Summary
Background
The SMIM1 protein carries the Vel blood group antigen, and homozygosity for a 17 bp deletion in the coding region of the SMIM1 gene represents the molecular basis of the Vel- blood group phenotype. We developed PCR-based methods for typing the SMIM1 17 bp (64-80del) gene deletion and performed a molecular screening for the Vel- blood type in German blood donors.
Methods
For SMIM1 genotyping, TaqMan-PCR and PCR-SSP methods were developed and validated using reference samples. Both methods were used for screening of donors with blood group O from southwestern Germany. Heterozygotes and homozygotes for the SMIM1 64-80del allele were serologically typed for the Vel blood group antigen. In addition, the rs1175550 SNP in SMIM1 was typed and correlated to the results of the phenotyping.
Results
Both genotyping methods, TaqMan-PCR and PCR-SSP, represent reliable methods for the detection of the SMIM1 64-80del allele. Screening of 10,598 blood group O donors revealed 5 individuals homozygous for the deletional allele. They were confirmed Vel- by serological typing. Heterozygotes for the 64-80del allele showed different antigen expressions ranging from very weak to regular positive.
Conclusion
Molecular screening of blood donors for the Vel- blood type is feasible and avoids the limitations of serological typing which might show false-negative results with heterozygous individuals. The identification of Vel- blood donors significantly contributes to the adequate blood supply of patients with anti-Vel.
Keywords: SMIM1 genotyping, Vel antigen, PCR-SSP, TaqMan-PCR, Molecular blood typing
Introduction
The Vel blood group antigen was first described in 1952 when a Vel- patient with anti-Vel developed an acute intravascular hemolytic reaction after transfusion of Vel+ red cells [1]. After further reports of acute hemolytic transfusion reactions and hemolytic disease of newborns of Vel- mothers, Vel was recognized as a clinically important blood group antigen [2,3]. The exact immunogenicity of the Vel antigen is not known but there are several requests and nation-wide searches for Vel- red cell concentrates every year in Germany. According to the report of the ISBT Working Party on Rare Donors, Vel- blood is one of the most difficult to obtain blood types in several countries (‘Global Definitions of Rare Donors’, WP Meeting ISBT Congress 2013, Amsterdam). Because of the clinical significance of anti-Vel that mostly leads to severe complement-mediated intravascular hemolysis, it is important to find enough Vel- blood donors. Up to date there are approximately 20-30 known Vel- blood donors and about 50 cryopreserved Vel- red cell concentrates in Germany. These numbers are not sufficient to provide compatible red cell units to all patients with anti-Vel. The prevalence of the Vel- blood type was found to be slightly different in the populations, i.e., 1 in 3,985 British individuals (0.025%), 1 in 2,500 Americans (0.04%) and 1 in 1,762 individuals (0.057%) in Sweden [1,3]. It was speculated whether or not the highest frequency of the Vel- blood type can be found in northern Scandinavia.
In 2013 different groups reported the genetic basis of the Vel- blood type [4,5,6]. A 17 bp frame-shift deletion (64-80del) in the coding region of the SMIM1 gene was homozygous in all Vel- samples. The reduced or weak expression of the Vel antigen was associated with a heterozygous 64-80del genotype [4,5]. The International Society of Blood Transfusion (ISBT) Working Party for Red Cell Immunogenetics and Blood Group Terminology assigned the blood group system name VEL and number 034 and promoted Vel from a genetically unresolved blood group antigen (ISBT number 212001) to the new system defined by the SMIM1 locus (antigen number 034001). No other null allele of the SMIM1 gene has been reported so far. Thus, molecular screening for the Vel- blood could be simply based on the specific detection of the SMIM1 64-80del allele.
Here, we describe PCR-based methods for genotyping the 17 bp deletion in the SMIM1 gene underlying the Vel- blood group phenotype. We screened 10,598 blood group O donors for the SMIM1 64-80del allele and confirmed the Vel- donors to be negative by serological testing.
Material and Methods
DNA Samples of Blood Donors
This study was performed in a blood donor cohort from the southwestern part of Germany. Donors gave written consent to provide blood samples for research purposes, and the use of blood samples for research purposes was approved by the ethics committee of the Heidelberg University, Medical Faculty Mannheim. DNA was isolated from EDTA-anticoagulated blood using a commercial system (QIAamp Blood DNA Mini Kit; Qiagen, Hilden, Germany).
