Abstract
Complex chromosome rearrangements (CCRs) are currently defined as structural genome variations that involve more than 2 chromosome breaks and result in exchanges of chromosomal segments. They are thought to be extremely rare, but their detection rate is rising because of improvements in molecular cytogenetic technology. Their population frequency is also underestimated, since many CCRs may not elicit a phenotypic effect. CCRs may be the result of fork stalling and template switching, microhomology-mediated break-induced repair, breakage-fusion-bridge cycles, or chromothripsis. Patients with chromosomal instability syndromes show elevated rates of CCRs due to impaired DNA double-strand break responses during meiosis. Therefore, the putative functions of the proteins encoded by ATM, BLM, WRN, ATR, MRE11, NBS1, and RAD51 in preventing CCRs are discussed. CCRs may exert a pathogenic effect by either (1) gene dosage-dependent mechanisms, e.g. haploinsufficiency, (2) mechanisms based on disruption of the genomic architecture, such that genes, parts of genes or regulatory elements are truncated, fused or relocated and thus their interactions disturbed - these mechanisms will predominantly affect gene expression - or (3) mixed mutation mechanisms in which a CCR on one chromosome is combined with a different type of mutation on the other chromosome. Such inferred mechanisms of pathogenicity need corroboration by mRNA sequencing. Also, future studies with in vitro models, such as inducible pluripotent stem cells from patients with CCRs, and transgenic model organisms should substantiate current inferences regarding putative pathogenic effects of CCRs. The ramifications of the growing body of information on CCRs for clinical and experimental genetics and future treatment modalities are briefly illustrated with 2 cases, one of which suggests KDM4C (JMJD2C) as a novel candidate gene for mental retardation.
Key Words: Complex chromosome rearrangements, DNA double-strand break, Haploinsufficiency, Mixed mutation mechanisms, Structural genome variations, Triplosufficiency
Currently, structural genome variations (SVs) are operationally defined as alterations in the organization of genomic elements involving at least 50 bp [Alkan et al., 2011]. The rate of occurrence of SVs is inversely proportional to their size, the number of breaks and the number of genes they involve. Thus, smaller SVs, those that involve only 1 or 2 breaks and those that do not affect any gene, occur more frequently than larger ones, those that involve more than 2 breaks or those that affect at least one gene. This indicates that the phenotypic impact of an SV may provoke purifying selection, which will limit its frequency and persistence in the general population. For instance, CNVs larger than 100 kb, which by definition involve 2 breaks, arise de novo in the general population at an estimated rate of ∼1.2 × 10−2 CNVs per meiosis [Itsara et al., 2010]. On the other hand, in ∼14-18% of children with developmental delay, a CNV larger than 400 kb may be phenotypically significant [Cooper et al., 2011; Hochstenbach et al., 2011]. Taken together, all classes of germline SVs occur more frequently than germline single nucleotide variations (SNVs), they affect more nucleotides and may have a greater phenotypic impact than SNVs [Stankiewicz and Lupksi, 2010; Campbell and Eichler, 2013].
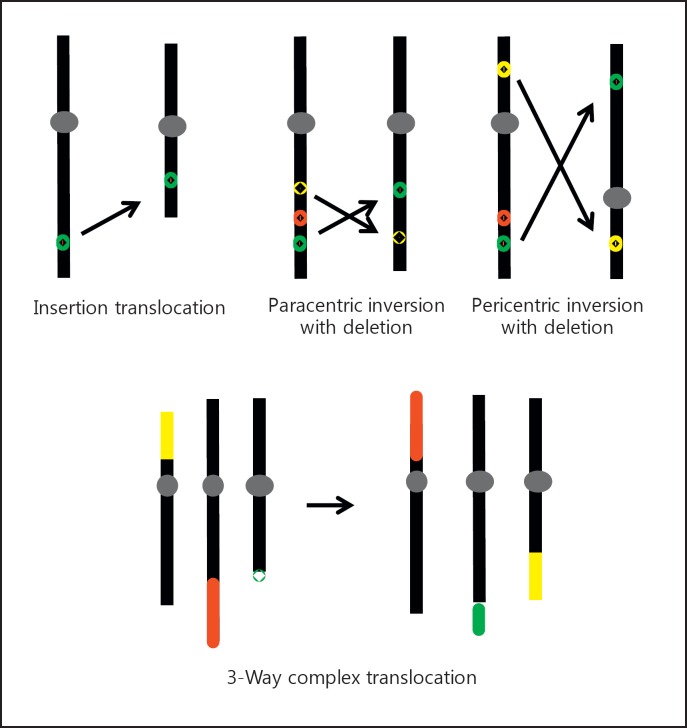
Complex chromosome rearrangements (CCRs) are a class of SVs that involve more than 2 chromosome breaks and result in exchanges of chromosomal segments. CCRs may be insertion-translocations, inversions associated with CNVs, translocations affecting more than 2 chromosomes, and combinations thereof (see fig. 1 for the major types of CCRs). Classically, CCRs were extremely rare events detected by karyotyping [Madan et al., 1997; Park et al., 2001]. The implementation of genome-wide assays for segmental aneuploidy, such as BAC, oligonucleotide and SNP arrays, flow karyotyping, and next-generation sequencing techniques, has revealed an increasing complexity of CCRs [Gribble et al., 2005; Fauth et al., 2006; De Gregori et al., 2007; Baptista et al., 2008; Higgins et al., 2008; Sismani et al., 2008; Schluth-Bolard et al., 2009; Gijsbers et al., 2010; Kang et al., 2010; Feenstra et al., 2011; Kloosterman et al., 2011; 2012; Nazaryan et al., 2014; Tabet et al., 2015]. Not only the detection rate of CCRs increased, they were also more often associated with CNVs than reciprocal translocations were [Feenstra et al., 2011]. An example of advancing insights into a CCR by progressively improving resolution is a case, which initially was reported as an ‘interstitial loss 6q14 contained within a de novo pericentric inversion 6(p11.2q15)’, but eventually became a 10-breakpoint CCR [Passarge, 2000; Poot et al., 2009; Kloosterman et al., 2012]. On the other hand, if genome aneuploidy screening is limited to arrays only, we may miss some of the CCRs or may misinterpret their nature and complexity [Hochstenbach et al., 2009; Brand et al., 2014]. Since the genome analysis method(s) used will affect the detection rate, estimates of population frequencies of CCRs are inherently subject to a technology-dependent ascertainment bias.
Fig. 1.
Schematic depictions of examples of CCRs including an insertion-translocation, 2 types of complex inversions, and a 3-way translocation. Top: chromosomes are in black, and centromeres are represented by gray ovals. The arrows indicate the direction of transposition of chromosomal loci and segments. Small colored symbols represent loci affected by the CCR. Red circles represent lost loci. Bottom: colored bars represent segments that are exchanged by the 3-way translocation.
CCRs have been classified according to their structure, the number of chromosome breaks detected, whether a single or several chromosomes were involved, and the mode of inheritance, i.e. de novo versus transmitted CCRs [Pai et al., 1980; Kausch et al., 1988; Kousseff et al., 1993; Batista et al., 1994; Park et al., 2001; Pellestor et al., 2011]. Heterozygous carriers of CCRs have an elevated risk for spontaneous abortion or chromosomally abnormal offspring. Empirical risk estimates range from 50 to 100% for spontaneous abortion [Batista et al., 1994] and from 20 to 90% for phenotypic abnormalities [Gorski et al., 1988]. However, lack of a detailed understanding of the origin and nature of a CCR renders medical genetic and reproductive counseling difficult. An additional compounding factor is that male carriers of CCRs are often subfertile or sterile due to arrest of spermatogenesis.
An appreciable number of CCRs were found among healthy individuals [Pellestor et al., 2011; de Pagter et al., 2015]. Thus, some CCRs may be nonpathogenic. Therefore, it is essential to obtain information allowing us to identify potentially pathogenic characteristics of CCRs. The prime pathogenic mechanism of a CCR is disruption of the genomic architecture either within a gene or between a gene and its regulatory elements [Yue et al., 2006; Klopocki and Mundlos, 2011]. If a CNV is associated with a CCR, haploinsufficiency or triplosufficiency of one or several genes may become an additional pathogenic mechanism [Yue et al., 2005; Poot et al., 2011a]. It is therefore cardinal to narrow down all breakpoint regions as much as possible, ideally to the single nucleotide level, and to determine whether CNVs are associated with a given CCR [Kloosterman et al., 2011, 2012]. Therefore, this review is limited to those CCRs that have been analyzed by molecular methods in sufficient detail regarding the sequences at their breakpoint regions. This information is crucial to address core questions in genetic counseling of carriers of CCRs: first, to infer the possible mechanism of origin and consequently recurrence risk and second, to determine whether disruption of gene-gene and gene-regulatory element architecture may be a possible pathogenic mechanism.
Definition and Classification of CCRs
CCRs have often been considered ‘curiosities of nature’, which, due to the limited resolution of classical karyotyping, were difficult to detect [Madan et al., 1997; Park et al., 2001]. In their pioneering study, Pai et al. [1980] referred to CCRs as ‘translocations that are more complex than the well described reciprocal translocations’. For decades, the definition and classification of CCRs were based upon the number of chromosome breaks and the visible structure of the rearrangements. Both depend upon the level of resolution of the detection method. Advances in molecular cytogenetic technology have dramatically improved both the detection rates and the resolution of CCRs. Consequently, the criteria for both definition and classification of CCRs have to be reconsidered.
Initially, CCRs were classified according to the outcome after meiosis [Kausch et al., 1988]. These authors distinguished 3 groups. First, 2 or more independent translocations leading to 2 or more meiotic quadrivalents of which each can segregate in a balanced or an unbalanced way. These CCRs are generally familial and can be transmitted from one generation to the next [Meer et al., 1981; Farrell et al., 1994; Zahed et al., 1998]. Second, ‘true’ CCRs involving ‘n’ chromosomes with ‘n’ breakpoints leading to the formation of a meiotic multivalent (2n). This leads to 2 possibilities of balanced segregation of chromosomes, as compared to several distinct unbalanced segregation products depending upon the number of chromosomes involved, and third, CCRs involving more breakpoints than chromosomes [Bass et al., 1985; Batanian and Eswara, 1998]. In such cases, additional insertions or inversions have taken place, producing even more complicated meiotic configurations leading to further possible unbalanced segregation outcomes. These rearrangements can be highly complex with as many as 7 derivative chromosomes [Kousseff et al., 1987] and up to 15 breakpoints [Tupler et al., 1992; Houge et al., 2003]. Most of these CCRs occur de novo. While this classification accommodates reciprocal, double reciprocal and more complex translocations, it does not include ‘simple’ inversions or insertion-translocations. The latter 2 types of rearrangements can only be detected unequivocally by molecular cytogenetic techniques.
Alternatively, CCRs have been classified according to the number of chromosome breaks involved [Kousseff et al., 1993]. These authors divided CCRs in 2 broad groups: one group ≤4 breaks and the other group >4 breaks. They observed that most familial CCRs belonged to the group with up to 4 breaks and have been transmitted by a balanced female carrier, whereas most of the de novo CCRs belonged to the group with more than 4 breaks and were of paternal origin [Kousseff et al., 1993; Batista et al., 1994; Joyce et al., 1999; Lee et al., 2002; Grasshoff et al., 2003; Kuechler et al., 2005]. The majority of the reported cases had 4 or less breaks [Pellestor et al., 2011]. This may either reflect reduced viability or reproductive fitness of individuals with CCRs made up by more than 4 breaks or an ascertainment bias due to technical limitations.
A third classification was based upon both the location and the distribution of breakpoints, thus distinguishing CCRs with intrachromosomal rearrangements (i.e. insertions, inversions, deletions, or duplications) from those involving several chromosomes at once [Lurie et al., 1994].
Meanwhile, several authors described complex inversions and insertion-translocations, which, depending on the definition applied, may or may not have been labeled CCRs [Madan and Menko, 1992; Bernardini et al., 2008; Jiang et al., 2008; Lybaek et al., 2008; de Vree et al., 2009; Poot et al., 2009; Kang et al., 2010]. A special case of a CCR involving a single chromosome is the inversion-duplication-distal deletion rearrangement [Hulick et al., 2009; van Binsbergen et al., 2012; Kowalczyk et al., 2013; Trachoo et al., 2013]. These studies indicated that rearrangements involving only a single chromosome can also be ‘complex’. Such ‘borderline CCRs’ prompted a reassessment of the definition of a CCR. Due to improved resolution of molecular cytogenetic techniques, more and more ‘simple’ rearrangements, such as inversions and reciprocal translocations, prove to be unexpectedly ‘complex’. Therefore, for the time being, the pragmatic approach of Madan [2013] may be the most appropriate. She advocated describing all possible CCRs as ‘a CCR involving that and that chromosome, being familial or de novo, apparently balanced or unbalanced as appropriate’. With this purely descriptive approach, all possible CCRs should prompt detailed molecular cytogenetic analyses, which may provide the information needed for proper genetic counseling of the concerned patients and their families.
Mechanisms of Origin of CCRs
During meiosis, 2 consecutive cell divisions, which involve each one round of chromosome segregation, without intervening DNA replication, produce haploid germ cells. During the first cell division, homologous chromosomes pair and form Holliday junctions, which are resolved by multiple recombination events between the chromosomes. Thereupon, the chromosomes are separated and pulled to opposite poles. After this division, each cell contains a haploid number of chromosomes. In the second equational division, the sister chromatids segregate from one another, such that haploid nuclei with chromosomes consisting of single chromatids result. Due to recombination events during the first meiotic division, some genes will be exchanged between the 2 parental chromosomes. These meiotic recombination events are initiated by Spo11-catalyzed DNA double-strand breaks (DSBs), which prompt subsequent recombinational repair for cells to remain viable [Richardson et al., 1998; Romanienko and Camerini-Otero, 2000; Keeney, 2008; Andersen and Sekelsky, 2010; Wahls and Davidson, 2010; Phadnis et al., 2011]. The finding that SPO11 is highly expressed in testes and ovaries and that spermatocytes and oocytes of Spo11−/− mice undergo cell cycle arrest, and elevated levels of apoptosis underscores that Spo11-catalyzed DSBs are the initial step required for meiosis [Romanienko and Camerini-Otero, 2000; Scott and Pandita, 2006].
Genes and Proteins
Although the biochemistry of DSB repair during meiosis is not completely understood, a number of human genetic disorders of defective DSB response may provide clues regarding the genes involved and their modes of action [O'Driscoll and Jeggo, 2006; Scott and Pandita, 2006]. Those are ataxia telangiectasia (OMIM 208900; gene: ATM), ataxia-telangiectasia-like disorders 1 and 2 (OMIM 604391 and 615919; genes: MRE11 and PCNA), Bloom syndrome (OMIM 210900; gene BLM), Nijmegen breakage syndrome - NBS - (OMIM 251260; gene: NBS1), Seckel syndrome (OMIM 210600; gene: ATR), Fanconi anemia/BRCA2 (OMIM 605724; gene: FANCD1/BRCA2), LIG4 syndrome (OMIM 606593; gene: LIG4), and Werner syndrome (OMIM 277700; gene WRN) [O'Driscoll and Jeggo, 2006; Scott and Pandita, 2006].
Some of these syndromes exhibit spontaneous chromosomal instability (ATR, BLM, FANCD1/BRCA2, NBS1, WRN) or elevated sensitivity to ionizing radiation (ATM, LIG4) and phenotypes such as hypogonadism and impaired spermatogenesis (ATM), primary ovarian failure (NBS1), hypospadias and cryptorchidism (ATR). Also male patients with Bloom syndrome (BLM) show azoospermia and cryptorchidism, while females present with an overall reduced fertility. Male patients with Werner syndrome (WRN) present with complete azoospermia, while females show reduced fertility [Epstein et al., 1966].
