Abstract
An international study was performed by 26 experienced PCR laboratories from 14 countries to assess the performance of duplex quantitative real-time PCR (qPCR) strategies on the basis of TaqMan probes for detection and quantification of parasitic loads in peripheral blood samples from Chagas disease patients. Two methods were studied: Satellite DNA (SatDNA) qPCR and kinetoplastid DNA (kDNA) qPCR. Both methods included an internal amplification control. Reportable range, analytical sensitivity, limits of detection and quantification, and precision were estimated according to international guidelines. In addition, inclusivity and exclusivity were estimated with DNA from stocks representing the different Trypanosoma cruzi discrete typing units and Trypanosoma rangeli and Leishmania spp. Both methods were challenged against 156 blood samples provided by the participant laboratories, including samples from acute and chronic patients with varied clinical findings, infected by oral route or vectorial transmission. kDNA qPCR showed better analytical sensitivity than SatDNA qPCR with limits of detection of 0.23 and 0.70 parasite equivalents/mL, respectively. Analyses of clinical samples revealed a high concordance in terms of sensitivity and parasitic loads determined by both SatDNA and kDNA qPCRs. This effort is a major step toward international validation of qPCR methods for the quantification of T. cruzi DNA in human blood samples, aiming to provide an accurate surrogate biomarker for diagnosis and treatment monitoring for patients with Chagas disease.
Chagas disease (CD), caused by the protozoan Trypanosoma cruzi, affects mostly the poor populations in 21 countries of the Americas, where close to 7 to 8 million people are infected, 25 million are at risk, and 10 thousand deaths are recorded annually (World Health Organization, www.who.int/mediacentre/factsheets/fs340/en, last accessed November 1, 2014).1 In recent years, this neglected tropical disease is becoming a global concern because of the increasing migration from Latin America to nonendemic countries from Europe and North America.2
Complex interactions between the genetic background of the parasite and the host and environmental and epidemiologic factors determine the outcome of the infection. In the acute phase of CD the symptoms are variable, and in most cases resolve spontaneously after some weeks. Appropriate treatment can eliminate the parasite during this phase, but the infection is only recognized in 1% to 2% of infected persons during the acute phase. In the chronic phase, approximately 70% of seropositive persons are asymptomatic, whereas 30% ultimately develop serious cardiac and/or digestive disorders several years or decades later, and necrotizing inflammatory injuries in the central nervous system in cases of CD reactivation under immunodepression. Each year, 2% to 3% of symptomatic persons start to present manifestations that can rapidly evolve to sudden death. However, the factors that govern the progression of chronic CD remain unknown, and no prognostic markers are available.3
Accurate diagnostics tools and surrogate markers of parasitologic response to treatment are priorities in CD research and development.4 To develop an accurate laboratory tool for diagnosis and treatment follow-up, several difficulties need to be addressed, such as the low and intermittent number of circulating parasites during the chronic phase of infection and parasite genotype diversity, because six discrete typing units (DTUs), TcI to TcVI, are unevenly distributed in different endemic regions.5 Quantitative real-time PCR (qPCR)-based assays may fill these gaps, but their application in the clinical practice requires prior analytical and clinical validation studies.6,7 So far, a few real-time PCR strategies have been developed for T. cruzi DNA detection and quantification in CD patients.8–11
As part of the Small Grants Programme (joined initiative of Communicable Diseases Research/Pan-American Health Organization) and The Special Programme for Research and Training in Tropical Diseases/United Nations Development Program/United Nations Children’s Fund/World Bank/World Health Organization, an international study was performed by 26 experienced PCR laboratories from 14 countries to assess the performance of duplex qPCR strategies on the basis of TaqMan probes for detection and quantification of the parasite loads in blood samples of CD patients.
Materials and Methods
Ethics Statement
The studies in which the samples were collected were approved by the ethical committees of the participating institutions, according to the principles expressed in the Declaration of Helsinki. Written informed consent forms were signed by the adult study subjects and from parents/guardians on behalf of all minor subjects. All samples were pre-existent at the time of this international study and were anonymized before being processed.
Spiked Blood Samples
Seronegative human blood samples were spiked with cultured epimastigotes of Sylvio X10 and CL-Brener stocks (TcId and TcVI, respectively) and were immediately mixed with one volume of guanidine hydrochloride 6 mol/L EDTA 0.2 mol/L buffer, pH 8.00 (GE).
Patients and Blood Specimens
Peripheral blood samples from 156 CD patients were distributed into eight groups according to their geographic origin, as follows. Group 1 (G1) included samples from four seropositive patients from Mexico, two patients with acute CD (G1a) and two patients with asymptomatic chronic CD (G1b). Group 2 (G2) included samples from two patients from French Guiana with acute CD acquired by oral transmission. One patient was positive and the other patient was negative for IgG serologic studies. Both patients experienced cardiac symptoms and were infected with TcI. Group 3 (G3) included samples from five seropositive patients from Bolivia; two patients with acute CD acquired by oral transmission (G3a) and three patients with asymptomatic chronic CD acquired from vectors or congenitally (G3b). Group 4 (G4) included samples from five seropositive patients from Venezuela with acute CD acquired by oral transmission. Two of these patients were infected with TcI. Group 5 (G5) included samples from 13 seropositive patients from Colombia with asymptomatic chronic CD acquired from vectors or congenitally. Five of these patients were infected with TcI. Group 6 (G6) included samples from 21 Bolivian seropositive patients, resident in Spain, with chronic CD acquired from vectors or congenitally. One patient experienced digestive symptoms and the others were asymptomatic. Group 7 (G7) included samples from 31 seropositive patients from Brazil with chronic CD and the following clinical manifestations: asymptomatic (n = 5), cardiac (n = 17), digestive (n = 2), and cardiodigestive (n = 7). Thirteen patients were infected with TcII. Group 8 (G8) included samples from 75 seropositive patients from Argentina with chronic CD and the following clinical manifestations: asymptomatic (n = 27), cardiac (n = 34), digestive (n = 1), and cardiodigestive (n = 13). Fifty-one patients were infected with TcV or TcVI or combinations of TcII, TcV, and TcVI.
In addition, samples from 50 persons from Argentina with negative serology for T. cruzi were included as negative controls to address the specificity of the procedures.
DNA Extraction
The blood samples were obtained and immediately mixed with an equal volume of GE (GEB). After 48 to 72 hours at room temperature GEB samples were boiled for 15 minutes (except for G3 and G5 groups and seronegative samples) and stored at 4°C for DNA extraction and PCR analysis.
GEB samples were processed with the High Pure PCR Template Preparation kit (Roche Diagnostics Corp., Indianapolis, IN) as described in Duffy et al.9 Those samples with cycle threshold (Ct) values lower than the Ct values for the most concentrated point of the standard curve were properly diluted in seronegative human blood treated with GE, and DNA extraction and qPCR procedures were repeated. To build the standard curves for quantification of parasitic loads, DNA from spiked blood samples were obtained in the same way as reported for the clinical samples. The DNA eluate was stored at −20°C until use in qPCR analysis.
