Abstract
Brassica oleracea and B. rapa are two important vegetable crops. Both are composed of dozens of subspecies encompassing hundreds of varieties and cultivars. Synthetic B. napus with these two plants has been used extensively as a research model for the investigation of allopolyploid evolution. However, the mechanism underlying the explosive evolution of hundreds of varieties of B. oleracea and B. rapa within a short period is poorly understood. In the present study, interspecific hybridization between B. oleracea var. alboglabra and B. rapa var. purpurea was performed. The backcross progeny displayed extensive morphological variation, including some individuals that phenocopied subspecies other than their progenitors. Numerous interesting novel phenotypes and mutants were identified among the backcross progeny. The chromosomal recombination between the A and C genomes and the chromosomal asymmetric segregation were revealed using Simple Sequence Repeats (SSR) markers. These findings provide direct evidence in support of the hypothesis that interspecific hybridization and backcrossing have played roles in the evolution of the vast variety of vegetables among these species and suggest that combination of interspecific hybridization and backcrossing may facilitate the development of new mutants and novel phenotypes for both basic research and the breeding of new vegetable crops.
Interspecific hybridization is an important driving force in plant evolution and speciation1,2,3. Following interspecific hybridization, there are two models for the subsequent evolution of plants: genome polyploidization and stabilization, and gene flow with ploidy maintenance4,5,6,7. It has been estimated that over 70% of flowering plants have undergone genome polyploidization2,8. Members of Brassicaceae, a family containing many important vegetable and oil crops, have undergone genomic duplication several times9. Crops such as radishes, cabbages, turnips, and Chinese cabbages are diploids. They have undergone ancient hexaploidization and subsequent fractionalization events10,11,12,13. The best-known recent genome polyploidization event is the interspecific hybridization of B. rapa (A genome), B. nigra (B genome), and B. oleracea (C genome) to form the allotetraploids B. juncea (AB genome), B. napus (AC genome), and B. carinata (BC genome)14. To date, artificial interspecific hybridization and allopolyploids have been primarily synthesized in the laboratory with the goal of exploring the mechanisms underlying polyploidization-dependent evolution and expanding genetic diversity of crops15,16,17,18,19,20,21,22. However, the evolutionary mechanisms underlying gene flow between homoploids have not been thoroughly investigated.
Brassica oleracea and B. rapa are two closely related taxa that contain the most important cruciferous vegetable crops. B. oleracea is composed of more than 15 subspecies, including B. oleracea var. alboglabra (Kai-lan, Chinese broccoli), B. oleracea var. acephala (Acephala, garden cultivars), B. oleracea var. botrytis (cauliflower and romanesco), B. oleracea var. capitata (cabbage), B. oleracea var. costata (tronchuda cabbage), B. oleracea var. gemmifera (Brussels sprout), B. oleracea var. gongylodes (kohlrabi), B. oleracea var. italica (broccoli), B. oleracea var. medullosa (marrow cabbage), B. oleracea var. oleracea (wild cabbage), B. oleracea var. palmifolia (palm cabbage), B. oleracea var. ramosa, B. oleracea var. sabauda (Savoy cabbage), B. oleracea var. sabellica (curly kale), and B. oleracea var. viridis23,24,25. B. oleracea var. alboglabra has been suggested to be a primitive type of kale crop and a possible ancestor of various cultivated B. oleracea vegetables23,24. B. rapa contains an even greater number of subspecies and varieties, such as B. rapa ssp. chinensis (pakchoi), B. rapa ssp. pekinensis (Chinese cabbage), B. rapa var purpurea (purple Cai-tai, B. compestris L. var purpurea was used in the past), B. rapa ssp. parachinensis (Cai xin), B. rapa ssp. dichotoma, B. rapa ssp. japonica, B. rapa ssp. Narinosa (wutacai), B. rapa ssp. Nipposinica (mizuna), B. rapa ssp. oleifera, B. rapa ssp. rapa (turnip), and B. rapa ssp. trilocularis23,24,26. The oldest members of B. rapa were planted thousands of years ago. However, most varieties of B. compestris cultivars, the hundreds of landraces of pakchoi in southern China and Chinese cabbage in northern China, emerged between the 14th and 17th centuries27. Though this evolutionary boom has attracted much attention, the mechanisms underlying it remain unclear.
In the present study, an interspecific hybrid of B. oleracea var. alboglabra and B. rapa var. purpurea (the primitive type of cultivated B. oleracea and B. rapa) was generated. After two rounds of backcrossing of the synthetic allotetraploid plant with B. rapa L. var. purpurea, the morphological variation and genetic composition of the progeny were investigated. The stereotypical traits of B. oleracea var. gemmifera (Brussels sprouts), B. oleracea var. gongylodes (kohlrabi), B. chinensis (pakchoi), and B. pekinensis (Chinese cabbage) were phenocopied or partially phenocopied in the BC2 plants, providing direct evidence in support of the hypothesis that interspecific hybridization and backcrossing played roles in the evolutionary expansion of vegetable varieties in B. oleracea and B. rapa. The intergenomic recombination and asymmetric segregation of chromosomes could be one of the principal mechanisms underlying this process. Many interesting mutants and previously unknown phenotypes were obtained, indicating that the combination of interspecific hybridization and backcrossing may facilitate the development of more mutants and novel phenotypes.
Results
Creation of hybrid plants by embryo rescue
A cytoplasmic male sterile line of B. oleracea var. alboglabra was hand-pollinated with pollen from an inbred line of B. rapa L. var. purpurea. On the 8th day after pollination, the young siliques were harvested, the surfaces were sterilized, and then ovules were dissected out and grown in embryo culture media. Three out of the ≈400 ovules developed into seedlings. Morphologically, the hybrid plants fall between their parents (Fig. 1). Chromosome analysis from the root tip cells revealed that the hybrid somatic cells contained 19 chromosomes (Fig. 1b). A series of codominant Simple Sequence Repeats (SSR) markers indicated ten of which were derived from B. rapa L. var. purpurea, and nine of which were derived from B. oleracea var. alboglabra (Fig. 1e). The hybrid plants were male sterile and produced no pollen. On the main stalk, the embryos were lethal when hand-pollinated with pollen from either B. rapa L. var. purpurea or a fertile B. oleracea var. alboglabra plant. However, some adventitious stalks generated from the bases of the plants produced several seeds by backcrossing with B. rapa L. var. purpurea. These partially fertile stalks were stout with shortened internodes that distinguished them morphologically from the original plants, indicating that these branches may underwent a spontaneous chromosomal doubling.
