Abstract
The growth and development in the last decade of accurate and reliable mass data collection techniques has greatly enhanced our comprehension of cell signaling networks and pathways. At the same time however, these technological advances have also increased the difficulty of satisfactorily analyzing and interpreting these ever-expanding datasets. At the present time, multiple diverse scientific communities including molecular biological, genetic, proteomic, bioinformatic, and cell biological, are converging upon a common endpoint, that is, the measurement, interpretation, and potential prediction of signal transduction cascade activity from mass datasets. Our ever increasing appreciation of the complexity of cellular or receptor signaling output and the structural coordination of intracellular signaling cascades has to some extent necessitated the generation of a new branch of informatics that more closely associates functional signaling effects to biological actions and even whole-animal phenotypes. The ability to untangle and hopefully generate theoretical models of signal transduction information flow from transmembrane receptor systems to physiological and pharmacological actions may be one of the greatest advances in cell signaling science. In this overview, we shall attempt to assist the navigation into this new field of cell signaling and highlight several methodologies and technologies to appreciate this exciting new age of signal transduction.
Keywords: Signaling, Network, Pathway, Phenotype, Receptor
1. Introduction
1.1. The Relentless Progression in Complexity
Many research scientists familiar with signal transduction research have in recent years realized that despite their enhanced output technologies, genomic, proteomic or metabolomic, they often consider themselves somewhat hampered by analytical techniques that do not seem able to adequately appreciate mass datasets. Our consideration of the nature of signal transduction systems has likely forever moved away from linear enzymatic cascades with near-Brownian modes of motion of individual signaling factors in intermediary metabolic systems. Current hypotheses, of at least receptor-mediated signal transduction pathways, include the presence of substate-specific isoforms of receptors coupled to preassembled signal transduction cascades consisting of subtype-specific, stable multiprotein signaling complexes that possess distinct subcellular targeting mechanisms (1, 2). Despite this conversion of thinking and the wider appreciation of the inherent increase in the complexity of signaling systems, the potential for hindrance of pharmacological research has not been seen, actually quite the reverse. The more subtle our appreciation of the intricate nature of receptor response mechanisms and their contextual variety, then the more selective and specific rationally designed pharmacotherapies may become (3, 4).
With the ability to rapidly and accurately measure multiple differences (genomic or proteomic) between either physiological or drug-induced states, our appreciation of the complex nature of biological processes has forced us to consider that often physiological disorders or drug responses are mediated by alterations in whole gene/protein networks, as opposed to simple activation or inhibition of a linear signal transduction pathway. A common phrase often used to describe this changing mindset in molecular biology is “pathways no longer exist, there are only networks.” This statement however does not negate the many years of prior signal transduction research but suggests perhaps that the delineation of discrete signaling pathways is likely an abstraction of the true hyper-complex signaling network due to our previous deficiencies in analytical technology. There are a huge variety of efficient and sensitive techniques which an investigator can use to assess genomic or proteomic differences in distinct pathophysiological or pharmacological scenarios, including fluorometric gene array analysis, genome-wide association screening and massive parallel sequencing, ChIP(chromatin immunoprecipitation)-on chip, antibody arrays, protein-binding microarrays, differential in-gel electrophoresis and quantitative mass spectrometry (MS). These techniques have been thoroughly discussed in recent years and therefore will not be repeated here. These era-changing technologies, however, often leave experimenters feeling lost in a mass of data that may or may not contain the specific scientific answers they are seeking. The application of biologically relevant mathematical processes to divine the eventual physiological meaning of these datasets will be the primary subject of this overview. We intend to provide a simple primer that researchers can use as a reference for interpretation of their complex datasets. The analytical tools and processes described will be applicable to both genomic and proteomic data and will hopefully facilitate a more holistic understanding of the creation and eventual pharmacological targeting of signal transduction networks. The primary goal of these bioinformatic analytical tools is the rational and biologically relevant condensation of these mass data lists into outputs that may predict the functional activities of the genes/proteins modulated between the control and test datasets. The clustering of gene/protein factors into functional groups or even signaling pathways will help to categorize characteristic gene/protein sets for future diagnostic and therapeutic use. Therefore in the future patient diagnosis, drug development, testing, and design may all take place initially at the signaling network level rather than at the single gene/protein measurement index level.