Duplex Real-Time PCR Procedures
Two duplex qPCR procedures were compared, Satellite DNA (SatDNA) qPCR and kinetoplastid DNA (kDNA) qPCR assays. The former targets the satellite sequence of the nuclear genome of the parasite and the sequence of an internal amplification control (IAC) as described in Duffy et al,9 and the latter is a modification of the method reported by Qvarnstrom et al11 which targets the conserved region of the minicircle parasite sequences with the addition of primers and TaqMan probe for the IAC.9 Both reactions were performed with 5 μL of re-suspended DNA, using FastStart Universal Probe Master Mix (Roche Diagnostics GmbHCorp., Mannheim, Germany) in a final volume of 20 μL.
Optimal cycling conditions for both qPCR assays were a first step of 10 minutes at 95°C, followed by 40 cycles at 95°C for 15 seconds and 58°C for 1 minute. The amplifications were performed using Rotor-Gene 6000 (Corbett Life Science, Cambridgeshire, United Kingdom) or ABI7500 (Applied Biosystems, Foster City, CA) devices.
Standard curves were plotted with 1/10 serial dilutions of total DNA obtained from a GEB-seronegative sample spiked with 105 parasite equivalents per milliliter of blood (par. eq./mL). TcId- and TcVI-DNA–based standard curves were used to quantify parasitic loads in G1a, G1b, G2, G3a, G4, and G5 and in G3b, G6, G7, and G8 samples, respectively.
Duplex Real-Time PCR Assays Performance
Terms
On the basis of the MICROVAL protocol,12 the analytical validation of both qPCR methods included the following parameters. i) Selectivity is defined as a measure of the degree of response from target and nontarget microorganisms and comprises inclusivity and exclusivity. Inclusivity is the ability of an alternative method (each qPCR assay in this case) to detect the target pathogen from different strains (DTUs in this case), and exclusivity is the lack of response from closely related but nontarget strains (other trypanosomatids in this case). ii) Reportable range is a set of values of measurands for which the error of a measuring instrument is intended to lie within specified limits. iii) Limit of detection (LOD) is the smallest amount that the method can reliably detect to determine the presence or absence of an analyte. iv) Precision is the closeness of agreement between independent test/measurement results obtained under stipulated conditions. v) Limit of quantification (LOQ) is the smallest amount that the method can reliably measure quantitatively.
The above-mentioned parameters were evaluated in the framework of the international study as described in the sections below.
Inclusivity
Both qPCR assays were tested with genomic DNA obtained from a panel of T. cruzi stocks belonging to the six different DTUs, plus TcI stocks representative of three different TcI Spliced Leader Intergenic Region (SL-IR)–based groups (TcIa, TcId, and TcIe), in concentrations that ranged from 0.0625 to 10 fg/μL tested on duplicates: TcI [K98 (TcIa SL-IR–based group), G (TcId group), and SE9V (TcIe group) stocks]13–16; TcII (Tu18 stock), TcIII (M5361 stock), TcIV (CanIII stock), TcV (PAH265 stock), and TcVI (CL-Brener stock).17
Exclusivity
Serial dilutions of purified DNAs from Trypanosoma rangeli and Leishmania major, Leishmania mexicana and Leishmania amazonensis that ranged from 0.1 fg/μL to 1000 pg/μL were assayed on duplicates by both qPCRs.
Reportable Range
A panel of GEB samples spiked with Sylvio X10 (TcId) and CL-Brener (TcVI) stocks, spanning 105 to 0.0625 par. eq./mL was prepared. After DNA purification, each dilution was amplified on triplicate by both qPCR assays. Assigned (theoretical) versus measured values were converted to Log10 par. eq./10 mL and plotted for linear regression analysis.
LOD
The LOD was calculated as the lowest parasitic load that gives ≥95% of qPCR detectable results, according to the Clinical and Laboratory Standards Institute guidelines.18 It was measured from two panels of GEB samples spiked with the CL-Brener stock; one panel was boiled for 15 minutes before preparing serial dilutions,7 and the other panel was diluted without prior boiling. For both panels, eight replicates from GEB dilutions that contained 0.125, 0.25, 0.5, and 1 par. eq./mL for SatDNA qPCR and 0.0625, 0.125, 0.25, and 0.5 par. eq./mL for kDNA qPCR were purified and amplified during five consecutive days. The LOD was determined by Probit regression analysis with Minitab 15 Statistical Software (Minitab Inc., State College, PA).
Precision
Precision experiments were performed with spiked GEB samples at concentrations of 0.5, 10, and 103 par. eq./mL (0.699, 2, and 4 Log10 par. eq./10 mL) for SatDNA qPCR and 0.25, 10, and 103 par. eq./mL (0.398, 2, and 4 Log10 par. eq./10 mL) for kDNA qPCR, assayed on duplicates during 20 consecutive experiments, one run per day, according to the Clinical and Laboratory Standards Institute guidelines.19 The estimates of within-laboratory precision SDs (St) were calculated with the following formula: , where B is the SD of the daily means and Sr is the estimate of repeatability SD (within-run precision).
LOQ
For both qPCR methods, the LOQ was derived from a 20% threshold value for the CV of measurements obtained in the precision experiments, as described in Schwarz et al.20 Assuming an exponential decrease in CV, a curve for the relation between CV and Log10 par. eq./10 mL was fitted with SigmaPlot 10.0 (Systat Software Inc., San Jose, CA).
Quality Controls for Analysis of Clinical Specimens
A negative control and two positive controls that contained different concentrations of T. cruzi DNA (namely a high-positive control and a low-positive control near the limit of detection) were included in every run as recommended.6
Internal Amplification Control
A pZErO-2 recombinant plasmid that contains an inserted sequence of Arabidopsis thaliana aquaporin was used as a heterologous extrinsic IAC.8
RNase P Assay
To check for DNA integrity, clinical samples were tested with TaqMan RNase P Control Reagents Kit (Applied Biosystems) in an ABI7500 Real-Time PCR device.
T. cruzi Genotyping
SatDNA qPCR quantifiable samples collected from laboratories that did not perform T. cruzi DTU typing (G1, G3, and G6) were genotyped with PCR-based strategies targeted to nuclear genomic markers, as described in Burgos et al.21
Evaluation of Parasitemia with Hemoculture
The 15 samples provided by the laboratory of Disciplina de Parasitología, Universidade Federal do Triângulo Mineiro (Uberaba, Brazil), were evaluated by hemoculture. The assay was performed according to the method described in Chiari et al.22 Immediately after collection, 30 mL of blood was centrifuged at 4°C to remove plasma. The packed cells were washed by centrifugation at 4°C in liver infusion tryptose medium. The sedimented erythrocytes were resuspended in 30 mL of liver infusion tryptose medium and uniformly distributed in six test tubes. Cultures were maintained at 28°C and homogenized weekly. The culture was microscopically examined 30, 60, and 90 days after culture in 10-μL aliquots of suspension.
