Abstract
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and is still one of the most fatal cancers. Hence, it needs to identify always new putative markers to improve its diagnosis and prognosis. The selenium is an essential trace mineral implicated as a key factor in the early stage of cancer and exerts its biological function through the selenoproteins. In the last years our group has been studying the involvement of some selenoproteins in HCC. However, no many data are reported in literature about the correlation between HCC and the glutathione peroxidases (GPXs), both selenium and non selenium-containing GPXs.
In this paper we have evaluated the GPX4 and GPX7 expression in some paraffin-embedded tissues from liver biopsy of patients with hepatitis C virus (HCV)-related cirrhosis and HCC by immunohistochemistry and RT-qPCR analysis. Our results evidenced that i) GPX4 and GPX7 had a statistically significant over-expression in HCC tissues compared to cirrhotic counterparts used as non tumor tissues, and ii) their expression was higher in grade III HCC tissues with respect to grade I-II samples. Therefore, we propose to use GPX4 and GPX7 as possible markers for improving HCC diagnosis/prognosis.
Key words: Hepatocellular carcinoma, GPX4, GPX7, immunohistochemistry, RT-qPCR
Introduction
Liver cancer is the second-leading cause of cancer mortality worldwide, accounting for approximately 600,000 cancer-related deaths annually.1 Hepatocellular carcinoma (HCC), the most common type of liver cancer, generally develops from chronic liver injury,2 and its risk factors are multiple, such as hepatitis B (HBV) or C virus (HCV) infection, alcohol-induced liver disease (ALD), nonalcoholic fatty liver disease (NAFLD), primary biliary cirrhosis, exposure to environmental carcinogens (particularly aflatoxin), or even type 2 diabetes and obesity.3 Despite recent advances in diagnosis and management, the median survival of HCC patients is less than 8 months, and the disease is still one of the most fatal cancers.4 Surgical resection, liver transplantation, and local ablation remain the only HCC curative modalities,5,6 and recurrence occurs in up to 70% of patients within 5 years after resection.7,8 Unfortunately, the molecular signaling mechanisms, which specifically lead to HCC, are shielded and perturbed by molecular signaling sustained by viral infection as well as other diseases such as, for instance, diabetes. Therefore, it is necessary to identify always new putative markers to improve the HCC prognosis. Recently some Authors evaluated the expression also of other two proteins, thymosin beta 4 (Tβ4) and thymosin beta 10 (Tβ10) in HCC tissues. This study showed the expression of both beta-thymosins in HCC with marked differences in their degree of expression and frequency of immunoreactivity. The higher incidence of Tβ10 expression and its higher reactivity in tumor cells involved in stromal invasion indicated a possible major role for Tβ10 in HCC progression.9
Some studies evidenced the role of selenium (Se) to assist cells in resisting to oxidative damage that is a major cause of cellular damage also because it was found implicated as a key factor in the early stage of cancer.10 In vivo, Se is primarily present as selenoproteins to maintain the balance of the cellular redox state, and in humans there are 25 selenoproteins.11 In the last years our group has been focusing on some selenoproteins and their involvement in HCC, also evaluating by immunohistochemistry (IHC) the expression of selenium binding protein-1 (SELENBP1), which is, in vivo, able to incorporate exogenously administered radioactive (75Se)-sodium selenite in the liver, as well as that of Selenoprotein M (SELM) in tissue samples of HCC patients.12-14 These studies provided evidence that SELENBP-1, as well as selenium, is down-regulated in the liver tissue of HCC patients and that its gradual loss is associated with an increased malignant grade.12,13 Moreover, we showed for the first time an increase of SELM expression in HCC liver tissues, and its correlation with their increased malignancy grade.14 Also, we have recently carried out the analysis of the global expression of the seleno-transcriptome protein family in two human hepatocellular carcinoma cell lines (HepG2 and Huh7) compared to the normal human hepatocytes by means of the RT-qPCR analysis.15,16 These studies have shown a signature of selenoprotein mRNAs specific for human hepatoma cells showing the genes that change their expression as a consequence of the liver cancer in the absence of any genetic mutations or viral infection, and, in particular, that in HepG2 and Huh7 cells there were three down-regulated and ten over-regulated genes, among which SELM, and two glutathione peroxidases such as GPX4 and GPX7.15,16
