Abstract
Malignant mesothelioma (MM) is an aggressive cancer of mesothelial cells of pleural and peritoneal cavities. In 85% of cases both pleural and peritoneal MM is caused by asbestos exposure. Although both are asbestos-induced cancers, the incidence of pleural MM is significantly higher (85%) than peritoneal MM (15%). It has been proposed that carcinogenesis is a result of asbestos-induced inflammation but it is not clear what contributes to the differences observed between incidences of these two cancers. We hypothesize that the observed differences in incidences of pleural and peritoneal MM are the result of differences in the direct response of these cell types to asbestos rather than to differences mediated by the in vivo microenvironment. To test this hypothesis we characterized cellular responses to asbestos in a controlled environment. We found significantly greater changes in genome-wide expression in response to asbestos exposure in pleural mesothelial cells as compared to peritoneal mesothelial cells. In particular, a greater response in many common genes (IL-8, ATF3, CXCL2, CXCL3, IL-6, GOS2) was seen in pleural mesothelial cells as compared to peritoneal mesothelial cells. Unique genes expressed in pleural mesothelial cells were mainly pro-inflammatory (G-CSF, IL-1β, IL-1α, GREM1) and have previously been shown to be involved in development of MM. Our results are consistent with the hypothesis that differences in incidences of pleural and peritoneal MM upon exposure to asbestos are the result of differences in mesothelial cell physiology that lead to differences in the inflammatory response, which leads to cancer.
Keywords: Mesothelioma, Mesothelial cells, Asbestos, RNA-Seq, Inflammation, Next Gen Sequencing
Malignant mesothelioma (MM) is an aggressive cancer of mesothelial cells causally related to asbestos exposure. Asbestos is a naturally occurring silicate mineral fiber and has been seen lodged in the lungs after inhalation. It is believed that after lodging into lungs, fibers move to locations such as the pleural cavity by unknown mechanisms and cannot be cleared from the body (reviewed in (Thompson, Westbom et al. 2014)). Asbestos fibers can directly or indirectly impact mesothelial cells resulting in MM development. Mesothelial cells are flat, thin cells that line pleural and peritoneal cavities, providing these cavities with a protective layer (Mutsaers 2004). These cells are very diverse in nature with numerous functions including injury/repair, migration and inflammation. Some of the mediators required for these functions are cytokines, chemokines, growth factors and matrix components secreted from mesothelial cells (Mutsaers 2004; Mossman, Shukla et al. 2013). It has also been reported that acute inflammatory response by asbestos fibers in lung parenchyma following asbestos inhalation may result in the flow of fluid and fibers directly into the pleural space thereby directly affecting mesothelial cells (Miserocchi 1997).
MM can develop either in the pleural or peritoneal cavity in response to asbestos exposure; however, the site of origin appears to be associated with different populations and outcomes (Kindler 2013). Pleural MM is more common than peritoneal MM (80–85% of MM incidents versus 10–15%, respectively), and patients with peritoneal MM are significantly younger than those with pleural MM (Rodriguez, Cheung et al. 2009). In addition, patients with peritoneal MM have a shorter median survival, and women with the same type of cancer live longer than men (Rodriguez, Cheung et al. 2009). These differences suggest that the pleural and peritoneal mesothelial cells are differentially susceptible to asbestos exposure in the development of MM.
Global gene expression studies have demonstrated that there are both similar and unique gene patterns in mesothelial cells originating from pleural and peritoneal cavities (Kanamori-Katayama, Kaiho et al. 2011). However, to our knowledge, no studies have indicated that pleural and peritoneal mesothelial cells have differential susceptibility to asbestos exposure. We hypothesize that mesothelial cell of different origin show differential responses to asbestos, which may in part be responsible for differences in incidences of pleural versus peritoneal MM. To test this hypothesis, we exposed two isolates of human peritoneal and pleural mesothelial cells each to crocidolite asbestos and assessed the global gene expression pattern by massive parallel sequencing (MPS).
Materials and Methods
Cell culture and asbestos exposure
Human primary pleural (HPM3, HPM4) and primary as well as immortalized peritoneal (HM3, LP9/hTERT) mesothelial cells were purchased from the Rheinwald lab (Brigham and Women’s Hospital, Harvard University, Boston, MA). Cell lines were validated by STR DNA fingerprinting using the Promega CELL ID System (Promega, Madison, WI) at the UVM DNA Analysis Facility. All cells were grown in 50:50 M199:MCDB106 medium (Invitrogen, Carlsbad, CA) supplemented with 15% FBS, 10 ng/mL EGF, 0.4 µg/mL hydrocortisone, 50 units/mL penicillin and 100 µg/mL streptomycin. All cells were incubated at 37°C in 5% CO2 and grown to 80–90% confluence before addition of asbestos or glass beads (GB). Crocidolite asbestos fibers were prepared and added (5 µg/cm2 or 75 × 106 µm2/cm2 for 8 h) to cell culture medium as previously described (Shukla, MacPherson et al. 2009). GB were used as negative control and added to cells at the same surface area concentration. One immortalized cell line (LP9) was included in the study as we had microarray data from this cell line (Shukla, MacPherson et al. 2009) and we wanted to validate those results by MPS.
The experimental design was a 2 × 2 design that included the factors Treatment and Cell line (Table 5). Treatment included biological duplicates of either 1) no application (control) or 2) application of asbestos. Glass beads were applied as a negative control for the application of a substance to the cells from the three non-immortalized cell lines. After initial analysis showed almost no effect of GB versus untreated (i.e. only one transcript for SNX16 passed a very liberal FDR of <0.2 and at least 2X fold change in the most variable sample set, HPM4, and only at 2.2X), these samples were omitted from the analysis.
Table 5.
Experimental design. Biological duplicates were created for all cell line x treatment samples, except for glass bead generated samples. Glass bead negative controls were only generated for the non-immortalized cell lines.
| Cell line | Cell source ocation |
Cell source gender |
Cell source age (years) |
Treatment |
|---|---|---|---|---|
| LP9 | Peritonium (immortalized) | Female | 26 | Control/Asbestos (no Glass Bead) |
| HM3 | Peritonium | Female | 29 | Control/Asbestos/Glass Bead |
| HPM3 | Pleural cavity | Male | 64 | Control/Asbestos/Glass Bead |
| HPM4 | Pleural cavity | Female | 65 | Control/Asbestos/Glass Bead |
One aim of this study was to examine the differential response of peritoneal and pleural mesothelial cells to asbestos exposure. However, reliable age and gender matched peritoneal and pleural cell lines were not available. The cell lines used in this study are confounded by age across cell sources, and thus cell source was considered as a correlate, but not explicitly built into the model. This is a limitation of the present study and a repeat will be planned as more age and gender matched primary cell lines become available from any source.
Cell viability assay
Following sterilization under ultraviolet light overnight to destroy microbial contaminants, asbestos was suspended in 1X Hanks’ Balanced Salt Solution (HBSS) at 1 mg/mL, sonicated for 15 min in a water bath sonicator, and triturated 5X through a 22-gauge needle. This suspension was added to cells in medium to achieve the desired surface area (SA)-based concentrations. After 24 h, cells were collected by trypsinization, and counted using a hemocytometer (Shukla, MacPherson et al. 2009). Trypan blue exclusion test was performed to show that cells were viable.
RNA preparation for MPS and qRT-PCR
For qRT-PCR, total RNA (1 µg) prepared by RNeasy plus kit (Qiagen) was reverse-transcribed with random primers using the Promega AMV Reverse Transcriptase kit (Promega, Madison, WI) according to the recommendations of the manufacturer as described previously (Shukla, MacPherson et al. 2009). Transcription was evaluated using the ΔΔCt method. Duplicate or triplicate assays were performed with RNA samples. The values obtained from cDNAs and hypoxanthine phosphoribosyl transferase (hprt) controls provided relative gene expression levels for the gene locus investigated. The primers and probes used were purchased from Applied Biosystems (Foster City, CA). For MPS, RNA was prepared by a Trizol extraction method by double DNAse treatment. RNA was quality checked by Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, CA) before subjection to MPS.
Massive Parallel Sequencing
RNA-Seq was performed using Illumina HiSeq 1000 platform in Advanced Genome Technologies Core, UVM. One µg of total RNA was subjected to library synthesis using the TruSeq V2 RNA-Seq kit. Briefly, 1 µg of total RNA was enriched for the Poly-A mRNA and reverse transcribed to double stranded cDNA. The cDNA was end repaired, adenylated, and ligated with Illumina indexes prior to 12 cycles of PCR. Sequencing was performed using 12pM hybridization to a 2×100 paired end flow cell. BAM files for the 19 samples analyzed in this report are available on GEO, accession GSE63966.