Statistical Analysis
The Cohen κ coefficient23 was used to compare the clinical sensitivity of SatDNA and kDNA qPCRs for the detection of T. cruzi DNA in samples from chronic CD patients. The unpaired t-test was used to compare the means of parasitic loads of quantifiable samples from acute versus chronic CD patients and asymptomatic versus symptomatic chronic CD patients for both qPCR methods. In addition, nonparametric analysis of variance was used to compare the parasitic loads of quantifiable samples grouped according to their T. cruzi genotypes for each qPCR assay. Bland-Altman bias plot6 was used to analyze the closeness of the agreement between the quantifiable results of both qPCR methods. Finally, the paired t-test was used to compare the means of IAC Ct values of both qPCR assays, and the Tukey criterion24 was used to detect samples with outlier Ct values of IAC (Ct > 75th percentile + 1.5× interquartile distance of median Ct), which indicated PCR inhibition or material loss during sample DNA extractions.
Results
Analytical Validation of qPCR Methods
The analytical validation results obtained for both qPCR assays are shown in Table 1. For the inclusivity, qPCR methods were assayed with DNA from strains that represented the six T. cruzi DTUs, plus TcI stocks representative of three different TcI SL-IR–based groups (TcIa, TcId, and TcIe).
Table 1.
Analytical Validation Results Obtained for qPCR Assays
| Validation Parameters | SatDNA qPCR* | kDNA qPCR |
|---|---|---|
| Inclusivity (detectable qPCR), fg/μL | ||
| TcIa | 0.0625 | 0.0625 |
| TcId | 0.25 | 0.0625 |
| TcIe | 1 | 0.0625 |
| TcII | 0.0625 | 0.0625 |
| TcIII | 0.0625 | 0.0625 |
| TcIV | 0.25 | 0.0625 |
| TcV | 0.0625 | 0.0625 |
| TcVI | 0.0625 | 0.0625 |
| Exclusivity (nondetectable qPCR), pg/μL | ||
| T. rangeli | 1 | 0.001 |
| L. major | 1000 | 1000 |
| L. mexicana | 1000 | 1000 |
| L. amazonensis | 1000 | 1000 |
| Reportable range, par. eq./mL | ||
| TcId | 105–1 | 105–1 |
| TcVI | 105–0.25 | 105–0.25 |
| Limit of detection, par. eq./mL | ||
| Boiled | 0.46 | 0.16 |
| Nonboiled | 0.70 | 0.23 |
| Precision, CV % | ||
| 0.25 par. eq./mL | ND | 31.98 |
| 0.5 par. eq./mL | 46.60 | ND |
| 10 par. eq./mL | 6.00 | 8.79 |
| 1000 par. eq./mL | 1.72 | 2.92 |
| Limit of quantification, par. eq./mL | 1.53 | 0.90 |
Data for SatDNA qPCR were taken from Duffy et al.9
kDNA, kinetoplastid DNA; ND, not done; par. eq./mL, parasite equivalents in 1 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
kDNA qPCR gave the same analytical sensitivity of 0.0625 fg/μL DNA, the lowest concentration tested, for all T. cruzi stocks analyzed. SatDNA qPCR gave an analytical sensitivity of 0.0625 fg/μL DNA for stocks representative of TcIa, TcII, TcIII, TcV, and TcVI; 0.25 fg/μL DNA for stocks belonging to TcId and TcIV; and 1 fg/μL DNA for the TcIe stock.
Exclusivity was assayed in T. rangeli and Leishmania spp. Both qPCR methods were nondetectable when up to 1000 pg/μL DNA, the highest concentration tested, from Leishmania stocks was analyzed. In the case of T. rangeli, 10 fg/μL DNA could be amplified by kDNA qPCR, whereas SatDNA qPCR required an input of at least 10 pg/μL T. rangeli DNA to obtain a detectable PCR result.
The reportable range was determined with 10 spiked GEB samples that contained serial dilutions of TcId- and TcVI-cultured epimastigotes. Linear regression analysis yielded the equation y =1.013x − 0.058 (R2 =0.992) and y =1.001x − 0.005 (R2 = 0.998) for SatDNA qPCR, and y = 0.813x + 0.824 (R2 =0.969) and y =1.011x − 0.048 (R2 =0.984) for kDNA qPCR and for TcId and TcVI representative stocks, respectively. Accordingly, the reportable range was from 105 to 1 par. eq./mL for TcId stock and from 105 to 0.25 par. eq./mL for TcVI stock, for both qPCR methods.
The LOD was determined for TcVI DTU. It was 0.16 par. eq./mL (95% CI, 0.13–0.24 par. eq./mL) and 0.23 par. eq./mL (95% CI, 0.18–0.35 par. eq./mL) for kDNA qPCR in boiled and nonboiled samples, respectively (P = 0.013). For SatDNA qPCR the LOD was 0.46 par. eq./mL (95% CI, 0.36–0.64 par. eq./mL) and 0.70 par. eq./mL (95% CI, 0.54–1.01 par. eq./mL) in boiled and nonboiled samples, respectively (P = 0.044). The LOD of kDNA qPCR was lower than that of SatDNA qPCR for both boiled (P = 0.013) and nonboiled samples (P = 0.044).
Estimates of precision were calculated for nonboiled GEB samples spiked with TcVI stock. The precision was higher for kDNA qPCR (CV = 31.98%) than for SatDNA qPCR (CV = 46.60%) at concentrations closer to the LOD, but it was higher for SatDNA qPCR (CV = 6.00% and 1.72%) than for kDNA qPCR (CV = 8.79% and 2.92%) for concentrations of 10 and 1000 par. eq./mL, respectively.
The LOQ was derived from a 20% threshold value of the CVs obtained in the precision experiments. Linear least squares regression for the equation, y = y0 + axe−bx, resulted in the best fit (R2 = 1.0) for both qPCRs. On the basis of the derived equation (y = 1.61 + 157.75xe−1.81x and y =1.79 + 43.39xe−0.91x), the absolute LOQ20%CV was estimated in 1.53 par. eq./mL and 0.90 par. eq./mL for SatDNA and kDNA qPCRs, respectively.
Comparison of qPCR Results in Blood Samples
The performance of both qPCR methods was compared with DNA obtained from 156 GEB samples that covered different epidemiologic and clinical settings. In addition, 50 GEB samples from persons with negative serology for T. cruzi were tested. No amplification was detected for any of these negative controls with the use of both SatDNA and kDNA qPCR methods (clinical specificity, 100%).
The results obtained for the different patients’ groups, including their geographic origin, routes of transmission, phase of CD, T. cruzi DTUs (only performed for samples with parasitic loads above SatDNA qPCR LOQ), qPCR positivity, and median parasitic loads, are summarized in Table 2.
Table 2.
