters.8 The mainstream of miRNAs transcription is aided by RNA polymerase II to yield long primary transcripts many of which are capped and polyadenylated.10 miRNA genes exhibit different location where some are either located in the intergeneic regions, or within introns of non-coding genes or exons of non-coding genes. It is speculated that around one third of miRNAs are intergenic and about one third of all miRNA loci contain clustered miRNAs.10,11
Figure 1 illustrates how miRNA could be classified according to their genomic locations relative to the positions of the exon and intron.11 Recent studies showed that most miRNA genes (≈70%) are located within a defined transcription unit rather than in intergenic regions as was previously thought.8
Figure 1.
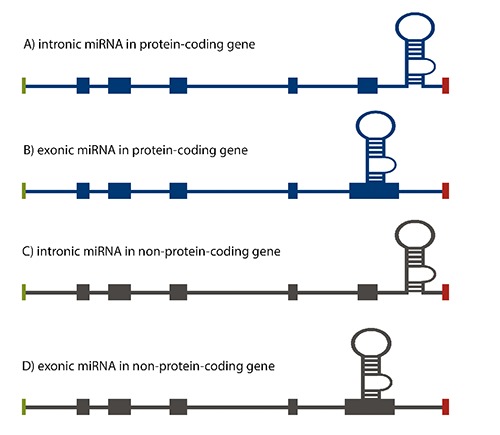
microRNA gene localization within the genome. A) intronic miRNA within the non-coding transcription unit, i.e., miR15a-16-1; B) exonic miRNA within the non-coding transcription unit, i.e., miR-155; C) intronic miRNA within the coding transcription unit, i.e., miR-25-93-106b; D) exonic miRNA within the coding transcription unit, i.e., miR-985.
Mammalian miRNA biogenesis and processing canonical maturation
miRNAs are transcribed in a more or less similar way, as the protein coding genes are. The majority of them if not all are transcribed by polymerase II demonstrated by experimental evidences.12,13 Lee and coworkers pointed out experimentally the role of polymerase II in transcribing miRNAs, by treating HeLa cells with α-amanitin at a concentration that inhibits the activity of polymerase II (50 μg/mL) it resulted in the reduction of many pri-miRNAs i.e., pri-miR-23a~27a~24-2 and pri-miR-30a. This emphasizes the role of polymerase II in the transcription of miRNA genes.12
MiRNAs biogenesis undergoes several activation steps involving both cellular compartments of nuclear and cytoplasm until its maturity. It starts with a primary transcript (pri-miRNA) production by the transcription of miRNA gene that is mediated by RNA polymerase II, which contains 7 methylgluanosine cap and poly (A)-tail.13 Stem loop structure is the major feature of the pri-miRNA to the subesequent processing reactions. The miRNA maturation is initiated when the stem-loop structure is cleaved in the nucleus by the action of RNase III Drosha that will release the pre-cursor from of the miRNA named as pre-miRNA. The Drosha enzyme along with its cofactor DiGeorge syndrome critical region gene 8 (DGCR8), forms a large complex of 650 KDa in human that act as a microprocessor complex. DGCR8 is composed of two double stranded RNA binding domains (dsRBDs) that is speculated to assist Drosha in recognizing its substrate although the biochemical mechanism is yet to be understood.13,14 After the production of the pre-miRNA in the nucleus it is transported into the cytoplasm by a process that is mediated by Exportin-5 (Exp-5) which is a RAN-GTP dependent nucleo/cytoplasmic cargo transporter, as the pre-miRNA processing is confined in the cytoplasm.3,8,15-17
In the cytoplasm, Dicer cleaves the pre-miRNA into a double stranded 22-nt miRNA, which is referred as miRNA/miRNA*. These 22-nt miRNAs are unwounded by the action of Argonaute proteins (helicases) producing a single mature strand. Where miRNA is the mature form or so called guide strand that is the most thermodynamically stable strand produced hence preferentially loaded into the RNA-induced silencing complex (RISC), whereas miRNA* [miRNA star] so called passenger strand, less stable thermodynamically, is thought to be degraded.13,18,19 However there are some evidences which show that both strands of the miRNA duplex exhibits functional roles and the mechanistic bases of selection are not yet understood.20 But deep sequencing studies data revealed that some passenger miRNAs hold functional roles in regulating miRNAs homeostasis as well as downstream effect on RNA and DNA transcription and translation19 (Figure 2). Particularly, the mature functional miRNA strand is incorporated in miRISC which is an effecter complex in association with Argonaute proteins. Though not much is known about the complex, it is found to be composed of Dicer and dsRNA-binding protein known as transactivating response RNA binding protein that is responsible for Ago-2 recruitment.18,21
Figure 2.