RNA-Seq analysis
RNA-Seq data were analyzed following the method described by Trapnell et al. (Trapnell, Roberts et al. 2012). Briefly, Illumina HiSeq 1000 reads were trimmed and clipped for quality control in Trimmomatic v0.27 (Bolger, Lohse et al. 2014). Read quality was checked for each sample using FastQC v0.10.1. High quality reads were then imported into TopHat v2.0.8 for alignment initially to the transcriptome (hg19, GRCh37.72), and then to the genome (hg19, hg19, GRCh37). BAM files were imported into the RNA-Seq pipeline of Partek Genomics Suite®, version 6.6 (Copyright © 2009, Partek Inc., St. Louis, MO, USA), and RPKM (reads per kilobase of transcript model per million mapped reads) counts for each of the 68,296 transcripts defined in RefSeq annotation file were calculated (release 2013-5–10). Exclusions included a minimum RPKM value of 1.0 in at least two of the 16 samples (potentially duplicates of one sample group). RPKM values were log2-transformed with an offset value of 1.0. Sequence duplication was in the 12–20% range, and the estimated library sizes ranged from 40–50 M, both of which were satisfactory. Approximately 80% of the reads aligned, suggesting that the reads adequately represented the biological system. The sequencing process resulted in reads of very high quality and quantity (phred score = 30 or better at p = 0.001). About 48% of the reads aligned to one location in the genome, either with one or both ends. The remainder aligned at more than one location in the genome (ambiguous reads). Based on the alignment and the RefSeq annotation, 37,893 expressed transcripts were identified in the data set. After filtering based on the minimum RPKM requirement, 20,185 transcripts were retained for downstream analyses.
Principal Component Analysis (PCA) was used to identify sources of variation in the absence of a model using the Partek Genomics Suite® software (PGS). Variation was assigned to components of a model that contained crossed (Treatment, Cell line) and nested (Cell line within Cell source) factors using the adonis procedure from the Bioconductor package vegan (Anderson 2001). While we cannot justify the assumptions required for permutational multivariate analysis of variance the decomposition nonetheless helps rank the sources of variation. Differential univariate expression statistics reflecting effect size (fold change, FC) and statistical significance (unadjusted p-value <0.05) were calculated using Analysis of Variance (ANOVA) as is implemented by PGS, and false positives were controlled for using the False Discovery Rate (FDR), or adjusted p-value, <0.05 based on the step-up method of Benjamini and Hochberg (Benjamini and Hochberg 2009).
Expression-based functional and pathway analyses were conducted using DAVID 6.7 (http://david.abcc.ncifcrf.gov) and Ingenuity Pathway Analysis (IPA; Ingenuity® Systems, www.ingenuity.com). For this analysis, the differential expression statistics were filtered for significance at p<0.05 and at least a 2X FC. IPA implements a Fisher test of the null hypothesis that the genes were differentially expressed due to chance.
IL-1β ELISA assay
The Quantikine Human IL-1β/IL-1f2 Immunoassay (R&D Systems, Minneapolis, MN, measures predominantly mature IL-1β) was used on concentrated cell medium. Medium (500 µL) was concentrated using Amcion® ultracentrifugal filters with a 10 K membrane (Millipore, Billerica, MA) by spinning at 14,000× g for 30 min. Columns were then reversed into new collection tubes and spun for 2 min at 1,000× g. IL-1β assay was performed on the concentrated medium according to the manufacturer’s instructions. A total of 500 µL of cell supernatant was concentrated. Two hundred µL samples with assay diluents were loaded into 96 well plates pre-coated with IL-1β antibody. Values are expressed as pg/mL of IL-1β from the original supernatant (non-concentrated).
Western blot analyses
Medium was collected and concentrated as described above. Sample buffer was added to the concentrated supernatant, and samples were boiled for 5 min. Western blot analyses were performed as previously described (Shukla, Barrett et al. 2006) on concentrated supernatants for IL-6 and IL-8. Antibodies were obtained from Abcam, Cambridge MA. As supernatant was processed for different protein analysis, no loading control was available. Ponceau staining of the membrane before probing for antibodies confirmed the equal loading (data not shown).
Results
Peritoneal mesothelial cells are more sensitive to asbestos-induced toxicity
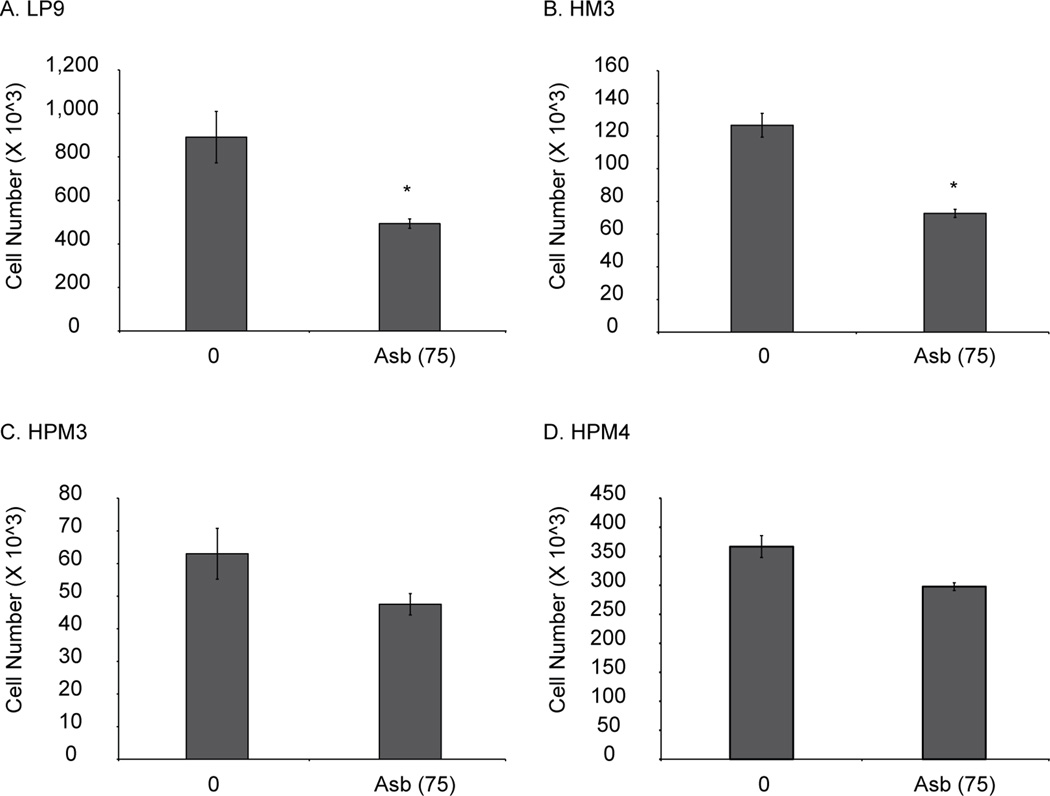
To assess sensitivity of pleural and peritoneal mesothelial cells to asbestos, we exposed two peritoneal (LP9, HM3) and two pleural (HPM3, HPM4) cell lines to 5µg/cm2 asbestos for 24 h. Figure 1 shows that both peritoneal lines (A, B) were more sensitive to asbestos-induced cell killing as compared to pleural lines (C, D), which showed no significant killing in response to asbestos. GB had no significant effect on any cell lines (data not shown).
Figure 1.
Asbestos is more cytotoxic to human peritoneal mesothelial cells compared to human pleural mesothelial cells. Two peritoneal (LP9 and HM3) and 2 pleural (HPM3 and HPM4) mesothelial cells were exposed to asbestos (5 µg/cm2 or 75) for 24 h and cells were counted using hemocytometer. (n=3) *p≤0.05 as compared to untreated control.
Responsiveness to asbestos exposure differs between pleural and peritoneal mesothelial cells
Multivariate analyses were used to evaluate the response of the system to treatment variable of interest (PERMANOVA), and to assess the ability of the data to reflect the biological factors of the design in the absence of a model (PCA).
Assignment of variance to model components (Table 1) suggests that cell sources is the largest contributor to variation, with a mean square error (MSE) twice that between cell lines. Each MSE (associated with treatment, cell source, and cell lines) was large (F > 11) compared with residual variation.
Table 1.