Figure 1. The F1 hybrid of B. oleracea var. alboglabra and B. rapa L. var. purpurea.
(a) Inflorescences and flowers; (b) Chromosome microscopy of root tip cells from the F1 hybrid plant, n = 19; (c) Plants at the vegetative growth stage; (d) Rosette leaves. (e) Chromosome specific SSR analysis of F1 plant, Ba, B. oleracea var. alboglabra; Br, B. rapa L. var. purpurea; C1–C9 and A1–A10 labeled on right side of lanes indicat corresponding chromosomes, bold alphanumeric characters below each gel indicate the SSR markers. Photographs were taken by Zhang X. and Li X.
Morphological divergence among BC1 (AAC) plants
A total of 72 seeds were harvested from the partially fertile F1 stalks that were hand pollinated with B. rapa var. purpurea. After sowing, 67 seedlings germinated and completed a life cycle. Within this population, morphological variations were observed in the size and shape of the leaves, the size and color of flowers and stalks, the structure of the inflorescence and the presence of surface wax (Fig. 2). White petals and surface wax were the dominant traits controlled by the genes from C genome, as indicated by the F1 plants (Figs 1 and 2). The occurrence of yellow petals and waxless surfaces suggests that some of the chromosome segments from the C genome had been lost from the BC1 plants.
Figure 2. Morphological variations in BC1 plants.
Panels from top to bottom show buds, flowers, young siliques, inflorescences, and rosette leaves. Photographs were taken by Zhang X.
Morphological traits of BC2 progeny
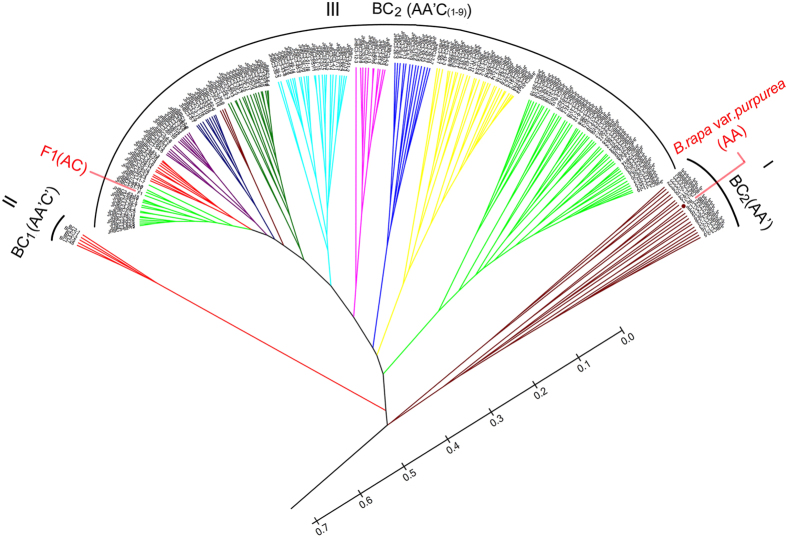
After hand pollination with B. rapa L. var. purpurea as the male parent, all of the 67 resulting BC1 plants were fertile. These 67 BC2 lines produced 935 seedlings. It was deduced that each plant in this generation contained a genome composed of AA′C(0–9) (A′ probably contains segments from C due to chromosomal recombination; C(0–9) contains 0–9 chromosomes from the C genome that probably contain segments from A). Flow cytometry showed that the chromatin content diverged significantly (Fig. 3). Morphologically, the BC2 plants bore a stronger resemblance to B. rapa L. var. purpurea, but the detailed traits diverged significantly among individuals within the population (Fig. 4). The plants were clustered based on 17 morphological traits, including plant height, architecture, stooling, stalk shape, stalk color, waxiness of the stalk, brawniness of the stalk, lateral stalk number, rosette leaf shape, rosette leaf color, rosette leaf crimp, waxiness of the leaf, petiole color, bolting time, flowering time, petal color and style structure. As shown in Fig. 5, 19 of the 274 (6.93%) BC2 plants clustered with B. rapa L. var. purpurea (group I), indicating that these plants were of the AA′ genotype, from which the chromosomes derived from the C genome had been eliminated. However, the individuals within this group were found to differ from each other significantly, indicating that recombination between chromosomes from the A and C genome occurred frequently during two rounds of meiosis, resulting in the replacement of many segments of the A′ genome with fragments from the C genome. Group II comprised BC1 plants. These plants were less diverse morphologically because they underwent only one round of recombination and contained an AA′C′ genome. The other plants were clustered according to nine major terms in group III, indicating the wide range of segregation events and complicated genetic interactions.
Figure 3. Flow cytometry analysis of parents and progeny.
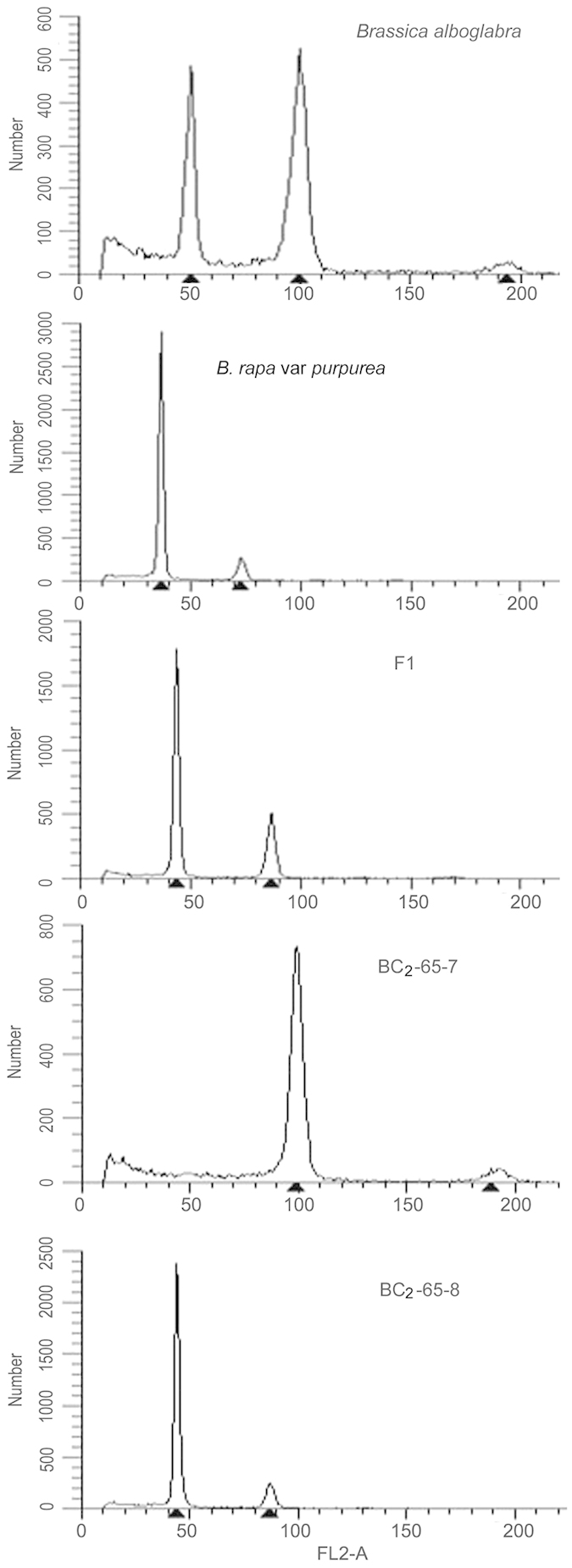
Leaf tissue cells were analyzed. The F1 plant contains chromatin content with the mean value of its parents. BC2-65-7 and BC2-65-8 are BC2 individuals showing significant differences in chromatin content.