Description of Clinical and Epidemiologic Data of the Different Groups of CD Patients Included in This Study, and Their Parasitic Loads Measured by SatDNA and kDNA qPCR Assays
| Group | Country | Transmission Route | CD Phase | Clinical Manifestation | T. cruzi DTU* | Boiled | Total (n) | SatDNA qPCR
|
kDNA qPCR
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detectable qPCR, n (%) | Quantifiable samples, n (%) | Median [IQR] (Log10 par. eq./10 mL) | Detectable qPCR, n (%) | Quantifiable samples, n (%) | Median [IQR] (Log10 par. eq./10 mL) | ||||||||
| G1a | Mexico | Unknown | Acute | Unknown | I | Yes | 2 | 2 (100) | 2 (100) | 8.41 [8.32–8.50] | 2 (100) | 2 (100) | 8.62 [8.55–8.69] |
| G1b | Chronic | Asymptomatic | Yes | 2 | 2 (100) | 1 (50) | 2.48 | 2 (100) | 1 (50) | 2.74 | |||
| G2 | French Guiana | Oral | Acute | Cardiac | I | Yes | 2 | 2 (100) | 2 (100) | 3.00 [2.64–3.35] | 2 (100) | 2 (100) | 4.17 [3.88–4.46] |
| G3a | Bolivia | Oral | Acute | Unknown | IV | No | 2 | 2 (100) | 2 (100) | 7.19 [7.18–7.20] | 2 (100) | 2 (100) | 6.76 [6.69–6.83] |
| G3b | Vectorial or congenital | Chronic | Asymptomatic | V/VI | No | 3 | 1 (33.3) | 0 (0) | 2 (66.7) | 0 (0) | |||
| G4 | Venezuela | Oral | Acute | Unknown | I | Yes | 5 | 5 (100) | 4 (80) | 3.13 [2.14–3.95] | 5 (100) | 5 (100) | 2.50 [2.43–4.46] |
| G5 | Colombia | Vectorial or congenital | Chronic | Asymptomatic | I | No | 13 | 13 (100) | 8 (61.5) | 2.43 [2.15–2.66] | 12 (92.3) | 8 (61.5) | 2.39 [2.03–2.92] |
| G6 | Spain (Bolivian immigrants) | Vectorial or congenital | Chronic | Asymptomatic | V/VI | Yes | 20 | 13 (65) | 2 (10) | 1.65 [1.63–1.66] | 17 (85) | 4 (20) | 1.49 [1.10–1.91] |
| Digestive | Yes | 1 | 1 (100) | 1 (100) | 2.06 | 1 (100) | 1 (100) | 1.84 | |||||
| G7 | Brazil | Unknown | Chronic | Asymptomatic | II | Yes | 5 | 4 (80) | 0 (0) | 4 (80) | 0 (0) | ||
| Cardiac | Yes | 17 | 16 (94.1) | 1 (5.9) | 1.79 | 14 (82.4) | 5 (29.4) | 1.04 [1.01–1.23] | |||||
| Digestive | Yes | 2 | 2 (100) | 0 (0) | 2 (100) | 1 (50) | 1.05 | ||||||
| Cardiodigestive | Yes | 7 | 5 (71.4) | 3 (42.9) | 1.93 [1.58–2.06] | 5 (71.4) | 5 (71.4) | 1.56 [1.09–2.03] | |||||
| G8 | Argentina | Unknown | Chronic | Asymptomatic | V/VI | Yes | 27 | 22 (81.5) | 7 (25.9) | 1.86 [1.61–2.24] | 21 (77.8) | 11 (40.7) | 1.57 [1.31–3.11] |
| Cardiac | Yes | 34 | 25 (73.5) | 10 (29.4) | 1.81 [1.42–2.58] | 29 (85.3) | 13 (38.2) | 1.59 [1.40–2.09] | |||||
| Digestive | Yes | 1 | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1.79 | ||||||
| Cardiodigestive | Yes | 13 | 12 (92.3) | 5 (38.5) | 1.74 [1.49–1.74] | 12 (92.3) | 6 (46.2) | 1.48 [1.37–2.14] | |||||
Genotyping was only performed for samples with parasitic loads above SatDNA qPCR limit of quantification (1.53 par. eq./mL).
CD, Chagas disease; DTU, discrete typing unit; IQR, interquartile range; kDNA, kinetoplastid DNA; par. eq./10 mL, parasite equivalents in 10 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
Acute CD
This group comprised 11 CD patients, 10 positives and 1 negative for serologic studies. All of them were qPCR detectable with the use of both SatDNA and kDNA qPCR methods (clinical sensitivity for acute CD, 100%). All cases also gave quantifiable parasitic loads by both qPCR methods, except one patient from Venezuela that was nonquantifiable with the use of SatDNA qPCR. The sample from the single seronegative patient from French Guiana presented 3.71 and 4.75 Log10 par. eq./10 mL for SatDNA and kDNA qPCRs, respectively. Most of these patients were infected by the oral route, and DNA from DTUs TcI or TcIV was detected in their samples. Only the patients from French Guiana presented cardiac symptoms.
Chronic CD
This group comprised 70 asymptomatic persons and 75 patients with cardiac and/or digestive syndromes. Except for the patients from Bolivia and Colombia who acquired the infection from vectors or congenitally, the transmission route in the remainder was unknown. Parasite genotyping revealed that the chronic CD patients were infected with all T. cruzi DTUs, except TcIII and TcIV.
For chronic CD patients, 117 and 122 of 145 samples were detectable with the use of SatDNA and kDNA qPCRs, clinical sensitivity equaled 80.69% and 84.14%, respectively; 113 of these samples were detectable by both qPCR methods, and 4 and 9 samples were detectable only by SatDNA or kDNA qPCRs, respectively; κ index was 0.691 (95% CI, 0.535–0.847). Moreover, 38 of 117 (32.48%) and 56 of 122 (45.90%) qPCR detectable samples were quantifiable with the use of SatDNA and kDNA qPCRs, respectively.
The details of the 13 samples with discordant qPCR results are shown in Table 3; because of the low parasitic loads of these samples, in each case the genotype was assumed to be that of the group to which the sample belonged.
Table 3.
Discordant Findings Using SatDNA and kDNA qPCR Assays to Evaluate Clinical Samples during the International Study
| Country | Sample ID | Clinical Manifestation | T. cruzi DTU* | Boiled | SatDNA qPCR
|
kDNA qPCR
|
||
|---|---|---|---|---|---|---|---|---|
| Ct | par. eq./mL | Ct | par. eq./mL | |||||
| Colombia | CE4 | Asymptomatic | I | No | 37.01 | NQ | ND | |
| Brazil | Fx230 | Cardiac | II | Yes | 34.04 | NQ | ND | |
| Fx2754 | 39.01 | NQ | ND | |||||
| Bolivia | M4 | Asymptomatic | V/VI | No | ND | 35.38 | NQ | |
| Spain (Bolivian immigrants) | 12,547 | Asymptomatic | V/VI | Yes | ND | 33.08 | NQ | |
| 19,592 | ND | 33.31 | NQ | |||||
| 27,259 | ND | 32.88 | NQ | |||||
| 34,749 | ND | 34.98 | NQ | |||||
| Argentina | PCH15 | Asymptomatic | V/VI | Yes | 39.69 | NQ | ND | |
| PCH43 | Cardiac | V/VI | Yes | ND | 35.07 | NQ | ||
| PCH58 | ND | 37.75 | NQ | |||||
| 437 | ND | 35.68 | NQ | |||||
| 919 | ND | 34.53 | NQ | |||||
The genotype of each sample was assumed to be that of the group to which the sample belonged.
