Abstract
We present data from our clinical department's experience with chromosomal microarray analysis (CMA) regarding the diagnostic utility of 1 or 2 long contiguous stretches of homozygosity (LCSHs) in an outbred population. The study group consisted of 2,110 consecutive patients from 2011 to 2014 for whom CMA was performed. The minimum cut-off size for defining a homozygous stretch was 5 Mb. To focus on cases with no parental consanguinity, we further studied only patients in whom the total length of homozygous stretches did not exceed 28 Mb or 1% of the autosomal genome length. We identified 6 chromosomal regions where homozygous stretches appeared in at least 3 patients and excluded these from further analysis. In 2 out of 120 patients with an isolated finding of 1 or 2 non-recurrent LCSHs, a plausible candidate gene associated with their phenotype was identified within the homozygous stretch. In both of these cases, a pathogenic mutation was detected, leading to diagnoses of pyruvate kinase deficiency and Marinesco-Sjögren syndrome. To clarify whether previously found homozygous stretches could be important for the interpretation of genome-wide sequencing data, we report 7 cases in which homozygous stretches not encompassing a clinically associated gene were first found on CMA, followed by the diagnostic whole-exome sequencing. The diagnostic utility of single LCSHs, unlikely to be caused by uniparental disomy, is discussed in detail.
Key Words: Autosomal recessive disease, Chromosomal microarray analysis, Clinical diagnostics, Long contiguous stretches of homozygosity, Variants of unclear clinical significance, Whole-exome sequencing
Chromosomal aberrations are a well-known cause of multiple congenital anomalies, intellectual disabilities (IDs), and autism spectrum disorders. Many centers use chromosomal microarrays (CMAs) as a first-tier diagnostic test for these indications because a remarkable proportion of causative genetic aberrations are in the submicroscopic range and, therefore, cannot be detected using conventional karyotyping methods [Hochstenbach et al., 2009; Miller et al., 2010; Vissers et al., 2010; Ahn et al., 2013]. In Estonia, CMAs have been funded as a first-tier diagnostic test by the Estonian Health Insurance Fund since 2011. All the diagnostic CMA analyses in Estonia are performed in 1 diagnostic laboratory (Department of Genetics, United Laboratories, Tartu University Hospital) using single nucleotide polymorphism (SNP) genotyping technology. An overview of the Estonian experience during the first years employing CMAs in clinical diagnostics was recently published by Zilina et al. [2014]. In contrast to array comparative genomic hybridization, SNP arrays also enable the detection of copy number neutral loss of heterozygosity regions, also known as regions of homozygosity or long contiguous stretches of homozygosity (LCSHs). The most widely used cut-off for the length of an LCSH is 5 Mb, as it was shown that LCSHs <4 Mb in length are common in an outbred European population [McQuillan et al., 2008]. LCSHs can result from ancestral homozygosity, uniparental disomy (UPD), or parental consanguinity [Kearney et al., 2011]. The degree of parental consanguinity can be reliably assessed based on the total length of LCSHs relative to the total autosomal genome length [Sund et al., 2013; Wang et al., 2015]. Parental consanguinity is very rare in Estonia having a small but outbred population of 1.3 million inhabitants.
In general, if well-known UPD syndromes are excluded, LCSH as a stand-alone finding rarely results in a definite diagnosis. However, the detection of an LCSH can lead to the selection of candidate genes within the stretch that cause an autosomal recessive disorder and, therefore, to further testing [Kearney et al., 2011; Sund et al., 2013; Wang et al., 2015]. A couple of published studies have investigated the diagnostic utility of LCSHs in a clinical setting, but they concentrated on UPD [Papenhausen et al., 2011] or parental consanguinity [Sund et al., 2013]. Recently, a large study of over 14,000 consecutive samples investigated the clinical utility of LCSHs caused by both UPD and parental relatedness [Wang et al., 2015]. In 9 cases, a clinically associated candidate gene was found in an LCSH, and in 7 of them, the pathogenic mutation was confirmed [Wang et al., 2015]. Whole-exome sequencing (WES) was used in another study to clarify the role of extended homozygosity in 50 patients with parental consanguinity, and it exhibited a diagnostic yield of 36% [Makrythanasis et al., 2014].