Sources of variation. The assignment of variation identified in the data to those biological treatment factors and latent sources of variation using MANOVA.
| Df | SumsOfSqs | MeanSqs | F.Model | R2 | |
|---|---|---|---|---|---|
| Treatment | 1 | 9048.824128 | 9048.824128 | 11.66710441 | 0.149499743 |
| Cell source | 1 | 20151.5438 | 20151.5438 | 25.98239972 | 0.33293283 |
| Cell line | 2 | 22795.55933 | 11397.77966 | 14.69573101 | 0.376615814 |
| Residuals | 11 | 8531.428359 | 775.5843963 | NA | 0.140951612 |
| Total | 15 | 60527.35561 | NA | NA | 1 |
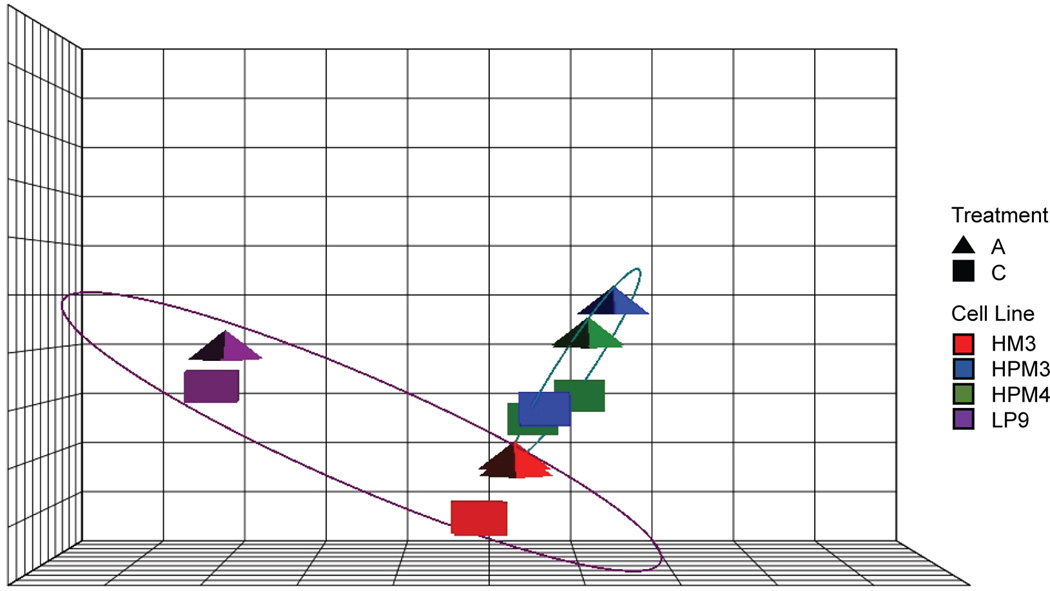
The PCA assessed sample group variation in the absence of a model, and reduced the data set from many dimensions, to a small number of vectors that represent the dominant sources of variation in the data. After RPKM normalization, PC components strongly reflected the sample groups, capturing 83% of the total variation in the first three components, clearly distinguishing Cell lines (PC1; x-axis) and Treatment (PC2; y-axis; Figure 2). The greatest variation lies in Cell source (PC1), like the MANOVA indicated, however the response to asbestos exposure (i.e. distance between treatment and control within cell lines) is in the same direction for each cell line, but to a greater extent in the pleural cells (PC2).
Figure 2.
Principal component analysis indicates common response to asbestos exposure. PCA plot reflects Treatment (triangles), Cell lines (colors), and Cell source (ellipses; red and purple = pleural; green and blue = peritoneal) in three components representing 83% of the overall variation. The x-axis differentiates the Cell source (purple vs. teal ellipses), and possibly immortalization based on the distance between the LP9 cell line as compared to the other three lines. The y-axis captures the response to Treatment, and is the same direction for all four cell lines, but to a greater extent in the pleural cell lines, as indicated by the distance between control (squares) and treated (triangle) samples (scalar data not shown).
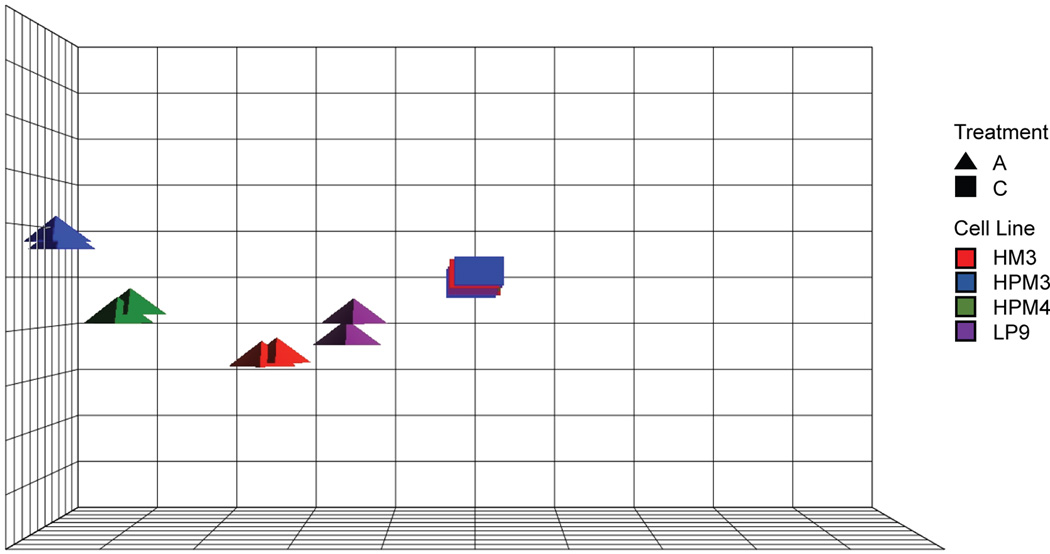
To reduce the number of factors in the model and further explore cell line to cell line response, the data were centered on the treatment controls. In centering on the controls, the control samples are averaged together and modeled as the reference sample for each cell line. One replicate of the HPM4 control sample set was an outlier for most transcripts (see Figure 2, green squares) and was not used in the centering of the HPM4 cell in response (replicate 2). Under this control-centered model, the first three principal components captured 78% of the overall variation (Figure 3). The resulting plot suggests largely the same response to asbestos across cell lines but with varying magnitudes across both Cell line and Cell source (PC1, x-axis, 61% of the variation). The second principal component also captures variation associated with difference in expression between the cell sources (PC2, y-axis, 9% of the variation). The progression along PCs 1 and 2 from immortalized peritoneal to primary peritoneal to pleural asbestos-treated cells provides evidence that the response to asbestos is larger in magnitude in the pleural cells as compared to peritoneal cells.
Figure 3.
Control-centered principal component analysis. Control-centered principal component analysis also indicates a common response to asbestos exposure but to a greater extent in the pleural cell lines. PCA plot centered on the Control samples for each cell line. Variation from the Control proceeds almost linearly from LP9 -> HM3 -> HPM4 -> HPM3, or peritoneum to pleural cavity.
While multivariate, sample-based PCA suggests a direction of common response in the absence of a model, a univariate analysis employing a linear model can identify the individual transcripts responsible for that response, and the biological pathway in which those transcripts are important components. Analysis of Variance (ANOVA) produced a transcript by transcript matrix of differential expression across sample groups. Statistical filters were applied, and the transcripts differentially expressed at an FDR <0.05 alone, and at p-value <0.05 and at least a 2X fold change, were counted (Figure 4, Table 2). The number of differentially expressed transcripts mirrored the distribution of the samples seen in the PCA plot under either filter, with HPM3 cells having the most differentially expressed transcripts from the controls (843), and immortalized LP9 cells the least (85). Collectively, pleural cell lines differential response was 2X or greater than that of the peritoneal cell lines.
Figure 4.
Transcripts commonly or uniquely differentially expressed in primary cell lines in response to asbestos exposure. A Venn diagram illustrating the number of transcripts commonly or uniquely differentially expressed based on the binary filter for the three primary cell lines.
Table 2.
Differentially expressed transcripts in response to asbestos exposure. Differentially expressed transcript counts for each contrast filtered by FDR alone, and a binary filter including unadjusted p-value and fold change.
| Cell source | Contrast | FDR <0.05 | FDR <0.05 unadjusted p<0.05 & 2X FC |
|---|---|---|---|
| Immortalized peritoneal mesothelium | LP9*A vs. LP9*C | 1231 | 85 |
| Primary peritoneal mesothelium | HM3*A vs. HM3*C | 3011 | 224 |
| Primary pleural mesothelium | HPM4*A vs. HPM4*C | 5846 | 481 |
| Primary pleural mesothelium | HPM3*A vs. HPM3*C | 8468 | 843 |
The difference in responsiveness between pleural and peritoneal mesothelial cells exposed to asbestos is a difference of magnitude, not direction
A Venn diagram of the differential expressed transcripts counts identified under the binary filter of p<0.05 and at least 2X fold change was created for the three primary cell lines (Figure 4). At this statistical threshold, 165 transcripts were shared in response to asbestos exposure, 29 were uniquely differentially expressed in HM3, and 121 and 473 were unique for HPM4 and HPM3, respectively. The 165 transcripts that are shared by cell line response to asbestos treatment include up-regulated IL-6 and 8, CXCL1, 2, and 3, ATF3, and NR4A1, and down-regulated ID1 (see Table 3A for partial list; for complete list see Additional file 1). Treatment samples are enriched for sequence-specific DNA binding proteins, including the CREB proteins, Jun/AP1 transcription factors, fos oncogene and fos-related proteins, NFkB, cell signaling, and inflammation response, based on DAVID functional analysis (p<0.001).
Table 3.