We shall consider the most commonly used techniques to extract functionally relevant and experimentally actionable information from mass data lists and then describe the most apt future uses of these paradigms. Even before more complex functional analysis can begin we shall discuss several important considerations with respect to the initial generation of the dataset and the relative merits and detractions of genomic/proteomic techniques.
1.2. Textual Definitions
In this overview, we shall consider both gene and protein datasets and will describe both as the same, that is, “dataset.” For most postexperimental analytical algorithms we find that the Gene Symbol nomenclature often provides the most reliable and flexible gene/protein annotation platform and therefore we shall primarily consider these in this overview. Individual genes or proteins will be individually and interchangeably described as “factors” in this overview.
2. Extracting Multiple Relevant Factors from Datasets
Since the advent of facile technologies that can generate large complex datasets, the primary goal of such experiments has been to identify many relevant factors (gene or protein) that may explain the pathophysiological outcome or drug response in the experimental paradigm. Typically, a single control and one or multiple test conditions are analyzed in a simple comparative manner. After the creation of the first readily available gene arrays, the primary data selection processes applied to these datasets were developed by classical statistical analysis (5). With respect to modern fluorometric gene arrays such as Illumina and also to quantitative proteomic techniques, the initial choices for data filtration are distinct due to the unique properties of either of the mass analytical techniques. Many of the analytical modes can be swapped between genomic or proteomic platforms but one must always take into account that often mass spectrometry is a discovery process while gene (and also antibody or protein) microarrays provide a standard reproducible platform for each experiment. The functional annotation of datasets provides an invaluable approach for divination of the physiological “meaning” of the output but specifically in the case of mass spectrometry proteomics provides a vital support for analysis of variability of function between experiments. This important aspect of functional annotation of proteomic data will be expanded upon in subsequent sections.
2.1. Fluorescent Microarrays
Using differential fluorescent dye attachment (typically Cy3 or Cy5) relative quantitative changes in mRNA expression are easily obtainable on a large scale (6, 7). As with most technologies based upon fluorescent dye usage, the presence of background residual signal can be problematical. Subtraction of such background intensity is achieved by statistically computing the average background intensity and using the standard deviation among this intensity to calculate a confidence interval, the upper limit of which is used for the subsequent background correction. To assist the comparison of multiple gene regulation profiles between microarray chips, normalization of the data is paramount. One of the most common methods employed for normalization of the respective gene fluorescent signal is the use of “housekeeping” genes. The valid employment of housekeeping genes to normalize biologically relevant fluctuating data on the array relies on the assumption that there is a set of standard genes whose expression does not change with experimental condition or ligand stimulation. However, with respect to our current thinking of physiological response/signal transduction networks, the concept of a nonchanging factor on the array unfortunately becomes less and less likely. Clearly, there will be a spectrum of perturbation of factors on the array and some genes may indeed be unperceivably altered and thus provide a de facto basis for normalization. It is likely though that in the next few years the reliance upon “housekeeping” factors will be an increasingly redundant concept even though it may be practically effective. Internal spotted standards of a control factor, for example, bovine serum albumin, can often provide an adequate control for the output from the assay chip instead of using an experimental sample. However, this merely controls for experimental detection process itself and not the differential factor data per se. An alternative approach though is the more reliable use of whole-array normalization. Typically, whole-array normalization is performed using linear or logarithmic regression techniques (8–12). The reliability of this process is likely to be affected by the network connectivity of the targets under study and the target selectivity of the experimental effect(s). This whole-array normalization also relies upon a potentially anachronistic assumption, that is, the majority of genes on the array are nondifferentially expressed between the experimental states, and that varying genes are not solely associated with one of the fluorescent labels. The latter assumption can be checked easily by dye-swapping paradigms in which fluorescent labels are reversed and experimental data obtained again. This can also be applied to quantitative proteomic technologies that we shall describe in later sections. As mentioned previously this assumption that there is only a minimal perturbation of genes on the array constructively reinforces our old concept of linear discrete signaling pathways. Practically, however, this technique may still yield the production of a de facto valid data set based on the “broadness” of the spectrum of variation in the response to the experimental actions (Fig. 1). To further prepare microarray data for functional analysis, it is typical to apply a log transformation to the fluorescent