Ct, cycle threshold; DTU, discrete typing unit; kDNA, kinetoplastid DNA; ND, nondetectable; NQ, nonquantifiable; par. eq./mL, parasite equivalents in 1 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
The nine kDNA qPCR detectable and SatDNA qPCR nondetectable samples belonged to four cardiac patients from Argentina and five asymptomatic cases from Bolivia, four of them living in Spain, infected with TcV, TcVI, or combinations of TcII, TcV, and TcVI. The four samples that were SatDNA qPCR detectable but nondetectable with the use of kDNA qPCR included samples from two cardiac patients from Brazil infected with TcII, one Colombian asymptomatic case infected with TcI, and one Argentinean asymptomatic case infected with TcV, TcVI, or combinations of TcII, TcV, and TcVI. Only two of these samples were boiled, the Bolivian sample that was kDNA qPCR detectable and SatDNA qPCR nondetectable and the Colombian sample that was SatDNA qPCR detectable and kDNA qPCR nondetectable. The parasitic loads of all these samples were below the LOQ of the corresponding qPCR detecting method; moreover, all four SatDNA qPCR detectable and kDNA qPCR nondetectable and five of the nine kDNA qPCR detectable and SatDNA qPCR nondetectable samples presented parasitic loads below the LOD of the corresponding qPCR assay (Table 1).
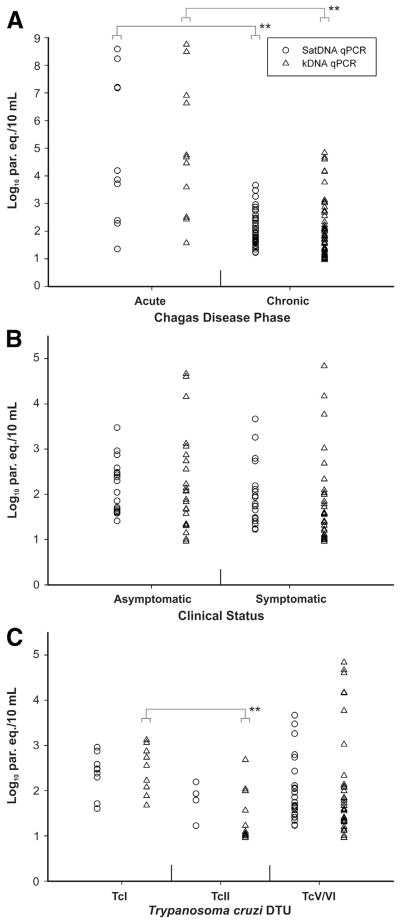
Figure 1 illustrates the comparative analysis of parasitic loads from quantifiable clinical samples on the basis of the CD phase of patients (Figure 1A), and the clinical status (Figure 1B) and T. cruzi DTUs (Figure 1C) of chronic CD patients, obtained by each qPCR assay.
Figure 1.
Comparative analysis of parasitic loads obtained by SatDNA (open circles) and kDNA (open triangles) qPCR assays for quantifiable samples from CD patients. Distribution of parasitic loads on the basis of the CD phase (A), the clinical status (B), and T. cruzi DTUs (C) of chronic CD patients. **P < 0.01. CD, Chagas disease; DTU, discrete typing unit; kDNA, kinetoplastid DNA; par. eq./10 mL: parasite equivalents in 10 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
Acute CD patients had higher parasitic loads (median, 4.03; interquartile range, 2.72 to 7.20 Log10 par. eq./10 mL) than chronic CD patients (1.90; 1.61 to 2.46 Log10 par. eq./10 mL) with the use of SatDNA qPCR (P = 0.0086). Likewise, with the use of kDNA qPCR the acute CD patients also had higher parasitic loads (4.67; 3.05 to 6.76 Log10 par. eq./10 mL) than the chronic CD patients (1.68; 1.31 to 2.28 Log10 par. eq./10 mL) (P = 0.0026) (Figure 1A).
The comparative analysis of the parasitic loads of chronic CD patients according to their clinical status (Figure 1B) did not show any significant differences between the parasitic loads of asymptomatic persons [(2.17; 1.65 to 2.48 Log10 par. eq./10 mL) and (1.97; 1.34 to 2.77 Log10 par. eq./10 mL)] and symptomatic patients [(1.77; 1.45 to 2.13 Log10 par. eq./10 mL) and (1.56; 1.21 to 2.01 Log10 par. eq./10 mL)] either for SatDNA (P = 0.2977) or kDNA (P = 0.1914) qPCRs, respectively.
Similarly, no significant differences were observed between the parasitic loads from chronic CD patients infected with TcI (2.47; 2.30 to 2.58 Log10 par. eq./10 mL), TcII (1.86; 1.65 to 2.00 Log10 par. eq./10 mL), and TcV, TcVI, or combinations of TcII, TcV, and TcVI (1.74; 1.49 to 2.11 Log10 par. eq./10 mL), with SatDNA qPCR. However, with the use of kDNA qPCR, the patients infected with TcI had higher parasitic loads (2.55; 2.08 to 2.87 Log10 par. eq./10 mL) than patients infected with TcV, TcVI, or combinations of TcII, TcV, and TcVI (1.59; 1.33 to 2.11 Log10 par. eq./10 mL) (not significant), and with TcII (1.09; 1.03 to 1.78 Log10 par. eq./10 mL) (P < 0.01) (Figure 1C).
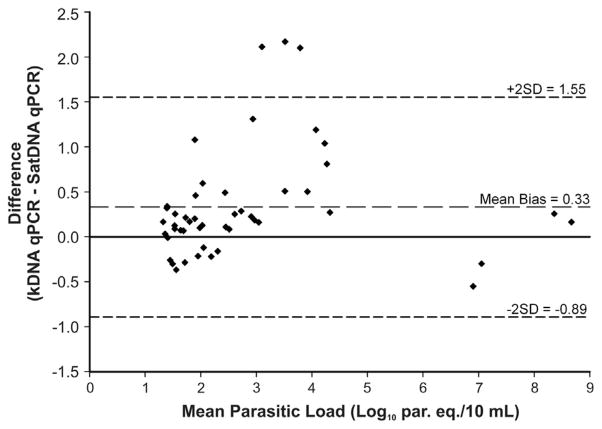
The degree of agreement between the quantifiable results obtained by both qPCR methods from clinical samples is represented in Figure 2 as a Bland-Altman bias (difference) plot. As shown, the mean bias was determined to be 0.33 Log10 par. eq./10 mL, indicating a systematic bias of 2.1-fold parasite equivalents per 10 mL between both methods. However, the bias was not statistically significant because the 95% CI (1.55 to −0.89 Log10 par. eq./10 mL), expressed in Figure 2 as bias ± 2 SD, contains zero (no difference).
Figure 2.