To our knowledge, no previous studies have examined the diagnostic utility of single LCSHs in patients without parental consanguinity (second cousin or closer). Therefore, the aim of this study was to fill this gap by investigating whether the detection of a single LCSH can aid in the discovery of phenotype-associated candidate genes, thereby leading to the confirmation of pathogenic mutations in routine clinical practice.
Materials and Methods
This study was approved by the Research Ethics Committee of the University of Tartu (approval date 1/13/2015 and number 243/T-3). The primary study group consisted of 2,110 consecutive patients who were analyzed with CMAs in our department from 2011 to 2014. No prenatal or non-affected parental samples were included in this study. The main indications for testing included - but were not restricted to - IDs, multiple congenital anomalies and autism spectrum disorders. In most of the cases, a CMA analysis was used as a first-tier diagnostic genetic test. Genomic DNA was extracted from peripheral blood samples. The CMA analysis was performed on all samples using HumanCytoSNP-12 BeadChips (Illumina Inc., San Diego, Calif., USA). Genotypes were called by GenomeStudio software v2010.3 (Illumina Inc.). The cnvPartition plugin (Illumina Inc.) for GenomeStudio was used to detect LCSHs, with the minimum region size set to 5 Mb. All chromosomes of each sample were also visually analyzed for quality control by a cytogeneticist, as well as to eliminate false-positive and false-negative calls.
The inclusion criteria for the subsequent analysis was the presence of 1 or 2 LCSHs with a minimal length of 5 Mb per LCSH and a total length not exceeding 28 Mb without the co-occurrence of pathogenic, likely pathogenic, or unclearly significant copy number variants. The upper limits of LCSH number and length were used to eliminate parental consanguinity, as 28 Mb equals ∼1% of the total autosomal genome, which is below the expected percentage for parents being second cousins [Sund et al., 2013]. Additionally, it is more likely that, in the case of parental consanguinity, there are multiple LCSHs on different chromosomes. We further excluded all LCSHs with lengths >25% of the whole chromosome on which they occurred to eliminate possible UPDs, as well as recurrent LCSHs present in at least 3 unrelated patients.
All the LCSHs included in the final study group were evaluated for clinical significance by searching for encompassed, autosomal recessive disease-associated genes. This was done using the web-based Genomic Oligoarray and SNP array evaluation tool v.2.0 [Wierenga et al., 2013]. The search criterion used was ‘OMIM genes with recessive inheritance pattern’. For every patient, the list of genes and associated phenotypes found by the software were compared with the patient's clinical information based on the referral documents. If a candidate gene matching the patient's phenotype was found, WES was performed to find the causative mutation. All probable pathogenic mutations identified by WES were confirmed by Sanger sequencing.
In a secondary analysis, we included all patients from the study group with LCSHs that did not encompass a good candidate gene and for whom WES was performed previously, although not as a part of this study. In case of a homozygous pathogenic mutation, the CMA data were reanalyzed to look for a <5-Mb LCSH in the region of the mutation.
Results
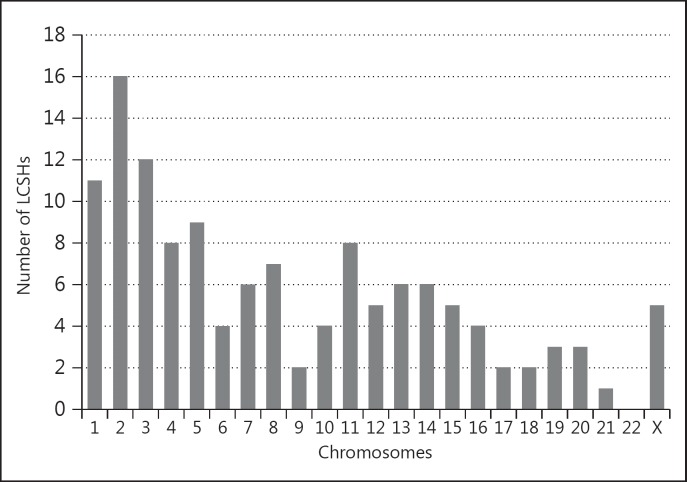
Out of 2,110 patients, 172 patients (8.2%) presented with an isolated finding of 1 or 2 LCSHs when taking into account the aforementioned inclusion and exclusion criteria (other than recurrence). A total of 183 LCSHs were detected, as 11 patients had 2 LCSHs. We identified 6 different recurrent LCSHs [52 (28.4%) in total] (table 1). After excluding all recurrent LCSHs, 120 patients (5.7%) with 129 LCSHs remained in the final study sample. The main characteristics of the detected non-recurrent LCSHs are listed in table 2, and their distribution among chromosomes is presented in figure 1.