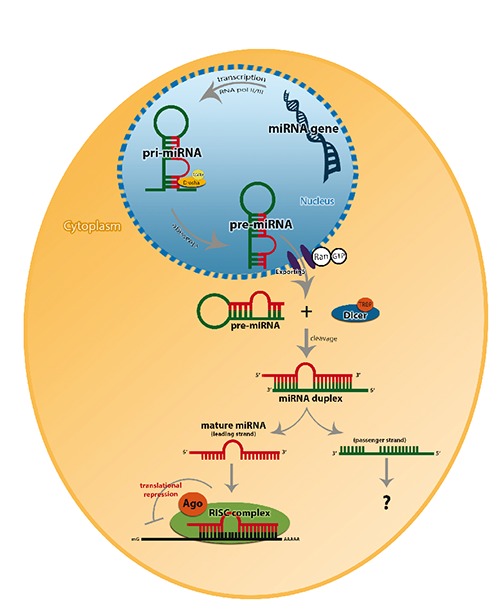
Canonical pathway of miRNA biogenesis. In the nucleous pri-miRNA is produced by poly-II or III then cleaved by Drosh/DGCR8 microprocessor complex resulting in a stem loop pre-miRNA. Pre-miRNA is then expeled to the cytoplasm by Exp-5/RAN-GTP and Dicer along with the transactivating response RNA binding protein (TRBP) cleaves the stem loop structure to a double stranded mature miRNA. Where the guide strand of miRNA along Ago2 protein are loaded into the RISC guiding it to its functional destination, whereas the passenger strand is reported to be either degraded or may hold functional roles in regulating miRNAs homeostasis.
Argonaute proteins and RNA-induced silencing complex loading in miRNA biological functions
Upon the miRNA:miRNA* duplex production, the mature miRNA strand is favored and associated with a member of the Argonaute proteins, which are the core components of RISC unit. Argonaute proteins along with the miRISC, act as the interface between the miRNAs and their target mRNA.22
miRNA act by two major mechanisms that occur post-transcriptionally either as translational repressors or mRNA degraders. The key role of their function is mediated by RISC which is guided by miRNAs to down-regulate gene expression.23,24
Gene target regulation
Translational repression
Translational repression is determined by the degree of complementarity based on Watson-Crick base pairing between miRNA seed sequence and the target mRNA.23 The translational repression occurs when the loading strand imperfectly pairs with the target mRNA directs RISC to bind to the 3’ un-translated region (3’UTR) of the target. The imperfect pairing causes a bulge in the duplex that by the proximate pairing of miRNA and the target mRNA. As a result of this bulging that targets mRNA, it is blocked from being translated and protecting the target mRNA from the endonucleases. This complex is then transported to a cytoplasmic compartment known as Processing bodies (P-bodies) which contain enzymes that are involved in mRNA degradation.25,26 The translational repression efficiency is affected by number of miRNA binding sites in a target mRNA.23,27
mRNA degradation
miRNA also causes target degradation, a result found where a perfect complementarity between RISC unit at 3’UTRs in mRNA and seed region of miRNA makes it sufficient to cause endonucleolytic cleavage and mRNA degradation.23 In the P bodies that contain the decapping enzymes along with the exonucleases to carry out the degradation process lead by the RISC, Argonaute proteins and miRNAs.28 In plants, extensive complementary between miRNA and target mRNA leads to mRNA cleavage rather than translational repression by RISC.23,27,29,30 Since the function of miRNAs depend on the seed sequence core region base pairing with its target, hence hundreds and thousands of potential targets could be regulated by single miRNA. However, many computational algorithms, that predict potential mRNA targets such as miRanda, TargetScan have been developed.31
MicroRNAs and tumorigenesis
Mounting evidence from cancer research provides insights into the role of miRNA in tumorigenesis.32 These researchers found that 50% of the human miRNA genes are located in the cancer associated regions or at the fragile sites of chromosomes.7,33 There are several studies showing the altered pattern of global miRNA expression. Although these studies are contradicting, some showed an up-regulation34 and others down-regulation35 in the miRNA expression profile but it is an disputed fact that miRNA expression is altered in cancers.