Bland-Altman bias (difference) plot analysis as a measure of the degree of agreement between the quantifiable results obtained by SatDNA and kDNA qPCR assays for samples from Chagas disease patients. kDNA, kinetoplastid DNA; par. eq./10 mL, parasite equivalents in 10 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
Analysis of Exogenous and Endogenous Amplification Controls
The IAC amplification was used for quality control throughout the procedure, from the DNA extractions to qPCR assays. Higher Ct values of IAC were obtained for the clinical samples with the use of SatDNA qPCR (19.05; 18.53 to 19.41) than kDNA qPCR (18.90; 18.40 to 19.25) (P < 0.0001); threshold Cts for outlier values were 20.72 and 20.52, respectively. However, the same three samples with IAC Ct outliers (24.89, 21.00, and 21.49, and 24.74, 20.98, and 21.45) were identified for both SatDNA and kDNA qPCRs, respectively. These three samples were detectable for T. cruzi DNA by both qPCRs, but their parasitic loads were below the LOQ of the corresponding qPCR assay (Table 1); except for the one with the lowest Ct values of IAC (21.00 and 20.98) which presented a parasitic load of 0.94 par. eq./mL (0.97 Log10 par. eq./10 mL) with the use of kDNA qPCR.
In addition, to evaluate DNA integrity of GEB samples, RNase P analysis was performed in a separate amplification reaction. All clinical samples were PCR detectable for this endogenous control with Ct values (22.32; 21.20 to 23.27) between cycle 19 and 27.
Comparison of qPCR Findings with Hemoculture
The parasitic loads of a subset of 15 GEB samples from Brazilian CD patients were compared with hemoculture results performed at the time the samples were collected (Table 4). Despite the time passed, an overall agreement was found between parasitic loads obtained by both qPCR methods and hemoculture results: the three samples with quantifiable parasitic loads corresponded to patients with five or six positive hemoculture results of a total of six replicates, whereas the three samples with nondetectable parasitic loads corresponded to patients with negative results by hemoculture assay, except one case with only one positive hemoculture result (Table 4).
Table 4.
Comparison of Hemoculture and qPCR Assays Results for a Subset of Fifteen Samples from Brazilian Chagas Disease Patients
| Sample ID | Year | Clinical Manifestation | Hemoculture, positive/total | SatDNA qPCR, par. eq./mL | kDNA qPCR, par. eq./mL |
|---|---|---|---|---|---|
| 87 | 1997 | Cardiac | 3/6 | NQ | NQ |
| 88 | Cardiodigestive | 4/6 | NQ | 1.24 | |
| 86 | 1998 | Cardiodigestive | 0/6 | ND | ND |
| 93F | 2000 | Asymptomatic | 0/6 | ND | ND |
| 90F | Asymptomatic | 1/6 | NQ | NQ | |
| 94F | Digestive | 1/6 | NQ | NQ | |
| 89F | Cardiodigestive | 5/6 | 1.68 | 3.67 | |
| 91F | Cardiodigestive | 6/6 | 8.58 | 10.80 | |
| 60 | 2002 | Cardiac | 1/6 | ND | ND |
| 01 | 2004 | Asymptomatic | 2/6 | NQ | NQ |
| 02 | 2006 | Asymptomatic | 6/6 | NQ | NQ |
| 72 | 2008 | Cardiac | 1/6 | NQ | 0.90 |
| 74 | Cardiac | 1/6 | NQ | NQ | |
| 69 | Cardiodigestive | 3/6 | NQ | 0.98 | |
| 66 | Cardiodigestive | 6/6 | 15.54 | 48.24 |
kDNA, kinetoplastid DNA; ND, nondetectable; NQ, nonquantifiable; par. eq./mL, parasite equivalents in 1 mL of blood; qPCR, quantitative real-time PCR; SatDNA, Satellite DNA.
Discussion
Analytical Validation of qPCR Methods in Different T. cruzi DTUs
This study presents the analytical validation and evaluation of two duplex qPCR methods on the basis of TaqMan probes designed for detection and quantification of T. cruzi DNA in human blood samples. The study was performed in the context of an international study organized in an attempt to establish standard operative procedures for quantification of parasitic loads in blood samples, using two methods ranked among the best ones by a previous qualitative PCR international study.7
This study involved the evaluation of DNA samples from parasite stocks representative of the different T. cruzi DTUs, including three distinct TcI SL-based groups. Analytical validation was performed with seronegative blood spiked with known numbers of cultured epimastigotes and treated with GE buffer. Furthermore, blood samples from diverse clinical settings of different geographic regions and harboring different parasite DTUs were assayed. The parasitic loads of these samples were compared with the same DNA extraction protocols, qPCR amplification procedures, master mixes, PCR thermocyclers, and quality controls in the same laboratory.
Analytical sensitivity was more uniform among the different T. cruzi DTUs for kDNA qPCR than for SatDNA qPCR, because this latter method was less sensitive for some TcI and TcIV strains, indicative of lower gene dosage in their genomes.8,25 Thus, in practice it would be advisable to construct standard qPCR curves with the use of regional strains representative of the prevailing DTUs in the affected population.
Parasitic Loads in Different Clinical Groups
As expected, parasitic loads in acute CD samples were higher than in chronic CD cases, reaching concentrations of up to 8 Log10 par. eq./10 mL for both qPCR methods. The four samples with the highest parasitic loads belonged to Mexican and Bolivian acute CD patients infected with TcI and TcIV, respectively. Accurate quantification of these samples was achieved after diluting them 1:10,000 in seronegative human GEB before doing the final DNA extractions and qPCR amplifications. One of the two acute CD samples from French Guiana belonged to a seronegative patient with suspicion of orally acquired T. cruzi infection on the basis of clinical and epidemiologic findings.26 The qPCR positivity of this case points to the usefulness of molecular diagnostic methods for detecting acute CD cases before seroconversion.
Parasitic loads in chronic CD patients ranged from nonquantifiable to values of 3.67 and 4.84 Log10 par. eq./10 mL with the use of SatDNA and kDNA qPCRs, respectively. We observed differences in the parasitic loads between chronic CD patients infected with TcI (eight asymptomatic persons from Colombia plus one from Mexico) and TcII (11 symptomatic patients from Brazil), when they were analyzed with kDNA qPCR. This finding is in agreement with previous observations obtained by Moreira et al27 with the use of a Sybr Green SatDNA qPCR assay in samples from Colombian and Brazilian symptomatic chronic CD patients recruited for the BENEFIT (Benznidazole Evaluation for Interrupting Trypanosomiasis) trial. Nevertheless, no differences were observed between the parasitic loads of asymptomatic and symptomatic chronic CD patients for the samples tested in our study. This is in agreement with previous observations that showed no correlation between T. cruzi parasitemia and clinical manifestations.28,29 However, studies in the murine model reported that T. cruzi reinfections leading to an increase of parasitemia could be related to the variability and severity of the clinical course of CD.30
Bland-Altman analysis was performed to summarize the agreement between parasitic loads obtained by both qPCR methods by calculating the bias and by estimating the mean difference and the SD of the differences. It is also common to determine the limits of agreement, which are by convention set at the 95% CI of the difference between the methods, usually specified as bias ± 2 SD. If the 95% CI for the mean difference includes zero, such as in our study, there is statistically no evidence of bias.6
Some samples presented discordant qPCR results by either of the methods tested; kDNA qPCR detected more samples than SatDNA qPCR, which is reasonable because the former method had higher analytical sensitivity (lower LOD). Moreover, in most of the discordant samples the parasitic load was below the LOD, and in all cases below the LOQ, of the corresponding qPCR assay that gave detectable results.