Table 1.
Location of 52 recurrent LCSHs found in this study
| Chromosomal bands | Maximal coordinates among the samples | Maximal size, Mb | Minimal overlapping stretch coordinates | Minimal overlapping region, Mb | No. of LCSHs (% of a total of 183) | First report of the recurrent LCSH |
|---|---|---|---|---|---|---|
| 3p21.33p21.1 | chr3:43,805,245 – 54,093,664 | 10.3 | chr3:48,787,219 – 50,483,437 | 1.7 | 17 (9.3%) | Wang et al., 2015 |
| 6p22.3p21.31 | chr6:23,404,279 – 33,887,363 | 10.4 | chr6:26,184,041 – 30,071,279 | 3.9 | 9 (4.9%) | this report |
| 7q11.21q11.22 | chr7:61,760,894 – 68,616,110 | 6.9 | chr7:61,834,214 – 66,904,395 | 5.1 | 5 (2.7%) | this report |
| 7q31.2q31.33 | chr7:116,266,792 – 124,008,463 | 7.7 | chr7:117,783,250 – 122,990,979 | 5.2 | 3 (1.6%) | this report |
| 11p11.2q12.1 | chr11:44,901,903 – 57,160,838 | 12.3 | chr11:46,342,376 – 51,274,692 | 4.9 | 11 (6%) | Wang et al., 2015 |
| Xq13.1q21.1 | chrX:71,595,785 – 83,584,844 | 12.0 | chrX:73,424,191 – 78,848,072 | 5.4 | 7 (3.8%) | this report |
All coordinates are according to the hg19 reference genome assembly.
Table 2.
Main characteristics of non-recurrent LCSHs detected in this study
| Number of patients with non-recurrent LCSH (% of a total of 2,110 patients) | 120 (5.7%) |
| Number of patients with 1 (2) LCSHs | 111 (9) |
| Non-recurrent LCSH count | 129 |
| Size range of single LCSHs | 5 – 28 Mb |
| Size range of the sum of 2 LCSHs in 1 patient | 11.1 – 25.1 Mb |
| Average size of a single LCSH | 7.7 Mb |
| Number of genes causing a recessive disorder in an LCSH (median) | 0 – 15(3) |
| Number of cases with a candidate recessive gene in an LCSH | 2a |
In both cases, the mutation was confirmed.
Fig. 1.
The number of non-recurrent LCSHs and their distribution among different chromosomes (total number = 129). The plot shows that homozygous stretches in our study group were located on all chromosomes except chromosome 22.
Searching for genes associated with recessive disorders revealed a median of 3 genes per LCSH (range 0 to 15). In 2 cases, an appropriate candidate gene was discovered (table 3). First, in a 1-year-old girl with transfusion-dependent hemolytic anemia, an LCSH on chromosome 1 encompassed the PKLR gene, which is known to cause pyruvate kinase deficiency leading to hemolysis and anemia. WES revealed a novel mutation in the PKLR gene that was predicted to cause a deletion of 1 amino acid. Although an enzyme analysis on the affected child was uninformative due to recurrent transfusions, the pathogenicity of the mutation was still confirmed by the detection of decreased pyruvate kinase activity in both heterozygous parents. Second, in a 6-year-old boy with growth failure, developmental delay, ataxia, and cerebellar atrophy, an LCSH on chromosome 5 was found to encompass the SIL1 gene which is associated with Marinesco-Sjögren syndrome. WES revealed a homozygous 1-bp duplication in the SIL1 gene that was predicted to cause a translational frameshift, thereby resulting in a truncated protein. The parents were confirmed to be heterozygous carriers.