data to make numerical manipulation more acceptable. Parametric tests used for statistical analysis of the factor variation are the most commonly utilized, as these tests are much more sensitive and require the data to be normally distributed. This is usually achieved by using log transformation of the spot intensities to achieve a Gaussian distribution of the data. To extract the actual differential expression profile of genetic factors from microarray data, a ratio of intensity (as a measure of expression level: z-ratio) between two samples is used. As with all biological experiments, replicates of array data are required if a fold-change cutoff of z-ratios is used to primarily filter the data set. Several model-based techniques have been developed that facilitate the assumption of multiplicative noise, and eliminate statistically significant outliers from the data (13). The typical parametric analytical methods applied to primary gene array data management include maximum-likelihood analysis, F-statistic, ANOVA (analysis of variance), and t-tests. The results of these tests are often improved by the log transformation of the primary data. Nonparametric tests used to analyze microarray data include Mann–Whitney tests (14) and Kruskal–Williams rank analysis (15). The primary goal of the initial statistical analysis of the array data is the calculation of significance values for gene expression, most commonly as a “p-value.” P-values, either fixed to 0.05 or 0.01 are then employed to reduce the dataset to significantly regulated gene lists before z-ratio/fold-change cutoffs are applied (typically ±1.5) as well as provisions for false data creation which are highly likely when large arrays are used. Protocols for the elucidation of random false results calculate the overall chance that at least one gene is a false-positive or -negative, that is, the family-wise error rate (16). Erroneous data discovery from arrays can also be assessed using the Bonferroni approach, that is, this technique multiplies the uncorrected p-value by the number of genes tested, treating each gene as an individual test. This protocol can increase significant data specificity by reducing the number of false-positives identified, but unfortunately attenuates the array sensitivity by increasing the number of false-negatives. A modification of the Bonferroni approach, the false-discovery rate (FDR), uses a random permutation while assuming each gene is an independent test. In addition, bootstrapping approaches can improve significantly on the Bonferroni approach, as they are less stringent (17). Resampling-based false discovery rate-controlling procedures can also be used (18). These array data extraction protocols can be applied to other array platforms, for example, antibody or protein arrays, as essentially the chip data can be easily analogized. However, one caveat is of course required, that is, the likelihood of high logarithmic increases in protein expression is highly unlikely as even a twofold change of protein expression may be sufficient to generate profound signaling actions, especially if the protein possesses enzymatic activity.
Fig. 1.
Contextuality of dataset housekeeping reliability. Accepting a high level of connectivity of signaling factors introduces the likelihood of disruption of potential “housekeeping” factors. In paradigm A where a relatively selective activation of a target that possesses only minimal connectivity with the greater network of factors does not perceptibly disrupt the chosen housekeeper and therefore creates a de facto housekeeping factor. However, in paradigm B where the target factor is multiply connected to other factors in the network an increased likelihood of the loss of housekeeper reliability is seen (a). The potential effects of the connectivity in the network of the target factor and the target selectivity of a biological perturbing action (α, highly selective acting on minimal targets, β moderately selective acting on several targets, γ poorly selective acting on multiple targets). Highly connected targets possess a greater chance of disrupting housekeeping reliability and perturbations to the network that are nonselective are also likely to disrupt housekeeping reliability (b).
2.2. Quantitative Mass Spectrometry
The primary contrast between proteomic datasets and those from array experiments is the expectation of inclusion of certain data-points, that is, proteins. Standard arrays provide a reproducible experimental platform while the recovery of the same protein between experiments is often unlikely. The use therefore of pathway bioinformatics, which can infer function from a variety of related proteins and not just based on individual identity, in such experiments may be paramount for the future use of proteomics. There are also recent advances in MS-based technologies that can be applied to mass spectrometers that can facilitate the accurate selection of protein species to be identified from a desired list (selective reaction monitoring, SRM; 19) in-part recreating the desired scanning pattern of an array. Such specific monitoring modes of MS may considerably slow down the rate of data retrieval and may only be suitable for experiments in which high levels of starting extract are available. In contrast to array technology though, the detection through SRM is still dependent on the ability of the MS to physically detect the specified peptides. This detection reliability is often more likely to demonstrate experiment to experiment variability than gene array platforms.
In this overview, our major focus is upon the functional interpretation of gene/protein datasets using bioinformatic approaches and therefore we shall focus upon the most commonly used current quantitative proteomic technique, that is, isobaric mass-tag labeling.