Transcripts commonly and uniquely differentially expressed in response to asbestos exposure. A) Transcripts known to be involved with MM that were significantly differentially expressed in all cell lines, and B) Transcripts uniquely differentially expressed between pleural and peritoneal mesothelial cells. These transcripts represent the interaction between Cell Source and Treatment ((p<0.05, FDR<0.05, 2X fold change).
| A. | |||||
|---|---|---|---|---|---|
| RefSeq ID | Gene symbol |
HPM3 Fold Change |
HPM4 Fold Change |
HM3 Fold Change |
LP9 Fold Change |
| NM_000584 | IL8 | 33.523 | 68.4999 | 12.6472 | 13.3992 |
| NM_001145033 | C11orf96 | 15.5014 | 68.4658 | 21.0523 | 2.87073 |
| NM_002089 | CXCL2 | 8.59856 | 59.7874 | 11.2084 | 4.85504 |
| NM_002090 | CXCL3 | 5.8495 | 57.1446 | 12.237 | 3.08622 |
| NM_004591 | CCL20 | 2.6658 | 41.6213 | 9.11657 | 1.69404 |
| NM_000600 | IL6 | 7.55715 | 34.5214 | 10.7801 | 1.97597 |
| NM_001030287 | ATF3 | 12.0644 | 31.31 | 25.8987 | 7.32549 |
| NM_001130046 | CCL20 | 4.0348 | 27.2693 | 6.23303 | 2.24153 |
| NM_015714 | G0S2 | 9.33613 | 20.1768 | 6.38625 | 1.79726 |
| NM_001206486 | ATF3 | 4.95986 | 17.7409 | 10.1859 | 5.9077 |
| NM_001674 | ATF3 | 2.88467 | 11.3234 | 7.8942 | 4.45283 |
| NM_173157 | NR4A1 | 9.26958 | 9.28196 | 6.07496 | 2.36559 |
| B. | |||
|---|---|---|---|
| RefSeq ID | Gene symbol | HMP3 vs. HM3 Fold Change |
HMP4 vs. HM3 Fold Change |
| NM_001145033 | C11orf96 | 2.1016 | 1.16537 |
| NM_004591 | CCL20 | 3.95134 | 1.84928 |
| NM_001130046 | CCL20 | 2.59972 | 1.24291 |
| NM_000758 | CSF2 | 3.37437 | 1.85269 |
| NM_172219 | CSF3 | 6.99657 | 3.669 |
| NM_000759 | CSF3 | 4.62554 | 2.09011 |
| NM_001178147 | CSF3 | 3.76625 | 2.13401 |
| NM_172220 | CSF3 | 3.21695 | 1.87829 |
| NR_033662 | CSF3 | 2.77213 | 1.67068 |
| NM_002089 | CXCL2 | 2.63689 | 1.14172 |
| NM_002090 | CXCL3 | 3.12556 | 1.44637 |
| NM_013409 | FST | 2.60138 | 2.01464 |
| NM_001191322 | GREM1 | 2.04879 | 1.78317 |
| NM_013372 | GREM1 | 1.78934 | 2.09912 |
| NM_000575 | IL1A | 4.64077 | 1.92285 |
| NM_000576 | IL1B | 3.84257 | 1.85831 |
| NM_000600 | IL6 | 2.1373 | 1.19435 |
| NM_001256339 | NKX3-1 | 2.19502 | 1.76747 |
| NM_006167 | NKX3-1 | 2.6504 | 1.27087 |
| NR_046072 | NKX3-1 | 2.29697 | 1.35888 |
| NM_002638 | PI3 | 2.14985 | 1.36983 |
| NM_003713 | PPAP2B | 2.65004 | 1.68872 |
| NM_000963 | PTGS2 | 2.27583 | 1.22729 |
| NM_002852 | PTX3 | 2.12739 | 1.05261 |
| NM_016321 | RHCG | 2.02976 | 1.27926 |
| NM_007115 | TNFAIP6 | 2.03878 | 1.40361 |
| NM_014350 | TNFAIP8 | 2.17025 | 1.30366 |
| NM_001190947 | TRAF1 | 2.03622 | 1.15353 |
| NM_006022 | TSC22D1 | 2.37599 | 1.75202 |
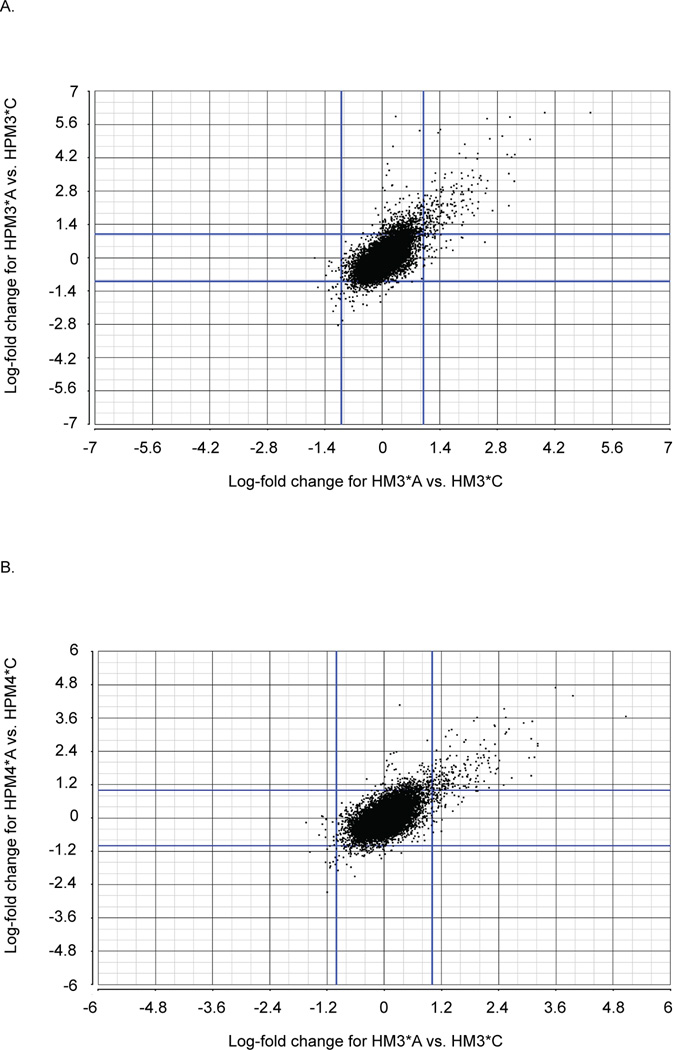
While a Venn diagram highlights the number of transcripts commonly and uniquely responding to treatment in each cell line under a specific filter, it 1) fails to inform as to the nature of the response, i.e. whether the shared differentially expressed transcripts are similarly up- or down-regulated, and 2) does not inform on instances where transcripts are similarly differentially expressed in two cell lines but do not meet the statistical filter parameters in one of them, potentially by a small margin. To address these limitations, scatter plots of the unfiltered transcript sets were created for each pairwise comparison using the log fold change (Figure 5, A and B). A diagonal line, lower left to upper right, versus crossed lines about zero, indicates that response to asbestos is similar and in the same direction (i.e. the same transcripts are mutually up-regulated versus up-regulated in one cell line and down-regulated in the other). The slope of the diagonal indicates the degree of similarity of the response, and can indicate when differential expression is shared yet not captured under an applied threshold, for example a 1.9X fold change is missed under a 2X fold change filter. The comparison between asbestos-exposed and control samples from either pleural cell line vs. HM3 demonstrates similarly up and down regulation, with a correlation coefficient of r = 0.7 for both comparisons. The distance off of the diagonal (slope > 1) is the result of two possible mechanisms, 1) cell lines present a mixed response, i.e. more cells respond in one cell line, or 2) cell lines present a differential response, i.e. one cell line is more sensitive. The consistency of the signal indicates that the pleural cell lines are more sensitive. A few differentially expressed genes lie within the “cross” region indicating up-regulation in HPM3 and HPM4 but not HM3 (for example CSF3, FST, and IL-1α), however, most of the genes in those regions were not statistically significant. There are no statistically significant transcripts incongruent across Cell source (upper left and lower right quadrants), again showing the general agreement in the direction of expression change across cell sources.
Figure 5.
Patterns of differential expression between pleural and peritoneal cell lines in response to asbestos exposure. Scatter plots of the log-fold changes for differentially expressed transcripts when A) pleural cell line HMP4 is compared to peritoneal cell line HM3, linear regression indicates a correlation coefficient of r = 0.70, and B) pleural cell line HPM3 is compared to peritoneal cell line HM3 (linear regression indicates a correlation coefficient of r = 0.72).