Table 3.
Confirmed pathogenic mutations found by whole-exome sequencing
| Patient No. | Phenotype | LCSH | Mutated gene | Affected transcript | Protein change | Disease | First report of the mutation |
|---|---|---|---|---|---|---|---|
| Patients in whom a pathogenic mutation was confirmed inside an LCSH | |||||||
| 1 | transfusion-dependent, non-immune hemolytic anemia | arr[hg19] 1q21.2q23.2(149,698,049 – 159,667,190)×2 hmz | PKLR | NM_000298.5: c.1137_1139del | NM_000298.5: p.(Lys380del) | pyruvate kinase deficiency of red cells (OMIM 266200) | this report |
| 2 | developmental delay, ataxia, cerebellar atrophy | arr[hg19] 5q31.1q31.2(132,669,425 – 139,522,328)×2 hmz | SIL1 | NM_022464.4: c.947dupT | NM_022464.4: p.(Arg317Glufs*35) | Marinesco-Sjögren syndrome (OMIM 248800) | Senderek et al., 2005 |
| Patients in whom a pathogenic mutation was confirmed outside an LCSH | |||||||
| 3 | intellectual disability, macrocephaly | arr[hg19] 1q25.3q32.1(182,559,510 – 200,136,582)×2 hmz, 14q13.3q21.2(37,290,124 – 44,790,527)×2 hmz | – | ||||
| in smaller LCSH: arr[hg19] 19q13.32q13.33(47,035,436 – 48,534,068)×2 hmz | KPTN | NM_007059.2: c.665dupA | NM_007059.2: p.(Ser223Glnfs* 18) | mental retardation, autosomal recessive 41 (OMIM 615637) | Pajusalu et al., 2015 | ||
| 4 | oligohydramnios, congenital renal insufficiency, hypotension, calvarial hypoplasia | arr[hg19] Xp22.1p21.2(23,811,075 – 29,602,016)×2 hmz | – | ||||
| outside LCSH in 1p32.1 | REN | NM_000537.3: c.[341C>T];[255G>C] | NM_000537.3: p.[(Ser114Phe)]; [(Gln85His)] | renal tubular dysgenesis (OMIM 267430) | this report | ||
Of the patients for whom WES had been ordered outside this study, 7 had previously been found to carry an LCSH by CMA analysis that did not encompass a good candidate gene, and 2 of them had a recurrent LCSH. Of these 7 patients, 2 received a definitive diagnosis by WES (table 3). First, WES in an adult man with ID and macrocephaly (reported in detail by us [Pajusalu et al., 2015]) revealed a homozygous 1-bp duplication in the KPTN gene (19q13.32) which is associated with such a phenotype [Baple et al., 2014]. The patient also has a sister with a similar phenotype who carried the same homozygous mutation. A previous CMA analysis found 2 LCSHs in the man (17.6 Mb in 1q25.3q32.1 and 7.5 Mb in 14q13.3q21.2), but only 1 of them (in 14q13.3q21.2) was also present in his sister. However, the KPTN gene is not located in either of these LCSHs. Reanalysis of the CMA data revealed a 1.5-Mb LCSH on chromosome 19 that encompasses the KPTN gene and which is shared by both sibs. This LCSH was not reported after the initial CMA analysis interpretation due to the 5-Mb cut-off length routinely used in our laboratory. The parents of the sibs did not report consanguinity but were born in the same parish. Second, in a girl carrying an LCSH on the X chromosome, compound heterozygous mutations were found in the REN gene (1p32.1), resulting in renal tubular dysgenesis.