Although the functions in meiosis of each of the genes involved in these syndromes is not yet fully understood, a picture begins to emerge. The ATM-encoded phosphatidylinositol 3-kinase cooperates with TEL1 to limit the number of DSB to one per pair of sister chromatid and one per quartet of chromatids [Zhang et al., 2011]. Atm−/− mice are infertile because meiosis is arrested at the zygotene/pachytene stage of prophase I as a result of abnormal chromosomal synapsis and subsequent chromosome fragmentation [Xu et al., 1996]. In Atm−/− mice ATR, a protein related to ATM, DMC1, a RAD51 family member, and RAD51 show reduced localization to developing synaptonemal complexes in spermatocytes [Barlow et al., 1998]. ATM appears to act as a monitor of the prophase I meiosis and also to control DSB formation [Barlow et al., 1998; Lange et al., 2011]. Cooperation of ATM with SPO11 is required for the obligate XY crossover and also appears to control autosomal crossovers and chromosome integrity [Barchi et al., 2008]. In Atm−/− Spo11+/− mice, ATR rescues spermatogenesis by phosphorylating H2AX in response to DNA DSBs, while folliculogenesis remains partially defective [Bellani et al., 2005; Di Giacomo et al., 2005]. Also DSBs harboring oocytes may progress through meiosis in the presence of an active ATM-dependent DSB control pathway, since they appear to lack a G2-phase type of DNA damage control mechanism [Marangos and Carroll, 2012].
Although the WRN and the BLM helicases belong both to the family of RecQ helicases, defects of either during DSB response may lead to opposing outcomes [Suhasini and Brosh, 2013; Croteau et al., 2014; Keijzers et al., 2014; Kitano, 2014]. Loss of WRN is associated with a variegated translocation mosaicism and a reduction in DNA recombination [Salk et al., 1981; Dhillon et al., 2007; Melcher et al., 2000]. Loss of BLM, in contrast, provokes elevated rates of sister chromatid exchanges during meiosis I [Bischof et al., 2001]. The WRN-encoded protein activates the ATR-CHK1-induced S-phase checkpoint in response to topoisomerase-I-DNA covalent complexes, recruits and stabilizes Rad51 and limits the activity of the MRE11-encoded exonuclease [Patro et al., 2011; Su et al., 2014]. In somatic cells, absence of the WRN-encoded 3′-5′ helicase-3′-5′ exonuclease causes a prolongation of the S phase of the cell cycle, hypersensitivity to DNA cross-linking and DSBs provoking agents, elevated frequencies of micronuclei and a reduction in recombinational DSB repair [Poot et al., 1992, 1999, 2001, 2002, 2004; Ogburn et al., 1997; Honma et al., 2002; Kamath-Loeb et al., 2004; Dhillon et al., 2007]. The extent of DSB ligation is normal in WRN deficient cells, albeit that more deletions are formed at the fusion sites [Rünger et al., 1994]. Cultured fibroblasts from patients with Werner syndrome show a typical and diagnostic variegated chromosomal translocation mosaicism and spontaneous deletion formation [Salk et al., 1981; Fukuchi et al., 1989; Oshima et al., 2002].
The BLM-encoded 3′-5′ helicase prevents elevated formation of sister chromatid exchanges and quadriradial exchanges between homologous chromosomes during the S phase of the cell cycle [Bartram et al., 1976; Bischof et al., 2001]. Thus, BLM prevents chromosomal recombination and CCR formation [Bartram et al., 1976; LaRocque et al., 2011]. BLM recruits Rad51 and, in cooperation with DNA topoisomerase III, dissolves displacement loops that form during DNA recombination, thus ensuring that recombination intermediates are completely resolved during meiosis [Wu and Hickson, 2003; Fasching et al., 2015; Kaur et al., 2015; Tang et al., 2015]. As part of a complex with either WRN or BLM, Rad51 searches the entire genome to locate a region of homology that can be fused to the broken chromosome. These regions of homology can be located on a sister chromatid, on a homologous chromosome, or on an unrelated chromosome. Rad51 then opens the intact double-stranded DNA template to allow strand invasion and formation of a 3-stranded displacement loop in which the single-stranded broken-end base pairs with its complementary strand of the intact duplex. At this point, the cell can follow one of several alternative pathways. The cell can proceed through a synthesis-dependent strand annealing pathway that copies the template to seal the break without an accompanying crossover. Alternatively, the cell can repair the DSB by formation of a branched intermediate, i.e. a double Holliday junction, which has to be resolved subsequently by producing crossovers between the homologs [Haber, 2015]. In somatic cells, such crossovers may lead to the loss of heterozygosity. During meiosis, such crossovers generate tension between paired homologs, which assures the proper disjunction of chromosomes during meiosis I. Failure to do so, results in elevated rates of sister chromatid exchanges, the hallmark of Bloom syndrome [Bartram et al., 1976].
Thus, the proteins encoded by the genes mutated in BLM and WRN perform critical, albeit distinct, functions required for proper chromatid disjunction [Shen and Loeb, 2000; Mohaghegh et al., 2001; Compton et al., 2008; Monnat, 2010; Rossi et al., 2010; Kamath-Loeb et al., 2012]. It is, however, unlikely that these well-recognizable and extremely rare autosomal recessive syndromes with reduced fertility may account for a significant number of cases with CCRs. On the other hand, WRN heterozygotes occur at a rate of 1:200 in the general population. Individuals who are heterozygous for WRN mutations, e.g. the parents of WRN patients, show normal fertility but an elevated sensitivity to DNA damage induced by DNA cross-linking agents [Ogburn et al., 1997; Poot et al., 1999]. This indicates that hemizygosity for WRN may be pathogenic through haploinsufficiency. A likely molecular mechanism underlying haploinsufficiency is the formation of protein complexes with altered stoichiometries of the contributing partners [Poot et al., 2011a]. In the case of WRN, these may be Rad51, topoisomerase I and MRE11, while for BLM these would be Rad51, topoisomerase III, RPA, Rad54 and Rmi1 [Su et al., 2014; Fasching et al., 2015; Kaur et al., 2015; Tang et al., 2015]. Thus, perturbation of the delicate balance between pro-recombination activity of the WRN-pathway versus anti-recombination activity of the BLM pathway may either produce variegated translocation mosaicism (WRN) or elevated rates of sister chromatid exchanges (BLM). In addition, imbalanced functioning or complete inactivity of ATM, ATR, MRE11, NBS1, PCNA, RAD51, etc. during meiosis may conceivably provoke the formation of CCRs.
In addition, the PCNA protein appears to facilitate resolution of structures resembling double Holliday junctions that were postulated as intermediates of double-strand break repair during meiosis [Giannattasio et al., 2014]. Loss of both alleles of the PCNA gene caused a defect in nucleotide excision repair, resulting in developmental delay, ataxia, sensorineural hearing loss, short stature, cutaneous and ocular telangiectasia, and photosensitivity [Baple et al., 2014]. Patients with mutations in LIG4 show a clinical syndrome closely resembling NBS. In contrast to NBS cells, those from patients with LIG4 syndrome showed normal cell cycle checkpoint responses but impaired DSB rejoining [O'Driscoll et al., 2001]. Patients with mutations in XRCC4, the obligate partners of LIG4 in the nonhomologous end joining (NHEJ) pathway of DSB repair, show extreme global growth failure in utero, microcephalic primordial dwarfism and severe combined immunodeficiency [Murray et al., 2015]. In a single family, a male and female adult patient with short stature, early-onset metabolic syndrome and gonadal failure were found to carry XRCC4 mutations [de Bruin et al., 2015]. By single-molecule fluorescence-resonance-energy-transfer analyses of the Ku/XRCC4/XLF/DNA ligase IV complex involved in NHEJ ligation, filament-forming proteins were found to bridge DSBs in vivo [Reid et al., 2015]. These filaments at either end of the DSB appeared to interact dynamically to achieve the optimal configuration and end-to-end positioning and ligation. Conceivably, a subtle defect in this process may lead to illegitimate repair of the DSB and subsequently to formation of a CCR. Overexpression of replication protein A1 (RPA1), for instance because of a recurrent duplication 17p13.3, provokes an abnormal S-phase transit, attenuated DSB-induced RAD51 retention, elevated rates of fused and triradial chromosomes, hydroxyurea-induced micronuclei, and increased sensitivity to camptothecin [Outwin et al., 2011]. These findings add to the growing genetic and phenotypic complexity of the biological pathways possibly involved in DSB response and CCR prevention.
Molecular Mechanisms: Fork Stalling and Template Switching, Microhomology-Mediated Break-Induced Repair and the Breakage-Fusion-Bridge Cycle
The above evidence suggests the involvement of these genes and proteins in assuring proper chromatid transmission during meiosis. Yet, no biological pathways leading to the formation of CCRs emerge. Based on studies of CNV formation, several mechanisms of CCR formation have been proposed [Hastings et al., 2009; Colnaghi et al., 2011; Liu et al., 2012]. These include the fork stalling and template switching mechanism (FoSTeS), microhomology-mediated break-induced repair (MMBIR) and the breakage-fusion-bridge cycle. The latter has been proposed after observations during classical karyotyping and involves NHEJ of the chromosomal pieces [Van Dyke et al., 1977; Colnaghi et al., 2011]. Finally, chromothripsis, i.e. shattering of chromosomes followed by randomly ‘stitching together’ of the pieces, has been proposed [Stephens et al., 2011]. Although it is impossible to observe these processes ‘in the act’ during meiosis, they leave specific patterns of sequence changes at the ‘joining points’, which may serve as ‘molecular signatures’ allowing us to infer the nature of the process that gave rise to the CCR in question.
In FoSTeS and MMBIR, active DNA replication forks are assumed. It is conceivable that similar processes may also occur during meiosis when Holliday junctions are being resolved [Hastings et al., 2009; Colnaghi et al., 2011]. Initially, FoSTeS assumed that if during DNA-lagging strand synthesis a DNA polymerase encountered a block, it would switch to a nearby active replication fork and continue nascent DNA synthesis [Hastings et al., 2009]. As a result, 2 simultaneously active replicons are fused in the nascent stretch of DNA. If this process would occur multiple times during attempts to resolve Holliday junctions, multiple crossovers, each involving a short stretch of DNA, would result. The eventual outcome of such a process would be a CCR. The molecular signature of such a FoSTeS-mediated CCR would be a set of closely interspersed joining points connecting multiple chromosomes or a set of interspersed duplications and triplications within a single chromosome, termed ‘chromoanasynthesis’ [Colnaghi et al., 2011; Hart and O'Driscoll, 2013; Plaisancié et al., 2014].
Alternatively, the DSB generated during meiosis may be subjected to MMBIR [Hastings et al., 2009; Colnaghi et al., 2011]. This model assumes that after exposure to high levels of endogenously generated DNA damage, e.g. nucleotide oxidation or DNA interstrand cross-linking, the DNA replication fork collapses. Thereupon, RAD51-mediated invasion of the 3′ end of the DSB into the double-stranded DNA template and 3′-5′ resection of one of the strands occurs. The WRN-encoded 3′-5′ helicase-3′-5′ exonuclease and the BLM-encoded 3′-5′ helicase may, together with the MRE11-encoded exonuclease, replication protein A and NBS1-encoded protein, be involved in such a process [Truong et al., 2013; Su et al., 2014]. On the one hand, the joining of 2 open DNA strands is based upon the microhomology between the 2 participating stretches of DNA. On the other hand, small deletions will be generated at the joining points during the exonuclease-mediated resection step [Colnaghi et al., 2011; Hart and O'Driscoll, 2013]. The molecular signature of MMBIR will be the concomitant presence of microhomologies and small deletions at the joining points of the thus generated CCRs.
Recently, several cases of ‘borderline CCRs’ consisting of duplications and triplications associated with segments of copy-number-neutral loss of heterozygosity have been reported [Carvalho et al., 2015]. Detailed analyses suggest that these have resulted from a postzygotic MMBIR-like process involving template switches between sister chromatids [Carvalho et al., 2015].
The third proposed mechanism for CCR-formation is the breakage-fusion-bridge cycle prompting NHEJ [Colnaghi et al., 2011; Hart and O'Driscoll, 2013]. DNA NHEJ is the predominant pathway of DSB repair, which involves heterodimers of Ku70 and Ku80, and DNA-dependent protein kinases, DNA-PKcs. Heterodimers of Ku bind to the DSBs, which in turn recruit DNA-PKcs. The activated DNA-PKcs complex subsequently recruits artemis, XRCC4, LIG4, and DNA polymerase μ. The nuclease activity of artemis prepares the DNA ends for ligation and gap filling by LIG4 and DNA polymerase μ. NHEJ functions throughout the mitotic cell cycle and is the predominant DSB-rejoining mechanism in the G1 phase of the cell cycle [Scott and Pandita, 2006]. The molecular signature of NHEJ will be the formation of small deletions at the joining points in the absence of microhomologies.
Chromothripsis as a Possible Cause of CCRs
Recently, a fourth mechanism of CCR formation has been proposed: chromosome shattering, also known as chromothripsis, followed by random stitching together of the resulting chromosomal segments. Chromothripsis occurs frequently in many tumor types, but it has also been invoked to explain a number of CCRs in the germline [Kloosterman et al., 2011, 2012; Stephens et al., 2011; Nazaryan et al., 2014]. Chromothripsis results from multiple DSBs arising simultaneously in close proximity to each other, often on several chromosomes at once. Also a few likely cases of chromothripsis involving only a single chromosome have been described [Lybaek et al., 2008; Poot et al., 2009; Kloosterman et al., 2012]. The initiating event may be uncontrolled overactivity of SPO11, but also environmental insults, such as free radicals or ionizing radiation, leading to an overwhelming number of DSBs have been suggested [Lieber, 2010]. Furthermore, covalent DNA modification or other damages interfering with proper resolution of Holliday junctions or defective cell cycle checkpoint function have been invoked [Maher and Wilson, 2012; Pellestor et al., 2014].
Interestingly, CCRs resulting from chromothripsis involve either one or a few, but never all chromosomes [Maher and Wilson, 2012]. A CCR involving exchanges between multiple chromosomes may arise if several Holliday junctions are in close proximity to each other at the same point in time [Hansen et al., 2010]. This may be the case during the pachytene stage of meiosis I. In contrast, a CCR affects only a single chromosome, when it is either isolated, e.g. in a micronucleus, or in a highly condensed state, probably during early meiosis I [Crasta et al., 2012; Maher and Wilson, 2012]. Thus, the number of chromosomes being affected by a CCR may indicate at what precise stage of meiosis I chromothripsis may have taken place.
Chromothripsis was originally discovered in dividing somatic cells of tumors [Stephens et al., 2011]. In contrast to this form of chromothripsis, the kind of chromothripsis giving rise to CCRs should be termed ‘constitutional’ or ‘germline chromothripsis’, as it arises during meiosis and harbors a set of highly specific characteristics [Ballarati et al., 2009; Poot et al., 2009; Gijsbers et al., 2010; Kloosterman et al., 2011, 2012; Chiang et al., 2012; Korbel and Campbell, 2013; Nazaryan et al., 2014]. Based on a series of cases with CCRs, chromothripsis was inferred for those, which had at least one nonreciprocal exchange in their karyotype and, upon genome-wide CNV analyses, showed association of either diploid or haploid segments with the rearrangement sites. In a case with an insertion-translocation, the inserted fragment occurred in the triploid state, as was predicted from the karyotype. In general, the CNVs in cases with chromothripsis are only losses and not duplications. Since the diploid segments of the genome had retained heterozygosity for SNP markers, chromothripsis most likely took place immediately before or during meiosis I.