Figure 4. Morphology of BC2 plants.
(a) Inflorescences vary in structure and color. (b) Purple or white patches on the tips of buds. (c) The pistils vary in color, length, and shape. (d) Colors of the siliques. (e) Differences in rosette leaves. (f) Poly and monostalks. (g) Differences in stalk in color and waxiness. (h) Variations in plant architecture. Photographs were taken by Zhang X.
Figure 5. Clustering of two generations of backcrossed progeny and their ancestor plants.
The UPGMA tree was constructed using MEGA 5 and morphological traits.
Phenocopying of Brassica vegetables by BC2 progeny
Some BC2 plants displayed traits characteristic of vegetables produced by plants other than their progenitors. As shown in Fig. 6, one BC2 plant generated enlarged corms that resemble kohlrabi (B. oleracea var. carlorapa) (Fig. 6a). Some BC2 plants produced axillary bud balls, which are characteristic of Brussels sprouts (B. oleracea L. var. gemmifera) (Fig. 6b). A number of BC2 plants displayed a phenotype nearly identical to several pakchoi landraces (B. rapa ssp. chinensis). For example, some BC2 plants phenocopied the “Paopaoqing,” a pakchoi landrace that produces characteristic dark green wrinkled rosette leaves (Fig. 6c). The leaves of the BC2 plants frequently folded inward (Fig. 6d), a fundamental developmental process that leads to the formation of the leafy head in cabbage and Chinese cabbage. This suggests that the leafy head formation if the petiole will be short and that the inner leaves will fall inward. These phenotypes morphologically recapitulate the evolutionary history of several Brassica vegetable varieties.
Figure 6. Phenocopying of other Brassica vegetables by BC2 progeny.
(a) A BC2 plant (BC2-15-10) generated enlarged bulbs (red arrow) that resemble kohlrabi. (b) A BC2 plant (BC2-55-5) produced axillary bud balls (red arrow) characteristic of Brussels sprouts. (c) BC2 plants (BC2-34-3 and BC2-29-13) with a phenotype nearly identical to landraces of pakchoi. (d) A BC2 plant (BC2-20-12) produced leaves that folded inward (red arrow), a fundamental trait for developing the leafy head found on Chinese cabbages and other cabbages. Photographs were taken by Zhang X. and Li X.
Interesting mutants and novel phenotypes
In addition to the evidence presented for morphological variation and evolutionary recurrence, many interesting mutants were generated at a relatively high frequency. For example, one mutant presented two cotyledons that merged into a monocotyledon (Fig. 7a). The backcross progeny segregated into mono and dicotyledonous individuals at a 1:1 ratio, indicating that the mutation is controlled by a single dominant gene. This mutant might be a good model for the study of cotyledon development and determination of the molecular basis of mono and dicotyledonous segregation, which is a fundamental question in the evolution and phylogeny of flowering plants. Another interesting mutant generated young plants from its underground root system (Fig. 7b). Similar schedules have been adopted as a primary method of reproduction by many plants. This mutant might be useful for studying mechanisms of this kind. Plants resembling bamboo shoots and Zingiber striolatum could be used as new vegetable cultivars.
Figure 7. Mutants generated from BC2 plants.
(a) WT, wild-type normal plant; M1, M2, and M3, monocotyledon mutants; M3 clearly shows a monocotyledon formed by this combination. (b) The mutants (left three panels) generated young plants from their underground root systems that resemble Zingiber striolatum Diels. Photographs were taken by Zhang X.
Intergenomic recombination and asymmetric segregation as indicated by SSR analysis
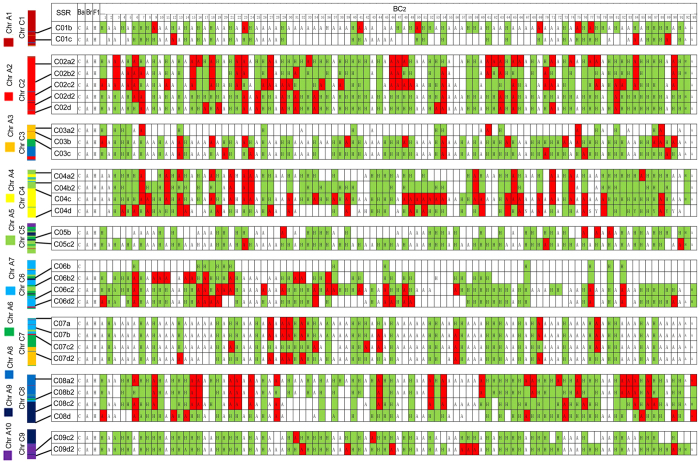
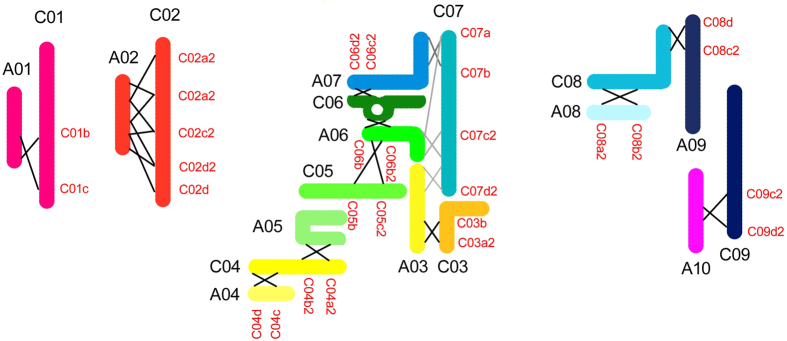
To investigate the genetic mechanism underlying the aforementioned morphological observations, 30 SSR markers (2–5 per chromosome, details in Table 1) were designed and used for genetic analysis of 93 BC2 plants, the F1, and their initial progenitors. As shown in Fig. 8, the BC2 plants contained 0–9 whole or partial C chromosomes from B. oleracea var. alboglabra. Intergenomic recombination between A and C chromosomes was detected between each pair of adjacent markers. Because both A and C genomes have been extensively rearranged since their divergence, the homeologous recombinations can result in asymmetric segregations with chromosome deletion and duplication, as demonstrated in Fig. 9.