Generally, when compared to normal, cancer is moderately featured by global down regulation of miRNA expression.35,36 However, deviation in any vital element of miRNA pathways, starting from biogenesis and ending with target function, contributes to cancer pathogenesis, progress and development suggesting the tight control of miRNA over the normal homeostasis. It is evident that miRNAs contribute to regulate tumorigenesis and tumor development by negatively regulating the expression genes involved in cell proliferation and apoptosis 32. At a molecular level, it was shown that some miRNAs act on tumor suppressor genes while others target oncogenes resulting in alterations that lead to tumorigenesis and cancer development. Additionally, at a physiological level, some miRNAs were found to regulate angiogenesis while others were found to contribute to metastasis.36 One of the first evidences linking the miRNAs with the human cancers came from chronic lymphocytic leukemia (CLL).37 Half of the patients with CLL have deletion in chromosome 13q14, however studies did not show any potential of the conventional genes around that region to correlate with the disease pathogenicity. Interestingly, miR-15 and miR-16-1 were found to be encoded in that region and their expression is aberrant in most of CLL cases. The inverse correlation between the expression of the miR-15 and miR-16-1 and the expression of the anti-apoptotic protein Bcl-2, suggests that as miR-15 and miR-16-1 posttranscriptional inhibition of Bcl2 induces apoptosis in leukemic cells.38,39 On the other hand, in vivo studies showed the role of some miRNAs in promoting metastasis. Tang et al. showed how miR-125b induces metastasis in MCF-7 and MDA-MB-231 breast cancer cell lines as it activates the STARD13 suggesting its role in pro-metastasis.40
Dual nature of miRNAs in cancer progression (tumor suppressor and oncogenic features)
Deregulation of microRNA molecules was found experimentally, using Northern Blotting, real-time PCR and miRNA microarray in many diseases. Additional studies reported that some miRNAs specifically regulate cell proliferation and apoptosis in cancer formation including solid cancers like breast, lung, liver, brain cancers. Zhang and coworkers showed that above 50% of miRNAs are localized in the range of human cancer genomic region, or in fragile sites or even near oncogenes or tumor suppressor genes which is not what was though previously.7,41,42 Evidence confirmed the dual function of the miRNAs in tumorigenesis. For examples, over expression of miR-31 in murine lung cancers was a feature; however when miR-31 was knocked down it substantially repressed the tumorigenecity of the lung cancer in a dose-dependent manner. Furthermore, miR-31, mRNA targets were identified and found to include tumor suppressive genes like PPP2R2A whose expression was amplified by the knockdown miR-31. This suggests its oncogenic role as it suppresses specific tumor suppressors.43 The first so called OncomiR was introduced in lymphoma tumorigenesis and best characterized transcript mir-17-92. Moreover, the first functional evidence of miRNA was demonstrated in this cluster of miRNAs where co-expression of miR-17-92, which is a truncated portion of miR-17-92, expedited the lymphomagenesis.42
On the other hand, many evidences demonstrating the role of let-7 in breast cancer and many other malignancies as tumor suppressor miRNA are accumulating. In a study trying to elucidate the role of let-7, Kim et al. associated the increase in the expression CCR7 and CCL21, which has been reported to play chemotactic and metastatic roles in breast cancer cell lines and malignant tissue than normal. They confirmed that the direct target of let-7a is the 3’UTR of CCR7 suggesting its role as a tumor suppressor. To support that, synthetic induction of let-7a decreased breast cancer cell proliferation, migration and invasion, whereas in MDA-MB-231 cell lines, the CCR7 protein expression was reduced, too.44 The significance of let-7 miRNA cluster was further highlighted by Hainz et al.’s group. They manipulated the expression of let-7c to ensure the role of let-7c as a tumor suppressor miRNA. In their study, let-7c expression was examined in breast cancer cell lines and tissues. They showed that there was a significant reduction in the expression of let-7c in tumor compared to their matched normal breast tissue. Moreover, they demonstrated that down-regulation of let-7c in breast cancer cell lines increased the cell proliferation. Interestingly, in xenograft models of breast cancer cells, let-7c was reconstituted and found to reduce the tumor burden.45
Other studies speculated that miR-15a and miR-16-1 exhibit tumor suppressing action as they are capable of down-regulating the anti-apoptotic protein Bcl-2. For instance, in CLL which is highly associated with loss of chromosomal region 13q14, miR-15a and miR-16-1cluster was found to localize in the same region hence loss of 68% of these miRNAs are feature in CLL patients.37,46,47
Kent et al., accentuated that to assign a definite mode of action for an altered miRNA either as an oncogene or a tumor suppressor gene one should be considered with caution due to the fact that miRNA expressions are highly cell type specific and cellular differentiation status dependent.47 Well-studied strong candidates of miRNAs either as tumor suppressive or oncogenic activities are summarized in Table 1.41,42,46-49
Table 1.