By paired-end and mate-pair sequencing the joining points of the rearrangements were investigated at nucleotide resolution [Kloosterman et al., 2011, 2012]. At these joining points, losses of a few base pairs up to several kb, and small insertions of non-templated sequences were found. In one case of a CCR caused by chromothripsis, 11 out of 12 joining points contained regions of microhomology, suggesting involvement of BLM and its partners [Truong et al., 2013; Nazaryan et al., 2014]. In another case, little or no microhomology was found, while in a series of cases, 45% of junctions displayed microhomologies, 29% no microhomology, and 26% had short insertions of 1-97 non-templated nucleotides [Kloosterman et al., 2011, 2012]. This indicates that after chromothripsis the chromosomal segments were either fused by NHEJ or by MMBIR [Liang et al., 1998; Holloway et al., 2010; Truong et al., 2013]. Remarkably, the segments appeared to be randomly joined in head-to-tail, tail-to-tail and head-to-head orientations [Kloosterman et al., 2012]. This means that the joining process operates in both the forward and the reverse direction, which differentiates it from FoSTeS [Hastings et al., 2009; Liu et al., 2012]. Another differentiating hallmark of chromothripsis is the close clustering of breaks, analogous to the clustering of mutations (kataegis), resulting from the activity of editing cytosine deaminases of the APOBEC superfamily [Kloosterman et al., 2012; Lada et al., 2015], which can only be fully ascertained by paired-end and mate-pair sequencing. Chromothripsis-based rearrangements affect only a single haplotype, which suggests that it occurred just before or during meiosis I and indicates a possible involvement of BLM and its partners [Bischof et al., 2001; Holloway et al., 2010].
Chromothripsis took place in the paternal germline in all 4 of the cases that could thus be evaluated [Kloosterman and Hochstenbach, 2014]. As a causative mechanism for this sex bias, the higher number of cell divisions of sperm cells versus oocytes has been proposed [Gribble et al., 2005; Hehir-Kwa et al., 2011; Kloosterman et al., 2012]. If this were true, the risk for chromothripsis increases with the age of the father, as has been found for SNVs and CNVs [Hehir-Kwa et al., 2011; O'Roak et al., 2012; Buizer-Voskamp et al., 2013]. Alternatively, an increased burden of free radical-mediated DNA damage or less active free radical defense or repair mechanisms may be responsible for the paternal bias for chromothripsis [Kloosterman et al., 2012; Pellestor et al., 2014]. No definitive evidence supporting this hypothesis has yet been published. A single study of the antioxidant defense of round spermatids of the rat is in agreement with this hypothesis, however [Den Boer et al., 1990]. In addition, higher levels of abortive apoptosis, telomere erosion, mitotic errors, micronuclei formation, and p53 inactivation in the male versus female germline have been proposed [Pellestor et al., 2014]. Interestingly, all these mechanisms have been identified as potential causes for reproductive failure and chromosomal abnormalities, thus linking chromothripsis to the human germline once again [Pellestor et al., 2014].
Recently, a novel mechanism of chromothripsis was proposed [Zhang et al., 2015]. In cultured retinal pigment epithelial cells, one or several missegregated chromosomes may become separated into a micronucleus. During the subsequent S phase of the cell cycle, the chromosome(s) in the micronucleus undergo underreplication, and the resulting chromosomal segments are fused by a microhomology-dependent mechanism, akin to MMBIR. The resulting cell nucleus then contains one or several chromosomes that have undergone chromothripsis. If the fusion process involves several chromosomes at once, a CCR may emerge. This mechanism, which requires one round of DNA replication, is in agreement with several of the described cases of chromothripsis [Kloosterman et al., 2011, 2012; Nazaryan et al., 2014]. Most micronuclei accumulate DSBs, which are associated with the phosphorylated histone H2AX [Crasta et al., 2012] and the RAD51 protein [Haaf et al., 1999]. The latter is mobilized by the BLM helicase to sites of DSBs [Wu and Hickson, 2003; Fasching et al., 2015; Kaur et al., 2015; Tang et al., 2015]. If the micronucleus does not contain sufficient BLM-encoded helicase, the balance between WRN- and BLM-dependent activities may become tilted towards the hypo-recombinogenic WRN pathway, and a variegated translocation in association with deletions may result [Salk et al., 1981; Fukuchi et al., 1989; Dhillon et al., 2007; Melcher et al., 2000]. Thus, these observations provide a novel molecular mechanism linking chromothripsis to CCRs.
The molecular details of processes such as FoSTeS, MMBIR, breakage-fusion-bridge cycle and chromothripsis in the germline have not yet been fully elucidated [Hastings et al., 2009; Liu et al., 2012; Korbel and Campbell, 2013]. In particular, a transgenic model for germline chromothripsis has not yet been described. For clinical purposes, such as to infer the mechanism of origin and recurrence risk of the CCR, rigorous criteria to distinguish FoSTeS, MMBIR, breakage-fusion-bridge cycle and chromothripsis are a prerequisite. Nevertheless, operational definitions based on molecular signatures can be obtained by paired-end or mate-pair analyses at the nucleotide level. In table 1, the molecular mechanisms of CCR formation, the potentially involved genes, and the molecular signatures of joining points are summarized.
Table 1.
Mechanisms of CCR formation, involved genes and molecular signatures of joining points
| FoSTeS | MMBIR | Breakage-fusion-bridge cycle | Chromothripsis | |
|---|---|---|---|---|
| Causes | stalled replication forks | DSBs | ? | overactivity of SPO11, free radicals or ionizing radiation, covalent DNA damage interfering with resolution of Holliday junctions or defective cell cycle checkpoints |
| Basic mechanism | strand invasion by lagging DNA replication strand | joining of 2 open DNA strands | ? | NHEJ and MMBIR |
| Enzymatic activities | DNA replication machinery | exonuclease-mediated resection | artemis-nuclease, gap-filling DNA polymerase, ligase 4 | not studied |
| Associated CNVs | deletions, duplications, triplications | none | none | only losses, no duplications; pachytene stage of meiosis I |
| DNA fragment orientation | head-to-tail | head-to-tail | head-to-tail | randomly joined in head-to-tail, tail-to-tail and head-to-head orientations |
| Junction sequences | ? | microhomologies, short deletions, non-templated insertions | small deletions due to NHEJ | losses of a few base pairs up to several kb, small insertions of non-templated sequences |
| Junction spacing | ? | ? | ? | closely spaced |
| Key references | Hastings et al., 2009; Colnaghi et al., 2011 | Hastings et al., 2009; Colnaghi et al., 2011 | Scott and Pandita, 2006 | Liang et al., 1998; Holloway et al., 2010; Truong et al., 2013 |
? = Not known.
Pathogenetic Mechanisms of CCRs
Roughly, 70% of all reported CCRs were found in healthy individuals [Pellestor et al., 2011]. This means that CCRs, although they affect large numbers of nucleotides, do not necessarily elicit a phenotypic effect. Hence the need to define a set of characteristics distinguishing phenotypically neutral from truly pathogenic CCRs. More than a decade of experience with CNVs may guide our attempts at understanding the possible phenotypic effects of CCRs. Based on this experience, there are mainly 3 types of pathogenic mechanism for an SV: First, gene dosage-dependent mechanisms which have been extensively explored in the context of CNVs [Poot et al., 2011a], and second, mechanisms based on disruption of genomic architecture such that genes, parts of genes, and regulatory elements are relocated and their interactions perturbed. The phenotypic effects of this mechanism are mediated by alteration of gene expression rather than changes in the structure of the encoded protein(s) [Reymond et al., 2007; Henrichsen et al., 2009a; Klopocki and Mundlos, 2011; Spielmann and Klopocki, 2013; Spielmann and Mundlos, 2013], third, mixed mutation mechanisms (MMMs) in which a CNV or a disrupted gene on one chromosome is paired with a different type of mutation on the other chromosome. Genetically this constellation appears as recessive, but the underlying mechanism is distinct from a homozygous or compound heterozygous pair of variants in a single gene.
Gene Dosage-Dependent Mechanisms
In a diploid genome, CNVs affecting the phenotype based on a change in dosage of a gene exert a dominant effect [Poot et al., 2011a; Hart and O'Driscoll, 2013]. Basically, changes in gene dosage may be phenotypically significant if the encoded protein interacts with other proteins [Veitia, 2010; Veitia and Birchler, 2010]. That means haploinsufficiency or triplosufficiency for a gene due to loss or gain CNV, respectively, may alter the stoichiometry of the proteins in a complex and thus the function of the latter [Veitia, 2010; Veitia and Birchler, 2010; Poot et al., 2011a]. A prerequisite for this mechanism is that the change in gene dosage is translated into altered levels of mRNA. Only in a few cases this has actually been demonstrated. Those include de novo CNVs in patients with atrial septal defect, 22q11 deletions in patients with the velocardiofacial syndrome, deletions in patients with the Williams Beuren syndrome, duplications of the 1q21.1 region, and both deletions and duplications of the 16p11.2 region [Merla et al., 2006; Harvard et al., 2011; Luo et al., 2012; Blumenthal et al., 2014]. In a study of 129 individuals of European and Yoruba descent, complex relationships between size and population frequencies of CNVs with gene expression level have been found [Schlattl et al., 2011]. The data supported a causal role of CNVs in expression of quantitative trait loci and revealed a dosage compensation mechanism of some transcript levels [Schlattl et al., 2011]. This large-scale study generalizes the observation of a correlation of a mouse expression of quantitative trait loci with corpus callosum thickness in patients with small deletions in the 1q44 region [Poot et al., 2011b]. Including fine-scale CNV information may thus complement association studies aimed at identifying phenotypically significant expression of quantitative trait loci [Schlattl et al., 2011].
Mechanisms Based on Disruption of the Genomic Architecture
A growing body of experimental evidence indicates that the genome consists of topologically associated domains (TADs), which are in the megabase range and separated by boundary regions [Dixon et al., 2012; Nora et al., 2012, 2013]. These TADs represent regulatory domains within which promoters and regulatory elements may interact with each other and protein-encoding genes [Spielmann and Klopocki, 2013; Spielmann and Mundlos, 2013]. Disruption of TADs by genomic rearrangements may then cause pathological interactions by perturbing gene expression [Spielmann and Klopocki, 2013; Spielmann and Mundlos, 2013; Lupiáñez et al., 2015]. In patients with skeletal dysmorphology (in particular of the hands), changes in noncoding elements have frequently been revealed by screening for CNVs [Klopocki et al., 2008, 2011; Benko et al., 2009; Dathe et al., 2009; Gordon et al., 2009, 2014; Kurth et al., 2009; Spielmann et al., 2012; Lohan et al., 2014; Tayebi et al., 2014]. This genomic mechanism is not necessarily limited to these disorders. Recently, losses of cis-regulatory elements, imbalance in coding and noncoding elements, illegitimate adoption of enhancer sequences and the like, resulting from CNVs have been proposed as a general mechanism of ‘genomic’ disease [Klopocki and Mundlos, 2011; Lettice et al., 2011; Poot and Kas, 2013; Spielmann and Klopocki, 2013; Gordon et al., 2014; Ibn-Salem et al., 2014; Lupiáñez et al., 2015]. CNVs may also cause ‘side effects’, i.e. they affect the level of expression of genes in the flanking regions [Reymond et al., 2007]. In mice, this effect extends to regions up to 250 kb for all tissues examined [Henrichsen et al., 2009b]. Systematic analysis of ENCODE data allowed establishing a map of regulatory domains consisting of sequences that enhance and/or inhibit the expression of CNV-flanking genes [Spielmann and Mundlos, 2013]. Changes in functioning of noncoding sequences as a mechanism of phenotypic effects of CCRs is a novel area of research for which the technological basis is still under construction [Lupiáñez et al., 2015].
A second mechanism by which disruption of the genomic architecture by CNVs or CCRs may affect organismal phenotypes is truncation or fusion of genes. Truncation of a gene may lead to expression of either a truncated mRNA or may, through loss of a legitimate transcription stop signal, activate nonsense-mediated mRNA decay [Wagner and Lykke-Andersen, 2002; Popp and Maquat, 2013]. A truncated mRNA may either be phenotypically inconsequential or exert an affect through a dominant-negative mode of action. Nonsense-mediated mRNA decay, on the other hand, will provoke a lowered level of gene expression, which amounts to a recessive mode of gene action, except for cases of gene haploinsufficiency or triplosufficiency [Poot et al., 2011a; Veitia, 2010; Veitia and Birchler, 2010]. In order to decide between these 2 possibilities, one needs to ascertain the level of transcription of the genes affected by the CCR.
If a CCR connects 2 genes with the same direction of transcription, a fusion transcript may result. In this way, the open reading frame of one gene, or part of a gene, will be fused with that of another gene or part of a gene. As a consequence, the level and the expression pattern of the fusion gene will come under the transcriptional control of one of the genes, which may lead to ectopic gene expression. To establish whether a CCR indeed provokes such a mode of perturbed gene expression, one needs to ascertain both the gene fusion points and whether the fusion gene is actually transcribed. In patients with autism and with schizophrenia, this mechanism as a result of deletions has already been demonstrated [Holt et al., 2012; Rippey et al., 2013]. Also duplications, either being tandem duplications, insertion-duplications, or triplications embedded within duplications, may cause disruption or fusion of genes [Newman et al., 2015]. Table 2 lists the currently reported CCRs, which produced gene fusions, together with the phenotypes of the patients, the involved chromosomal regions, and the affected genes. In 9 out of the 10 reported cases, 1-3 fusion transcripts were actually detected. In one case, such a fusion transcript was not detected, presumably because the direction of transcription of the 2 affected genes were opposite to each other [Malli et al., 2014]. In a patient with schizophrenia, the CCR apparently activated an alternative transcription mode, since the authors found 3 fusion transcripts [Eykelenboom et al., 2012]. While in 8 cases a fusion transcript was successfully demonstrated, other causes of the patient's phenotypes, such as haploinsufficiency of truncated genes, altered expression of unrelated genes, or additional CNVs, still have to be considered [Nothwang et al., 2001; Yue et al., 2005; Mansouri et al., 2006; Borsani et al., 2008; Backx et al., 2011; Eykelenboom et al., 2012; Kloosterman et al., 2012; Di Gregorio et al., 2013; Malli et al., 2014]. In 2 cases, regions of homology have been identified at the fusion points, such that for the other 8 cases NHEJ seems to be the most likely mechanism of fusion.
Table 2.