Discussion
During the last decade, studies demonstrated the great utility of CMA analysis in clinical diagnostics as a first-tier diagnostic cytogenetic test for patients with ID, autism spectrum disorders and multiple congenital anomalies [Hochstenbach et al., 2009; Miller et al., 2010; Vissers et al., 2010; Ahn et al., 2013]. The primary goal of CMA analysis is to find a causative microdeletion or microduplication. SNP arrays have the advantage of also revealing genotype information, which allows the detection of copy number neutral chromosomal aberrations associated with an increased risk of autosomal recessive disorders. Multiple LCSHs on different chromosomes that indicate close parental consanguinity, as well as LCSHs that cover the majority of single chromosomes and are caused by UPD, have been studied during the last few years, and their clinical implications are, therefore, better understood [Papenhausen et al., 2011; Wang et al., 2015]. Single LCSHs in patients without parental consanguinity are not infrequently found during routine diagnostics in centers where SNP array technology is in use, but their clinical significance remains unclear in most of the cases [Zilina et al., 2014]. Reporting a variant of uncertain significance (VUCS) can cause anxiety in patients and frustration in referring doctors who are not specialists in the field of medical genetics [Coughlin et al., 2012; Zilina et al., 2014]. Therefore, it is of great importance to work toward minimizing the number of reported VUCSs.
One of the most straightforward ways to decrease the number of VUCSs is to identify recurrent LCSHs and to interpret them as benign polymorphisms, which can be left unreported depending on laboratory policies. In this study, we identified recurrent LCSHs in 28.9% of all isolated LCSH findings. Thus, we can clarify the clinical significance of nearly one-third of all LCSHs. Nevertheless, caution is still needed in classifying recurrent LCSHs as benign, because there is the possibility that a shared haplotype could be mutated in both parents. Interestingly, out of 6 chromosomal regions where we identified recurrence of an LCSH, only 2 matched with the 5 regions reported by Wang et al. [2015]. This could be attributed to different study populations, which clearly indicates the need for population-specific LCSH databases.
Although homozygosity of a chromosomal region can be intuitively attributed to an increased risk of autosomal recessive disorders, the clinical significance of single LCSHs remains unclear. In this study, only patients with 1 or 2 LCSHs, with total length not exceeding 28 Mb (i.e. 1% of the autosomal genome), were analyzed. As suspected, because only a very small proportion of the whole genome was covered, a candidate gene well-associated with a patient's phenotype was very rarely identified. Nevertheless, if a well-matched candidate gene is found, the confirmation of a pathogenic mutation is likely, since we found 2 patients with a plausible candidate gene, mutations in which were confirmed in both of them. Wang et al. [2015] reported 9 patients with good candidate genes, and a pathogenic mutation was detected in 7 of them. Although the study sample of Wang et al. [2015] was almost 7 times larger than ours, and patients with parental consanguinity were also included, we can conclude that our results are consistent with theirs due to the very small number of mutation-confirmed cases and the associated probability of a large statistical error. Despite the low success rate in our study, we can still recommend looking through detected, non-recurrent LCSHs for candidate genes, as this can lead to a molecular diagnosis. Additionally, if software tools such as the Genomic Oligoarray and SNP array evaluation tool [Wierenga et al., 2013] are used, the evaluation of LCSH regions is not very time-consuming, as the median number of genes causing a recessive disorder per LCSH was only 3 in our study group.
We also referred to a previously published case, in which a shared LCSH was discovered in an adult man and his sister, and in which subsequent WES led to the discovery of a homozygous pathogenic mutation outside the shared LCSH [Pajusalu et al., 2015]. Nevertheless, reevaluating the CMA data of the sibs revealed a small 1.5-Mb LCSH in the region of the pathogenic mutation. Therefore, if there is a strong indication for recessive inheritance and distant consanguinity or common ancestry is likely, lowering the cut-off of the LCSH size can aid in the identification of candidate genes. For example, the LCSH minimum cut-off length of 1 Mb was used in a study that focused on familial recessive IDs in outbred families [Schuurs-Hoeijmakers et al., 2011]. However, one should not presume that the finding of homozygosity is a general rule, as we also demonstrated that in a girl with a distinct phenotype and an LCSH on the X chromosome, compound heterozygous mutations causing her disease were detected outside the LCSH.
In conclusion, we have reported the results of a study based on a routine clinical diagnostic laboratory's CMA data from a 4-year period. This publication will bring more attention to single LCSHs in patients with no parental consanguinity, as more studies with large sample sizes are needed to broaden the understanding of the diagnostic utility of single LCSHs.