In this study, correlation between parasitic loads and frequency of positive hemocultures was also observed, strengthening the notion that detectable PCR results are indicative of live parasites.31
Use of Internal Amplification Controls
Previous studies for screening and quantification of parasitic loads of T. cruzi with the use of different real-time PCR approaches included a host DNA sequence, such as RNase P human gene, as an internal control.8,10,27 This type of endogenous control is useful for qualitative purposes, as in the validation of the use of archival samples. In the present study, for instance, the RNase P assay was useful for checking the DNA integrity of GEB samples stored for >10 years. Nevertheless, we do not recommend the use of endogenous controls for quantitative purposes because the content of human blood cells can be highly variable, depending on the nutritional, metabolic, and immunologic status of the persons.9 Therefore, a normalized amount of DNA of a plasmid that contains a heterologous sequence was added before DNA extractions and was used as an internal amplification control to monitor the whole procedure. Indeed, in our study, the IAC was useful to detect three samples with outlier Ct values.
Application of qPCR Methods in Different Scenarios of T. cruzi Infection
Comparison of the analytical parameters for both qPCR methods suggests that kDNA qPCR possesses higher sensitivity for detection and quantification of samples with low parasitic loads. However, in some Central and South American countries, such as Venezuela, Guatemala, Panama, Colombia, El Salvador, and some regions of Brazil, where T. rangeli might cause a false-positive diagnosis of T. cruzi infection,32 SatDNA and not kDNA qPCR should be the qPCR-based method of choice (Table 1). Furthermore, qualitative and quantitative SatDNA real-time PCR approaches were recently used in clinical trials with new antiparasitic drugs and proved to be useful to detect treatment failure.33,34
Clinical sensitivities of both qPCR methods when tested in samples from chronic CD patients (80.69% and 84.14% for SatDNA and kDNA qPCRs, respectively) were similar to those obtained by the four best performing methods selected in a previous international PCR study for T. cruzi detection.7 However, these sensitivities are not good enough for the application of PCR as confirmatory testing of blood donors or clinic patients who are serologically positive. Future prospective studies must be conducted to determine the optimal real-time PCR-based algorithm for diagnosis of T. cruzi infections in other epidemiologic and/or clinical scenarios, such as in early detection of congenital or oral transmission, and reactivation of infection in immunocompromised patients due to organ transplantation or HIV coinfection. Finally, the availability of these standardized and validated qPCR methods opens up new possibilities to monitoring patients in clinical trials with trypanocidal drugs,27,33,34 contributing to improve the quality of life of CD patients.
Acknowledgments
Supported by CDR/Small Grant Programme PAHO/WHO (A.G.S.) and partially by CONICET grant PIP 112-200801-02915 and the National Agency of Science and Technology grant PICT 33955.
We thank the Instituto de Biología y Medicina Experimental (IBYME), Instituto Nacional de Parasitología “Dr. Mario Fatala Chabén”, and Instituto de Investigaciones en Ingeniería Genética y Biología Molecular (INGEBI) (Buenos Aires, Argentina) for supplying facilities and equipment and Life Technologies Argentina for providing the ABI StepOne Real-Time PCR device.
Samples from Mexico, French Guiana, Bolivia, Venezuela, Colombia, Spain, Brazil, and Argentina were provided by the Laboratorio Estatal de Salud Pública (Guerrero, Mexico), the Laboratoire Hospitalier et Universitaire-CH Andrée Rosemon (Cayenne, French Guiana), the Laboratorio de Parasitología y Biología Molecular, Instituto Nacional de Laboratorios en Salud (La Paz, Bolivia), the Instituto de Medicina Tropical, Universidad Central de Venezuela (Caracas, Venezuela), the Centro de Investigaciones en Microbiología y Parasitología Tropical, Universidad de los Andes (Bogota, Colombia), the Laboratorio de Parasitología Molecular, Pontificia Universidad Javeriana (Bogota, Colombia), and Centro Nacional de Microbiología, Instituto de Salud Carlos III (Madrid, Spain), the Laboratório de Biología Molecular e Doenças Endêmicas, Instituto Oswaldo Cruz/Fiocruz (Rio de Janeiro, Brazil), Universidade Federal do Triângulo Mineiro (Uberaba, Brazil), the Universidade Federal do Rio Grande do Norte (Natal, Brazil), the Instituto Nacional de Parasitología “Dr. Mario Fatala Chabén” (Buenos Aires, Argentina), and the Instituto de Patología Experimental, CONICET-Universidad Nacional de Salta (Salta, Argentina), respectively. Samples from seronegative persons were provided by Dr. Cecilia Irurtia (Hospital Posadas, Buenos Aires, Argentina).
We thank Drs. Patricio Diosque (IPE, CONICET-Universidad Nacional de Salta, Argentina) for PAH265, Tu18, CanIII, and M5631 strains; Aldo Solari (Universidad de Chile, Santiago, Chile) for SE9V isolate; Jose Franco Da Silveira (EPM, Sao Paulo, Brazil) for G isolate; Juan David Ramirez and Felipe Guhl (CIMPAT, Universidad de los Andes, Colombia) for T. rangeli DNA; and Alexandre Dasilva (CDC, Atlanta, GA) for Leishmania spp. DNAs.
Footnotes
The findings and conclusions of this manuscript are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention (Atlanta, GA).
Disclosures: A.G.S. is a research member of CONICET. Z.E.Y. is Regional Advisor on Communicable Diseases Research and Health Analysis, Pan American Health Organization. B.A.d.N., A.R., S.S.-E., F.G., I.R., and A.G.S. are members of NHEPACHA Network. Life Technologies Argentina provided access to an ABI StepOne Real-Time PCR machine.