Reported cases of gene fusion as a result of a CCR
| Major patient phenotypes | Karyotype | Fusion transcripts: 5’ gene, exons; 3’ gene, exons | 5’ gene breakpoint | 3’ gene breakpoint | Truncated gene products | Additional SV, SNVs, or transcription changes | Fusion mechanism | Parental origin | Reference |
|---|---|---|---|---|---|---|---|---|---|
| DD, ataxia, brain atrophy | t(1;19)(q21.3;q13.2)dn | PAFAH1B3, exons 1–5; CLK2, exons 1–12 | in intron 4 (PAFAH1B3) | 5’ of exon 1 (CLK2) | PAFAH13B exons 1–5 | ? | AluSP repeats | ? | Nothwang et al., 2001 |
| DD, delay in expressive language, macrocephaly, kyphoscoliosis | t(7;10)(q33;q23)dn | SEC8L1, exons 1–11; PTEN, exons 3–9 | in intron 11 (SEC8L) | in intron 2 (PTEN) | none | 7-Mb deletion in 7q33q34 (PTEN, TPK1) | ? | Paternal germline | Yue et al., 2005 |
| Hypospadias, anal atresia, rectourethral fistula, hypoplastic kidney | t(6;17)(p21.31;q11.2) | 182-FIP, exon 1; LHFP5, exon 4 | in intron 1 (FIP1) | in intron 3 (LHFP5) | ? | increased transcription of SRPK1 and TAOK1 | 2-bp deletion and a 7-bp duplication | ? | Mansouri et al., 2006 |
| Left renal agenesis, neutropenia, recurrent pulmonary infections, long bone diaphysis broadening, growth and DD | t(2;7)(p13;p12)dn | EXOC6B, exon 1; TNS3, exon 16 and TNS3, exon 15; EXOC6B, exon 2 | in intron 1 (EXOC6B) | in intron 15 (TNS3) | none | none; haploinsufficiency for both TNS3 and EXOC6B | NHEJ? | ? | Borsani et al., 2008 |
| DD + agenesis of the corpus callosum | t(6;14)(q25.3;q13.2)dn | ARID1B, exons 1–5; MRPP3, exons 5–8; MRPP3, exons 1–4; MRPP3, exons 6–19 | in intron 5 (ARID1B) | in intron 4 (MRPP3) | none | none; haploinsufficiency for both ARID1B and MRPP3 | 1. NHEJ? 2. non-templated insert | ? | Backx et al., 2011 |
| Schizophrenia | t(1;11)(q42.1;q14.3)dn | DISC1, exons 1–8; DISC1FP1, exons 4–7b; DISC1, exons 1–8; DISC1FP1, exons 3a–7b; DISC1, exons 1b–2; DISCFP1, exons 9–13 | in exons 8 and 2 (DISC1) | in exons 4, 3a, and 9 (DISC1FP1) | none | haploinsufficiency for DISC1 | NHEJ? | ? | Eykelenboom et al., 2012 |
| Psycho motor retardation, speech delay, facial dysmorphisms, preaxial polydactyly of the thumbs | t(1;12;7)(p21;q14;p21)dn | DPYD, exons 1–3; ETV1, exons 10–14; FOXP1, exons 1–11; unknown, exons 1–2 | in intron 3 (DPYD) | in intron 9 (ETV1) | FOXP1, exons 1–11; DPYD, exons 1–3 | decreased expression of FOXP1 and DPYD | NHEJ? | ? | Kloosterman et al., 2012; van Heesch et al., 2014 |
| Psycho motor retardation, cerebellar hypoplasia | t(X;8)(q25;q24)dn | PTK2, exon 1; THOC, exons 2–36 | in 5’ UTR (PTK2) | in intron 1 (THOC) | none | ? | MER4/AluJ on 8q24 and SVA on Xq25 | ? | Di Gregorio et al., 2013 |
| DD, speech impairment, dysmorphisms | t(5;6)(q12.3;q25.3)dn | no fusion transcripts expressed | in intron 10 (ADAMTS6) | in intron 2 (ARID1B) | none | haploinsufficiency for ARID1B and ADAMST6 | NHEJ? | ? | Malli et al., 2014 |
| GTS, OCD, ADHD + comorbidities | t(3;9)(q25.1;q34.3)mat | OLFM1, exon 7; TCONS_12_00019929; OLFM1, exon 7; TCONS_12_00019930 | in exon 7 (OLFM1) | in TCONS_12_00019929/30 | OLFM1, exons 1–7 | haploinsufficiency for OLFM1 | NHEJ? | maternally inherited | Bertelsen et al., 2015 |
ADHD = Attention deficit hyperactivity disorder; DD = developmental delay; GTS = Gilles de la Tourette syndrome; OCD = obsessive-compulsive disorder; ? = not known.
Mixed Mutation Mechanisms
Disruption or deletion of one allele of a gene may also exert a phenotypic effect via a recessive mechanism. This occurs if, for the same locus as the deletion, an SNV or CNV residing on the chromosome from the other healthy parent is transmitted. This ‘unmasking’ of a recessive allele constitutes a mixed mutation mechanism (MMM). In table 3, all well-documented cases of ‘unmasking’ are compiled. Of the 24 cases with a deletion, 11 involved a known recurrent CNV. This is in agreement with frequent suggestions in the literature that the phenotypic variability of patients with a recurrent CNV may be due to an MMM, such as unmasking [Mefford et al., 2008; Hannes et al., 2009; Kumar et al., 2009; Bedeschi et al., 2010; Kunishima et al., 2013; McDonald-McGinn et al., 2013]. For 5 of the 11 de novo CNVs, the parental origin of the rearrangement could be determined: 4 occurred in the germ line of the father. Interestingly, in 2 cases a second overlapping deletion on the chromosome of the other healthy parent was discovered [Masurel-Paulet et al., 2010; Hochstenbach et al., 2012]. Thus far, only 2 cases of unmasking due to a de novo CCR have been reported. The phenotypes of 10 of the 11 cases with unmasking due to a de novo deletion could be classified as ‘double syndromes’, i.e. the patients exhibit features pertaining to 2 syndromes simultaneously [Flipsen-ten Berg et al., 2007]. No SNVs were unmasked in a series of 20 patients with random assortments of phenotypes, but in one case, an overlapping deletion was found in a genomic footprint of 1.53 Mb [Hochstenbach et al., 2012]. This is in agreement with the relative prevalence of potentially pathogenic SNVs and CNVs in the human genome [Stankiewicz and Lupksi, 2010; Campbell and Eichler, 2013]. The patients with a transmitted deletion and a concomitantly inherited SNV displayed a seemingly random assortment of phenotypes, not reminiscent of any Mendelian disorder. In contrast, genes associated with a Mendelian disease are less tolerant to phenotypically significant SNVs than genes that do not cause any known disease [Petrovski et al., 2013]. The loss of gene function due to gene disrupting mutations such as nonsense, coding indels, and splice acceptor/donor site mutations have been found at significantly elevated frequencies in the genomes of patients with severe ID, epilepsy and atrial septal defect [Petrovski et al., 2013]. As of yet, no clear phenotypic or genetic pattern allowing us to pinpoint an unmasking mechanism in patients with a CNV or a CCR has as yet emerged.
Table 3.
Literature on unmasking of second SNVs or CNVs by inherited and de novo CNVs and CCRs
a.
Published cases of inherited recessive mutations unmasked by hemizygosity due to a de novo deletion
| Phenotype of proband | Unmasked gene | Gene location | SNV | Parental origin of SNV | Chromosomal location | Deletion size, Mb | Parental origin of de novo deletion | Reference |
|---|---|---|---|---|---|---|---|---|
| Prader-Willi syndrome; oculocutaneous albinism | OCA2 | 15q12-q13.1 | p. Val443Ile; c. 1327G>A | mother is heterozygous carrier | del(15)(q11.2q13.1) | nd | paternal | Lee et al., 1994 |
| VCF/DiGeorge syndrome; Bernard-Soulier syndrome | GP1BB | 22q11.21 | c. −133C>G | nd | del(22)(q11.2q11.2) | nd | nd | Ludlow et al., 1996 |
| MR, retinoblastoma + Wilson disease | ATP7B | 13q14.3 | nd | maternal | del(13)(q14.2q21.1) | nd | paternal | Riley et al., 2001 |
| Smith-Magenis syndrome; sensorineural hearing loss | MYO15A | 17p11.2 | p. Thr2205Ile; c. 6614C>T | mother is heterozygous carrier | del(17)(p11.2p11.2) | nd | paternal | Liburd et al., 2001 |
| Angelman syndrome; oculocutaneous albinism | OCA2 | 15q12-q13.1 | c. 647-?_807+?del (deletion of exon 7) | father is heterozygous carrier | del(15)(q11.2q13.1) | ∼5 | maternal | Fridman et al., 2003 |
| Williams syndrome; chronic granulomatous disease | NCF1 | 7q11.23 | p. Tyr26fs; c. 75-76del | nd | del(7)(q11.23q11.23) | nd | nd | Kabuki et al., 2003 |
| Sotos syndrome; reduced coagulation factor 12 activity | F12 | 5q35.3 | c. −4C>T (rs1801020) | nd | del(5)(q35.3q35.3) | 2.2 | nd | Kurotaki et al., 2005 |
| 22q11 del syndrome; psychotic disorder | COMT | 22q11.21 | p. Val158Met; c. 472G>A | nd | del(22)(q11.2q11.2) | nd | nd | Gothelf et al., 2005 |
| Wolf-Hirschhorn syndrome; Wolfram syndrome | WFS1 | 4p16.1 | p. Gln366X; c. 1096C>T | mother is heterozygous carrier | inv dup del(4) (:p15.33→p16.1::p16.1→qter) | 8.3 | paternal | Flipsen-ten Berg et al., 2007 |
| 22q13 del syndrome; metachromatic leukodystrophy | ARSA | 22q13.33 | p. Pro426Leu c. 1277C>T | nd | del(22)(q13.2qter).ish cgh dup(13)(q22qter) | ∼8 | nd | Bisgaard et al., 2009 |
| Werner syndrome | WRN | 8p12 | p. Tyr57X; c. 171C>G | nd | del (8)(p12p12) | 0.553 | nd | Friedrich et al., 2010 |
b.
Published cases of inherited recessive mutations unmasked by hemizygosity due to a deletion inherited from a healthy parent
| Phenotype of proband | Unmasked gene | Gene location | SNV | Parental origin of SNV | Chromosomal location | Deletion size, Mb | Parental origin of deletion | Reference |
|---|---|---|---|---|---|---|---|---|
| MR; sensorineural hearing loss | probably USH1E | 21q21 | nd | nd | del(21)(q11.2q21.3) | ∼14 | maternal | Wakui et al., 2002 |
| Peters Plus syndrome | B3GALTL | 13q12.3 | p.?; c.660 + 1G>A | father is heterozygous carrier | del(13)(q12.3q13.1) | ∼1.5 | maternal | Lesnik Oberstein et al., 2006 |
| Growth retardation; autism; mild dysmorphic signs | DIAPH3 | 13q21.2 | p. Pro614Thr; c. 1840C>A | father is heterozygous carrier | del(13)(q21.1q21.31) | ∼10 | maternal | Vorstman et al., 2011 |
| DD/MR; ataxia; areflexia; macrocephaly; giant axonal neuropathy | GAN | 16q23.2 | p. Glu486Lys; c. 1456G>A | father is heterozygous carrier | del(16)(q23.2q23.2) | 0.057–0.131 | maternal | Buysse et al., 2010 |
| Van den Ende-Gupta syndrome | SCARF2 | 22q11.21 | c. 854 + 1G 1 T (intron 4) | Maternal | del(22)(q11.2) | 2.56 | paternal | Bedeschi et al., 2010 |
| Oculocutaneous albinism | OCA2 | 15q12q13.1 | p. Gly651Glu; c. 1952G>A | father is heterozygous carrier | del(15)(q13.1q13.1) | 0.184 (intragenic deletion) | maternal | Rooryck et al., 2011 |
| Oculocutaneous albinism | OCA2 | 15q12q13.1 | p. AlaA334Val; c. 1001C>T | Nd | del(15(q13.1q13.1) | 0.2 | maternal | Rooryck et al., 2011 |
| Cockayne syndrome | ERCC6 | 10q11.2 | ? frameshift | Maternal | del(10q11.2) | 5.0 | paternal | Ghai et al., 2011 |
| Cohen syndrome | COH1 | 8q22.2 | p. Arg1696X; c. 5086C>T | father is heterozygous carrier | del(8)(q22.2q22.2) | 0.067 (intragenic deletion) | maternal | Rivera-Brugués et al., 2011 |
| Cohen syndrome | COH1 | 8q22.2 | p. Lys3835fsX43, c. 11505delA | mother is heterozygous carrier | del(8)(q22.2q22.2) | 0.193 (intragenic deletion) | paternal | Rivera-Brugués et al., 2011 |
| Cohen syndrome | COH1 | 8q22.2 | p. Thr1289Ser c. 3866C>G + p. Asp3942_Gly3943insAsp; c. 11827_11828insATG | father is heterozygous carrier | del(8)(q22.2q22.2) | 0.315 (intragenic deletion) | maternal | Rivera-Brugués et al. 2011 |
| DD, retinitis pigmentosa + juvenile neuronal ceroid lipofuscinosis | CLN3 | 16p11.2 | ? | ? | del(16)(p11.2) | 220 kb (distal deletion) | ? | Pebrel-Richard et al., 2014 |
c.
Published cases of CNVs or SNVs unmasked by a CCR
| Phenotype of proband | Unmasked gene | Gene location | CNV or SNV | Parental origin of SNV | Chromosomal location | Deletion size, Mb | Parental origin of deletion | Reference |
|---|---|---|---|---|---|---|---|---|
| Primary amenorrhea, hypergonadotropic hypogonadism, disturbed folliculogenesis | FSHR | 2p16.3 | p. Pro587His; c. 1760C4A | Paternally inherited | t(2;8)(p16.3or21;p23.1) | ? | maternally inherited | Kuechler et al., 2010 |
| Short stature + congenital pulmonary alveolar proteinosis | CSF2RA | Yp11.32 | gene deletion | maternally inherited | Xp22.33p22.2 | ? | de novo paternal | Auger et al., 2013 |
d.
Published cases of 2 overlapping deletions transmitted by healthy parents
| Phenotype of proband | Affected gene | CNV location | CNV type | Parental origin | CNV location | CNV type | Parental origin | Reference |
|---|---|---|---|---|---|---|---|---|
| Severe epileptic encephalopathy, retinopathy, ASD and choreoathetosis | CHRNA7 | 15q13.3 | recurrent deletion | maternally inherited | del(15)(q13.3) | recurrent deletion | paternally inherited | Masurel-Paulet et al., 2010 |
| Severe MR, lack of speech, cheilognathopalatoschisis, microcephaly and bilateral hearing loss | HSBP1 | 16q23.3 | unique deletion of 16 kbp | paternally inherited | del(16)(q23.3q24) | unique deletion of 2.166 Mb | maternally inherited | Hochstenbach et al., 2011 |
ASD = Atrial septal defect; DD = developmental delay; MR = mental retardation; nd = not determined; VCF = velocardiofacial syndrome; ? = not known.
Multiple Possibly Pathogenic Mechanisms Provoked by CCRs: Two Cases
Since, by definition, a CCR is composed of multiple genomic alterations occurring simultaneously, it inherently is likely to provoke multiple pathogenic mechanisms, all operating at once in a single patient. It is a major challenge to determine to what extent each of these mechanisms may explain (part of) the patient's clinical phenotype(s). This is illustrated by the following cases.
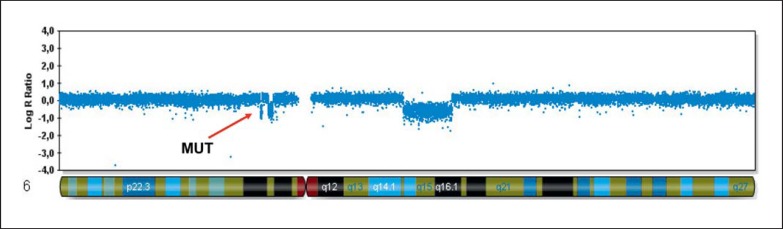
The first case was a de novo CCR involving multiple deletions in association with a pericentric inversion of chromosome 6 [Passarge, 2000; Poot et al., 2009; Kloosterman et al., 2012]. Initially, this patient was described as a case with a pericentric inversion of 6p11.2;q15 with an additional deletion within 6q14, which upon SNP-array analysis was found to harbor 3, instead of 1, deletions within the de novo pericentric inversion (fig. 2) [Passarge, 2000; Poot et al., 2009]. The patient's phenotype consists of a dysmorphic face, nonprogressive deficit of motor control, lack of speech development, and a strongly reduced sensitivity to pain [Passarge, 2000; Poot et al., 2009]. Following a gene-centric approach, only haploinsufficiency of deleted genes or the disruption of the MUT gene by the leftmost breakpoint of deletion 1 (table 4) would be considered [Poot and Hochstenbach, 2010; Poot et al., 2011a]. In view of the ENCODE data, also regulatory domains consisting of sequences that enhance and/or inhibit the expression of breakpoint-flanking genes should be taken into account [Spielmann and Mundlos, 2013]. Thus, loss or disruption of the long noncoding RNA LOC101927048, binding sites for the transcription factors GATA1, GATA2, GATA3, p300 and c-fos, and the histone methylation marks of active promoter regions H3K4Me1 and H3K27AC (table 4), should also be considered when trying to explain the patient's phenotype.