Table 1. Characteristics of the 30 analyzed simple sequence repeat (SSR) markers.
| SSR | primer-F | primer-R | Band in A genome (bp) | Band in C genome (bp) | Locus on A genome | Locus on C genome |
|---|---|---|---|---|---|---|
| C01b | GCAACACTTGCTGTTTTGGA | GCTGAGGTAGGAAGGGAAGG | 207 | 233 | A01_15917060 | C01_20708918 |
| C01c | ATCAACGAGGATGTTGGAGA | TGACCCAATCAGGTCAAGAA | 189 | 183 | A01_20992040 | C01_31003906 |
| C02a2 | GATCGACCTTCCATCACGTC | CCTTGTGCAAGAAGGTGTTG | 199 | 176 | A02_4660427 | C02_6784924 |
| C02b2 | ATATACACATGGCCGGTGCT | TTGGTGGAGTTTCCGAGTTT | 114 | 100 | A02_11831311 | C02_16112719 |
| C02c2 | TTGGCTTCAACGTGTCTGAG | CCACTTAGGCGATGGTGTTT | 179 | 146 | A02_15538343 | C02_24870903 |
| C02d2 | TGGGTTCGAATAAGTCGGAT | CTTGGCCCATCATCAAAACT | 278 | 193 | A02_21212404 | C02_37480057 |
| C02d | GTCAGGGCCGGTTGGTAT | CCAAAACCGAACTAAACGGA | 278 | 266 | A02_26805121 | C02_42253466 |
| C03a2 | GGACTAGGCCCAGAAAGGTC | ATGAGAATCACACGCGTCAC | 181 | 141 | A03_5096072 | C03_5782559 |
| C03b | CCGGACCATAAATTATCGCA | GCTCCTCCTCCTCCATCTTC | 165 | 174 | A03_9810560 | C03_11586692 |
| C03c | AGCCGTTGAATCATAATGGC | AATGACAAACGCCGGTCTAC | 142 | 100 | A06_16840545 | C03_29211252 |
| C04a2 | AAAGTCAGGCGTCTTTCTGC | TTGTTGTTGTATAGACCAGACA | 269 | 137 | A05_3393634 | C04_6311603 |
| C04b2 | CATCACCACCATCTACACGC | ATACAAACTGCCGAGACGCT | 153 | 130 | A05_7742417 | C04_17130510 |
| C04c | GCAAAGACCTTTTTGCAAGC | ACCGACCAAAGGTGAACAAC | 213 | 253 | A04_9337281 | C04_28980301 |
| C04d | CGAAGAAATGGGCAATGAGT | CCATGGTCGGACAATCTCTT | 123 | 93 | A04_18399294 | C04_40018307 |
| C05b | CTCGCATTGAAGGAGTGTCA | ATCTCCAGCCTTTTCCAGGT | 263 | 255 | A06_6560520 | C05_13746011 |
| C05c2 | AAAAGCAACCAAAGCGCTAA | TCGTTTCCTTGATTGCTTCC | 139 | 114 | A06_8543638 | C05_21938996 |
| C06b | CGTTGTTTGTCGTCTGCATC | GCCGAATCAGAACCGTTAGA | 246 | 238 | A06_1811353 | C06_13619390 |
| C06b2 | TGGAATCACTTCAGCGTTCA | ACGAGTTTTCTCGGCTTTCA | 265 | 72 | A06_14493472 | C06_17947219 |
| C06c2 | TCAGTATTCTCATCGCCCAA | AGGTCGACCAAAGACGAAAA | 188 | 146 | A07_15806647 | C06_28866205 |
| C06d2 | GCTTCATTGGATCCCACATC | GGGTTCGTGATTGATGGTAAA | 143 | 117 | A07_19411829 | C06_35244684 |
| C07a | ATCCGACCAGTAACGTCCAG | GGGGGATTACCTTCATCTTTT | 217 | 208 | A06_24653900 | C07_463379 |
| C07b | CCCGTAGTTAGAACACCCGA | GCGCAGACCTAGCCTTATTG | 248 | 265 | A07_8821086 | C07_16930026 |
| C07c2 | TGATGGGTAAAATGCTCCAA | CTGCGTCTTTTCTTCCAGTG | 160 | 125 | A06_23763438 | C07_27826196 |
| C07d2 | CGCGTAGAAGTAGTTCCAGTG | TCATCAAACGTGGAGGTCAA | 167 | 131 | A03_23672505 | C07_41422404 |
| C08a2 | TGTTTTCAGCACCTTTTCAAGA | CAAGCAGTGCAACCACAAAG | 241 | 129 | A08_5763859 | C08_5598895 |
| C08b2 | TGCTGCTAAGTCTAGTCCACAA | CCTCAAGATCCACAATGCCT | 248 | 178 | A08_19250863 | C08_18384496 |
| C08c2 | ACTCTCAACAATGGCAAGGG | GCTGTTTTCCATATTCGGCT | 215 | 159 | A09_31431992 | C08_32596668 |
| C08d | CATCGGCCGTAACAAAGAAT | CAGAAACGGGATTCGATCAT | 253 | 231 | A09_38876771 | C08_41482344 |
| C09c2 | TAGCCAACTAACATGGCACG | GCCCACATCTTTCAAAACAAA | 177 | 143 | A10_9735500 | C09_28720170 |
| C09d2 | AAGCTACTTGCCTCACTATCCT | TGATTTCACATAAGGTCCCCA | 250 | 159 | A10_12909906 | C09_35598537 |
Figure 8. Chromosome recombination and segregation as indicated by SSR markers.