Aberrantly expressed miRNAs in different types of cancer and their potential function.
| miRNA | Suggestive role | Cancer type | References |
|---|---|---|---|
| miR-15a~16-1 | TS | CLL, prostate, multiple myeloma | 46-48 |
| Let-7 | TS | Breast, lung, cervix, ovary, urothelial, colon | 41,46-48 |
| miR-143, 145 | TS | Colorectal, bladder | 41,47,48 |
| miR-155 | ON | Hematopoietic, breast, lung, pancreas, colon | 41,46-48 |
| miR-17-92 | ON | B-cell lymphoma, lung | 42,46-48 |
| miR-21 | ON | Breast, colon, lung, pancreas, stomach, prostate and glioblastoma | 47,48 |
| miR-372,373 | ON | Testicular germ cell tumors | 41,49 |
TS, tumor suppressor; CLL, chronic lymphocytic leukemia; ON, oncogene.
MicroRNAs and breast cancer: current knowledge of relevant miRNAs to breast cancer
After the first report on deregulation of miRNA in human lymphocytic leukemia,37 many other studies showed aberrant expression profiles of miRNA in breast cancer. Generally, miRNA expression is down-regulated in poorly differentiated breast tumors and miRNAs has been associated with specific pathological features like estrogen receptor (ER), progesterone receptor (PR), lymph node status, vascular invasion, proliferation and tumor stage.50,51 Iorio et al., demonstrated experimentally aberration of miRNA in breast cancer tissue samples. They compared normal versus breast cancer tissue samples and found 29 miRNAs whose expression was distinctively deregulated that could separate the normal form tumor samples. The most significantly deregulated miRNAs found were miR-125b, miR-145, miR-21 and miR-155. The first two were down-regulated and were believed to have tumor suppressive effect, while the remaining two exhibited oncogenic effect, and were up-regulated in the breast tumor tissues.50 Deregulation in miRNA expression in breast cancer has been also associated in many other studies. To name a few, let-7d, miR-210 and miR-221 were found to be down-regulated in ductal carcinoma in situ (DCIS) however up-regulated in the invasive carcinoma in a study aiming at revealing the role of miRNAs in breast cancer progression from DCIS to invasive ductal carcinoma.52 In relation to clinico-pathological features of breast cancer patients, Let-7 was found to be down-regulated with presence of positive lymph node metastasis of high proliferative index suggesting its association with poor prognosis an observation that was also reported in human lung cancer.50,53 Moreover, miR-30 found to be down-regulated in both ER and PR negative suggesting that its expression is regulated by these hormones.50
In triple negative cell that have been challenged therapeutically, a reduction in miR-342 was observed but its elevation was associated with the luminal B breast cancer subtype. Moreover, down-regulation of miR-520g in ER and PR positive tumors indicated that these deregulations in miRNA expression can serve as a maker for poor prognosis.51 In another study with triple negative breast cancer, when compared with normal tissue, miR-10b, miR-145, miR-205 and miR-122 were found to be down-regulated. Although it turned not to be statistically significant, miR-21, miR-210, miR-221 and miR-222 exhibited a higher expression with both patients disease free and overall survival.54 Moreover, miR-21 is found to be associated with high proliferative index, advanced tumor stage, and lymph node involvement. It is also involved in disease progression and metastasis in pregnancy-associated breast cancer55 (Table 2).48,56,57
Table 2.