Serological Vel Typing
The serological testing for the Vel antigen was performed using the indirect antiglobulin test in the gel technique (ID cards, Bio-Rad Cressier, Switzerland; and ScanGel, BioRad, Marnes-la-Coquette, France) using a polyclonal anti-Vel serum from an immunized patient. To enhance the reactions bithermal incubation (15 min at 37 °C and additional 15 min at room temperature) was applied. The gel cards were centrifuged for 10 min at 85 × g and visually inspected for the agglutination. The serological testing was followed by re-testing of the phenotyped blood using PCR with sequence-specific primers (PCR-SSP) to verify the SMIM1 genotype.
SMIM1 Genotyping
For genotyping the 17 bp deletion in SMIM1 specific for the Vel blood group we developed a PCR-SSP and a PCR with fluorescent allele-specific TaqMan probes. In addition, a PCR-SSP method was developed for genotyping the rs1175550 SNP in SMIM1 that was reported to be associated with expression levels of the Vel antigen [6]. The sequences of primers and probes are listed in table 1. Both methods were validated using phenotyped samples including two samples with the Vel- blood group phenotype.
Table 1.
Oligonucleotides for SMIM1 genotyping
| Name | Allele specificity | Direction | Sequence (5'–3') | Amplicon size, bp |
|---|---|---|---|---|
| PCR-SSP | ||||
| SMIMlwt-F | wild type | sense | GCAGCAGGGACGGAGTCA | 216 |
| SMIM1del-F | 64-80del | sense | GCAGCAGGGACGGAGTCC | 199 |
| SMIM1-R | SMIM1 | antisense | CCAAAGGCTGCGGTTTGCTG | |
| Rs1175550-F1a | Major A | sense | TCAGGGGCTGCAGCCTAGA | 160 |
| Rs1175550-F1g | minor G | sense | TCAGGGGCTGCAGCCTAGG | 160 |
| Rs1175550-R1 | SMIM1 | antisense | CTCAGGCCAAGCCCTGACC | |
| TaqMan PCR | ||||
| SMIM1-F1 | SMIM1 | sense | GCCACGTCCACTATAGTAGG | 142/125 |
| SMIM1-R1 | SMIM1 | antisense | GGAGACACCAGCCTGCTATG | |
| SMIM1wt-P | wild type | sense | VIC-CTAGGGGCTGTGTC-MGB* | |
| SMIM1del-P | 64-80del | sense | FAM-ACGGAGTCCAGCACAG-MGB* |
TaqMan probe modifications: FAM, VIC, minor groove binding (MGB).
For PCR-SSP part of the HBB gene (β-globin) was co-amplified as an internal control PCR fragment (536 bp) using primers β-glob-F (5ʹ-GGTTGGCCAATCTACTCCCAGG-3ʹ) and β-glob-R (5ʹ-GCTCACTCAGTGTGGCAAAG-3ʹ). Genomic DNA (10-50 ng) was subjected to 10 μl PCR reactions including 1 μmol/l SMIM1wt-F primer (for the SMIM1 wild-type allele) or SMIM1del17-F (for the SMIM1 64-80del allele), 1 µmol/l SMIM1-R primer, 0.2 μmol/l each internal control primer, 10 mmol/l Tris-HCl (pH 8.3), 50 mmol/l KCl, 1.5 mmol/l MgCl2, 200 μmol/l each dNTP, and 0.5 units Taq DNA polymerase (Bioron GmbH, Ludwigshafen, Germany). The cycling conditions were: 2 min initial denaturation at 95 °C, followed by 10 cycles with 20 s denaturation at 95 °C and 1 min annealing/extension at 65 °C, followed by 20 cycles with 20 s denaturation at 95 °C, 1 min annealing at 61 °C and 30 s extension at 72 °C. Amplification products were separated on 2 % agarose gels containing GelRed DNA stain (Biotium, Hayward, CA, USA). Results were documented using a UV gel documentation device (UVP Inc., Upland, CA, USA). The same protocol was used for the rs1775550 genotyping.
For TaqMan-PCR, 20-50 ng genomic DNA was mixed with 0.9 µmol/l each of the SMIM1-F1 and SMIM-R1 primers, 0.2 µmol/l each of the TaqMan probes in a final volume of 7.5 µl. After addition of 7.5 µl HotRox mastermix (Bioron GmbH) the background fluorescence (pre-read) was detected using a RealTime PCR cycler (ABI 7000; Applied Biosystems, Waltham, MA, USA). Cycling was performed in a standard PCR cycler using the following program: 10 min at 95 °C, 40 cycles with 15 s at 92 °C and 1 min at 60 °C. After cycling the fluorescence signals were detected in a post-read and allocated with the pre-read data. Values indicating heterozygous and homozygous del17 alleles were determined using reference samples.