Fig. 2.
SNP-array data of chromosome 6 of the proband with a de novo 46,XX,der(6)(pter/p12.3::p12.1→p12.1::q14.3→p12.1::p12.3→p12.2::q16.1→qter) [Passarge, 2000; Poot et al., 2009]. The arrow indicates the gene disrupted by the breakpoint.
Table 4.
Breakpoints identified by SNP-array analysis of the proband with a de novo 46,XX,der(6)(pter→p12.3::p12.1→p12.1::q14.3→p12.1::p12.3→p12.2::q16.1→qter)
| Deletiona | Breakpoint | Chromosome band | Defining SNPs |
Disrupted gene | Breakpoint in gene | Histone modificationb | DNAse I hypersensitivity | CpG island | DNA methylation sites | Transcription factor binding sites | Replication timingc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| left | right | |||||||||||
| 1 | left | 6p12.3 | rs6458687 | rs6458690 | MUT | exon 12-intron 11 | no | no | 1 | 0 | no | S2-S3 |
| right | rs6905366 | rs700002 | LOC101927048 | no | no | no | 0 | 0 | no | S3-S4 | ||
| 2 | left | 6p12.2 | rs1342622 | rs10498786 | no | no | H3K4me1H3K27ac | strong | 1 | 1 | GATA3, p300 | G2 |
| right | rs9463802 | rs2063643 | no | no | H3K4me1 H3K27ac | weak | 1 | 1 | GATA1, GATA2, p300, c-fos | G1b-S1 | ||
| 3 | left | 6q14.2-6q12.3 | rs2324476 | rs4707016 | no | no | no | no | 0 | no | no | S4-G2 |
| right | rs2799633 | rs2745650 | no | no | no | no | no | no | no | S4-G2 | ||
Refers to the deletion regions shown in figure 2.
H3K27ac and H3K4me1 marks are often found near regulatory elements.
Replication timing data according to Hansen et al. [2010]. SNPs in bold are diploid; SNPs in normal font are deleted. Genes, motives or sites of interest within 1 kbp of the breakpoint regions [Passarge, 2000; Poot et al., 2009].
The second case was initially described as a balanced insertion (9;12) translocation, which a mother transmitted such that it resulted in a trisomy of 9p22p24 in the proband [de Pater et al., 2002]. The mother, the proband and 2 other children, who inherited the balanced insertion (9;12) translocation all showed mental retardation. Therefore, the authors attributed the mental retardation in this 2-generation family to gene disruption brought about by the insertion (9;12) translocation [de Pater et al., 2002].
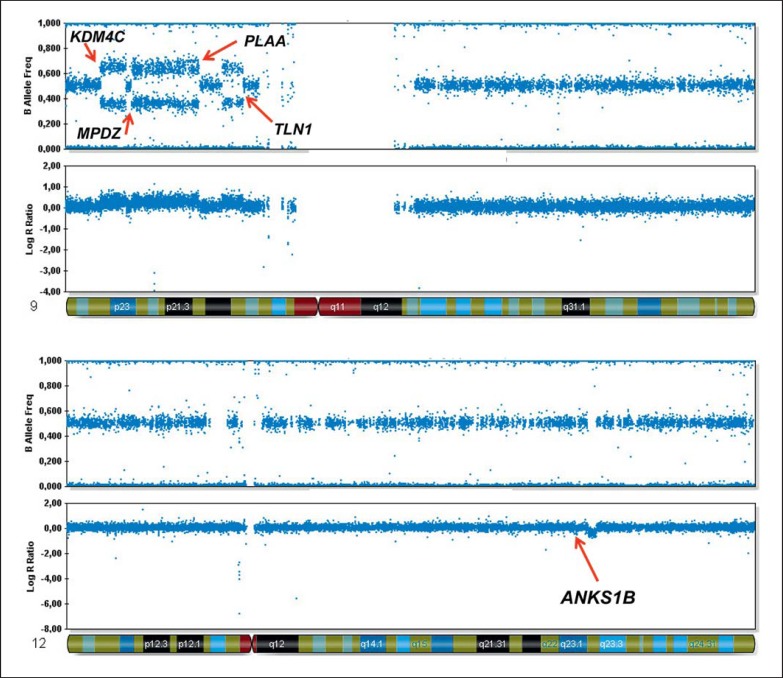
Upon genome-wide SNP-array analysis the patient's karyotype was described as: 46,XY,der(12)dir ins(12;9)(q23;p21p23)mat arr dup(9)(p21p23)(rs4740872×2,rs7860449×3,rs946452×3,rs1339284×2)(rs11790910×2,rs11793993×3,rs7038314×3,rs7863476×2)(rs10120052×2,rs13285154×3,rs2295797×3,rs3763630×2) arr del(12)(q23.1; q23)(rs11615348×2,rs10777975×1,rs10860431×1,rs7301622×2)(rs7965233×2,rs1007916×1,rs11111423×1,rs4764936×2), which involved 10 breakpoints (fig. 3). These breakpoints disrupted 5 candidate genes: KDM4C, MPDZ, PLAA, TLN1 and ANKS1B, and may affect binding sites of the GATA3, CTCF, RFX5, NFYB, POLR2A and MAFK transcription factors (table 5). By mate-pair sequencing of the mother, a grand total of 23 fusion points were detected, of which 16 were transmitted to the son with the inherited mental retardation and the de novo dysmorphisms [Kloosterman et al., 2012]. The mate-pair data confirmed the breakpoints found by SNP-array genotyping of the son but also revealed breaks in regions 6q22.31 (disrupting TRDN), 7p14.2 (disrupting AOAH), 9p21.2 (disrupting ITF74), and in 12q24.21 (disrupting TRPV4) in the genome of the mother.
Fig. 3.
SNP-array data of chromosomes 9 and 12 of the proband, who inherited a der(12)dir ins(12;9)(q23;p21p23)mat [de Pater et al., 2002]. The arrows mark the breakpoints identified by SNP-array and mate-pair sequencing analysis. The disrupted genes are indicated.
Table 5.
Breakpoint regions identified by SNP-array analysis of the 46,XY,der(12)dir ins(12;9)(q23;p21p23)mat case
| Breakpointa | Chromosome band | Defining SNP |
Disrupted gene | Breakpoint in gene | Histone modificationb | DNAse I hypersensitivity | CpG island | DNA methylation sites | Transcription factor binding sites | Replication timingc | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| left | right | ||||||||||
| 1 | rs4740872 | rs7860449 | KDM4C | intron 17 | no | no | no | no | no | S3-S4 | |
| 2 | rs946452 | rs1339284 | no | no | no | Strong | no | no | no | S4-G2 | |
| 3 | 9p23 | rs11790910 | rs11793993 | MPDZ | intron 3 | no | no | no | no | GATA3 | S1-S2 |
| 4 | 9p23 | rs7038314 | rs7863476 | PLAA | exon 14 | no | no | no | no | no | S1-S2 |
| 5 | 9p23 | rs10120052 | rs13285154 | no | no | no | no | no | no | no | S3-S4 |
| 6 | 9p23 | rs2295797 | rs3763630 | TLN1 | intron 1 | H3K27ac | Strong | no | no | CTCF, RFX5, NFYB, POLR2A | G1b |
| 7 | 12q23.1 | rs11615348 | rs10777975 | ANKS1B | intron 12 | no | no | no | no | no | S4-G2 |
| 8 | 12q23.1 | rs10860431 | rs7301622 | ANKS1B | intron 11 | no | no | no | no | MAFK | S4-G2 |
| 9 | 12q23.2 | rs7965233 | rs1007916 | no | no | no | no | no | no | no | S3-S4 |
| 10 | 12q23.2 | rs11111423 | rs4764936 | no | no | no | no | no | no | no | S4-G2 |
Refers to the deletion regions shown in figure 3.
H3K27ac mark is often found near regulatory elements.
Replication timing data according to Hansen et al. [2010]. SNPs in bold are diploid, italicized are duplicated and SNPs in normal font are deleted. Genes, motives or sites of interest within 1 kbp of the breakpoint regions [de Pater et al., 2002].
The breakpoint 1 in region 9p21 disrupts KDM4C, which is a histone demethylase of the Jumonji domain 2 (JMJD2) family. KDM4C, together with the histone demethylases of the same subclass exhibit distinct and combinatorial functions in control of the embryonal stem cell state [Das et al., 2014]. KDM4C is targeted to H3K4me3-positive transcription start sites, where it contributes to transcriptional regulation [Pedersen et al., 2014]. KDM4A/ C-mediated control of histone methylation regulates intrinsic factors and signaling factors and may thus provide a novel control mechanism of lineage decision [Cascante et al., 2014]. Given this role in brain development, disruption of one copy of KDM4C may be involved in the transmitted form of mental retardation in this family.
Apart from detecting additional breakpoints beyond those seen by classical karyotyping, SNP-array genotyping and mate-pair sequencing revealed an additional level of complexity. In addition to CNVs and gene disruption, side effects and changes in noncoding sequences, e.g. DNA and histone methylation, and transcription factor binding sites may all occur simultaneously (table 5). Thus, the improved resolution of mate-pair sequencing showed that these 2 CCRs were even more complex than already revealed by SNP-array genotyping. Therefore, these CCRs must be considered as potential cases of MMMs operating simultaneously.
Diagnostic, Medical Genetic and Experimental Challenges
Facing patients with a complex set of phenotypes, not clearly fitting into a Mendelian syndrome, medical geneticists generally decide to first perform genome-wide segmental aneuploidy screening [Miller et al., 2010; Kearney et al., 2011]. This choice appears justified by the relatively high yield of potentially pathogenic CNVs in cohorts of patients with intellectual delay and neurodevelopmental disorders [Hochstenbach et al., 2009, 2011]. However, apparently ‘balanced’ SVs, which represent a small but significant number of cases, will thus escape detection [De Gregori et al., 2007; Hochstenbach et al., 2009]. With improving resolution of the CNV detecting arrays, the number of truly ‘balanced’ cases of SVs decreases, since the breakpoint regions of these SVs are increasingly found to contain small CNVs [Gribble et al., 2005; Fauth et al., 2006; De Gregori et al., 2007; Baptista et al., 2008; Higgins et al., 2008; Sismani et al., 2008; Schluth-Bolard et al., 2009; Gijsbers et al., 2010; Kang et al., 2010; Feenstra et al., 2011; Kloosterman et al., 2011, 2012; Tabet et al., 2015]. Also cases with multiple de novo or transmitted CNVs raise the suspicion of a CCR [Houge et al., 2003; Lybaek et al., 2008; Ballarati et al., 2009; Poot et al., 2009, 2010a; Schluth-Bolard et al., 2009; Tzschach et al., 2010]. Such cases should be further investigated by classical karyotyping or by paired-end or mate-pair sequencing [Korbel et al., 2007; Kloosterman et al., 2011, 2012; Hooper et al., 2012; Nazaryan et al., 2014].
The latter, highly laborious technique reveals the full complexity of the CCRs and will also allow determining the mode(s) of fusion of the chromosome breaks at nucleotide resolution [Kloosterman and Hochstenbach, 2014]. This information may contain clue(s) regarding possible mechanism(s) of origin of the CCR (see table 1). If these fusion points involve small deletions, microhomologies or non-templated insertions, this points towards biological pathways in which genes such as ATM, BLM, WRN, RAD51, MRE11, ATR, or NBS1 participate. In such cases, the pedigree of the patient should be scrutinized for individuals with either CCRs, impaired fertility or recurrent miscarriages. Medical geneticists may then attempt to trace possible ‘risk alleles’, which cosegregate with such phenotypes in these pedigrees. Such transmitted risk alleles are by themselves not sufficient to cause CCRs, but may represent susceptibility factors, which may confer some degree of vulnerability towards CCR formation. If the fusion points do not contain any molecular signatures such as microhomologies, deletions or insertions, the CCR may have been stitched together by NHEJ. For the latter, no empirical or population data-based recurrence risk estimate is currently available.
A second aspect of the fusion point sequences is the respective orientation of the fused chromosomal fragments. If those are not exclusively head-to-tail, but also tail-to-tail, head-to-head, etc., they may have resulted from chromothripsis [Kloosterman et al., 2011, 2012]. This may indicate that male individuals in the family of the patient may harbor elevated levels of oxidative stress, defective checkpoint functions, such as p53 activity, higher levels of abortive apoptosis, telomere erosion, mitotic errors, or micronucleus formation [Pellestor et al., 2014]. These suggested mechanisms of germline chromothripsis need further scrutiny before they can be incorporated into routine medical genetic counseling.
Since roughly 70% of all reported CCRs were found in healthy individuals, this type of SV does not necessarily elicit a phenotypic effect [Pellestor et al., 2011]. Hence, the need to distinguish phenotypically neutral from truly pathogenic CCRs. Again, obtaining information on the breakpoints at nucleotide resolution is a pivotal first step in identifying the possible pathogenic effects of a CCR. It will also allow ascertaining which genes and which additional architectural relationships within the genome are disrupted. The 2 cases described above illustrate this problem and show that CCRs inherently harbor multiple plausible mutational mechanisms, each of which need separate scrutiny (tables 4, 5).
As a first step, databases, such as ECARUCA (http://umcecaruca01.extern.umcn.nl:8080/ecaruca/ecaruca.jsp), DECIPHER (https://decipher.sanger.ac.uk/), ISCA (http://clinicalgenome.org/), and PubMed should be consulted for each of the disrupted genes to determine whether such a gene disruption has previously been discovered in a patient with a similar phenotype [Feenstra et al., 2006; Firth et al., 2009; Kaminsky et al., 2011; South and Brothman, 2011; de Leeuw et al., 2012; Riggs et al., 2012]. The Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home), on the other hand, summarizing data from 62 studies of healthy individuals, reports 3,024,212 CNVs and 2,335 inversions [MacDonald et al., 2014]. These databases are mainly ‘populated’ by CNVs, such that the reported SVs are biased towards those that contain dosage-sensitive genes, which may affect the individual's phenotype via haploinsufficiency or triplosufficiency [Poot et al., 2011a]. A recently developed database contains 2,643 individually curated breakpoints of congenital and somatic chromosomal rearrangements (dbCRID, http://dbCRID.biolead.org) [Kong et al., 2011]. Since there is no clinical need to karyotype randomly selected, healthy individuals, the number of true CCR carriers is underestimated, and no database of ‘phenotypically neutral’ CCR breakpoints, similar to the Database of Genomic Variants for CNVs, exists. In view of the recent advances in paired-end and mate-pair sequencing, there is clearly a need for such a more comprehensive database [Kloosterman and Hochstenbach, 2014].