Aberrantly expressed miRNAs in breast cancer and their suggestive roles.
| miRNA | Suggestive role | Biological function | References |
|---|---|---|---|
| miR-21 | ON | Invasion and metastasis | 56 |
| miR-10b | ON | Invasion and metastasis | 48,56,57 |
| miR-155 | ON | Cell invasion and metastasis | 56 |
| miR-27a | ON | Cell cycle progression | 48 |
| miR-206 | TS | ER signaling | 48,56 |
| miR-125a/b | TS | Migration and invasion | 48,56 |
| Let-7 family | TS | Proliferation and metastasis | 56,57 |
ON, oncogene; TS, tumor suppressor; ER, estrogen receptor.
miRNAs as diagnostic and therapeutic tools in breast cancer
Due to the accumulating knowledge about the breast cancer heterogeneity, it was always considered that breast cancer with its molecular and pathological types as separate entities. Hence, with the advanced genomic and proteomics technology, therapy is thought to be tailored accordingly and identification of new target anticancer therapies that work at different levels of the breast cancer biology as signal transduction, cell cycle, apoptotic, pro-angiogenic and metastatic pathways.58 Kim’s lab used synthetic let-7 to target C-C chemokine receptor type 7 (CCR7) in MDA-MB-231 cells and observed a decreased level of cell proliferation, migration and invasion. This suggests the role of let-7 in targeting CCR7 is a valid approach to suppress migration and invasion of human breast cancer and this serves the therapeutic potential of let-7 as an anti-metastatic in breast cancer.44
It is now established that miRNAs play a major role in cancer progression and many studies substantiate a distinctive expression profiles within cancerous tissues.34,59 These findings open the gate for new therapeutic modalities targeting miRNAs associated with disease progression. Recently, miRNA therapeutic companies are taking potential miRNAs into clinical trials. For examples, miravirsen is a drug, which is designed to inhibit miR-122 which is found abundantly in liver accounting for functional infection of hepatitis C virus (HCV). Although the real mechanism of how miR-122 aids the replication of the virus is not yet known, it is though that miR-122 interaction with the seed sequence of the 5’ non coding region of the viral transcript that induces its target rather than inhibiting the function as the classical role of miRNA. When treating chronic HCV-infected chimpanzees with miravirsen, interestingly it leads to HCV suppression with no side-effects.10
Other approach replacing miRNA rather than inhibiting miRNAs have been also developed. In a pre-clinical set up, mice cancer models were injected with miR-34a mimics showed a potent anti-tumor effects.60 Apart from the therapeutic potential that these miRNAs hold, miRNAs have been also proven their potentiality in helping in disease diagnosis. In many studies, it has been well illustrated how does these miRNAs are capable of differentiating between tumor profiles. For instance, in 17 poorly differentiated tumors, the miRNAs expression profile analysis was capable of classifying the correct diagnosis of these tumors than the mRNA profile.35
Despite these achievements, there are several facts that we have to take into consideration. For instance, the fact that some miRNAs hold dual function as oncogene or tumor suppressor increase the awareness that artificial regulation may result in unexpected antagonistic effects.31,61. Additionally, the impact of the therapeutics may become affected by the fact the some single miRNA may target hundreds or thousands of genes.1 Therefore, realizing and having a better understanding of the miRNA function, target recognition and their role in specific types of cancerous cells is necessary to develop therapeutic agents that target specific tumor types, and reduce or eliminate the potential for adverse effects of these drugs.