Acknowledgements
We are grateful to all the patients and their families, as well as to the referring doctors. Additionally, we acknowledge the staff in the Estonian Biocenter and Estonian Genome Center at the University of Tartu for the fruitful collaboration. This work was supported by Estonian Research Council grant PUT355.
References
- 1.Ahn JW, Bint S, Bergbaum A, Mann K, Hall RP, Ogilvie CM. Array CGH as a first line diagnostic test in place of karyotyping for postnatal referrals - results from four years' clinical application for over 8,700 patients. Mol Cytogenet. 2013;6:16. doi: 10.1186/1755-8166-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baple EL, Maroofian R, Chioza BA, Izadi M, Cross HE, et al. Mutations in KPTN cause macrocephaly, neurodevelopmental delay, and seizures. Am J Hum Genet. 2014;94:87–94. doi: 10.1016/j.ajhg.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coughlin CR, 2nd, Scharer GH, Shaikh TH. Clinical impact of copy number variation analysis using high-resolution microarray technologies: advantages, limitations and concerns. Genome Med. 2012;4:80. doi: 10.1186/gm381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochstenbach R, van Binsbergen E, Engelen J, Nieuwint A, Polstra A, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009;52:161–169. doi: 10.1016/j.ejmg.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Kearney HM, Kearney JB, Conlin LK. Diagnostic implications of excessive homozygosity detected by SNP-based microarrays: consanguinity, uniparental disomy, and recessive single-gene mutations. Clin Lab Med. 2011;31:595–613. doi: 10.1016/j.cll.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Makrythanasis P, Nelis M, Santoni FA, Guipponi M, Vannier A, et al. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum Mutat. 2014;35:1203–1210. doi: 10.1002/humu.22617. [DOI] [PubMed] [Google Scholar]
- 7.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pajusalu S, Reimand T, Õunap K. Novel homozygous mutation in KPTN gene causing a familial intellectual disability-macrocephaly syndrome. Am J Med Genet A. 2015;167:1913–1915. doi: 10.1002/ajmg.a.37105. [DOI] [PubMed] [Google Scholar]
- 10.Papenhausen P, Schwartz S, Risheg H, Keitges E, Gadi I, et al. UPD detection using homozygosity profiling with a SNP genotyping microarray. Am J Med Genet A. 2011;155A:757–768. doi: 10.1002/ajmg.a.33939. [DOI] [PubMed] [Google Scholar]
- 11.Schuurs-Hoeijmakers JH, Hehir-Kwa JY, Pfundt R, van Bon BW, de Leeuw N, et al. Homozygosity mapping in outbred families with mental retardation. Eur J Hum Genet. 2011;19:597–601. doi: 10.1038/ejhg.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, et al. Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 2005;37:1312–1314. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 13.Sund KL, Zimmerman SL, Thomas C, Mitchell AL, Prada CE, et al. Regions of homozygosity identified by SNP microarray analysis aid in the diagnosis of autosomal recessive disease and incidentally detect parental blood relationships. Genet Med. 2013;15:70–78. doi: 10.1038/gim.2012.94. [DOI] [PubMed] [Google Scholar]
- 14.Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010;47:289–297. doi: 10.1136/jmg.2009.072942. [DOI] [PubMed] [Google Scholar]
- 15.Wang JC, Ross L, Mahon LW, Owen R, Hemmat M, et al. Regions of homozygosity identified by oligonucleotide SNP arrays: evaluating the incidence and clinical utility. Eur J Hum Genet. 2015;23:663–671. doi: 10.1038/ejhg.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wierenga KJ, Jiang Z, Yang AC, Mulvihill JJ, Tsinoremas NF. A clinical evaluation tool for SNP arrays, especially for autosomal recessive conditions in offspring of consanguineous parents. Genet Med. 2013;15:354–360. doi: 10.1038/gim.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilina O, Teek R, Tammur P, Kuuse K, Yakoreva M, et al. Chromosomal microarray analysis as a first-tier clinical diagnostic test: Estonian experience. Mol Genet Genomic Med. 2014;2:166–175. doi: 10.1002/mgg3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]