In general, the GPXs belong to a family of phylogenetically related enzymes, and GPX4 and GPX7 are strictly correlated because the Cys-containing GPX7 is evolved from a GPX4-like ancestor.17 Actually, these two proteins present a percentage of sequence identity of about 50% and the same fold topology of an alpha-beta 3-layer sandwich type.18 GPX4 has some role in the regulation of apoptosis, whereas GPX7 is reported to be involved in the protein folding.17 Also, GPX4 was found over-regulated at the protein level in human colon carcinoma tissue and the impaired expression of its gene in peripheral blood mononuclear cells was proposed as a biomarker of increased breast cancer risk.19,20 Further, GPX4 has been significantly associated with breast cancer survival among the patients with the highest Native American (NA) ancestry whereas its variants resulted to be correlated with the risk of lethal prostate cancer, and able to modify the relation between γ-tocopherol and prostate cancer survival.21,22
Recently, GPX4 has been found to play an essential role in the hepatitis C virus (HCV) life cycle. In fact, the modulation of the oxidative stress, which specifically targets GPX4 in chronic hepatitis C (CHC), may decrease the virions infectivity preventing the hepatocarcinogenesis, principally in patients who remain difficult to be treated in the new era of interferon-free regimens.23 On the other hand, it is also reported that the dysfunction of GPX7 in oesophageal cells increases the levels of reactive oxygen species (ROS) and oxidative DNA damage, which are common risk factors for Barrett oesophagus and oesophageal adenocarcinoma.24 Hence, in this paper we report the evaluation of the GPX4 and GPX7 expression in tissue samples of patients with HCC by IHC and RT-qPCR analysis to understand if these two GPXs could be used as new markers for improving HCC diagnosis/prognosis.
Materials and Methods
Tissue sample
Paraffin-embedded HCC tissues obtained by biopsy from thirty patients were subjected to IHC and to reverse transcription-qPCR. In this study we used the cirrhotic counterparts of all tumor tissue sections as non tumor tissue controls. All patients in this study provided informed consent, and the study was approved by the Second University of Naples Ethics Committee. The clinic-pathological assessment of patients are listed in Table 1. In details, all patients had HCV-related cirrhosis and included 10 HCC with grade I, 11 HCC with grade II, and 9 HCC with grade III. No information related to follow-up data of these patients is known.
Table 1.
Hepatocellular carcinoma patients clinical-pathological characteristics.
| Number of patients | |
|---|---|
| Age | |
| ≤70 | 14 |
| ≥70 | 16 |
| Gender | |
| Female | 13 |
| Male | 17 |
| Grading | |
| I | 10 |
| II | 11 |
| III | 9 |
| Hepatitis C Virus (HCV) RNA | |
| Positive | 30 |
| Negative | None |
| Tumor size | |
| <2 | 6 |
| 2-5 | 16 |
| >5 | 8 |
Tissue immunohistochemistry
Briefly, xylene dewaxed and alcohol rehydrated paraffin sections were placed in Coplin jars filled with a 0.01 M trisodium citrate solution and microwaved. After heating, slides were thoroughly rinsed in cool running water for 5 min. Sections were immersed in 3% H2O2 at room temperature for 30 min to block any endogenous peroxidase activity. They were then washed in Tris-buffered saline (TBS) pH 7.4 before incubating at 4°C overnight with rabbit anti-human polyclonal GPX4 (LifeSpan BioSciences Inc., Seattle, WA, USA), diluted 10 µg/mL and with mouse anti-human monoclonal GPX7 (LifeSpan BioSciences Inc), diluted 10 µg/mL. After incubation with the primary antibody, tissue sections were stained with species-specific biotinylated secondary antibodies, followed by peroxidase labelled streptavidine (Dako, Glostrup, Denmark); the signal was developed by using diaminobenzidine (DAB) chromogen (Dako) as substrate. Mayer’s Hematoxylin solution was used as a nuclear counterstaining. Incubations with diluent antibody buffer omitting the specific antibody (for GPX4 and GPX7) were used as negative controls (Supplementary Figure 1).
Figure 1.