Results
Molecular Screening for the Vel- Blood Type
The PCR-SSP and TaqMan-PCR methods revealed the expected results from the two Vel- reference samples, i.e., the SMIM1 wild-type allele was absent, and only the 64-80del allele could be detected. In addition, all samples positive for the Vel antigen were homozygous or heterozygous for the SMIM1 wild-type allele.
Screening for the SMIM1 64-80del allele causing the Vel- phenotype by using the PCR-SSP method was performed in a two-step procedure (fig. 1). In the first step the DNA samples were analyzed only using the primers specific for the 64-80del allele. In the second step only the positive DNA samples from the first step were re-analyzed using both the primers for the wild-type allele and the 64-80del allele. Because the TaqMan-PCR method detects both alleles simultaneously, it determines the genotype of a DNA sample by a single reaction (fig. 2).
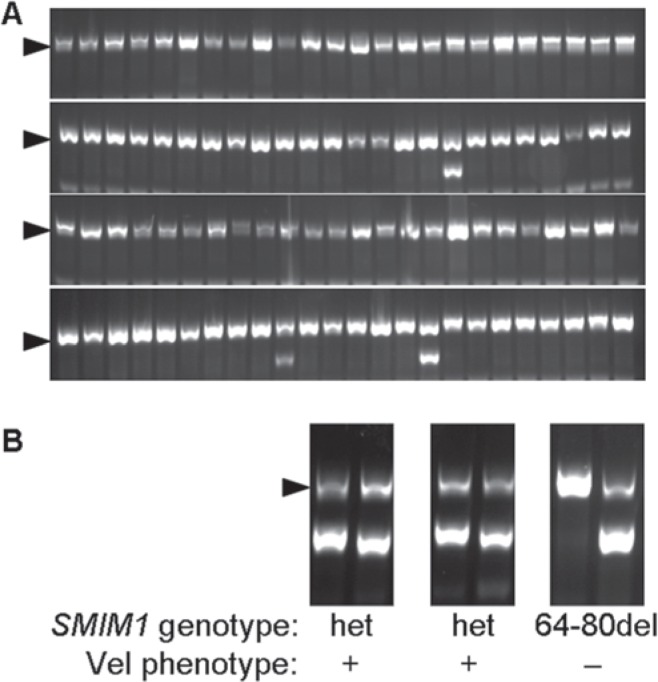
Fig. 1.
A Representative results from screening of 96 DNA samples for the SMIM1 64-80del allele by using PCR-SSP. Most DNA samples were negative because only the internal control fragment (arrow head) was amplified. B Three positive DNA samples were re-analyzed using both the primers for the wild-type allele and the 64-80del allele. One of the three DNA samples was negative for the wild-type allele indicating a homozygous 64-80del genotype.
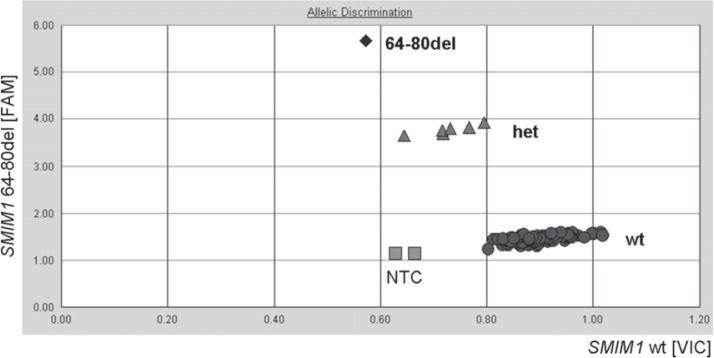
Fig. 2.
Representative results from screening of 94 DNA samples for the SMIM1 genotype by using TaqMan-PCR. The simultaneous detection of the wild-type allele in the VIC channel (X-axis) and the 64-80del allele in the FAM channel (Y-axis) enables the discrimination of the three genotypes: homozygous wild type (circles), heterozygotes (triangles), homozygous 64-80del (diamonds). NTC = No template control.
Both methods were used to screen for 64-80del homozygotes in 10,598 blood donors (table 2). The observed genotype frequencies showed no significant deviation from the Hardy-Weinberg equilibrium. However, based on the frequency of the 64-80del allele of 1.81% (383 of 21,196 SMIM1 alleles analyzed) we would expect a prevalence for the homozygous genotype of 1 in 3,063 blood donors in our study cohort. We identified 5 donors (0.05%) with the homozygous 64-80del genotype, indicating a prevalence of 1 in 2,120 donors.
Table 2.