A second way to assess the potential pathogenicity of CCRs is functional enrichment analyses of cohorts of patients with similar phenotypes. This approach has allowed us to relate phenotypes such as intellectual delay, autism, or attention deficit/hyperactivity disorder to certain biological pathways [Marshall et al., 2008; Webber et al., 2009; Hehir-Kwa et al., 2010; Pinto et al., 2010; Poot et al., 2010b; Webber, 2011; Noh et al., 2013; Andrews et al., 2015; Hawi et al., 2015]. To associate CCRs, CNVs, genes, and biological pathways with phenotypes, these have to be categorized systematically in so-called phenotype-ontologies [Robinson and Webber, 2014; Deans et al., 2015]. Next, the phenotypes of model organisms with disruptions of the orthologous genes, and features of breakpoint regions of the human CCRs, such as LINE elements, segmental duplications, ‘side effects’ on transcription levels of genes in the vicinity of the breakpoints, markers of active promoters (e.g. H3K4Me1, H3K27Ac), and adoption of enhancers or insulators have to be taken into consideration [Reymond et al., 2007; Hehir-Kwa et al., 2010; Spielmann and Mundlos, 2013; Ibn-Salem et al., 2014]. All these sophisticated methods of bioinformatic predictions notwithstanding, experimental confirmation of gene disruption or alteration of mRNA levels in a phenotypically relevant tissue of the patient remains necessary [Kloosterman and Hochstenbach, 2014]. The above highlights the need for both detailed clinical investigations of individuals with CCRs and complementary experimental studies to grasp the full impact of CCRs in the human genome. Induced pluripotent stem cells from patients with the 22q11.2 deletion syndrome, Down syndrome, and Prader-Willi syndrome have provided novel insights into the pathology of these disorders [Yang et al., 2010; Mou et al., 2012; Shcheglovitov et al., 2013; Paşca et al., 2014]. Transgenic animal models, such as mice and zebrafish, have allowed studying the effects of SVs at the organismal level and during development [Carvalho et al., 2014; Kloosterman et al., 2014; Portmann et al., 2014; Yang et al., 2015]. The novel genome editing CRISPR/Cas9 technology has opened an entirely new avenue for testing hypotheses on the functional impact of CNVs and CCRs, such as disruption of topological chromatin domains [Lupiáñez et al., 2015]. In addition, gene-gene interactions and novel treatment modalities can be studied in these models [Leblond et al., 2012; Schmeisser et al., 2012; Poot, 2013; Shcheglovitov et al., 2013]. In this way, the study of transgenic animals provides not only ‘the proof of the pudding’, it also opens up novel treatment avenues for otherwise difficult to study, rare disorders [Poot, 2013]. Eventually, investigation of CCRs may help to improve our insights into proper and disordered functioning and of our genome as a whole.
URLs of websites
Database of Genomic Variants: http://dgv.tcag.ca/dgv/app/home.
DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER): https://decipher.sanger.ac.uk/.
European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA): http://umcecaruca01.extern.umcn.nl:8080/ecaruca/ecaruca.jsp.
International Standards for Cytogenomic Arrays Consortium (ISCA): http://clinicalgenome.org/.
Acknowledgment
The authors wish to thank Prof. emer. E. Passarge, Drs. J. de Pater and P.F. Ippel for allowing us to investigate their respective patients.
References
- 1.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen SL, Sekelsky J. Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays. 2010;32:1058–1066. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews T, Meader S, Vulto-van Silfhout A, Taylor A, Steinberg J, et al. Gene networks underlying convergent and pleiotropic phenotypes in a large and systematically-phenotyped cohort with heterogeneous developmental disorders. PLoS Genet. 2015;11:e1005012. doi: 10.1371/journal.pgen.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auger J, Bonnet C, Valduga M, Philippe C, Bertolo-Houriez E, et al. De novo complex X chromosome rearrangement unmasking maternally inherited CSF2RA deletion in a girl with pulmonary alveolar proteinosis. Am J Med Genet A. 2013;161A:2594–2599. doi: 10.1002/ajmg.a.36097. [DOI] [PubMed] [Google Scholar]
- 5.Backx L, Seuntjens E, Devriendt K, Vermeesch J, Van Esch H. A balanced translocation t(6;14)(q25.3;q13.2) leading to reciprocal fusion transcripts in a patient with intellectual disability and agenesis of corpus callosum. Cytogenet Genome Res. 2011;132:135–143. doi: 10.1159/000321577. [DOI] [PubMed] [Google Scholar]
- 6.Ballarati L, Recalcati MP, Bedeschi MF, Lalatta F, Valtorta C, et al. Cytogenetic, FISH and array-CGH characterization of a complex chromosomal rearrangement carried by a mentally and language impaired patient. Eur J Med Genet. 2009;52:218–223. doi: 10.1016/j.ejmg.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Baple EL, Chambers H, Cross HE, Fawcett H, Nakazawa Y, et al. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J Clin Invest. 2014;124:3137–3146. doi: 10.1172/JCI74593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, et al. Breakpoint mapping and genome-wide array analysis in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, Jasin M. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, et al. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development. 1998;125:4007–4017. doi: 10.1242/dev.125.20.4007. [DOI] [PubMed] [Google Scholar]
- 11.Bartram CR, Koske-Westphal T, Passarge E. Chromatid exchanges in ataxia telangiectasia, Bloom syndrome, Werner syndrome, and xeroderma pigmentosum. Ann Hum Genet. 1976;40:79–86. doi: 10.1111/j.1469-1809.1976.tb00166.x. [DOI] [PubMed] [Google Scholar]
- 12.Bass HN, Sparkes RS, Lessner MM, Fox M, Phoenix B, Bernar J. A family with thress independent autosomal translocations associated with 7q32→7qter syndrome. J Med Genet. 1985;22:59–63. doi: 10.1136/jmg.22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batanian JR, Eswara MS. De novo apparently balanced complex chromosomes rearrangement (CCR) involving chromosome 4, 18, and 21 in a girl with mental retardation: report and review. Am J Med Genet. 1998;78:44–51. [PubMed] [Google Scholar]
- 14.Batista DA, Pai GS, Stetten G. Molecular analysis of a complex chromosomal rearrangement and a review of familial cases. Am J Med Genet. 1994;53:255–263. doi: 10.1002/ajmg.1320530311. [DOI] [PubMed] [Google Scholar]
- 15.Bedeschi MF, Colombo L, Mari F, Hofmann K, Rauch A, et al. Unmasking of a recessive SCARF2 mutation by a 22q11.12 de novo deletion in a patient with Van den Ende-Gupta syndrome. Mol Syndromol. 2010;1:239–245. doi: 10.1159/000328135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 17.Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 18.Bernardini L, Palka C, Ceccarini C, Capalbo A, Bottillo I, et al. Complex rearrangement of chromosomes7q21.13-q22.1 confirms the ectrodactyly-deafness locus and suggests new candidate genes. Am J Med Genet A. 2008;146A:238–244. doi: 10.1002/ajmg.a.32093. [DOI] [PubMed] [Google Scholar]
- 19.Bertelsen B, Melchior L, Jensen LR, Groth C, Nazaryan L, et al. A t(3;9)(q25.1;q34.3) translocation leading to OLFM1 fusion transcripts in Gilles de la Tourette syndrome, OCD and ADHD. Psychiatry Res. 2015;225:268–275. doi: 10.1016/j.psychres.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 20.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bisgaard AM, Kirchhoff M, Nielsen JE, Kibaek M, Lund A, et al. Chromosomal deletion unmasking a recessive disease: 22q13 deletion syndrome and metachromatic leukodystrophy. Clin Genet. 2009;75:175–179. doi: 10.1111/j.1399-0004.2008.01113.x. [DOI] [PubMed] [Google Scholar]
- 22.Blumenthal I, Ragavendran A, Erdin S, Klei L, Sugathan A, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borsani G, Piovani G, Zoppi N, Bertini V, Bini R, et al. Cytogenetic and molecular characterization of a de-novo t(2p;7p) translocation involving TNS3 and EXOC6B genes in a boy with a complex syndromic phenotype. Eur J Med Genet. 2008;51:292–302. doi: 10.1016/j.ejmg.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Brand H, Pillalamarri V, Collins RL, Eggert S, O'Dushlaine C, et al. Cryptic and complex chromosomal aberrations in early-onset neuropsychiatric disorders. Am J Hum Genet. 2014;95:454–461. doi: 10.1016/j.ajhg.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buizer-Voskamp JE, Blauw HM, Boks MP, van Eijk KR, Veldink JH, et al. Increased paternal age and the influence on burden of genomic copy number variation in the general population. Hum Genet. 2013;132:443–450. doi: 10.1007/s00439-012-1261-4. [DOI] [PubMed] [Google Scholar]
- 26.Buysse K, Vergult S, Mussche S, Ceuterick-de Groote C, Speleman F, et al. Giant axonal neuropathy caused by compound heterozygosity for a maternally inherited microdeletion and a paternal mutation within the GAN gene. Am J Med Genet A. 2010;152A:2802–2804. doi: 10.1002/ajmg.a.33508. [DOI] [PubMed] [Google Scholar]
- 27.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho CM, Vasanth S, Shinawi M, Russell C, Ramocki MB, et al. Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am J Hum Genet. 2014;95:565–578. doi: 10.1016/j.ajhg.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho CM, Pfundt R, King DA, Lindsay SJ, Zuccherato LW, et al. Absence of heterozygosity due to template switching during replicative rearrangements. Am J Hum Genet. 2015;96:555–564. doi: 10.1016/j.ajhg.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cascante A, Klum S, Biswas M, Antolin-Fontes B, Barnabé-Heider F, Hermanson O. Gene-specific methylation control of H3K9 and H3K36 on neurotrophic BDNF versus astroglial GFAP genes by KDM4A/C regulates neural stem cell differentiation. J Mol Biol. 2014;426:3467–3477. doi: 10.1016/j.jmb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44:390–397. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colnaghi R, Carpenter G, Volker M, O'Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22:875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and Holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das PP, Shao Z, Beyaz S, Apostolou E, Pinello L, et al. Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol Cell. 2014;53:32–48. doi: 10.1016/j.molcel.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dathe K, Kjaer KW, Brehm A, Meinecke P, Nürnberg P, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am J Hum Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deans AR, Lewis SE, Huala E, Anzaldo SS, Ashburner M, et al. Finding our way through phenotypes. PLoS Biol. 2015;13:e1002033. doi: 10.1371/journal.pbio.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Bruin C, Mericq V, Andrew SF, van Duyvenvoorde HA, Verkaik NS, et al. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J Clin Endocrinol Metab. 2015;100:E789–E798. doi: 10.1210/jc.2015-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, et al. Cryptic deletions are a common finding in ‘balanced’ reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, Carter NP, Feuk L, et al. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012;33:930–940. doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Den Boer PJ, Poot M, Verkerk A, Jansen R, Mackenbach P, Grootegoed JA. Glutathione-dependent defence mechanisms in isolated round spermatids from the rat. Int J Androl. 1990;13:26–38. doi: 10.1111/j.1365-2605.1990.tb00957.x. [DOI] [PubMed] [Google Scholar]
- 44.de Pagter MS, van Roosmalen MJ, Baas AF, Renkens I, Duran KJ, et al. Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am J Hum Genet. 2015;96:651–656. doi: 10.1016/j.ajhg.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Pater JM, Ippel PF, van Dam WM, Loneus WH, Engelen JJ. Characterization of partial trisomy 9p due to insertional translocation by chromosomal (micro)FISH. Clin Genet. 2002;62:482–487. doi: 10.1034/j.1399-0004.2002.620611.x. [DOI] [PubMed] [Google Scholar]
- 46.de Vree PJ, Simon ME, van Dooren MF, Stoevelaar GH, Hilkmann JT, et al. Application of molecular cytogenetic techniques to clarify apparently balanced complex chromosomal rearrangements in two patients with abnormal phenotype: case report. Mol Cytogenet. 2009;2:15. doi: 10.1186/1755-8166-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, et al. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 48.Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci USA. 2005;102:737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Gregorio E, Bianchi FT, Schiavi A, Chiotto AM, Rolando M, et al. A de novo X;8 translocation creates a PTK2-THOC2 gene fusion with THOC2 expression knockdown in a patient with psychomotor retardation and congenital cerebellar hypoplasia. J Med Genet. 2013;50:543–551. doi: 10.1136/jmedgenet-2013-101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 52.Eykelenboom JE, Briggs GJ, Bradshaw NJ, Soares DC, Ogawa F, et al. A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum Mol Genet. 2012;21:3374–3386. doi: 10.1093/hmg/dds169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farrell SA, Summers AM, Gadner HA, Uchida IA. Balanced complex chromosome rearrangement ascertained through prenatal diagnosis. Am J Med Genet. 1994;52:360–361. doi: 10.1002/ajmg.1320520322. [DOI] [PubMed] [Google Scholar]
- 54.Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. Top3-Rmi1 dissolve Rad51-mediated D-loops by a toposiomerase-based mechanism. Mol Cell. 2015;57:595–606. doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fauth C, Gribble SM, Porter KM, Codina-Pascual M, Ng BL, et al. Micro-array analyses decipher exceptional complex familial chromosomal rearrangement. Hum Genet. 2006;119:145–153. doi: 10.1007/s00439-005-0103-z. [DOI] [PubMed] [Google Scholar]
- 56.Feenstra I, Fang J, Koolen DA, Siezen A, Evans C, et al. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur J Med Genet. 2006;49:279–291. doi: 10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- 57.Feenstra I, Hanemaaijer N, Sikkema-Raddatz B, Yntema H, Dijkhuizen T, et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet. 2011;19:1152–1160. doi: 10.1038/ejhg.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flipsen-ten Berg K, van Hasselt PM, Eleveld MJ, van der Wijst SE, Hol FA, et al. Unmasking of a hemizygous WFS1 gene mutation by a chromosome 4p deletion of 8.3 Mb in a patient with Wolf-Hirschhorn syndrome. Eur J Hum Genet. 2007;15:1132–1138. doi: 10.1038/sj.ejhg.5201899. [DOI] [PubMed] [Google Scholar]
- 60.Fridman C, Hosomi N, Varela MC, Souza AH, Fukai K, Koiffmann CP. Angelman syndrome associated with oculocutaneous albinism due to an intragenic deletion of the P gene. Am J Med Genet A. 2003;119A:180–183. doi: 10.1002/ajmg.a.20105. [DOI] [PubMed] [Google Scholar]
- 61.Friedrich K, Lee L, Leistritz DF, Nürnberg G, Saha B, et al. WRN mutations in Werner syndrome patients: genomic rearrangements, unusual intronic mutations and ethnic-specific alterations. Hum Genet. 2010;128:103–111. doi: 10.1007/s00439-010-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghai SJ, Shago M, Shroff M, Yoon G. Cockayne syndrome caused by paternally inherited 5 Mb deletion of 10q11.2 and a frameshift mutation of ERCC6. Eur J Med Genet. 2011;54:272–276. doi: 10.1016/j.ejmg.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21:884–892. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gijsbers AC, Bosch CA, Dauwerse JG, Giromus O, Hansson K, et al. Additional cryptic CNVs in mentally retarded patients with apparently balanced karyotypes. Eur J Med Genet. 2010;53:227–233. doi: 10.1016/j.ejmg.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Gothelf D, Eliez S, Thompson T, Hinard C, Penniman L, et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 67.Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- 68.Gordon CT, Attanasio C, Bhatia S, Benko S, Ansari M, et al. Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence. Hum Mutat. 2014;35:1011–1020. doi: 10.1002/humu.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorski JL, Kistenmacher ML, Punnett HH, Zackai EH, Emanuel BS. Reproductive risks for carriers of complex chromosome rearrangements: analysis of 25 families. Am J Med Genet. 1988;29:247–261. doi: 10.1002/ajmg.1320290202. [DOI] [PubMed] [Google Scholar]
- 70.Grasshoff U, Singer S, Liehr T, Starke H, Fode B, et al. A complex chromosomal rearrangement with a translocation 4;10;14 in a fertile male carrier: ascertainment through an offspring with partial trisomy 14q24→1q22 and partial monosomy 4q27→q28. Cytogenet Genome Res. 2003;103:17–23. doi: 10.1159/000076282. [DOI] [PubMed] [Google Scholar]