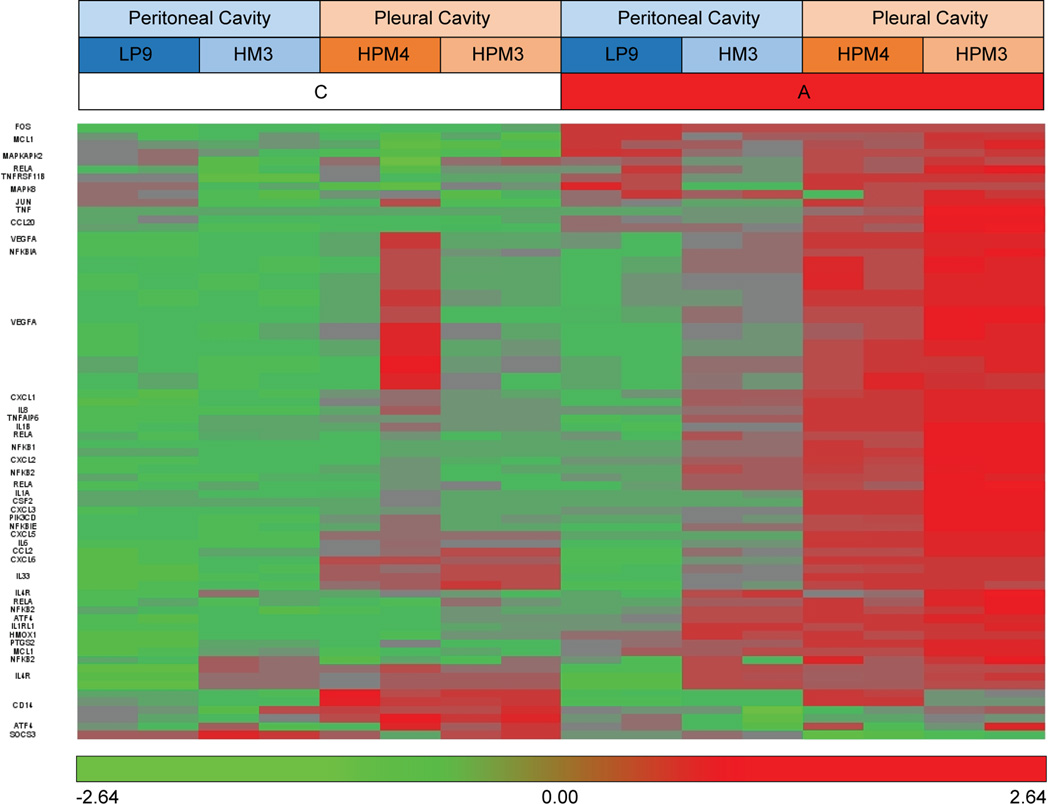
For a broader systems biological analysis of the response to asbestos, the differentially expressed transcripts from each cell line were analyzed independently. IPA indicated Cellular Growth and Proliferation as the most enriched molecular and cellular functions, Cancer as the most enriched disease, and IL-17 and IL-6 signaling as the most significantly enriched pathways in all primary cell lines (p<0.05), again emphasizing the similarity in the response across cell lines and cell sources. IL-10 signaling was also enriched. Seventy transcripts representing 35 genes were involved in IL-17, IL-6, and IL-10 signaling (Table 4, Figure 6).
Table 4.
Transcripts involved in IL17, IL6, and IL10 signaling based on IPA canonical pathways annotation. IL17 signaling pathways were the most significantly enriched pathways in response to asbestos exposure in all four cell lines. IL6 and IL10 signaling pathways were also highly enriched and included many of the same transcripts. Bolded transcripts are in all three signaling pathways.
| RefSeq ID | Gene symbol | RefSeq ID | Gene symbol |
|---|---|---|---|
| NM_182810 | ATF4 (IL17) | NM_001165412 | NFKB1 |
| NM_001675 | ATF4 (IL17) | NM_003998 | NFKB1 |
| NM_002982 | CCL2 (IL17) | NM_002502 | NFKB2 |
| NM_001130046 | CCL20 (IL17) | NM_001261403 | NFKB2 |
| NM_004591 | CCL20 (IL17) | NM_001077494 | NFKB2 |
| NM_001040021 | CD14 (IL6 and IL10) | NR_048560 | NFKB2 |
| NM_001174104 | CD14 (IL6 and IL10) | NM_020529 | NFKBIA |
| NM_001174105 | CD14 (IL6 and IL10) | NM_004556 | NFKBIE |
| NM_000591 | CD14 (IL6 and IL10) | NM_005026 | PIK3CD (IL17 and IL6) |
| NM_000758 | CSF2 (IL17) | NM_000963 | PTGS2 (IL17) |
| NM_001511 | CXCL1 (IL17) | NM_001243984 | RELA |
| NR_046035 | CXCL1 (IL17) | NM_021975 | RELA |
| NM_002089 | CXCL2 (IL17) | NM_001145138 | RELA |
| NM_002090 | CXCL3 (IL17) | NM_001243985 | RELA |
| NM_002994 | CXCL5 (IL17) | NM_003955 | SOCS3 (IL6 and IL10) |
| NM_002993 | CXCL6 (IL17) | NM_000594.5 | TNF (IL6 and IL10) |
| NM_005252 | FOS | NM_007115 | TNFAIP6 (IL6) |
| NM_002133 | HMOX1 (IL10) | NM_002546 | TNFRSF11B (IL6) |
| NM_000575 | IL1A (IL6 and IL10) | NM_001171622 | VEGFA (IL6) |
| NM_000576 | IL1B | NM_001171630 | VEGFA (IL6) |
| NM_003856 | IL1RL1 (IL6 and IL10) | NM_001025369 | VEGFA (IL6) |
| NM_033439 | IL33 (IL6 and IL10) | NM_001171627 | VEGFA (IL6) |
| NM_001199640 | IL33 (IL6 and IL10) | NM_001204384 | VEGFA (IL6) |
| NM_001199641 | IL33 (IL6 and IL10) | NM_001204385 | VEGFA (IL6) |
| NM_000418 | IL4R (IL10) | NM_001025370 | VEGFA (IL6) |
| NM_001257406 | IL4R (IL10) | NM_001171628 | VEGFA (IL6) |
| NM_001257997 | IL4R (IL10) | NM_001033756 | VEGFA (IL6) |
| NM_001257407 | IL4R (IL10) | NM_001171629 | VEGFA (IL6) |
| NM_000600 | IL6 | NM_001025368 | VEGFA (IL6) |
| NM_000584 | IL8 (IL17 and IL6) | NM_001171626 | VEGFA (IL6) |
| NM_002228 | JUN (IL6 and IL10) | NM_001025367 | VEGFA (IL6) |
| NM_139046 | MAPK8 | NM_001171625 | VEGFA (IL6) |
| NM_139047 | MAPK8 | NM_001171624 | VEGFA (IL6) |
| NM_004759 | MAPKAPK2 (IL6) | NM_003376 | VEGFA (IL6) |
| NM_032960 | MAPKAPK2 (IL6) | NM_001025366 | VEGFA (IL6) |
| NM_001197320 | MCL1 (IL6) | ||
| NM_021960 | MCL1 (IL6) | ||
| NM_182763 | MCL1 (IL6) |
Figure 6.
Transcripts from the IL-17, IL-6, IL-10 signaling pathways commonly differentially expressed in response to asbestos. Both were identified by pathway analysis as the most enriched in all cell lines.
Unique genes expressed in pleural mesothelial cells were mainly pro-inflammatory and have been shown to be involved in development of MM
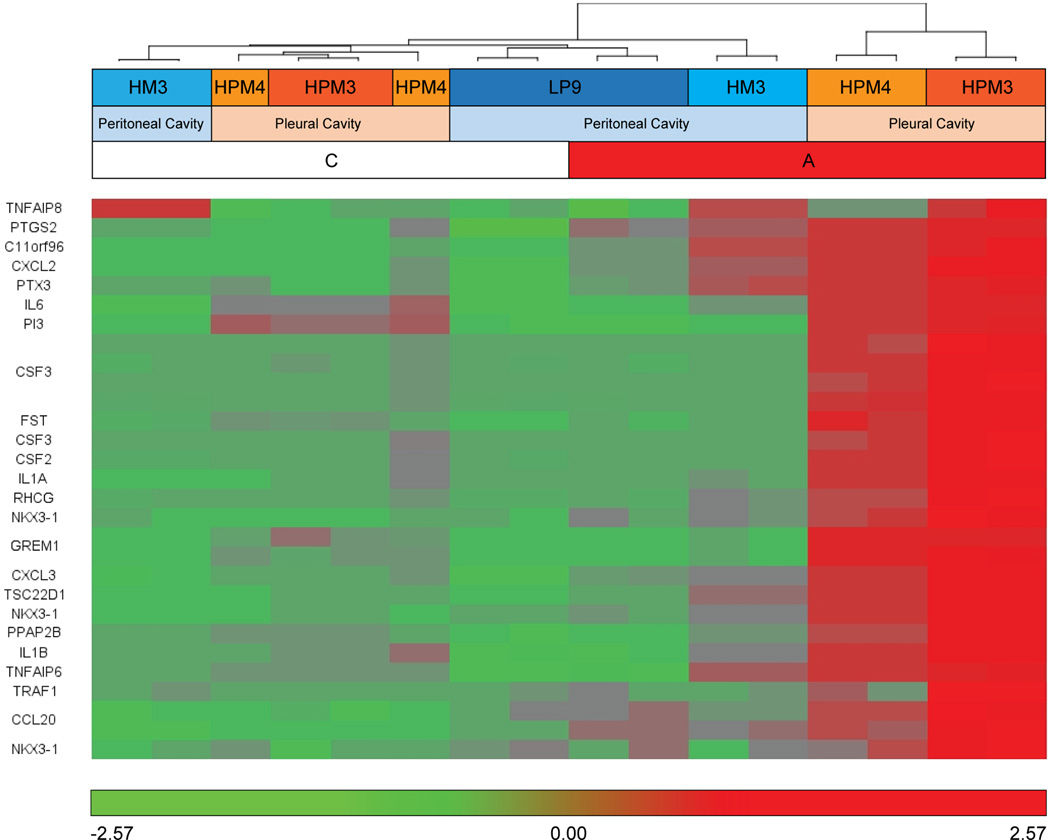
Despite significant agreement in response to asbestos, when the interaction is modeled, i.e. the difference of difference between each pleural cell line treatment versus control, versus the HM3 cell line treatment versus control, a collection of transcripts for which differential expression in response to asbestos exposure was unique to pleural cell lines was identified as significantly differentially expressed (p<0.05 and at least 2X fold change; see fold differences in Table 3B or heat map in Figure 7) including CSF3, FST, IL-1α, IL-1β and GREM1. Based on these transcript profiles, the most significantly differentially enriched clusters between the collective pleural and the peritoneal primary cell line in response to asbestos were extracellular matrix, inflammatory and immune response, and cytokine activity (p<0.01, FCR<0.05). Uniquely expressed transcripts responsive to asbestos in pleural mesothelial cells are mostly pro-inflammatory genes and have been shown to be involved in the development of MM.