The C chromosomes are shown at the left. The segments homologous to A01–A10 chromosomes are indicated with ten different colors. The chromosome loci of the SSRs are given as solid lines. C, A, and H indicate the C chromosomes, A chromosomes, and hybrid type bands. The detection of C chromosome was highlighted in green, the loss of C chromosome segments resulted from intergenomic recombination are highlighted in red.
Figure 9. Homeologous chromosome recombination model.
A sketch of homeologous chromosome recombination between the A and C genomes constructed with the events detected by SSR analysis. A01-A10, B.rapa chromosomes. C01-C09, B. oleracea chromosomes. Red alphanumeric characters indicate the SSR markers. Cross lines indicate the recombination events.
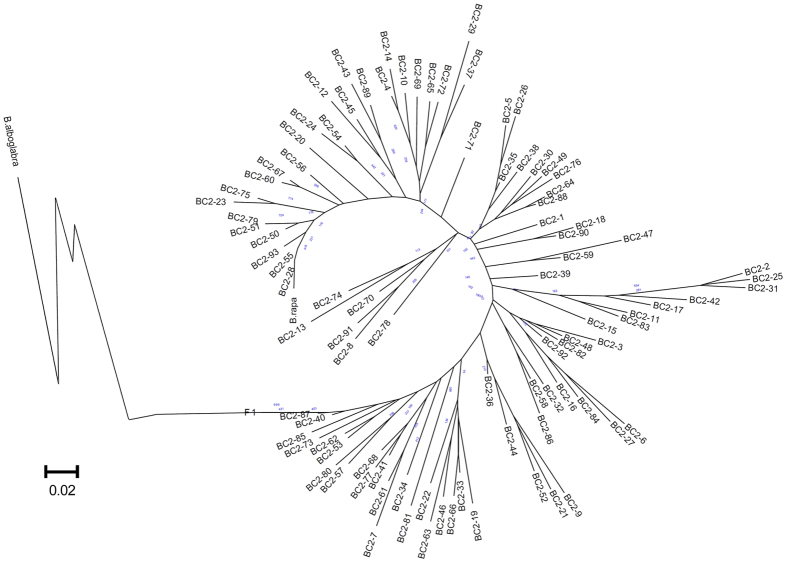
A neighbor-joining (NJ) tree was generated based on this set of SSR data (Fig. 10). The BC2 plants also clustered in nine major branches, which are in accordance with the tree constructed with morphological traits (Fig. 5). Due to two generations of backcrossing with B. rapa L. var. purpurea, it is not surprising that the B. oleracea var. alboglabra outbranched far from the BC2 groups. The F1 and B. rapa L. var. purpurea clustered separately in two outside branches, which were again consistent with the morphological tree. One of these two branches contained the BC2 plants harboring nearly the whole set of C chromosomes and the other those that had lost nearly all of the C chromosomes. The inner branches were composed of those harboring many different numbers of C chromosomes. These molecular data confirmed the morphological observations.
Figure 10. Neighbor-joining tree.
The tree was constructed with 30 SSR markers using MEGA 5. The blue numbers indicate the bootstraps.
InDel mutations detected in BC2 plants
The SSR analysis also detected four novel bands in the BC2 plants (Fig. 11). All of these four mutations are short stretch insertions. The mutation rate was 0.15%, much higher than the spontaneous mutation rate, indicating the minor genomic alteration such as short stretch insertion was induced by interspecific hybridization.
Figure 11. InDel mutations detected by PCR.
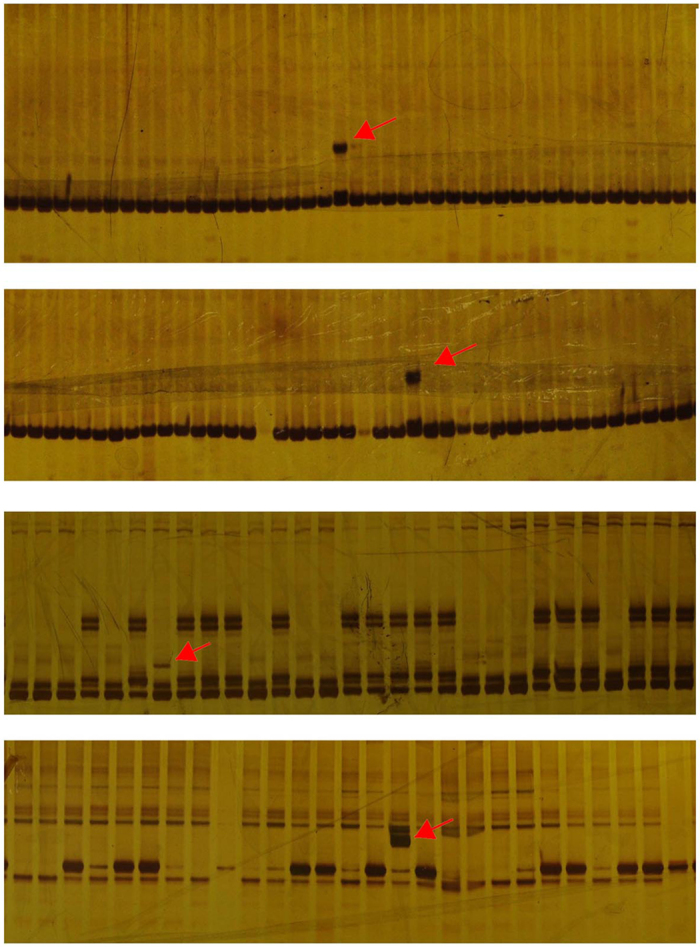
The red arrows point the mutational bands.