- 71.Gribble SM, Prigmore E, Burford DC, Porter KM, Ng BL, et al. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet. 2005;42:8–16. doi: 10.1136/jmg.2004.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haaf T, Raderschall E, Reddy G, Ward DC, Radding CM, Golub El. Sequestration of mammalian Rad51-recombination protein into micronuclei. J Cell Biol. 1999;144:11–20. doi: 10.1083/jcb.144.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haber JE. TOPping off meiosis. Mol Cell. 2015;57:577–581. doi: 10.1016/j.molcel.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hart L, O'Driscoll M. Causes and consequences of structural genomic alterations in the human genome. eLS Wiley. 2013 http://www.els.net. [Google Scholar]
- 77.Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, et al. Understanding the impact of 1q21.1 copy number variant. Orphanet J Rare Dis. 2011;6:54. doi: 10.1186/1750-1172-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hawi Z, Cummins TD, Tong J, Johnson B, Lau R, et al. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol Psychiatry. 2015;20:289–297. doi: 10.1038/mp.2014.183. [DOI] [PubMed] [Google Scholar]
- 80.Hehir-Kwa JY, Wieskamp N, Webber C, Pfundt R, Brunner HG, et al. Accurate distinction of pathogenic from benign CNVs in mental retardation. PLoS Comput Biol. 2010;6:e1000752. doi: 10.1371/journal.pcbi.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hehir-Kwa JY, Rodríguez-Santiago B, Vissers LE, de Leeuw N, Pfundt R, et al. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J Med Genet. 2011;48:776–778. doi: 10.1136/jmedgenet-2011-100147. [DOI] [PubMed] [Google Scholar]
- 82.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009a;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 83.Henrichsen CN, Vinckenbosch N, Zöllner S, Chaignat E, Pradervand S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009b;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 84.Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hochstenbach R, van Binsbergen E, Engelen J, Nieuwint A, Polstra A, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009;52:161–169. doi: 10.1016/j.ejmg.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 86.Hochstenbach R, Buizer-Voskamp JE, Vorstman JA, Ophoff RA. Genome arrays for the detection of copy number variations in idiopathic mental retardation, idiopathic generalized epilepsy and neuropsychiatric disorders: lessons for diagnostic workflow and research. Cytogenet Genome Res. 2011;135:174–202. doi: 10.1159/000332928. [DOI] [PubMed] [Google Scholar]
- 87.Hochstenbach R, Poot M, Nijman IJ, Renkens I, Duran KJ, et al. Discovery of variants unmasked by hemizygous deletions. Eur J Hum Genet. 2012;20:748–753. doi: 10.1038/ejhg.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holt R, Sykes NH, Conceição IC, Cazier JB, Anney RJL, et al. CNVs leading to fusion transcripts in individuals with autism spectrum disorder. Eur J Hum Genet. 2012;20:1141–1147. doi: 10.1038/ejhg.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Honma M, Tadokoro S, Sakamoto H, Tanabe H, Sugimoto M, et al. Chromosomal instability in B-lymphoblasotoid cell lines from Werner and Bloom syndrome patients. Mutat Res. 2002;520:15–24. doi: 10.1016/s1383-5718(02)00144-4. [DOI] [PubMed] [Google Scholar]
- 91.Hooper SD, Johansson AC, Tellgren-Roth C, Stattin EL, Dahl N, et al. Genome-wide sequencing for the identification of rearrangements associated with Tourette syndrome and obsessive-compulsive disorder. BMC Med Genet. 2012;13:123. doi: 10.1186/1471-2350-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Houge G, Liehr T, Schoumans J, Ness GO, Solland K, et al. Ten years follow up of a boy with a complex chromosomal rearrangement: going from a >5 to 15-breakpoint CCR. Am J Med Genet A. 2003;118A:235–240. doi: 10.1002/ajmg.a.10106. [DOI] [PubMed] [Google Scholar]
- 93.Hulick PJ, Noonan KM, Kulkarni S, Donovan DJ, Listewnik M, et al. Cytogenetic and array-CGH characterization of a complex de novo rearrangement involving duplication and deletion of 9p and clinical findings in a 4-month-old female. Cytogenet Genome Res. 2009;126:305–312. doi: 10.1159/000251966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ibn-Salem J, Köhler S, Love MI, Chung HR, Huang N, et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 2014;15:423. doi: 10.1186/s13059-014-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang YH, Martinez JE, Ou Z, Cooper ML, Kang SH, et al. De novo and complex imbalanced chromosomal rearrangements revealed by array CGH in a patient with an abnormal phenotype and apparently ‘balanced’ paracentric inversion of 14(q21q23) Am J Med Genet A. 2008;146A:1986–1993. doi: 10.1002/ajmg.a.32408. [DOI] [PubMed] [Google Scholar]
- 97.Joyce CA, Cabral de Almeida JC, Santa Rose AA, Correia P, Moraes L, et al. A de novo complex chromosomal rearrangement with nine breakpoints characterised by FISH in a boy with mild mental retardation, developmental delay, short stature and microcephaly. Clin Genet. 1999;6:86–92. doi: 10.1034/j.1399-0004.1999.560113.x. [DOI] [PubMed] [Google Scholar]
- 98.Kabuki T, Kawai T, Kin Y, Joh K, Ohashi H, et al. A case of Williams syndrome with p47-phox-deficient chronic granulomatous disease (in Japanese) Nihon Rinsho Meneki Gakkai Kaishi. 2003;26:299–303. doi: 10.2177/jsci.26.299. [DOI] [PubMed] [Google Scholar]
- 99.Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J Biol Chem. 2004;279:55499–5505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 100.Kamath-Loeb A, Loeb LA, Fry M. The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLoS One. 2012;7:e30189. doi: 10.1371/journal.pone.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang SH, Shaw C, Ou Z, Eng PA, Cooper ML, et al. Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results. Am J Med Genet A. 2010;152A:1111–1126. doi: 10.1002/ajmg.a.33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaur H, DeMuyt A, Lichten M. Top3-Rmi1 DNA single-stranded decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell. 2015;57:583–594. doi: 10.1016/j.molcel.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kausch K, Haaf T, Köhler J, Schmid M. Complex chromosomal rearrangement in a woman with multiple miscarriages. Am J Med Genet. 1988;31:415–420. doi: 10.1002/ajmg.1320310221. [DOI] [PubMed] [Google Scholar]
- 105.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST, Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 106.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keijzers G, Maynard S, Shamanna RA, Rasmussen LJ, Croteau DL, Bohr VA. The role of RecQ helicases in non-homologous end-joining. Crit Rev Biochem Mol Biol. 2014;49:463–472. doi: 10.3109/10409238.2014.942450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kitano K. Structural mechanisms of human RecQ helicases WRN and BLM. Front Genet. 2014;5:366. doi: 10.3389/fgene.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kloosterman WP, Hochstenbach R. Deciphering the pathogenic consequences of chromosomal aberrations in human genetic disease. Mol Cytogenet. 2014;7:100. doi: 10.1186/s13039-014-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 111.Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 112.Klopocki E, Mundlos S. Copy-number variations, noncoding sequences, and human phenotypes. Annu Rev Genomics Hum Genet. 2011;12:53–72. doi: 10.1146/annurev-genom-082410-101404. [DOI] [PubMed] [Google Scholar]
- 113.Klopocki E, Ott CE, Benatar N, Ullmann R, Mundlos S, Lehmann K. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J Med Genet. 2008;45:370–375. doi: 10.1136/jmg.2007.055699. [DOI] [PubMed] [Google Scholar]
- 114.Klopocki E, Lohan S, Brancati F, Koll R, Brehm A, et al. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am J Hum Genet. 2011;88:70–75. doi: 10.1016/j.ajhg.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kong F, Zhu J, Wu J, Peng J, Wang Y, et al. dbCRID: a database of chromosomal rearrangements in human diseases. Nucleic Acids Res 39(Database issue) 2011;D895-D900 doi: 10.1093/nar/gkq1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;52:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 117.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kousseff BG, Nichols P, Essig YP, Miller K, Weiss A, Tedesco TA. Complex chromosome rearrangements and congenital anomalies. Am J Med Genet. 1987;26:771–782. doi: 10.1002/ajmg.1320260403. [DOI] [PubMed] [Google Scholar]
- 119.Kousseff BG, Papenhausen P, Essig YP, Torres MP. Complex chromosome rearrangement with ankyloblepharon filiform adnatum. J Med Genet. 1993;30:167–170. doi: 10.1136/jmg.30.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kowalczyk M, Tomaszewska A, Podbioł-Palenta A, Constantinou M, Wawrzkiewicz-Witkowska A, et al. Another rare case of a child with de novo terminal 9p deletion and co-existing interstitial 9p duplication: clinical findings and molecular cytogenetic study by array-CGH. Cytogenet Genome Res. 2013;139:9–16. doi: 10.1159/000342165. [DOI] [PubMed] [Google Scholar]
- 121.Kuechler A, Ziegler M, Blank C, Rommel B, Bullerdiek J, et al. A highly complex chromosomal rearrangement between five chromosomes in a healthy female diagnosed in preparation for intracytoplasmatic sperm injection. J Histochem Cytochem. 2005;53:355–357. doi: 10.1369/jhc.4B6437.2005. [DOI] [PubMed] [Google Scholar]
- 122.Kuechler A, Hauffa BP, Köninger A, Kleinau G, Albrecht B, et al. An unbalanced translocation unmasks a recessive mutation in the follicle-stimulating hormone receptor (FSHR) gene and causes FSH resistance. Eur J Hum Genet. 2010;18:656–661. doi: 10.1038/ejhg.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumar RA, Marshall CR, Badner JA, Babatz TD, Mukamel Z, et al. Association and mutation analyses of 16p11.2 autism candidate genes. PLoS One. 2009;4:e4582. doi: 10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kunishima S, Imai T, Kobayashi R, Kato M, Ogawa S, Saito H. Bernard-Soulier syndrome caused by a hemizygous GPIbβ mutation and 22q11.2 deletion. Pediatr Int. 2013;55:434–437. doi: 10.1111/ped.12105. [DOI] [PubMed] [Google Scholar]
- 125.Kurotaki N, Shen JJ, Touyama M, Kondoh T, Visser R, et al. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet Med. 2005;7:479–483. doi: 10.1097/01.gim.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- 126.Kurth I, Klopocki E, Stricker S, van Oosterwijk J, Vanek S, et al. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat Genet. 2009;41:862–863. doi: 10.1038/ng0809-862. [DOI] [PubMed] [Google Scholar]
- 127.Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, et al. Disruption of transcriptional coactivator Sub1 leads to genome-wide re-distribution of clustered mutations induced by APOBEC in active yeast genes. PLoS Genet. 2015;11:e1005217. doi: 10.1371/journal.pgen.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, et al. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci USA. 2011;108:11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee MH, Park SY, Kim YM, Kim JM, Han JY, et al. Prenatal diagnosis of a familial complex chromosomal rearrangement involving chromosomes 5, 10, 16 and 18. Prenat Diagn. 2002;22:102–104. doi: 10.1002/pd.224. [DOI] [PubMed] [Google Scholar]
- 132.Lee ST, Nicholls RD, Bundey S, Laxova R, Musarella M, Spritz RA. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N Engl J Med. 1994;330:529–534. doi: 10.1056/NEJM199402243300803. [DOI] [PubMed] [Google Scholar]
- 133.Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, et al. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet. 2006;79:562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- 135.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- 137.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lohan S, Spielmann M, Doelken SC, Flöttmann R, Muhammad F, et al. Microduplications encompassing the Sonic hedgehog limb enhancer ZRS are associated with Haas-type polysyndactyly and Laurin-Sandrow syndrome. Clin Genet. 2014;86:318–325. doi: 10.1111/cge.12352. [DOI] [PubMed] [Google Scholar]
- 140.Ludlow LB, Schick BP, Budarf ML, Driscoll DA, Zackai EH, et al. Identification of a mutation in a GATA binding site of the platelet glycoprotein Ibbeta promoter resulting in the Bernard-Soulier syndrome. J Biol Chem. 1996;271:22076–22080. doi: 10.1074/jbc.271.36.22076. [DOI] [PubMed] [Google Scholar]
- 141.Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, et al. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lurie IW, Wulfsberg EA, Prabhakar G, Rosenblum-Vos LS, Supovitz KR, Cohen MM. Complex chromosomal rearrangements: some breakpoints may have cellular adaptive significance. Clin Genet. 1994;46:244–247. doi: 10.1111/j.1399-0004.1994.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 144.Lybaek H, Øyen N, Fauske L, Houge G. A 2.1 Mb deletion adjacent but distal to a 14q21q23 paracentric inversion in a family with spherocytosis and severe learning difficulties. Clin Genet. 2008;74:553–559. doi: 10.1111/j.1399-0004.2008.01072.x. [DOI] [PubMed] [Google Scholar]
- 145.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42(Database issue) 2014;D986-D992 doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Madan K. What is a complex chromosome rearrangement? Am J Med Genet A. 2013;161A:1181–1184. doi: 10.1002/ajmg.a.35834. [DOI] [PubMed] [Google Scholar]
- 147.Madan K, Menko FH. Intrachromosomal insertions: a case report and a review. Hum Genet. 1992;89:1–9. doi: 10.1007/BF00207032. [DOI] [PubMed] [Google Scholar]
- 148.Madan K, Nieuwint AW, van Bever Y. Recombination in a balanced complex translocation of a mother leading to a balanced reciprocal translocation in the child. Review of 60 cases of balanced complex translocations. Hum Genet. 1997;99:806–815. doi: 10.1007/s004390050453. [DOI] [PubMed] [Google Scholar]
- 149.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Malli T, Duba HC, Erdel M, Marschon R, Kranewitter W, et al. Disruption of the ARID1B and ADAMTS6 loci due to a t(5;6)(q12.3;q25.3) in a patient with developmental delay. Am J Med Genet A. 2014;164A:3126–3131. doi: 10.1002/ajmg.a.36738. [DOI] [PubMed] [Google Scholar]
- 151.Mansouri MR, Carlsson B, Davey E, Nordenskjöld A, Wester T, et al. Molecular genetic analysis of a de novo balanced translocation t(6;17)(p21.31;q11.2) associated with hypospadias and anorectal malformation. Hum Genet. 2006;119:162–168. doi: 10.1007/s00439-005-0122-9. [DOI] [PubMed] [Google Scholar]
- 152.Marangos P, Carroll J. Oocytes progress beyond prophase in the presence of DNA damage. Curr Biol. 2012;22:989–994. doi: 10.1016/j.cub.2012.03.063. [DOI] [PubMed] [Google Scholar]
- 153.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Masurel-Paulet A, Andrieux J, Callier P, Cuisset JM, Le Caignec C, et al. Delineation of 15q13.3 microdeletions. Clin Genet. 2010;78:149–161. doi: 10.1111/j.1399-0004.2010.01374.x. [DOI] [PubMed] [Google Scholar]
- 155.McDonald-McGinn DM, Fahiminiya S, Revil T, Nowakowska BA, Suhl J, et al. Hemizygous mutations in SNAP29 unmask autosomal recessive conditions and contribute to atypical findings in patients with 22q11.2DS. J Med Genet. 2013;50:80–90. doi: 10.1136/jmedgenet-2012-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meer B, Wolff G, Back E. Segregation of a complex rearrangement of chromosomes 6, 7, 8 and 12 through three generations. Hum Genet. 1981;58:221–225. doi: 10.1007/BF00278717. [DOI] [PubMed] [Google Scholar]
- 157.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Melcher R, von Golitschek R, Steinlein C, Schindler D, Neitzel H, et al. Spectral karyotyping of Werner syndrome fibroblast cultures. Cytogenet Cell Genet. 2000;91:180–185. doi: 10.1159/000056841. [DOI] [PubMed] [Google Scholar]
- 159.Merla G, Howald C, Henrichsen CN, Lyle R, Wyss C, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Monnat RJ., Jr Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semin Cancer Biol. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mou X, Wu Y, Cao H, Meng Q, Wang Q, et al. Generation of disease-specific induced pluripotent stem cells from patients with different karyotypes of Down syndrome. Stem Cell Res Ther. 2012;3:14. doi: 10.1186/scrt105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Murray JE, van der Burg M, IJspeert H, Carroll P, Wu Q, et al. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am J Hum Genet. 2015;96:412–424. doi: 10.1016/j.ajhg.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nazaryan L, Stefanou EG, Hansen C, Kosyakova N, Bak M, et al. The strength of combined cytogenetic and mate-pair sequencing techniques illustrated by a germline chromothripsis rearrangement involving FOXP2. Eur J Hum Genet. 2014;22:338–343. doi: 10.1038/ejhg.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Newman S, Hermetz KE, Weckselblatt B, Rudd MK. Next-generation sequencing of duplication CNVs reveals that Mmst are tandem and some create fusion genes at breakpoints. Am J Hum Genet. 2015;96:208–220. doi: 10.1016/j.ajhg.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noh HJ, Ponting CP, Boulding HC, Meader S, Betancur C, et al. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 2013;9:e1003523. doi: 10.1371/journal.pgen.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nothwang HG, Kim HG, Aoki J, Geisterfer M, Kübart S, et al. Functional hemizygosity of PAFAH1B3 due to a PAFAH1B3-CLK2 fusion gene in a female with mental retardation, ataxia and atrophy of the brain. Hum Mol Genet. 2001;10:797–806. doi: 10.1093/hmg/10.8.797. [DOI] [PubMed] [Google Scholar]