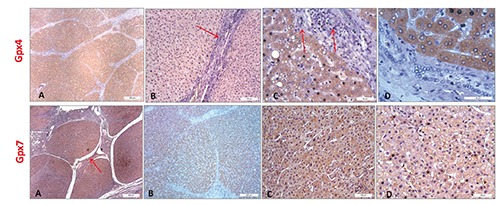
GPX4 and GPX7 staining in cirrhotic tissues. The immunohistochemical observation of the human GPX4 and GPX7 expressions at 40x (A), 200x (B), 400x (C) and 630x (D) magnifications for GPX4 and at 40x (A), 100x (B), 200x (C) and 400x magnifications for GPX7 in cirrhotic tissues. In the GPX4 staining it can be observed a portal lobule richly infiltrated by non-immunoreactive lymphocytic elements (B, red arrow), as well as bile ducts (C, red arrows) whose epithelia show no reactivity to this protein; more details were visible in panel D (630x). In the GPX7 staining the hepatic parenchyma is organized into macronodular areas (A, red arrow), visible at higher magnification in the panel B (100x).
Scoring methods proposed by Sinicrope et al.25 were utilised in the evaluation of immunoreactivity for both staining intensity and percentage positive of stained tumor cells. The percentage of positive cells that revealed stronger staining intensity in respect to the adjacent hepatocyte cells were scored as follows: 0 (if 0-4% of tumor cells were stained); 1 (if 5-25% of tumor cells were stained); 2 (if 26-50% of tumor cells were stained); 3 (if 51-75% of tumor cells were stained); and 4 (if more than 75% of tumor cells were stained). In order to determine the staining intensity, these categories were sub-classified as follows: 0, no expression, 1, extremely weak; 2, weak; 3, moderate; and 4, strong expression. Associations between immunohistochemical scores and clinicopathological characteristics of tissue specimens were evaluated by Pearson correlation coefficients and P<0.05 was considered statistically significant.
RNA preparation and RT-qPCR analysis
Total RNA was extracted from cirrhotic and tumoral liver FFPE (formalin-fixed paraffin embedded) tissue sections equivalent to 60 µm by RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies-Ambion, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extracted RNA was dissolved in diethylpyrocarbonatetreated water, and its concentration and purity were assessed by measurement of optical density at 260⁄280 nm. RNA samples were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Two microgram of total RNA of each sample was reverse-transcribed with SuperScript VILO cDNA Synthesis kit (Life Technologies-Ambion) according to the manufacturer’s instructions and subsequently diluted with nuclease-free water (Life Technologies-Ambion).
Sequences for mRNAs from the nucleotide data bank (National Center for Biotechnology Information, USA) were used to design primer pairs for RT-qPCR (Primer Express, Applied Biosystems, CA, USA). Oligonucleotides were obtained from Sigma Aldrich. The primer sequences are provided in Table 2. An appropriate region of 18S rRNA was used as control. RT-qPCR assays were run on an Step-One Real Time PCR System (Applied Biosystem). Two µL of cDNA were amplified in a total volume of 25 µL containing 2X SYBR Green PCR Master Mix (Applied Biosystem) and 300 nM of forward and reverse primers. The thermal cycling conditions were as follows: 5 min of denaturation at 95°C followed by 44 cycles of a two-step program (denaturation at 95°C for 30 sec and annealing/extension at 60°C for 1 min). For each target the primer sequences and the melting temperature are reported in Table 2. Dilutions of standards and test samples were run in duplicate. Each reaction was repeated at least three times. To show that only one PCR product has been formed using the designed GPX4 and GPX7 primers, we report as example four curves and the related separation of PCR products on a gel (Supplementary Figures 2 and 3). Expression levels of each target gene in HCC tissue were compared with cirrhotic liver tissue. Data were normalized using the 18S rRNA as housekeeping gene which has been previously used in our recent paper.15-16 The 2 x-fold change between expression levels in HCC and cirrhotic liver tissues was chosen as the threshold for significance of target genes. Statistical analyses (paired Student’s t-test) were performed using Prism software (Graphpad Software, La Jolla, CA, USA). Significant differences in relative gene expression between HCC and cirrhotic liver tissue are marked by *(P<0.05), and ** (P<0.01).
Table 2.
Parameters for the RT-qPCR analysis.
| Gene | Temperature (°C) | Sequence (5’ -> 3’) |
|---|---|---|
| GPX4 | 59.8 | AGAGATCAAAGAGTTCGCCGC (21) |
| TCTTCATCCACTTCCACAGCG (21) | ||
| GPX7 | 57.9 | TTGGTCCCATCATTCTTGTGG (21) |
| GGCTGGTGATTCACTGGTCAA (21) | ||
| CTGCCCTATCAACTTTCGTG (20) | ||
| 18S | 60 | GTAGTTTCTCAGGCTCCCTCTC (22) |
Figure 2.