Discussion
Many interspecific hybrids have been produced from various types of wild and cultivated A and C genome plants15,16,17,18,19,20,21,22. However, this is the first report of the hybridization of the two flower stalk vegetables, B. oleracea var. alboglabra and B. rapa L. var. purpurea. Morphological analysis, chromosome counting, flow cytometry and SSR analysis demonstrated that hybridization was successful. The hybrid plants showed a relatively high degree of spontaneous chromosome doubling. Because the initial progenitors were highly homologous, the morphological divergence among the BC1 plants could not have been derived from genetic segregation between sister chromosomes. Mechanisms such as recombination between homoeologous chromosomes and alterations in the expression pattern and DNA methylation status of genes have been proven to lead to rapid phenotypic changes in newly synthesized allopolyploids16,17,18,19,20,21,22,28,29,30,31. The DNA methylation and expression level were not analyzed in these newly synthesized hybrids. However, homoeologous chromosome recombination was found to be active in this system. The changes in qualitative traits, such as flower color, indicate the non-homologous chromosomal recombination and gene loss took place during meiosis in the F1 plants32,33. The relatively low fertility rate of the first round of backcrosses indicated that non-homologous chromosomal recombination occurred extensively, resulting in female gamete lethality due to chromosome disorder during meiosis. This is consistent with the observation that newly synthesized allopolyploids undergo consistent chromosome crossover and recombination17,34,35.
Chromosome recombination requires homologous sequences for DNA pairing. Sequencing of cabbages and Chinese cabbages has shown that the A and C genomes share many homologous blocks10,13, presenting opportunities for recombination. Both the A and C genomes underwent two rounds of genome duplication and produced three subgenomes9,36. This type of genome redundancy provides considerable plasticity for non-homologous recombination between A and C chromosomes. These facts may be the reason for the extensive homeologous recombination displayed in the hybrid plants, which leaded to chromosome deletion, duplication and rearrangement in high frequency and finally resulted in the numerous phenotypes observed in the BC2 progenies. On the other hand, our backcrossing strategy also contributing to the high frequency homeologous recombination observed, because a complete single set of A chromosomes always obtained from the recurrent parent, which will rescue the lethal chromosome rearrangements in the maternal plants.
One well-known example of interspecific hybrid evolution is the U’s triangle model, in which three tetraploids, B. napus, B. juncea, and B. carinata, were proposed to have evolved from hybrids of any two of the three diploids B. rapa, B. nigra, and B. oleracea14. Each of the three major Brassica vegetable species, B. rapa, B. oleracea, and B. juncea, have a large number of subspecies and varieties and various crop cultivars that produce a diverse variety of edible organs. Previous studies have suggested that these plants were domesticated independently in China, Europe, and the Middle East37,38,39. Primitive types, such as kale and turnip, may have been cultivated more than 5,000 years. However, historical records indicate that most other types emerged suddenly between 800–400 years before present27. The evolutionary history of the expansion of this cultivar is still a mystery; the natural variation and selection model cannot explain this rapid speciation. Recent re-sequencing and comparison between three subspecies of B. apa, a turnip, a rapid cycling, and the reference genome of Chinese cabbage estimated the date of divergence among the three morphotypes at approximately 250,000 YA, long predating the date of domestication40. One explanation for this paradox is that the genetic variation may have come from interspecific hybridization. The present study provides solid evidence that interspecific hybridization and backcrossing have the potential to generate multiple crop types within the B. rapa and B. oleracea clans. It is here proposed that the primitive crop types of B. rapa and B. oleracea were domesticated independently in China and Europe for more than two thousand years and evolved slowly and smoothly until the 12–14th centuries, when the crops were introduced to each other between China and the Mediterranean via the Maritime Silk Road. These interspecific crossings were spontaneous and occurred in quotidian settings such as kitchen gardens. Backcrossing to B. rapa and B. oleracea crops took place in China and the Mediterranean, respectively, due to the population proportion effect. Genetic rearrangement and morphological variation manifested in the progeny of these crosses. Then, various traits were selected according to the selector’s preferences and were consequently refined and stabilized in later generations to form the diverse varieties and cultivars known today. This scenario is also supported by historical biogeographical data that all of the varieties of B. rapa were first generated in southern China, where only one Chinese native kale crop, B. oleracea var. alboglabra, coexists with pakchoi cultivars, including B. rapa L. var. purpurea27,41. Even the Chinese cabbage, which is currently planted through much of northern China, Korea, and Japan, originated in southern China and was later distributed elsewhere27. In the current model, it was not possible confirm that the two plants used in this study were the initial ancestors of current cultivars, but a trend toward a polychronism was observed, suggesting that multiple interspecific hybridization events took place and that many Brassica plants, including allotetraploid crops, could have participated in them.
Mutants are important resources for genetic research and for plant breeding. Natural mutation is rare and the mutants are difficult to obtain. Several methods, including EMS and γ-radiation, have been used to induce mutations42. The majority of these methods induce point mutations, creating loss-of-function mutants. The results of the present work showed distant crossing to be a powerful method for producing mutations, including valuable gain-of-function mutants. In distant hybrids, mutants can be generated by chromosome fragment deletion and gene conjunction, which is called “genome reshuffling,” and by activated transposable elements, which induce mutations termed “genome shock”16,17,43. It is not easy to use linkage-map-based cloning on these types of mutants, so they have not been widely used for functional genetic analysis. Currently, with the rapid development of sequencing technology and the accompanying dramatic reduction in cost, these mutants may be more frequently utilized in genetic studies.
Beside the large chromosome fragment rearrangements, minor genomic alteration such as short stretch elimination and insertion also play important roles in the evolution of interspecific hybrid plants30. Though only four InDel mutations have been detected in this study, it is far from reflecting the true mutation level in these plants. Because SSR is not an effective mutation detecting tool and it cannot detect the point mutations.
The use of distant cross technology in the plant breeding industry have a long and glorious history. Many important agronomic traits, such as male sterility, disease resistance, and stress tolerance have been introduced to cultivars from wild and interspecific relatives by distant crossing44,45,46,47,48. However, crop diversity is currently in sharp decline, and current vegetable innovation cannot match that of the pre-industrial age. One reason for this trend is that commercial seeds are highly homogeneous; thus, new traits are not likely to occur nor to be adopted to form the new varieties. The present study shows that artificial distant hybridization and backcrossing can generate new traits at a relatively high frequency. Some of the new traits have the potential to breed new vegetable cultivars. For this reason, it is here proposed that breeders use distant hybridization and backcrossing strategies to generate new types of crops for the enrichment of the human diet rather than limiting themselves to the transfer of existing traits from one plant to another for the minor modification of commercial cultivars.
Among the BC3 plants, typical traits such as enlarged corms, bud balls, wrinkled rosette leaves, leaf folding, mono-cotyledon, and root-born seedlings were also observed, indicating that these traits are heritable rather than epigenetic or due to phenotypic plasticity. However, because the maternal progenitor B. oleracea var. alboglabra is a cytoplasmic male sterile line, their progeny cannot self, so the traits cannot be reproduced stably. After generations of backcrossing, the traits are gradually lost and the phenotype finally approaches that of the recurrent parent B. rapa L. var. purpurea. This study is to be repeated using a fertile system, and further stabilize and manifest the traits by selfing and sister crossing, which will provide stronger support for the hypothesis raised in this study.