References
- 1.Organización Panamericana de la Salud (OPS) Estimación cuantitativa de la enfermedad de Chagas en las Américas. Montevideo, Uruguay: OPS; 2006. OPS/HDM/CD/425-06. [Google Scholar]
- 2.Schmunis GA, Yadon ZE. Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 2010;115:14–21. doi: 10.1016/j.actatropica.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Prata A. Clinical and epidemiological aspects of Chagas disease. Lancet Infect Dis. 2001;1:92–100. doi: 10.1016/S1473-3099(01)00065-2. [DOI] [PubMed] [Google Scholar]
- 4.WHO/TDR Disease Reference Group on Chagas Disease, Human African Trypanosomiasis and Leishmaniasis. Research Priorities for Chagas Disease, Human African Trypanosomiasis and Leishmaniasis. Geneva, Switzerland: WHO; 2012. WHO Technical Report Series (No. 975) [PubMed] [Google Scholar]
- 5.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12:240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Burd EM. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5:e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, Levin MJ, Favaloro RR, Freilij H, Schijman AG. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in Chagas disease patients. PLoS Negl Trop Dis. 2009;3:e419. doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, Parrado R, Bello ZD, Velazquez E, Munoz-Calderon A, Juiz NA, Basile J, Garcia L, Riarte A, Nasser JR, Ocampo SB, Yadon ZE, Torrico F, de Noya BA, Ribeiro I, Schijman AG. Analytical performance of a multiplex real-time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis. 2013;7:e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piron M, Fisa R, Casamitjana N, Lopez-Chejade P, Puig L, Verges M, Gascon J, Gomez i Prat J, Portus M, Sauleda S. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 11.Qvarnstrom Y, Schijman AG, Veron V, Aznar C, Steurer F, da Silva AJ. Sensitive and specific detection of Trypanosoma cruzi DNA in clinical specimens using a multi-target real-time PCR approach. PLoS Negl Trop Dis. 2012;6:e1689. doi: 10.1371/journal.pntd.0001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Committee for Standardization. Protocol for the validation of alternative methods (EN ISO 16140) Paris, France: European Committee for Standardization; 2002. Microbiology of food and animal feeding stuffs. [Google Scholar]
- 13.Cura CI, Mejia-Jaramillo AM, Duffy T, Burgos JM, Rodriguero M, Cardinal MV, Kjos S, Gurgel-Goncalves R, Blanchet D, De Pablos LM, Tomasini N, da Silva A, Russomando G, Cuba CA, Aznar C, Abate T, Levin MJ, Osuna A, Gurtler RE, Diosque P, Solari A, Triana-Chavez O, Schijman AG. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int J Parasitol. 2010;40:1599–1607. doi: 10.1016/j.ijpara.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Herrera C, Bargues MD, Fajardo A, Montilla M, Triana O, Vallejo GA, Guhl F. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions in Colombia. Infect Genet Evol. 2007;7:535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Herrera C, Guhl F, Falla A, Fajardo A, Montilla M, Adolfo Vallejo G, Bargues MD. Genetic variability and phylogenetic relationships within Trypanosoma cruzi I isolated in Colombia based on miniexon gene sequences. J Parasitol Res [Internet] 2009;2009 doi: 10.1155/2009/897364. http://dx.doi.org/10.1155/2009/897364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. Second Satellite Meeting: A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 18.National Committee flor Clinical Laboratory Standards (NCCLS) Protocols for Determination of Limits of Detection and Limits of Quantification: Approved Guideline. Wayne, Pennsylvania: NCCLS; 2004. [Google Scholar]
- 19.National Committee flor Clinical Laboratory Standards (NCCLS) Evaluation of Precision Performance of Quantitative Measurement Methods: Approved Guideline-Second Edition. Wayne, Pennsylvania: NCCLS; 2004. [Google Scholar]
- 20.Schwarz G, Baumler S, Block A, Felsenstein FG, Wenzel G. Determination of detection and quantification limits for SNP allele frequency estimation in DNA pools using real time PCR. Nucleic Acids Res. 2004;32:e24. doi: 10.1093/nar/gnh020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Chiari E, Dias JC, Lana M, Chiari CA. Hemocultures for the parasitological diagnosis of human chronic Chagas’ disease. Rev Soc Bras Med Trop. 1989;22:19–23. doi: 10.1590/s0037-86821989000100004. [DOI] [PubMed] [Google Scholar]
- 23.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 24.Burns MJ, Nixon GJ, Foy CA, Harris N. Standardisation of data from real-time quantitative PCR methods - evaluation of outliers and comparison of calibration curves. BMC Biotechnol. 2005;5:31. doi: 10.1186/1472-6750-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, Miles MA. Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am J Trop Med Hyg. 2009;81:1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchet D, Breniere SF, Schijman AG, Bisio M, Simon S, Veron V, Mayence C, Demar-Pierre M, Djossou F, Aznar C. First report of a family outbreak of Chagas disease in French Guiana and posttreatment follow-up. Infect Genet Evol. 2014;28:245–250. doi: 10.1016/j.meegid.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Moreira OC, Ramirez JD, Velazquez E, Melo MF, Lima-Ferreira C, Guhl F, Sosa-Estani S, Marin-Neto JA, Morillo CA, Britto C. Towards the establishment of a consensus real-time qPCR to monitor Trypanosoma cruzi parasitemia in patients with chronic Chagas disease cardiomyopathy: a substudy from the BENEFIT trial. Acta Trop. 2013;125:23–31. doi: 10.1016/j.actatropica.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Castro C, Prata A, Macedo V. The influence of the parasitemia on the evolution of the chronic Chagas’ disease. Portuguese Rev Soc Bras Med Trop. 2005;38:1–6. doi: 10.1590/s0037-86822005000100001. [DOI] [PubMed] [Google Scholar]
- 29.Pereira JB, Wilcox HP, Coura JR. The evolution of chronic chagasic cardiopathy. I. The influence of parasitemia. Portuguese Rev Soc Bras Med Trop. 1992;25:101–108. doi: 10.1590/s0037-86821992000200003. [DOI] [PubMed] [Google Scholar]
- 30.Bustamante JM, Novarese M, Rivarola HW, Lo Presti MS, Fernandez AR, Enders JE, Fretes R, Paglini-Oliva PA. Reinfections and Trypanosoma cruzi strains can determine the prognosis of the chronic chagasic cardiopathy in mice. Parasitol Res. 2007;100:1407–1410. doi: 10.1007/s00436-006-0425-3. [DOI] [PubMed] [Google Scholar]
- 31.Lages-Silva E, Crema E, Ramirez LE, Macedo AM, Pena SD, Chiari E. Relationship between Trypanosoma cruzi and human chagasic megaesophagus: blood and tissue parasitism. Am J Trop Med Hyg. 2001;65:435–441. doi: 10.4269/ajtmh.2001.65.435. [DOI] [PubMed] [Google Scholar]
- 32.Guhl F, Jaramillo C, Carranza JC, Vallejo GA. Molecular characterization and diagnosis of Trypanosoma cruzi and T. rangeli Arch Med Res. 2002;33:362–370. doi: 10.1016/s0188-4409(02)00380-6. [DOI] [PubMed] [Google Scholar]
- 33.Molina I, Gomez i Prat J, Salvador F, Trevino B, Sulleiro E, Serre N, Pou D, Roure S, Cabezos J, Valerio L, Blanco-Grau A, Sanchez-Montalva A, Vidal X, Pahissa A. Randomized trial of posaconazole and benznidazole for chronic Chagas’ disease. N Engl J Med. 2014;370:1899–1908. doi: 10.1056/NEJMoa1313122. [DOI] [PubMed] [Google Scholar]
- 34.Pinazo MJ, Thomas MC, Bua J, Perrone A, Schijman AG, Viotti RJ, Ramsey JM, Ribeiro I, Sosa-Estani S, Lopez MC, Gascon J. Biological markers for evaluating therapeutic efficacy in Chagas disease, a systematic review. Expert Rev Anti Infect Ther. 2014;12:479–496. doi: 10.1586/14787210.2014.899150. [DOI] [PubMed] [Google Scholar]