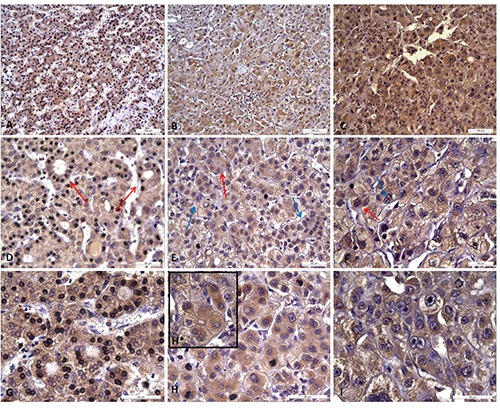
GPX4 staining. Immunohistochemical observation of the human GPX4 expression in HCC tissues with grades I, II and III, respectively, at 200x (A, B, C), 400x (D, E, F) and 630x (G, H, box in H, I) magnifications. In particular, it was possible observe in HCC of grade I some acinar cell variants (D, red arrows, and higher magnification in G), in HCC of grade II (E, and higher magnification in H and box in H, respectively) a rosette-like structure (red arrow) and mono and bi-layered sheets (blue arrows), and in HCC of grade III (F, and higher magnification in I) a strong stain in highly undifferentiated (red arrow) and binuclear hepatocytes (blue arrow).
Figure 3.
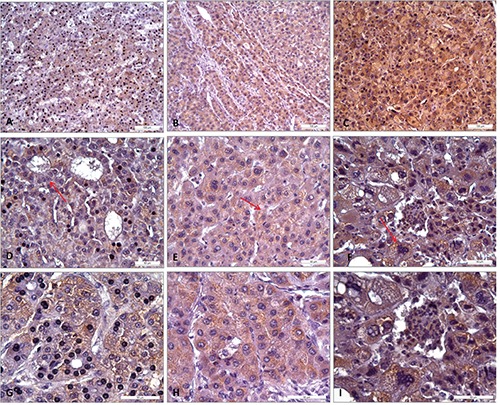
GPX7 staining. Immunohistochemical at 200x (A, B, C) 400x (D, E, F) and 630x (G, H, I) magnifications. In particular, it was possible to observe in HCC of grade I acinar cell variant (D, red arrow, and higher magnification in G), in grade II HCC a multi-layered sheet (E, red arrow, and higher magnification in H) and in grade III HCC atypical hepatocytes with monstrous nuclei (F, red arrow, and higher magnification in I).
Results and Discussion
GPX4 and GPX7 expression in HCC tissues by immunohistochemistry
Collected tissues were subjected to GPX4 and GPX7 staining by IHC. GPX4 staining in cirrhotic tissues showed a mild and specific cytoplasmic reactivity of the hepatocytes (Figure 1A). In Figure 1 B,C,D it has been possible to observe a portal lobule richly infiltrated by non-immunoreactive lymphocytic elements, and some bile ductules whose epithelia showed no reactivity to GPX4. In HCC tissues, the hepatocytes showed at 200x magnification a cytoplasmic positivity with stroma immunoreactivity and inflammatory elements interposed between the cirrhotic nodules as visible at 400x and 630 magnifications (Figure 2). In details, the cytoplasm of the hepatocytes showed a specific immunoreactivity for GPX4, gradually grown up from acinar cell variants in HCC tissues with a well-differentiated grade I (Figure 2D) to form a diffuse poorly differentiated grade III (Figure 2F). A diffuse positivity and a marked intensity of the cytoplasmic staining was observed in HCC samples with grade I, in a similar way to those with grade II (Figure 2 D,E). In particular, for the grade II HCC tissues we noted an asymmetrical arrangement of the hepatocytes, which are organized in rosette-like structures and in mono and bi-layered sheets (Figure 2 E,H, and box in panel H). On the other hand, the HCC tissues with grade III showed a more intense cytoplasmic staining when compared to those with grades I and II. Moreover, the hepatocytes were distributed in multi-layered sheets and their greater cell volume was associated both to an increased nuclear polymorphism and to the presence of bi-nucleate hepatocytes (Figure 2 F,I). After GPX7 staining in cirrhotic tissues, the hepatic parenchyma was organized into macro-nodular areas (Figure 1 A,B) and the hepatocytes expressed a mild cytoplasmic immuno-reactivity. GPX7 showed a weak and diffuse immunoreactivity in grade I HCC tissues similarly to those with grade II whereas the staining became more widespread and intense in grade III HCC tissues (Figure 3). In grade I and II HCC tissues we observed GPX7 plurifocal and moderate positivity (Figure 3A, 3 D,G,B,E,H). In particular, in grade II HCC tissues there was a strong architectural disorder with hepatocytes cellular distribution in multi-layered sheets (Figure 3 E,H). In grade III HCC the hepatocytes were atypical with monstrous nuclei and prominent nucleoli of variable sizes (Figure 3 F,I).