Figure 7.
Twenty-nine transcripts uniquely differentially expressed between pleural and peritoneal mesothelial cells. These transcripts represent the interaction between Cell Source and Treatment. Samples and transcripts were clustered based on Euclidean distance.
Validation of RNA-Seq results at protein and RNA levels
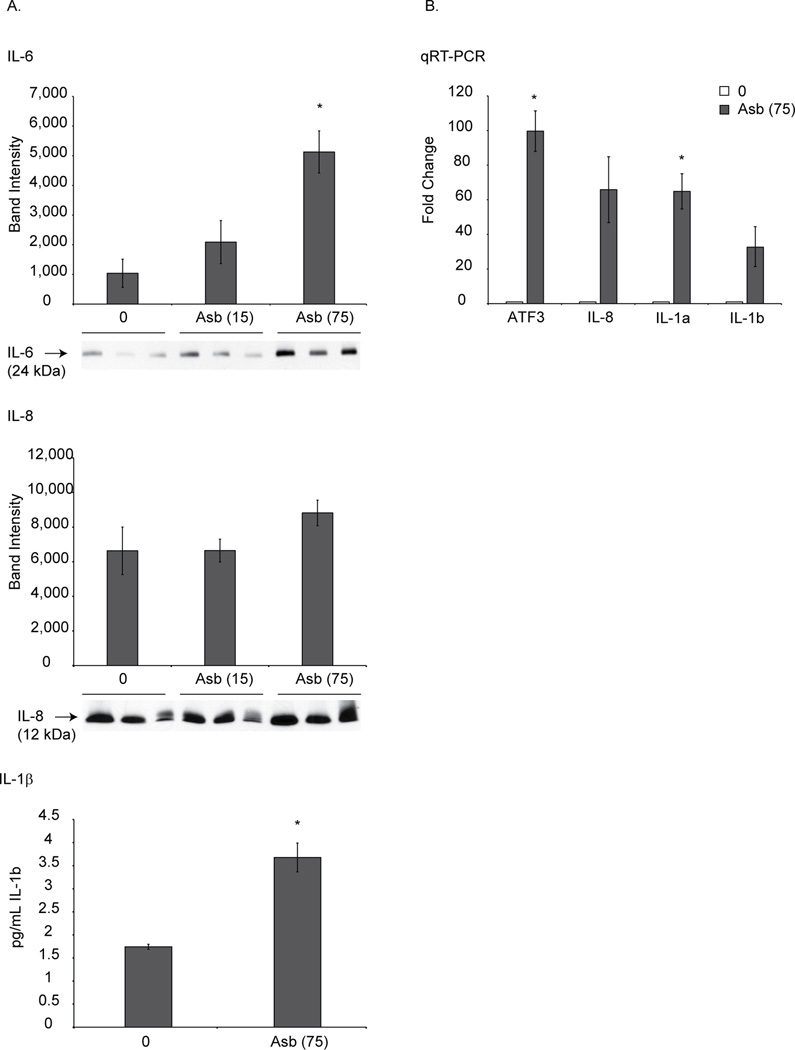
We assessed the protein levels of highly expressed cytokines like IL-6, IL-8 and IL-1β in HPM3 to validate RNA-Seq data (Figure 8). Medium from asbestos-exposed HPM3 cells at 8 h was collected and concentrated as discussed above. Either ELISA (IL-1β) or Western blot analysis (IL-8, IL-6) was performed on concentrated medium. As shown in Figure 8A asbestos exposure caused increased protein levels of IL-6, IL-8 and IL-1β from HPM3 cells. Results were validated at the RNA level also using qRT-PCR technique (Figure 8B).
Figure 8.
RNA-Seq expression data was validated at the protein level. (A) HPM3 cells were exposed to asbestos (1(15 × 106) or 5 (75 × 106) µg/cm2) for 8 h. Medium was collected and concentrated as described in the method section. Concentrated medium was analyzed for IL-1β (ELISA assay), IL-6 or IL-8 (Western blot analysis). N=3 samples/group. *p≤0.05 as compared to untreated control. (B) Validation of some highly expressing genes by qRT-PCR.
Validation of previously published microarray data in LP9 cells
We have previously published microarray data on LP9 cells exposed to asbestos (Shukla, MacPherson et al. 2009) (5 µg/cm2 for 8 h). In the present study we included LP9 cells exposed to the same concentration of asbestos for 8 h to validate our microarray results. MPS data validated previously published microarray data in LP9 cells, indicating up-regulation, for example, of ATF3, FOSB, TFPI2, IL-8, and NR4A2 in response to asbestos exposure using both platforms [8].
Discussion
Malignant mesothelioma (MM) is an aggressive cancer of the pleural or peritoneal cavities, caused by asbestos exposure. Although the cell of origin for both pleural and peritoneal MM is the mesothelial cell and the causative factor is asbestos, the incidence rate for pleural MM is 85% and while that for peritoneal MM is only 10–15%. In addition, there are many other differences between these two types of MMs, as discussed in the introduction section (Kindler 2013), however, it is not clear what contributes to these differences. We hypothesized that differences in response could be linked to different susceptibility of pleural and peritoneal mesothelial cells to asbestos exposure. To verify our hypothesis, in the present study we performed a global gene expression profile on asbestos exposed human pleural and peritoneal mesothelial cells. These are the first published RNA-Seq results for mesothelial cells exposed to controlled doses of asbestos. The results validate previous results inferred from microarray data generated from the same exposure using only an immortalized peritoneal cell line (Shukla, MacPherson et al. 2009). In addition, they provide greater resolution of genome-wide expression changes and preliminary data on the differential response of pleural and peritoneal cavities.
Multivariate and univariate analysis performed on RNA-Seq data suggested greater magnitude of asbestos-response in pleural mesothelial cells as compared to peritoneal mesothelial cells under comparable conditions of asbestos exposure. Detailed analysis showed that in response to asbestos a higher number of total genes were changing in pleural mesothelial cells as compared to peritoneal mesothelial cells, but with a commonality of response. Next we compared fold changes of top 10 common genes (IL-8, IL-6, ATF3, CXCL2, CXCL3, NR4A1, GOS2, CCL20, CHAC1 and ID1) between two different types of mesothelial cells. Many of these genes have been reported previously, by our group or others, to be overexpressed in response to asbestos exposure (Shukla, MacPherson et al. 2009) and involved in asbestos-induced MM development. IL-8, for example, is released from mesothelial and epithelial cells in response to asbestos exposure (Boylan, Ruegg et al. 1992; Hillegass, Miller et al. 2013; Duncan, Cook et al. 2014). Furthermore, IL-8 is present in higher levels in the serum and peritoneal lavage fluid (PLF) of MM tumor bearing humans (Davidson, Vintman et al. 2004; Eikawa, Ohue et al. 2010; Corradi, Goldoni et al. 2013; Vlaeminck-Guillem, Bienvenu et al. 2013) and mice (Shukla, Miller et al. 2013), and has been used as a diagnostic marker in combination with other cytokines. Other common cytokines and chemokines highly expressed by pleural cells in comparison to peritoneal cells were IL-6 and MIP-2 (CXCL2). MM is known as an IL-6 secreting tumor (Adachi, Aoki et al. 2006; Hillegass, Shukla et al. 2010) and IL-6 levels can be used as biomarkers of asbestos exposure (Amati, Tomasetti et al. 2008). Thus, IL-6 can serve as a target molecule for designing treatment for MMs (Adachi, Yoshio-Hoshino et al. 2010). MIP-2 has also been reported by us (Shukla, MacPherson et al. 2009) and others (Hill, Mangum et al. 2003) to be secreted by mesothelial cells in response to asbestos exposure and may contribute to inflammation. This study of common genes identified in the analysis demonstrated that although mesothelial cells of both origins express important shared genes, the magnitude of increase in response to asbestos in most of the genes was significantly higher in pleural mesothelial cells as compared to peritoneal mesothelial cells. This provides us with the first clue that pleural mesothelial cells may be more responsive to asbestos exposure.