Recent studies on the effect of neo-adjuvant chemotherapy on miRNAs expression
Of the early reports, a study on the effect of cisplatin and 5-fluorouracil on esophageal carcinoma cell lines, where a panel of miRNAs expression changes were detected to correlate with treatment induction and suggest their involvement in cell survival after treatment.62
Preliminary results from Baldoni et al., work showed that neo-adjuvant chemotherapy (NAC) induced alteration at the miRNA levels in gastric cancer patients compared to normal gastric tissue, where they have identified 14 differential miRNAs expression in gastric cancer patients. As further analyses are needed, they are speculating that association of these miRNAs with clinico-pathological features of the patients could determine some predictable values of miRNAs.63 Recent study on the effect of NAC in circulating miRNAs in breast cancer revealed 25 plasma miRNAs were altered by NAC among which miR-34a and miR-122 were highly up-regulated, particularly in patients with pathological partial response with aggressive breast cancer.64
Many ongoing studies emphasized the effect of NAC on miRNAs profile and expression of which some are summarized in Table 3.62,65,66
Table 3.
Summary of some studies on the effect of chemotherapy treatment on miRNA expression.
| miRNA | Cancer type | Chemotherapy agent | Notes and suggestive target effect | References |
|---|---|---|---|---|
| miR-199a-5p, miR-302f, miR-320a, miR-342-3p, miR-425, miR-455-3p, miR-486-3p, miR-519c-5p, miR-548d-5p, miR-617, miR-758, miR-766, miR-1286 | Esophageal carcinoma cell lines | Cisplatin or 5-fluorouracil | Cellular response to a cytotoxic stimulus | 62 |
| miR-125b | Breast cancer | Taxane-anthracycline | Validated protein targets of miR-125b are involved in apoptotic pathways and cell cycle control | 65 |
| miR-122a, miR-145 and miR-205 | MDA-MB-453 triple-negative breast cancer cells | Capecitabine and ixabepilone | - | 66 |
Conclusions
The study of miRNA role in disease in particular cancer is rapidly evolving field in the recent few years. There is a greater understanding of these molecules in terms of their identification and function. Moreover, their contribution in RNA regulation and protein synthesis will aid our understanding to their contribution in disease status in particular carcinogenesis. The expression of miRNA in tissue and serum may contribute to the identification of biomarkers of prognostic and predictive significance.
References
- 1.Di Leva G, Calin GA, Croce CM. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res Part C 2006;78:180-9. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Yang P, Wang X-F. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 2014;24:153-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Planell-Saguer D, Mariã N, Rodicio MC. Detection methods for microRNAs in clinic practice. Clin Biochem 2013;46:869-78. [DOI] [PubMed] [Google Scholar]
- 4.Molnár V, Tamási V, Bakos B, et al. Changes in miRNA expression in solid tumors: An miRNA profiling in melanomas. Semin Cancer Biol 2008;18:111-22. [DOI] [PubMed] [Google Scholar]
- 5.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res Fund Mol Mechan Mutagen 2011;717:1-8. [DOI] [PubMed] [Google Scholar]
- 7.Fabbri M, Croce C, Calin G. MicroRNAs. Cancer J 2008;14:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [DOI] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11. [DOI] [PubMed] [Google Scholar]
- 10.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Research 2013;2:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126-39. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Kim M, Ham J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarnieri DJ, DiLeone RJ. MicroRNAs: A new class of gene regulators. Ann Med 2008;40:197-208. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom K-H., et al. , The Drosha-DGCR8 complex in primary microRNA processing. Genes Develop 2004;18:3016-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund E, Güttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science 2004;303:95-8. [DOI] [PubMed] [Google Scholar]
- 16.Tétreault N, De Guire V. miRNAs: their discovery, biogenesis and mechanism of action. Clin Biochem 2013;46:842-5. [DOI] [PubMed] [Google Scholar]
- 17.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature Rev Mol Cell Biol 2014;15:509-24. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biol 2009;11:228-34. [DOI] [PubMed] [Google Scholar]
- 19.Bernardo BC, Charchar FJ, Lin RCY, McMullen JR. A MicroRNA guide for clinicians and basic scientists: background and experimental techniques. Heart Lung Circulation 2011;21:131-42. [DOI] [PubMed] [Google Scholar]
- 20.Hu HY, Yan Z, Xu Y, et al. Sequence features associated with microRNA strand selection in humans and flies. BMC Genom 2009;10:413-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell 2007;26:611-23. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Zhang Z, O’Loughlin E, et al. Quantitative functions of argonaute proteins in mammalian development. Genes Develop 2012;26:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol 2007;171:728-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel D. MicroRNAs genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 2005;7:719-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker R, Sheth U. Bodies and the control of mRNA translation and degradation. Mol Cell 2007;25:635-46. [DOI] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [DOI] [PubMed] [Google Scholar]