These evaluations have suggested that the gradual raise of GPX4 and GPX7 expression were associated with an increased malignant grade. Actually, the cells percentage and the expression intensity increased from better differentiated histopathological forms to those poorly differentiated. In particular, using scoring methods proposed by Sinicrope et al.25 for GPX4 staining we can underline that: i) seven HCC tissue samples displayed a weak expression (with score 2); ii) fourteen revealed a moderate expression (score 3); and iii) nine evidenced a strong expression (score 4). For GPX7 staining we can underline that: i) six HCC tissue samples displayed a weak expression (score 2); ii) sixteen revealed a moderate expression (score 3); and iii) eight evidenced a strong expression (score 4). A significant correlation was detected between the immunohistochemical scores and cancer progression/grading (P<0.05).
Analysis of GPX4 and GPX7 expression in HCC/cirrhotic tissues by RT-qPCR analysis
GPX4 and GPX7 gene expression levels in human HCC tissues were evaluated by RT-qPCR compared to the cirrhotic liver tissue. No significant change in GPX4 and GPX7 expression levels between the grade I and grade II HCC samples was observed therefore we treat together their gene expression levels. In details, Figure 4 shows that: i) GPX4 and GPX7 had a statistically significant over-expression in HCC tissues; and ii) their expression was higher in grade III HCC tissues with respect to grade I-II samples. It highlighted that GPX4 and GPX7 expression was associated with an increased malignancy grade. However, these gene data agreed with the protein GPX4 and GPX7 over-expression obtained by IHC and with our gene data obtained on two HCC cell lines, HepG2 and Huh7, when compared to normal human hepatocyte cells.16 Then, we evaluated the correlation coefficient between the GPX4 and GPX7 expression levels and the clinical-pathological data of HCC patients. A significant correlation was detected between the GPX4 and GPX7 gene expression levels and tumor grading (P=0.012 and P=0.043, respectively) whereas no correlation has been found between GPX4 and GPX7 expression and age/gender/tumor size.
Figure 4.
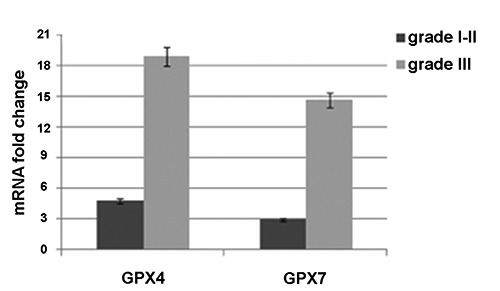
mRNA fold change evaluated between HCC tissues and cirrhotic liver tissues used as control for the expression levels of GPX4 and GPX7.
In conclusion in this paper i) we evaluated the GPX4 and GPX7 expression in HCC tissue samples by IHC and RT-qPCR; ii) we verified that both these GPXs had a statistically significant over-expression; iii) this over-expression was associated with an increased malignancy grade. Furthermore, this paper highlighted for the first time that the GPX4 and GPX7 over-expression in human HCC tissues can be easily and precisely detected by the simple IHC or RT-qPCR techniques.
Acknowledgments
The authors thank Marilina Russo for assistance in figure preparation.