Material and Methods
Plant material and growth conditions
B. oleracea var. alboglabra 4E286 is a cytoplasmic male sterile line. The B. rapa L. var. purpurea HCT3 is an inbred line. The seeds of these plants and their progeny were surface-sterilized in 1% NaClO and germinated on glass Petri dishes at 25 °C. After germination, seedlings were transplanted into a mixture of peat soil (peat:moss:perlite:vermiculite soil = 3:2:1:1). The seedlings were transplanted to a greenhouse after 2 weeks and were watered and fertilized regularly.
Distant hybridization and embryo rescue
The B. oleracea var. alboglabra cytoplasmic male sterile line 4E286 served as the maternal parent. The B. rapaL. var. purpurea inbred line HCT3 served as the paternal parent. The inflorescence was bagged and hand-pollinated one day later. Eight days after pollination, the siliques were harvested and surface-sterilized in 70% ethanol for 30 s followed by 1% NaClO for 15 min and washed three times in purified, sterile water. The siliques were dissected under a stereomicroscope on a superclean bench. The embryos were cultivated in a solidified MS medium containing 2 mg/L 6-benzyladenine (6-BA) and 0.2 mg/L 1-naphthaleneacetic acid (NAA) and then placed in a tissue culture room at 25 °C with a 16:8 light-dark cycle49. The germinated seedlings were propagated on the same medium and then transplanted to a root-induction medium composed of solidified MS + 0.2 mg/L NAA. The rooted plants were transplanted to soil, kept humid for a week, and then cultivated in a greenhouse.
Morphological observation and cluster analysis
The morphological traits recorded include shape, color, the crimp and wax of the cotyledons, rosette-leaves, stem-leaves, petioles, stalks, flowers, and siliques. The plant height, architecture, bolting time, stooling and brawniness of stalk, number of lateral stalks, and flowering time were observed and measured at the seedling, vegetative, bolting, flowering, and fruiting stages. The morphological data were used for cluster analysis, and the UPGMA tree was constructed using MEGA 550.
Chromosome observation
Root tips (1 mm) were cut and incubated in 2 mM 8-hydroxyquinoline for 4 h. The tissues were fixed in Carnoy’s Fluid (3:1 ethanol:acetic acid, v/v) for 24 h, then stored in 70% ethanol at 4 °C. The tips were digested in 1 M HCl at 60 °C for 10 min and washed twice in distilled water. Then the apical meristems (i.e., the white spots at the tips) were sliced and squashed in a drop of 10% Carbol fuchsin. The chromosome spreads were observed under a microscope.
Flow cytometry analysis
Rosette leaves (1 cm2) were diced with a razor blade in 1 mL of Galbraith’s buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-(N-morpholino) propanesulfonic acid [MOPS], 0.1% (v/v) Triton X, pH = 7)51. Then the diced leaves were mixed with another 1 mL of Galbraith’s buffer and filtered with a nylon mesh (50 μm aperture). Cells were harvested from the solution in a centrifuge at 1000 rpm for 5 min, then suspended in 200 mL of 50 mg/L propidium iodide (PI) and incubated in the dark for 20 min. The DNA content was assayed on a flow cytometer (BD FACSCalibur, NJ, U.S.) with excitation at 488 nm.
SSR analysis
The SSR markers were designed based on comparison between the genomes of B. oleracea10 and B. rapa13. The genome sequences were downloaded from (ftp://bradata:zhl410ivf@brassicadb.org/Brassica_oleracea/Bol_Chromosome_V1.1/BOL.seq.lst.new.chr20110802_check.fa.gz) and (ftp://bradata:zhl410ivf@brassicadb.org/Brassica_rapa/Bra_Chromosome_V1.5/Brapa_sequence_v1.5.fa.gz). The SSRs were firstly identified in B. rapa genome using MIcroSAtellite identification tool-MISA (http://pgrc.ipk-gatersleben.de/misa/). Then the primers were design on primer 352. Subsequently, the primers were mapped on B. oleracea and B. rapa genome using Electronic PCR (e-PCR)53. The SSR primers which produced fragments of more than six nucleotides difference between the two plants and also have only one local in each genome were retained. From which, 66 primer pairs equidistributed on the A genome were manual selected and experimentally tested via PCR analysis on B. oleracea var. alboglabra and B. rapa L. var. purpurea. The 30 SSR primers (Table 1) generate clear distinguishable bands were used for further analysis. Genomic DNA was extracted from leaf tissues using the cetyl trimethylammonium bromide method. PCR amplifications were performed in a 20 μL volume containing 20–50 ng template DNA, 0.5 pmol primers, 0.5 U Taq enzyme and 1× PCR reaction buffer. Reactions were performed with an initial denaturation step of 3 min at 95 °C; followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. The PCR products were then separated on an 8% polyacrylamide gel electrophoresis and visualized with silver staining.
Additional Information
How to cite this article: Zhang, X. et al. Interspecific hybridization, polyploidization, and backcross of Brassica oleracea var. alboglabra with B. rapa var. purpurea morphologically recapitulate the evolution of Brassica vegetables. Sci. Rep. 6, 18618; doi: 10.1038/srep18618 (2016).
Acknowledgments
Many thanks to Anne-Marie Chèvre for reading this manuscript and for her valuable suggestions. This work was supported by grants from the National Key Technology R & D Program of the Ministry of Science and Technology of China (2013BAD01B04-2), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS) and the Beijing Research Station of Vegetable Crop Gene Resource and Germplasm Enhancement.
Footnotes
Author Contributions X.Z. and X.L. conceived this work, interpreted the data and written the manuscript. X.Z., T.L., M.D., J.W., Y.Q., H.W., J.S. and D.S. performed the experiments. All authors reviewed the manuscript.