We and others have shown that mesothelial cells are very sensitive to asbestos exposure and that they undergo apoptosis or necrosis when exposed to asbestos (Shukla, MacPherson et al. 2009). Here we compared relative sensitivity of pleural and peritoneal mesothelial cells to asbestos-induced cell killing. Twenty-four hour exposure to a high dose of asbestos caused significant cell killing in two peritoneal mesothelial cells but not in pleural mesothelial cells. Although this observation needs to be verified by many more cell lines in both groups, it indicates that asbestos could be more mesotheliomagenic to pleural mesothelial cell as it is not cytotoxic to them.
Assessment of unique gene patterns in two mesothelial cell types also shed some light on possible increased reactivity of pleural mesothelial cells to asbestos. A unique gene list was generated at equal or more than two fold differential expression and a conservative FDR. The top genes comprising the unique gene list for pleural mesothelial cells included CSF3/GCSF, IL-β, IL-1α, GREM1, and others. These are mostly pro-inflammatory genes and have been shown to be involved with pathogenesis of asbestos-induced diseases including MM. We and others have demonstrated that asbestos exposure of mesothelial cells can cause secretion of IL-1β that is inflammasome-dependent (Hillegass, Miller et al. 2013). The secreted IL-1β can then affect mesothelial cells in an autocrine manner, thereby regulating many other pro-inflammatory cytokines including IL-8, IL-6 and G-CSF (Hillegass, Miller et al. 2013). Shannahan et al. (Shannahan, Ghio et al. 2012) have also reported increased IL-1β transcript in lung tissue of Libby asbestos exposed rats. IL-1β is a pro-inflammatory cytokine and has been demonstrated to play significant role(s) in potentiation of asbestos-induced transformation, inflammation and injury of mesothelial cells (Choe, Tanaka et al. 1998; Choe, Zhang et al. 1999; Wang, Faux et al. 2004). Biological effects of IL-1β can be exerted alone or by regulation of other pro-inflammatory cytokines like IL-8, IL-6, VEGF or G-CSF (Tanaka, Kanai et al. 2000; Weber, Wasiliew et al. 2010; Hillegass, Miller et al. 2013). Elevated concentrations of IL-1β and G-CSF are reported in serum and pleural fluid derived from MM patients (Soderblom, Pettersson et al. 1999; Yoshimoto, Kasahara et al. 2005) suggesting importance of these cytokines in MM pathogenesis. Another IL-1 family member cytokine highly expressed uniquely by pleural mesothelial cells in response to asbestos was IL-1α. Although less well studied than IL-1β, reports show that IL-1α may play important roles in regulating fibrinolytic system of mesothelial cells (Mandl-Weber, Haslinger et al. 2001). In addition, Griffith et al. (Griffith, Miller et al. 1994) have shown that IL-8 released in response to asbestos exposure of human pleural mesothelial cells, may in part be mediated by IL-1α. Unique genes in peritoneal mesothelial cells included IL-33, RCAN1, GK, 2NF331 and others. Little is known about the relationship of these genes to asbestos-induced MM.
RNA-Seq data analysis puts emphasis on two points, 1) the fold increases in most of the common genes in response to asbestos, many of which have been shown to be involved in pathogenesis of mesothelioma, were significantly higher in pleural mesothelial cells as compared to peritoneal mesothelial cells, and 2) some of the unique genes highly up-regulated in response to asbestos in pleural mesothelial cells have been shown to be involved in pathogenesis of MM, whereas such a link between unique genes in peritoneal mesothelial cells and MM is not clear. In addition to the genome-wide expression data, our cell viability data suggest that pleural mesothelial cells are more resistant to asbestos-induced toxicity as compared to peritoneal mesothelial cells.
IL-17 signaling is indicated as the most significantly enriched pathway in all mesothelial cell lines tested in the present study. Interleukin-17, a pro-inflammatory cytokine, can promote tumorigenesis by enhancing inflammation and prevent cancer cells from immune surveillance, and is considered to promote colorectal cancer progression (Wu, Wu et al. 2013). IL-17 is also known to promote tumor growth and metastasis by inducing neo-angiogenesis by VEGF pathway (Zarogoulidis, Katsikogianni et al. 2014). A literature search revealed some important links between pro-inflammatory cytokines being released from mesothelial cells in response to asbestos exposure and IL-17. Our present findings, as well as a recent published report (Hillegass, Miller et al. 2013), suggested that asbestos exposure of mesothelial cells can cause release of IL-1 cytokines and these cytokines are known to promote IL-17 production from Th17 and γδ T cells and play important roles in host protective immunity to infection (Mills, Dungan et al. 2013). Another growth factor, G-CSF, up-regulated in response to asbestos exposure to mesothelial cells (Hillegass, Miller et al. 2013) has been shown to play a role in IL-17-mediated peritoneal polymorphonuclear leukocyte accumulation (Witowski, Ksiazek et al. 2007) and thereby inflammation. These reports indicate that factors released by asbestos exposure to mesothelial cells may exert their biological/pathological effects via IL-17. Moreover, in addition to indirect role of asbestos in IL-17 stimulation, there are two recent reports indicating that both erionite and amphibole asbestos induce the production of IL-17 in mice after 7–8 months of exposure (Ferro, Zebedeo et al. 2014; Zebedeo, Davis et al. 2014). No evidence of IL-17 gene expression in response to asbestos exposure in the present experiment could be attributed due to the short time point (8 h) of asbestos exposure as compared to 7–8 months in the published literature. Taken together it may be possible that cytokines and growth factors released early in response to asbestos exposure exert their biological effects via modulating IL-17 later. In addition to IL-17 enriched pathway, IL-10 and IL-6 enriched pathways were also shown to be significant pathways modulated by asbestos by IPA analysis. Importance of IL-6 in asbestos-induced responses has already been discussed. Studies from the literature also show the modulated levels of IL-10 in asbestos exposed population and MM patients suggesting a role of IL-10 in the pathobiology of asbestos-induced diseases (Brunetti, Delmastro et al. 2003; Miura, Nishimura et al. 2006; Maeda, Miura et al. 2008).
To conclude we can say that while we clearly see a similar pattern of response across cell lines and cell sources, we also see a clear difference in the extent of the response between cell lines and cell sources. The pleural mesothelial cells clearly show similar but increased magnitude of response in important cytokines involved with the development of asbestos-induced diseases including MM. In addition, unique genes expressed by pleural mesothelial cells are all reported to be heavily involved in fibrosis and cancer. The increased responsiveness of the pleural cell lines could provide a mechanism with which to explain the greater frequency of pleural mesothelioma as compared to peritoneal mesothelioma. Because of the unavailability of sufficient numbers of gender and age matched samples, this study has limitations. In the future we plan to repeat the study with age-matched samples if and when they become available.
Supplementary Material
Additional file 1. The 165 transcripts commonly differentially expressed across cell lines in response to asbestos exposure (p<0.05, FDR<0.05, and 2X fold change).
Acknowledgments
We thank Dr. Jeffery Bond for his intellectual input in the experimental design, data analysis and critical review of the manuscript. We thank Dr. Jim Vincent of the Vermont Genetics Network for valuable feedback as well. We also acknowledge Dr. Jill Miller’s help with the experiment. Help from the UVM AGTC-DNA core facility is duly acknowledged. This work is supported by the VCC/LCCRO and an NIH RO1 grant ES021110.