Results from Vel genotyping of 10,598 blood donors
|
SMIM1 alleles |
SMIM1 genotypes |
Vel phenotype* |
|||||
|---|---|---|---|---|---|---|---|
| wt | 64-80del | wt | het | 64-80del | Vel+ | Vel− | |
| Numbers | 20,813 | 383 | 10,220 | 373 | 5 | 10,593 | 5 |
| % | 98.19 | 1.81 | 96.43 | 3.52 | 0.05 | 99.95 | 0.05 |
Deduced from genotype.
Vel Phenotyping
Expression of the Vel blood group antigen was determined in 45 genotyped samples using a gel-based agglutination assay. The phenotype was clearly Vel+ in homozygotes for the SMIM1 wild-type allele (n = 2) and clearly Vel- in homozygotes for the 64-80del allele (n = 4). Among the 39 SMIM1 heterozygotes, we found a broad range of the Vel antigen strength in the agglutination assay (fig. 3). Using ID gel cards our anti-Vel serum failed to detect the Vel antigen in 11 of the 39 (28.2%) 64-80del heterozygotes. In the more sensitive ScanGel cards all heterozygotes showed at least a very weak agglutination. The DNA marker rs1175550 is a SNP (A>G) in the non-coding region of the SMIM1 gene and is significantly associated with weak expression of the Vel antigen [6]. We genotyped rs1175550 in the 45 Vel-phenotyped samples and could confirm a significantly higher expression of the Vel antigen in carriers (n = 5) of the minor G allele of rs1175550 (fig. 3).
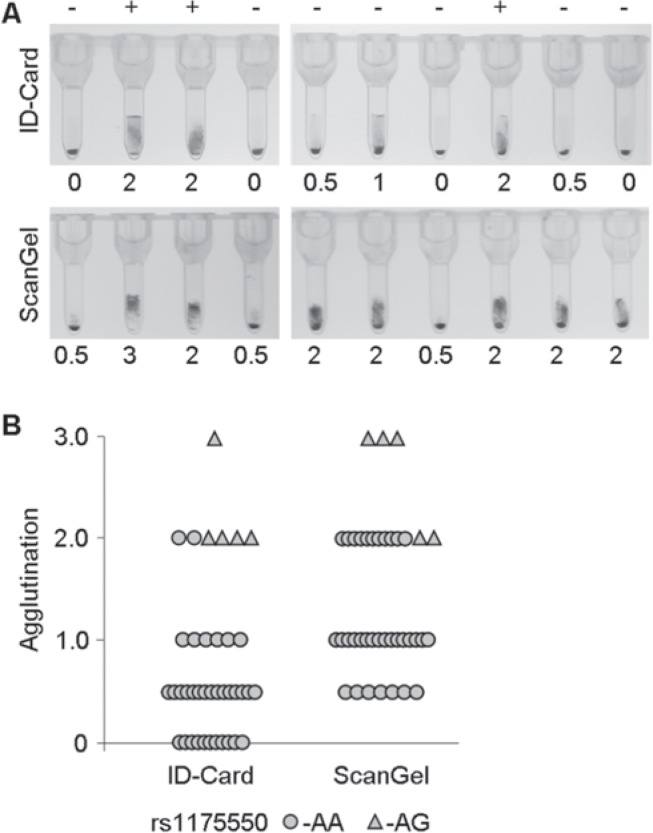
Fig. 3.
Serological typing Vel of blood donors heterozygous for the SMIM1 64-80del allele. A Representative results from phenotyping using an anti-Vel patient serum and gel-based agglutination. The same samples were analyzed using ID-Cards (upper panel) and ScanGel cards (lower panel). Agglutination was categorized from negative (0) to very weak (0.5), weak (1.0), moderate (2.0) and regular (3.0) positive. Absence (-) or presence (+) of the minor G-allele of the rs1175550 SNP is indicated above. B Summary of the agglutination of 39 SMIM1 64-80del heterozygous blood donors in the two gel cards and under consideration of the rs1175550 genotype. The minor G-allele (genotype AG) was found in 5 of the samples, 34 samples were homozygous for the major A-allele. The G-allele was significantly associated with a stronger agglutination. None of the samples were negative in the ScanGel cards indicating a higher sensitivity in Vel phenotyping.
Discussion
As expected, the wild-type allele of the SMIM1 gene defining the Vel blood group antigen was highly prevalent in our study cohort of 10,598 blood donors. More than 99.9% of the donors were either homozygous or heterozygous for the wild-type allele. In our approach using PCR-SSP or TaqMan-PCR the SMIM1 wild-type allele was detected by the absence of the 64-80 deletion. We cannot exclude other deletions or point mutations; however, no other genetic mechanism for the Vel- blood type than the SMIM1 64-80del allele has been reported so far. Therefore, it is most likely that all donors in our study homozygous or heterozygous for the SMIM1 wild-type allele express the Vel antigen. Five donors were homozygous for the 17 bp deletion, and we could confirm their Vel- phenotype, except for one donor who was not available for a fresh blood sample any more. To the best of our knowledge and based on the donor registry data the 5 donors were not related.