References
- 1.International Agency for Research on Cancer. GLOBOCAN 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012. International Agency for Research on Cancer, Lyon; 2013. [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76. [DOI] [PubMed] [Google Scholar]
- 3.Costantini S, Capone F, Guerriero E, Marfella R, Sorice A, Maio P, et al. Cytokinome profile of patients with type 2 diabetes and/or chronic hepatitis C infection. PLoS One 2012;7:e39486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology 2004;127:S27-S34. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Llovet JM. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology 2002;35:519-24. [DOI] [PubMed] [Google Scholar]
- 6.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology 2005;42:1208-36. [DOI] [PubMed] [Google Scholar]
- 7.Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol 2003;38:200-7. [DOI] [PubMed] [Google Scholar]
- 8.Llovet JM, Fuster J, Bruix J. Intention-to-treat analysis of surgical treatment for early hepatocellular carcinoma: resection versus transplantation. Hepatology 1999;30:1434-40. [DOI] [PubMed] [Google Scholar]
- 9.Theunissen W, Fanni D, Nemolato S, Di Felice E, Cabras T, Gerosa C, et al. Thymosin beta 4 and thymosin beta 10 expression in hepatocellular carcinoma. Eur J Histochem 2014;58:2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 2006;160:1-40. [DOI] [PubMed] [Google Scholar]
- 11.Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J 2009;422:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raucci R, Colonna G, Guerriero E, Capone F, Accardo M, Castello G, et al. Structural and functional studies of the human selenium binding protein-1 and its involvement in hepatocellular carcinoma. Biochim Biophys Acta 2011;1814:513-22. [DOI] [PubMed] [Google Scholar]
- 13.Di Stasio M, Volpe MG, Colonna G, Nazzaro M, Polimeno M, Scala S, et al. A possible predictive marker of progression for hepatocellular carcinoma. Oncol Lett 2011;2: 1247-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerriero E, Accardo M, Capone F, Colonna G, Castello G, Costantini S. Assessment of the Selenoprotein M (SELM) over-expression on human hepatocellular carcinoma tissues by immunohistochemistry. Eur J Histochem 2014;58:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantini S, Di Bernardo G, Cammarota M, Castello G, Colonna G. Gene expression signature of human HepG2 cell line. Gene 2013;518:335-345. [DOI] [PubMed] [Google Scholar]
- 16.Guariniello S, Di Bernardo G, Colonna G, Cammarota M, Castello G, Costantini S. Evaluation of the selenotranscriptome expression in two hepatocellular carcinoma cell lines. Anal Cell Pathol (Amst) 2015;2015:419561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta 2013;1830:3289-303. [DOI] [PubMed] [Google Scholar]
- 18.Scheerer P, Borchert A, Krauss N, Wessner H, Gerth C, Höhne W, et al. Structural basis for catalytic activity and enzyme polymerization of phospholipid hydroperoxide glutathione peroxidase-4 (GPX4). Biochemistry 2007;46:9041-9. [DOI] [PubMed] [Google Scholar]
- 19.Yagublu V, Arthur JR, Babayeva SN, Nicol F, Post S, Keese M. Expression of selenium-containing proteins in human colon carcinoma tissue. Anticancer Res 2011;31:2693-8. [PubMed] [Google Scholar]
- 20.Bermano G, Smyth E, Goua M, Heys SD, Wahle KW. Impaired expression of glutathione peroxidase-4 gene in peripheral blood mononuclear cells: a biomarker of increased breast cancer risk. Cancer Biomark 2010;7:39-46. [DOI] [PubMed] [Google Scholar]
- 21.Pellatt AJ, Wolff RK, John EM, Torres-Mejia G, Hines LM, Baumgartner KB, et al. SEPP1 influences breast cancer risk among women with greater native american ancestry: the breast cancer health disparities study. PLoS One 2013;8:e80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Blarigan EL, Ma J, Kenfield SA, Stampfer MJ, Sesso HD, Giovannucci EL, et al. Plasma antioxidants, genetic variation in SOD2, CAT, GPX1, GPX4, and prostate cancer survival. Cancer Epidemiol Biomarkers Prev 2014;23:1037-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brault C, Lévy P, Duponchel S, Michelet M, Sallé A, Pécheur EI, et al. Glutathione peroxidase 4 is reversibly induced by HCV to control lipid peroxidation and to increase virion infectivity. Gut 2014;65:144-54. [DOI] [PubMed] [Google Scholar]
- 24.Peng DF, Hu TL, Soutto M, Belkhiri A, El-Rifai W. Glutathione peroxidase 7 suppresses bile salt-induced expression of pro-inflammatory cytokines in Barrett’s carcinogenesis. J Cancer 2014;5:510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinicrope FA, Ruan SB, Cleary KR, Stephens LC, Lee JJ, Levin B. Bcl-2 and p53 oncoprotein expression during colorectal tumorigenesis. Cancer Res 1995;55:237-41. [PubMed] [Google Scholar]