References
- Adachi Y, Aoki C, et al. Interleukin-6 induces both cell growth and VEGF production in malignant mesotheliomas. Int J Cancer. 2006;119(6):1303–1311. doi: 10.1002/ijc.22006. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Yoshio-Hoshino N, et al. VEGF targeting in mesotheliomas using an interleukin-6 signal inhibitor based on adenovirus gene delivery. Anticancer Res. 2010;30(6):1947–1952. [PubMed] [Google Scholar]
- Amati M, Tomasetti M, et al. Assessment of biomarkers in asbestos-exposed workers as indicators of cancer risk. Mutat Res. 2008;655(1–2):52–58. doi: 10.1016/j.mrgentox.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 2009;57(1):289–300. Series B (Methodological) [Google Scholar]
- Bolger AM, Lohse M, et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan AM, Ruegg C, et al. Evidence of a role for mesothelial cell-derived interleukin 8 in the pathogenesis of asbestos-induced pleurisy in rabbits. J Clin Invest. 1992;89(4):1257–1267. doi: 10.1172/JCI115710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti G, Delmastro M, et al. Immunocytological and mineralogical study of bronchoalveolar lavage in a group of subjects exposed to asbestos. G Ital Med Lav Ergon. 2003;25(2):152–160. [PubMed] [Google Scholar]
- Choe N, Tanaka S, et al. Asbestos fibers and interleukin-1 upregulate the formation of reactive nitrogen species in rat pleural mesothelial cells. Am J Respir Cell Mol Biol. 1998;19(2):226–236. doi: 10.1165/ajrcmb.19.2.3111. [DOI] [PubMed] [Google Scholar]
- Choe N, Zhang J, et al. Asbestos exposure upregulates the adhesion of pleural leukocytes to pleural mesothelial cells via VCAM-1. Am J Physiol. 1999;277(2 Pt 1):L292–L300. doi: 10.1152/ajplung.1999.277.2.L292. [DOI] [PubMed] [Google Scholar]
- Corradi M, Goldoni M, et al. YKL-40 and mesothelin in the blood of patients with malignant mesothelioma, lung cancer and asbestosis. Anticancer Res. 2013;33(12):5517–5524. [PubMed] [Google Scholar]
- Davidson B, Vintman L, et al. Heparanase and basic fibroblast growth factor are co-expressed in malignant mesothelioma. Clin Exp Metastasis. 2004;21(5):469–476. doi: 10.1007/s10585-004-3150-2. [DOI] [PubMed] [Google Scholar]
- Duncan KE, Cook PM, et al. In vitro determinants of asbestos fiber toxicity: effect on the relative toxicity of Libby amphibole in primary human airway epithelial cells. Part Fibre Toxicol. 2014;11:2. doi: 10.1186/1743-8977-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikawa S, Ohue Y, et al. Enrichment of Foxp3+ CD4 regulatory T cells in migrated T cells to IL-6- and IL-8-expressing tumors through predominant induction of CXCR1 by IL-6. J Immunol. 2010;185(11):6734–6740. doi: 10.4049/jimmunol.1000225. [DOI] [PubMed] [Google Scholar]
- Ferro A, Zebedeo CN, et al. Amphibole, but not chrysotile, asbestos induces anti-nuclear autoantibodies and IL-17 in C57BL/6 mice. J Immunotoxicol. 2014;11(3):283–290. doi: 10.3109/1547691X.2013.847510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith DE, Miller EJ, et al. Interleukin-1-mediated release of interleukin-8 by asbestos-stimulated human pleural mesothelial cells. Am J Respir Cell Mol Biol. 1994;10(3):245–252. doi: 10.1165/ajrcmb.10.3.8117443. [DOI] [PubMed] [Google Scholar]
- Hill GD, Mangum JB, et al. Soluble ICAM-1, MCP-1, and MIP-2 protein secretion by rat pleural mesothelial cells following exposure to amosite asbestos. Exp Lung Res. 2003;29(5):277–290. doi: 10.1080/01902140303788. [DOI] [PubMed] [Google Scholar]
- Hillegass JM, Miller JM, et al. Asbestos and erionite prime and activate the NLRP3 inflammasome that stimulates autocrine cytokine release in human mesothelial cells. Part Fibre Toxicol. 2013;10:39. doi: 10.1186/1743-8977-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillegass JM, Shukla A, et al. Inflammation precedes the development of human malignant mesotheliomas in a SCID mouse xenograft model. Ann N Y Acad Sci. 2010;1203:7–14. doi: 10.1111/j.1749-6632.2010.05554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori-Katayama M, Kaiho A, et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS One. 2011;6(10):e25391. doi: 10.1371/journal.pone.0025391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler HL. Peritoneal mesothelioma: the site of origin matters. Am Soc Clin Oncol Educ Book. 2013;33:182–188. doi: 10.14694/EdBook_AM.2013.33.182. [DOI] [PubMed] [Google Scholar]
- Maeda M, Miura Y, et al. Immunological changes in mesothelioma patients and their experimental detection. Clin Med Circ Respirat Pulm Med. 2008;2:11–17. doi: 10.4137/ccrpm.s577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl-Weber S, Haslinger B, et al. Heat-killed microorganisms induce PAI-1 expression in human peritoneal mesothelial cells: role of interleukin-1alpha. Am J Kidney Dis. 2001;37(4):815–819. doi: 10.1016/s0272-6386(01)80131-1. [DOI] [PubMed] [Google Scholar]
- Mills KH, Dungan LS, et al. The role of inflammasome-derived IL-1 in driving IL-17 responses. J Leukoc Biol. 2013;93(4):489–497. doi: 10.1189/jlb.1012543. [DOI] [PubMed] [Google Scholar]
- Miserocchi G. Physiology and pathophysiology of pleural fluid turnover. Eur Respir J. 1997;10(1):219–225. doi: 10.1183/09031936.97.10010219. [DOI] [PubMed] [Google Scholar]
- Miura Y, Nishimura Y, et al. Involvement of IL-10 and Bcl-2 in resistance against an asbestos-induced apoptosis of T cells. Apoptosis. 2006;11(10):1825–1835. doi: 10.1007/s10495-006-9235-4. [DOI] [PubMed] [Google Scholar]
- Mossman BT, Shukla A, et al. New insights into understanding the mechanisms, pathogenesis, and management of malignant mesotheliomas. Am J Pathol. 2013;182(4):1065–1077. doi: 10.1016/j.ajpath.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsaers SE. The mesothelial cell. Int J Biochem Cell Biol. 2004;36(1):9–16. doi: 10.1016/s1357-2725(03)00242-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Cheung MC, et al. Malignant abdominal mesothelioma: defining the role of surgery. J Surg Oncol. 2009;99(1):51–57. doi: 10.1002/jso.21167. [DOI] [PubMed] [Google Scholar]
- Shannahan JH, Ghio AJ, et al. Transcriptional activation of inflammasome components by Libby amphibole and the role of iron. Inhal Toxicol. 2012;24(1):60–69. doi: 10.3109/08958378.2011.633942. [DOI] [PubMed] [Google Scholar]
- Shukla A, Barrett TF, et al. Transcriptional up-regulation of MMP12 and MMP13 by asbestos occurs via a PKCdelta-dependent pathway in murine lung. FASEB J. 2006;20(7):997–999. doi: 10.1096/fj.05-4554fje. [DOI] [PubMed] [Google Scholar]
- Shukla A, MacPherson MB, et al. Alterations in gene expression in human mesothelial cells correlate with mineral pathogenicity. Am J Respir Cell Mol Biol. 2009;41(1):114–123. doi: 10.1165/rcmb.2008-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Miller JM, et al. Extracellular signal-regulated kinase 5: a potential therapeutic target for malignant mesotheliomas. Clin Cancer Res. 2013;19(8):2071–2083. doi: 10.1158/1078-0432.CCR-12-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderblom T, Pettersson T, et al. High pleural fluid hyaluronan concentrations in rheumatoid arthritis. Eur Respir J. 1999;13(3):519–522. doi: 10.1183/09031936.99.13351999. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kanai H, et al. Induction of VEGF gene transcription by IL-1 beta is mediated through stress-activated MAP kinases and Sp1 sites in cardiac myocytes. J Mol Cell Cardiol. 2000;32(11):1955–1967. doi: 10.1006/jmcc.2000.1228. [DOI] [PubMed] [Google Scholar]
- Thompson JK, Westbom CM, et al. Malignant mesothelioma: development to therapy. J Cell Biochem. 2014;115(1):1–7. doi: 10.1002/jcb.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaeminck-Guillem V, Bienvenu J, et al. Intraperitoneal cytokine level in patients with peritoneal surface malignancies. A study of the RENAPE (French Network for Rare Peritoneal Malignancies) Ann Surg Oncol. 2013;20(8):2655–2662. doi: 10.1245/s10434-013-2933-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Faux SP, et al. Interleukin-1beta and tumour necrosis factor-alpha promote the transformation of human immortalised mesothelial cells by erionite. Int J Oncol. 2004;25(1):173–178. [PubMed] [Google Scholar]
- Weber A, Wasiliew P, et al. Interleukin-1 (IL-1) pathway. Sci Signal. 2010;3(105):cm1. doi: 10.1126/scisignal.3105cm1. [DOI] [PubMed] [Google Scholar]
- Witowski J, Ksiazek K, et al. Role of mesothelial cell-derived granulocyte colony-stimulating factor in interleukin-17-induced neutrophil accumulation in the peritoneum. Kidney Int. 2007;71(6):514–525. doi: 10.1038/sj.ki.5002082. [DOI] [PubMed] [Google Scholar]
- Wu D, Wu P, et al. Interleukin-17: a promoter in colorectal cancer progression. Clin Dev Immunol. 2013;2013:436307. doi: 10.1155/2013/436307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto A, Kasahara K, et al. Granulocyte colony-stimulating factor-producing malignant pleural mesothelioma with the expression of other cytokines. Int J Clin Oncol. 2005;10(1):58–62. doi: 10.1007/s10147-004-0432-2. [DOI] [PubMed] [Google Scholar]
- Zarogoulidis P, Katsikogianni F, et al. Interleukin-8 and interleukin-17 for cancer. Cancer Invest. 2014;32(5):197–205. doi: 10.3109/07357907.2014.898156. [DOI] [PubMed] [Google Scholar]
- Zebedeo CN, Davis C, et al. Erionite induces production of autoantibodies and IL-17 in C57BL/6 mice. Toxicol Appl Pharmacol. 2014;275(3):257–264. doi: 10.1016/j.taap.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The 165 transcripts commonly differentially expressed across cell lines in response to asbestos exposure (p<0.05, FDR<0.05, and 2X fold change).