The frequency of the SMIM1 64-80del allele of 1.81% in our study cohort corresponds to the observations of approximately 1.6% in other European populations [6,7]. The highest allele frequencies were reported for Scandinavian countries, e.g. 2.9% in Swedish blood donors [5], whereas African blacks and Asian donors revealed allele frequencies of only 0.56 and 0.6%, respectively [7]. In all populations, the homozygous 64-80del genotype correlated with the Vel- phenotype.
We found individual differences in the Vel antigen expression in donors heterozygous for the 64-80del allele ranging from regular positive to very weak positive (+/-). In general, phenotyping very much depends on the quality of anti-Vel serum and on the sensitivity of the test technique. Because anti-Vel sera of immunized patients mostly contain a high IgM fraction, the results in the ScanGel cards which were known to be very sensitive for IgM antibodies gave positive reactions with all heterozygous Vel samples even though the widely used ID cards were negative with some of them. Furthermore, genetic factors such as the rs1175550 SNP in the non-coding region of SMIM1 can affect the expression of the Vel antigen [6,7]. We could confirm the association of the minor G allele of rs1175550 with a significantly stronger antigen expression in 5 blood donors, who were presumably compound heterozygous for 64-80del and the rs1175550-G alleles. However, this additional genetic factor could not explain all variability in Vel antigen expression of 64-80del heterozygotes.
In summary, the molecular screening for Vel- blood donors can be performed by fast and robust PCR methods such as PCR-SSP or TaqMan-PCR. It is more reliable than serological screening and contributes significantly to supplying compatible blood for patients with anti-Vel. In our cohort we found a prevalence of 1 in approximately 2,000 donors of the Vel- genotype. Because of the significant clinical relevance of the Vel antigen, it is highly recommended to screen for and to register Vel- blood donors, irrespective of the size of the blood bank. Of course, cost issues for molecular or serological screening need to be considered and balanced against costs of cryopreservation and storage of Vel- RBC units.
Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Sussman LN, Miller EB. New blood factor: Vel. Rev Hematol. 1952;7:368–371. [PubMed] [Google Scholar]
- 2.Sussman LN. Current status of the Vel blood group system. Transfusion. 1962;2:163–171. doi: 10.1111/j.1537-2995.1962.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 3.Daniels G. Human Blood Groups. Blackwell Science Ltd: Oxford; 2002. [Google Scholar]
- 4.Ballif BA, Helias V, Peyrard T, Menanteau C, Saison C, Lucien N, Bourgouin S, Le Gall M, Cartron JP, Arnaud L. Disruption of SMIM1 causes the Vel- blood type. EMBO Mol Med. 2013;5:751–761. doi: 10.1002/emmm.201302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storry JR, Jöud M, Christophersen MK, Thuresson B, Åkerström B, Sojka BN, Nilsson B, Olsson ML. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat Genet. 2013;45:537–541. doi: 10.1038/ng.2600. [DOI] [PubMed] [Google Scholar]
- 6.Cvejic A, Haer-Wigman L, Stephens JC, Kostadima M, Smethurst PA, Frontini M, van den Akker E, Bertone P, Bielczyk-Maczyńska E, Farrow S, Fehrmann RS, Gray A, de Haas M, Haver VG, Jordan G, Karjalainen J, Kerstens HH, Kiddle G, Lloyd-Jones H, Needs M, Poole J, Soussan AA, Rendon A, Rieneck K, Sambrook JG, Schepers H, Silljé HH, Sipos B, Swinkels D, Tamuri AU, Verweij N, Watkins NA, Westra HJ, Stemple D, Franke L, Soranzo N, Stunnenberg HG, Goldman N, van der Harst P, van der Schoot CE, Ouwehand WH, Albers CA. SMIM1 underlies the Vel blood group and influences red blood cell traits. Nat Genet. 2013;45:542–545. doi: 10.1038/ng.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haer-Wigman L, Stegmann TC, Solati S, Ait Soussan A, Beckers E, van der Harst P, van Hulst-Sundermeijer M, Ligthart P, van Rhenen D, Schepers H, de Haas M, van der Schoot CE. Impact of genetic variation in the SMIM1 gene on Vel expression levels. Transfusion. 2015;55:1457–1466. doi: 10.1111/trf.13014. [DOI] [PubMed] [Google Scholar]