- 28.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nature Cell Biol 2005;7:633-6. [DOI] [PubMed] [Google Scholar]
- 29.Czech B, Hannon GJ. Small RNA sorting: matchmaking for argonautes. Nat Rev Genet 2011;12:19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [DOI] [PubMed] [Google Scholar]
- 31.Croce C. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med 2013;5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volinia S, Calin G, Liu C, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, Getz G, Miska E, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [DOI] [PubMed] [Google Scholar]
- 36.Jiang RLY. Regulation of miRNA pathway and roles of micrornas in tumorigenesis and metastasis. Human Genet Embryol 2013;S2:007. [Google Scholar]
- 37.Calin GA, Dumitru CD, Shimizu M., et al. , Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci 2002;99:15524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirnezami AHF, Pickard K, Zhang L, et al. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol (EJSO) 2009;35:339-47. [DOI] [PubMed] [Google Scholar]
- 39.Liu C-G, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A 2004;101:9740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang F, He Y, Zou M, et al. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One 2012;7:e35435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Develop Biol 2007;302:1-12. [DOI] [PubMed] [Google Scholar]
- 42.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Develop 2006;16:4-9. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Investig 2010;120:1298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S-J, Shin J-Y, Lee K-D, et al. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res 2012;14:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hainz YX. Let-7 microRNA: tumour suppression activity in breast cancer. Am J BioMed 2014;2:512-26. [Google Scholar]
- 46.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell 2009;136:586-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 2006;25:6188-96. [DOI] [PubMed] [Google Scholar]
- 48.Shenouda S, Alahari S. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metast Rev 2009;28:369-78. [DOI] [PubMed] [Google Scholar]
- 49.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006;124:1169-81. [DOI] [PubMed] [Google Scholar]
- 50.Iorio M, Ferracin M, Liu C, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70. [DOI] [PubMed] [Google Scholar]
- 51.Lowery A, Miller N, Devaney A, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 2009;11:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volinia S, GalassoM Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci 2012;109:3024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [DOI] [PubMed] [Google Scholar]
- 54.Radojicic J, Zaravinos A, Vrekoussis T, et al. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011;10:507-17. [DOI] [PubMed] [Google Scholar]
- 55.Walter BA, Valera VA, Sobel M, Merino MJ. miR-21 expression in pregnancy-associated breast cancer: a possible marker of poor prognosis. J Cancer 2011;2:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res 2010;12:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh R, Mo Y-Y. Role of microRNAs in breast cancer. Cancer Biol Ther 2013;14:201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serpico D, Molino L, Di Cosimo S. microRNAs in breast cancer development and treatment. Cancer Treat Rev 2014;40:595-604. [DOI] [PubMed] [Google Scholar]
- 59.Calin G, Croce C. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [DOI] [PubMed] [Google Scholar]
- 60.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 2011;19:1116-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett 2011;585:2087-99. [DOI] [PubMed] [Google Scholar]
- 62.Hummel R, Wang T, Watson D, et al. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol Rep 2011;26:1011-7. [DOI] [PubMed] [Google Scholar]
- 63.Baldoni MMB, Camillo CMC, Begnami MDF. MicroRNA expression signature of gastric carcinoma patients treated with neoadjuvant chemotherapy. Cancer Res 2014;74:abstract 4672. [Google Scholar]
- 64.Freres P, Josse C, Bovy N, et al. Neoadjuvant chemotherapy in breast cancer patients induces expression of miR-34a and miR-122. Eur J Cancer 2014;50:53-4. [DOI] [PubMed] [Google Scholar]
- 65.Zheng Y, Boohaker RJ, Liu X, et al. A microRNA expression signature in taxane-anthracycline-based neoadjuvant chemotherapy response. J Cancer 2015;6:671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao Y, Chen S, Zhou XIN, et al. 5-FU and ixabepilone modify the microRNA expression profiles in MDA-MB-453 triple-negative breast cancer cells. Oncology Lett 2014;7:541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