- 171.O'Driscoll M, Jeggo PA. The role of double-strand break repair-insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 172.O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 173.Ogburn CE, Oshima J, Poot M, Chen R, Hunt KE, et al. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 174.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Oshima J, Huang S, Pae C, Campisi J, Schiestl RH. Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res. 2002;62:547–551. [PubMed] [Google Scholar]
- 176.Outwin E, Carpenter G, Bi W, Withers MA, Lupski JR, O'Driscoll M. Increased RPA1 gene dosage affects genomic stability potentially contributing to 17p13.3 duplication syndrome. PLoS Genet. 2011;7:e1002247. doi: 10.1371/journal.pgen.1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pai GS, Thomas GH, Mahoney W, Migeon BR. Complex chromosome rearrangements. Clin Genet. 1980;18:436–444. doi: 10.1111/j.1399-0004.1980.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 178.Park SY, Kim JW, Kim YM, Kim JM, Lee MH, et al. Frequencies of fetal chromosomal abnormalities at prenatal diagnosis: 10 years experiences in a single institution. J Korean Med Sci. 2001;16:290–293. doi: 10.3346/jkms.2001.16.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Paşca SP, Panagiotakos G, Dolmetsch RE. Generating human neurons in vitro and using them to understand neuropsychiatric disease. Annu Rev Neurosci. 2014;37:479–501. doi: 10.1146/annurev-neuro-062012-170328. [DOI] [PubMed] [Google Scholar]
- 180.Passarge E. A distinctive phenotype associated with an interstitial deletion 6q14 contained within a de novo pericentric inversion 6 (p11.2q15) Cytogenet Cell Genet. 2000;91:192–198. doi: 10.1159/000056843. [DOI] [PubMed] [Google Scholar]
- 181.Patro BS, Frøhlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pebrel-Richard C, Debost-Legrand A, Eymard-Pierre E, Greze V, Kemeny S, et al. An unusual clinical severity of 16p11.2 deletion syndrome caused by unmasked recessive mutation of CLN3. Eur J Hum Genet. 2014;22:369–373. doi: 10.1038/ejhg.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pedersen MT, Agger K, Laugesen A, Johansen JV, Cloos PA, et al. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol Cell Biol. 2014;34:1031–1045. doi: 10.1128/MCB.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, et al. Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update. 2011;17:476–494. doi: 10.1093/humupd/dmr010. [DOI] [PubMed] [Google Scholar]
- 185.Pellestor F, Gatinois V, Puechberty J, Geneviève D, Lefort G. Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil Steril. 2014;102:1785–1796. doi: 10.1016/j.fertnstert.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 186.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Phadnis N, Hyppa RW, Smith GR. New and old ways to control meiotic recombination. Trends Genet. 2011;27:411–421. doi: 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Plaisancié J, Kleinfinger P, Cances C, Bazin A, Julia S, et al. Constitutional chromoanasynthesis: description of a rare chromosomal event in a patient. Eur J Med Genet. 2014;57:567–570. doi: 10.1016/j.ejmg.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 190.Poot M. Towards identification of individual etiologies by resolving genomic and biological conundrums in patients with autism spectrum disorders. Mol Syndromol. 2013;4:213–226. doi: 10.1159/000350041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Poot M, Hochstenbach R. A three-step workflow procedure for the interpretation of array-based comparative genome hybridization results in patients with idiopathic mental retardation and congenital anomalies. Genet Med. 2010;12:478–485. doi: 10.1097/GIM.0b013e3181e3914a. [DOI] [PubMed] [Google Scholar]
- 192.Poot M, Kas MJ. Antisense may make sense of 1q44 deletions, seizures, and HNRNPU. Am J Med Genet A. 2013;161A:910–912. doi: 10.1002/ajmg.a.35770. [DOI] [PubMed] [Google Scholar]
- 193.Poot M, Hoehn H, Rünger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 194.Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- 195.Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 196.Poot M, Gollahon KA, Emond MJ, Silber JR, Rabinovitch PS. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–758. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- 197.Poot M, Jin X, Hill JP, Gollahon KA, Rabinovitch PS. Distinct functions for WRN and TP53 in a shared pathway of cellular response to 1-β-D-arabinofuranosylcytosine and bleomycin. Exp Cell Res. 2004;296:327–336. doi: 10.1016/j.yexcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 198.Poot M, van't Slot R, Leupert R, Beyer V, Passarge E, Haaf T. Three de novo losses and one insertion within a pericentric inversion of chromosome 6 in a patient with complete absence of expressive speech and reduced pain perception. Eur J Med Genet. 2009;52:27–30. doi: 10.1016/j.ejmg.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 199.Poot M, Beyer V, Schwaab I, Damatova N, Van't Slot R, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010a;11:81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 200.Poot M, Eleveld MJ, van ‘t Slot R, Ploos van Amstel HK, Hochstenbach R. Recurrent copy number changes in mentally retarded children harbour genes involved in cellular localization and the glutamate receptor complex. Eur J Hum Genet. 2010b;18:39–46. doi: 10.1038/ejhg.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Poot M, van der Smagt JJ, Brilstra EH, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011a;135:228–240. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]
- 202.Poot M, Badea A, Williams RW, Kas MJ. Identifying human disease genes through cross-species gene mapping of evolutionary conserved processes. PLoS One. 2011b;6:e18612. doi: 10.1371/journal.pone.0018612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Portmann T, Yang M, Mao R, Panagiotakos G, Ellegood J, et al. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci USA. 2015;112:E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reymond A, Henrichsen CN, Harewood L, Merla G. Side effects of genome structural changes. Curr Opin Genet Dev. 2007;17:381–386. doi: 10.1016/j.gde.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 207.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Riggs ER, Church DM, Hanson K, Horner VL, Kaminsky EB, et al. Towards an evidence-based process for the clinical interpretation of copy number variation. Clin Genet. 2012;81:403–412. doi: 10.1111/j.1399-0004.2011.01818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Riley D, Wiznitzer M, Schwartz S, Zinn AB. A 13-year-old boy with cognitive impairment, retinoblastoma, and Wilson disease. Neurology. 2001;57:141–143. doi: 10.1212/wnl.57.1.141. [DOI] [PubMed] [Google Scholar]
- 210.Rippey C, Walsh T, Gulsuner S, Brodsky M, Nord AS, et al. Formation of chimeric genes by copy-number variation as a mutational mechanism in schizophrenia. Am J Hum Genet. 2013;93:697–710. doi: 10.1016/j.ajhg.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rivera-Brugués N, Albrecht B, Wieczorek D, Schmidt H, Keller T, et al. Cohen syndrome diagnosis using whole genome arrays. J Med Genet. 2011;48:136–140. doi: 10.1136/jmg.2010.082206. [DOI] [PubMed] [Google Scholar]
- 212.Robinson PN, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 214.Rooryck C, Morice-Picard F, Lasseaux E, Cailley D, Dollfus H, et al. High resolution mapping of OCA2 intragenic rearrangements and identification of a founder effect associated with a deletion in Polish albino patients. Hum Genet. 2011;129:199–208. doi: 10.1007/s00439-010-0913-5. [DOI] [PubMed] [Google Scholar]
- 215.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rünger TM, Bauer C, Dekant B, Möller K, Sobotta P, et al. Hypermutable ligation of plasmid DNA ends in cells from patients with Werner syndrome. J Invest Dermatol. 1994;102:45–48. doi: 10.1111/1523-1747.ep12371730. [DOI] [PubMed] [Google Scholar]
- 217.Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 218.Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21:2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schluth-Bolard C, Delobel B, Sanlaville D, Boute O, Cuisset JM, et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009;52:291–296. doi: 10.1016/j.ejmg.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 220.Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 221.Scott SP, Pandita TK. The cellular control of DNA double-strand breaks. J Cell Biochem. 2006;99:1463–1475. doi: 10.1002/jcb.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sismani C, Kitsiou-Tzeli S, Ioannides M, Christodoulou C, Anastasiadou V, et al. Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol Cytogenet. 2008;1:15. doi: 10.1186/1755-8166-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.South ST, Brothman AR. Clinical laboratory implementation of cytogenomic microarrays. Cytogenet Genome Res. 2011;135:203–211. doi: 10.1159/000331425. [DOI] [PubMed] [Google Scholar]
- 226.Spielmann M, Klopocki E. CNVs of noncoding cis-regulatory elements in human disease. Curr Opin Genet Dev. 2013;23:249–256. doi: 10.1016/j.gde.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 227.Spielmann M, Mundlos S. Structural variations, the regulatory landscape of the genome and their alteration in human disease. Bioessays. 2013;35:533–543. doi: 10.1002/bies.201200178. [DOI] [PubMed] [Google Scholar]
- 228.Spielmann M, Brancati F, Krawitz PM, Robinson PN, Ibrahim DM, et al. Homeotic arm-to-leg transformation associated with genomic rearrangements at the PITX1 locus. Am J Hum Genet. 2012;91:629–635. doi: 10.1016/j.ajhg.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 230.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Su F, Mukherjee S, Yang Y, Mori E, Bhattacharya S, et al. Nonenzymatic role for WRN in preserving nascent DNA strands after replication stress. Cell Rep. 2014;9:1387–1401. doi: 10.1016/j.celrep.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Suhasini AN, Brosh RM., Jr Disease-causing missense mutations in human DNA helicase disorders. Mutat Res. 2013;752:138–152. doi: 10.1016/j.mrrev.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tabet AC, Verloes A, Pilorge M, Delaby E, Delorme R, et al. Complex nature of apparently balanced chromosomal rearrangements in patients with autism spectrum disorder. Mol Autism. 2015;6:19. doi: 10.1186/s13229-015-0015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tang S, Wu MKY, Zhang R, Hunter N. Pervasive and essential roles of Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell. 2015;57:607–621. doi: 10.1016/j.molcel.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tayebi N, Jamsheer A, Flöttmann R, Sowinska-Seidler A, Doelken SC, et al. Deletions of exons with regulatory activity at the DYNC1I1 locus are associated with split-hand/split-foot malformation: array CGH screening of 134 unrelated families. Orphanet J Rare Dis. 2014;9:108. doi: 10.1186/s13023-014-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Trachoo O, Assanatham M, Jinawath N, Nongnuch A. Chromosome 20p inverted duplication deletion identified in a Thai female adult with mental retardation, obesity, chronic kidney disease and characteristic facial features. Eur J Med Genet. 2013;56:319–324. doi: 10.1016/j.ejmg.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 237.Truong LN, Li Y, Shi LZ, Hwang PY, He J, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci USA. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tupler R, Maraschio P, Gerardo A, Mainieri R, Lanzi G, Tiepolo L. A complex chromosome rearrangement with 10 breakpoints tentative assignement of the locus for Williams syndrome to 4q33→q35.1. J Med Genet. 1992;29:253–255. doi: 10.1136/jmg.29.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tzschach A, Menzel C, Erdogan F, Istifli ES, Rieger M, et al. Characterization of an interstitial 4q32 deletion in a patient with mental retardation and a complex chromosome rearrangement. Am J Med Genet A. 2010;152A:1008–1012. doi: 10.1002/ajmg.a.33343. [DOI] [PubMed] [Google Scholar]
- 240.van Binsbergen E, Hochstenbach R, Giltay J, Swinkels M. Unstable transmission of a familial complex chromosome rearrangement. Am J Med Genet A. 2012;158A:2888–2893. doi: 10.1002/ajmg.a.35580. [DOI] [PubMed] [Google Scholar]
- 241.Van Dyke DL, Weiss L, Logan M, Pai GS. The origin and behavior of two isodicentric bisatellited chromosomes. Am J Hum Genet. 1977;29:294–300. [PMC free article] [PubMed] [Google Scholar]
- 242.van Heesch S, Simonis M, van Roosmalen MJ, Pillalamarri V, Brand H, et al. Genomic and functional overlap between somatic and germline chromosomal rearrangements. Cell Rep. 2014;9:2001–2010. doi: 10.1016/j.celrep.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 243.Veitia RA. A generalized model of gene dosage and dominant negative effects in macromolecular complexes. FASEB J. 2010;24:994–1002. doi: 10.1096/fj.09-146969. [DOI] [PubMed] [Google Scholar]
- 244.Veitia RA, Birchler JA. Dominance and gene dosage balance in health and disease: why levels matter! J Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- 245.Vorstman JA, van Daalen E, Jalali GR, Schmidt ER, Pasterkamp RJ, et al. A double hit implicates DIAPH3 as an autism risk gene. Mol Psychiatry. 2011;16:442–451. doi: 10.1038/mp.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wagner E, Lykke-Andersen J. mRNA surveillance: the perfect persist. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- 247.Wahls WP, Davidson MK. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wakui K, Toyoda A, Kubota T, Hidaka E, Ishikawa M, et al. Familial 14-Mb deletion at 21q11.2-q21.3 and variable phenotypic expression. J Hum Genet. 2002;47:511–516. doi: 10.1007/s100380200076. [DOI] [PubMed] [Google Scholar]
- 249.Webber C. Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet Genome Res. 2011;135:277–285. doi: 10.1159/000331670. [DOI] [PubMed] [Google Scholar]
- 250.Webber C, Hehir-Kwa JY, Nguyen DQ, de Vries BB, Veltman JA, Ponting CP. Forging links between human mental retardation-associated CNVs and mouse gene knockout models. PLoS Genet. 2009;5:e1000531. doi: 10.1371/journal.pgen.1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 252.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 253.Yang J, Cai J, Zhang Y, Wang X, Li W, et al. Induced pluripotent stem cells can be used to model the genomic imprinting disorder Prader-Willi syndrome. J Biol Chem. 2010;285:40303–40311. doi: 10.1074/jbc.M110.183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yang M, Mahrt EJ, Lewis F, Foley G, Portmann T, et al. 16p11.2 deletion syndrome mice display sensory and ultrasonic vocalization deficits during social interactions. Autism Res (2015), Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 255.Yue Y, Grossmann B, Holder SE, Haaf T. De novo t(7;10)(q33;q23) translocation and closely juxtaposed microdeletion in a patient with macrocephaly and developmental delay. Hum Genet. 2005;117:1–8. doi: 10.1007/s00439-005-1273-4. [DOI] [PubMed] [Google Scholar]
- 256.Yue Y, Stout K, Grossmann B, Zechner U, Brinckmann A, et al. Disruption of TCBA1 associated with a de novo t(1;6)(q32.2;q22.3) presenting in a child with developmental delay and recurrent infections. J Med Genet. 2006;43:143–147. doi: 10.1136/jmg.2004.029660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zahed L, Der Kaloustian V, Batanian JR. Familial complex chromosome rearrangement giving rise to balanced and unbalanced recombination products. Am J Med Genet. 1998;79:30–34. [PubMed] [Google Scholar]
- 258.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhang L, Kim KP, Kleckner NE, Storlazzi A. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci USA. 2011;108:20036–20041. doi: 10.1073/pnas.1117937108. [DOI] [PMC free article] [PubMed] [Google Scholar]