References
- Jiao Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- Soltis P. S. & Soltis D. E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 97, 7051–7057 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H. The role of hybridization in evolution. Mol. Ecol. 10, 551–568 (2001). [DOI] [PubMed] [Google Scholar]
- Adams K. L. & Wendel J. F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 (2005). [DOI] [PubMed] [Google Scholar]
- Adams K. L. Evolution of duplicate gene expression in polyploid and hybrid plants. J Hered 98, 136–141 (2007). [DOI] [PubMed] [Google Scholar]
- Otto S. P. The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- Baack E. J. & Rieseberg L. H. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424 (1994). [DOI] [PubMed] [Google Scholar]
- Schranz M. E., Lysak M. A. & Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542 (2006). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H. et al. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 21, 481–490 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe G. D. et al. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant cell 26, 1925–1937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Geneti. 43, 1035–1039 (2011). [DOI] [PubMed] [Google Scholar]
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot. 7, 389–452 (1935). [Google Scholar]
- Jesske T., Olberg B., Schierholt A. & Becker H. C. Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L.: genetic diversity and hybrid yield. Theor. Appl. Genet. 126, 1053–1065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski E. et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186, 102–112 (2010). [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E. & Osborn T. C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens L. N. et al. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140, 336–348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M. E. & Osborn T. C. Novel flowering time variation in the resynthesized polyploid Brassica napus. J. Hered. 91, 242–246 (2000). [DOI] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K. & Osborn T. C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92, 7719–7723 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhong L., Wu X., Fang X. & Wang J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229, 471–483 (2009). [DOI] [PubMed] [Google Scholar]
- Szadkowski E. et al. Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol. 191, 884–894 (2011). [DOI] [PubMed] [Google Scholar]
- Song K., Osborn T. C. & Williams P. H. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) : 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestns). Theor. Appl. Genet. 79, 497–506 (1990). [DOI] [PubMed] [Google Scholar]
- Song K. M., Osborn T. C. & Williams P. H. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) : 2. Preliminary analysis of subspecies within B. rapa (syn. campestris) and B. oleracea. Theor. Appl. Genet. 76, 593–600 (1988). [DOI] [PubMed] [Google Scholar]
- von Bothmer R., Gustafsson M. & Snogerup S. Brassica sect.Brassica (Brassicaceae). Genet. Resour. Crop Ev. 42, 165–178 (1995). [Google Scholar]
- Zhao J. et al. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 110, 1301–1314 (2005). [DOI] [PubMed] [Google Scholar]
- Ye J. The development and evolution of the Chinese Cabbage in Ming and Qing Dynasties. Agricultural History of China 1, 53–60 (1991). [Google Scholar]
- Li X. et al. Instability of chromosome number and DNA methylation variation induced by hybridization and amphidiploid formation between Raphanus sativus L. and Brassica alboglabra Bailey. BMC Plant Biol. 10, 207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. Genetic and epigenetic interactions in allopolyploid plants. Plant mol. biol. 43, 387–399 (2000). [DOI] [PubMed] [Google Scholar]
- Comai L. et al. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell 12, 1551–1568 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Rapid genetic and epigenetic alterations under intergeneric genomic shock in newly synthesized Chrysanthemum morifolium x Leucanthemum paludosum hybrids (Asteraceae). Genome Biol. Evol. 6, 247–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S. D. et al. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175, 487–503 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S. D., Monod H., Eber F., Chevre A. M. & Jenczewski E. Non-random distribution of extensive chromosome rearrangements in Brassica napus depends on genome organization. Plant J. 70, 691–703 (2012). [DOI] [PubMed] [Google Scholar]
- Suay L. et al. Crossover rate between homologous chromosomes and interference are regulated by the addition of specific unpaired chromosomes in Brassica. New Phytol. 201, 645–656 (2014). [DOI] [PubMed] [Google Scholar]
- Leflon M. et al. Crossovers get a boost in Brassica allotriploid and allotetraploid hybrids. Plant Cell 22, 2253–2264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014). [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I. A. The genera of Brassiceae (Cruciferae: Brassicaceae) in the southeastern United States. Journal of the Arnold Arboretum 66, 279–351 (1985). [Google Scholar]
- Hedge I. C. In The biology and chemistry of the Cruciferae (eds Vaughn J. G., MacLeod A. J., & Jones B. M. G. ) 1–45 (London Academic Press, 1976). [Google Scholar]
- Arias T., Beilstein M. A., Tang M., McKain M. R. & Pires J. C. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 101, 86–91 (2014). [DOI] [PubMed] [Google Scholar]
- Lin K. et al. Beyond genomic variation–comparison and functional annotation of three Brassica rapa genomes: a turnip, a rapid cycling and a Chinese cabbage. BMC Genomics 15, 250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. The introduction, cultivation and evolution of Brassica oleracea vegetables in China. Studies in the History of Natural Sciences 5, 247–255 (1986). [Google Scholar]
- Shirasawa K., Hirakawa H., Nunome T., Tabata S. & Isobe S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 10.1111/pbi.12348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T. & Pires J. C. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol 186, 18–28 (2010). [DOI] [PubMed] [Google Scholar]
- Sigareva M. A. & Earle E. D. Direct transfer of a cold-tolerant Ogura male sterile cytoplasm into cabbage (Brassica oleracea ssp. capitata) via protoplast fusion. Theor. Appl. Genet. 94, 213–220 (1997). [Google Scholar]
- Sharma T. R. & Singh B. M. Transfer of resistance to Alternaria brassicae in Brassica juncea through interspecific hybridization among Brassica. J. Genet. Breed. 46, 373–378 (1992). [Google Scholar]
- Wei K., Pang S., Yang J. & Wei Z. Enhancement of cadmium tolerance and accumulation by introducing Perilla frutescens (L.) Britt var. frutescens genes in Nicotiana tabacum L. plants. Environ. Sci. Pollut. Res. Int. 22, 5405–5416 (2015). [DOI] [PubMed] [Google Scholar]
- Rygulla W. et al. Broadening the genetic basis of Verticillium longisporum resistance in Brassica napus by interspecific hybridization. Phytopathology 97, 1391–1396 (2007). [DOI] [PubMed] [Google Scholar]
- Lelivelt C. L., Leunissen E. H., Frederiks H. J., Helsper J. P. & Krens F. A. Transfer of resistance to the beet cyst nematode (Heterodera Schachtii Schm.) from Sinapis alba L. (white mustard) to the Brassica napus L. gene pool by means of sexual and somatic hybridization. Theor. Appl. Genet. 85, 688–696 (1993). [DOI] [PubMed] [Google Scholar]
- Murashige T. & Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plantarum 15, 473–479 (1962). [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- Galbraith D. W. et al. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051 (1983). [DOI] [PubMed] [Google Scholar]
- Untergasser A. et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D. Sequence mapping by electronic PCR. Genome Res. 7, 541–550 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]