Mass-tag labeling (Fig. 2), for example, iTRAQ (isobaric tag for relative and absolute quantitation), SILAC (stable incorporation of labeled amino acids in culture) or SILAM (stable incorporation of labeled amino acids in mammals), allows the rapid ratiometric analysis of multiple peptides separated by multidimensional cation-exchange liquid chromatography (LC) identified with either time-of-flight (TOF) or linear ion-trap tandem mass spectrometry (LC-MS2) with modified dissociation techniques such as PQD (20) and HCD (21). These instruments, and the diverse workflows they support, have in common that they both generate up to thousands of fragment ion spectra per hour of data acquisition. The assignment of these fragment ion spectra to peptide sequences, the inference of the proteins represented by the identified peptides and the determination of their abundances in the analyzed sample present complex computational and statistical challenges. It is important for the future use of MS and proteomics in metabolic signaling analysis to develop technological solutions to these issues that provide accurate and reproducible quantitative differential protein expression data. To this end, one of the major advances will be the application of accurate functional annotation and categorization into metabolic pathways of the protein sets created. As MS generally does not provide a factor identification process as reliable as microarrays, the physiological and rational prediction of the signaling consequences of the protein streams will facilitate experiment to experiment comparison.
Fig. 2.
Principle of isobaric mass-tags in quantitative mass spectrometry. (a) Several combinations of different-sized reporters of iTRAQ tags facilitate quantification of up to 8 different samples (masses from 113 to 121, excluding 120 as this corresponds to phenylalanine). Quantitative information is obtained from relative intensities of reporter ions in MS/MS spectrum. TMT (tandem mass tag: Thermo Electron Corporation) has the same property with iTRAQ but has different reporter and balancer chemistry. (b) In SILAC, isobaric amino acids are metabolically incorporated into all the cellular proteins. Animals can be fed and bred through multiple generations using feed with differential amino acid composition [SILAM: 60]. The equal amount of samples are combined and then applied to LC-MS/MS analysis. Quantitative information is obtained from relative intensities of light- and heavy-peptide ions in MS spectrum. (c) A representative analytical procedure of quantitative MS. In the bottom-up approach, complex peptide mixtures are fractionated through strong cation-exchange chromatography (SCX), which is essential for reducing sample complexity and increasing the number of identified peptides. Each fraction is analyzed through reverse-phase (RP) LC-MS/MS. For the nonisotopic study, quantitative information is obtained through peak intensity of specific peptides in ion chromatogram and more widely through counting finally matched MS/MS spectra and statistical manipulation. In case of using isobaric-tags, differentially labeled samples are combined before SCX chromatography. Quantitative information is obtained from MS or MS/MS spectrum, dependent on the property of isobaric tag. (d) Modes of sample preparation, labeling, and mixing for MS analysis. For mass-tag labeling procedures such as iTRAQ the individual extraction of proteins, then peptides from each sample is followed by individual mass-tag labeling and then mixing for single-run MS analysis. For stable isotope incorporation procedures, sufficient cell passages or animal generations in the presence of differential isotopes is required before mixing for single-run MS analysis.
In contrast to array-based technologies, the primary concerns for MS-based dataset creation approaches involves the actual accurate identification of the proteins in the sample, for example, control versus test. For TOF and LC-MS2 the identification of proteins in the sample is based upon fragmentation ion spectrum (MS2-spectrum) of a specific peptide ion that is broken down into its constituent components in a gas-filled collision cell. Due to the enormous complexity of peptides composed of 20 amino acids, however, a large number of MS/MS spectra do not contain sufficient identity information to allow error-free peptide definition. In order to minimize false identification, a strict filtering criterion is required, which can be enforced, for example, by searching retrieved MS/MS spectra against a composite of both “target” and “decoy” (often reverse peptide alignments) sequence database (22). Much of the statistical manipulation used for protein datasets has focused upon the actual generation of the identified protein list rather than on the bioinformatic/pathway structure of the resultant data list itself. In recent years, however, with the advent of sophisticated automated identification software more attention is now paid to the physiological relevance of the mass datasets. The correct correlation and attribution of an MS2-spectrum to its originating peptide sequence followed by eventual protein matching and identification is the first and central step in proteomic data processing. Numerous computational approaches and software tools have been developed to automatically assign candidate peptide sequences to fragment ion spectra, for example, SEQUEST, MASCOT, ProteinProspector, or ProbID (23–26). These computational approaches can involve database searching, where peptide sequences are identified by correlating acquired fragment ion spectra with theoretical spectra predicted for each peptide contained in a protein sequence database, or by correlating acquired fragment ion spectra with libraries of experimental MS2 spectra identified in previous experiments. In addition de novo sequencing can also be used, where peptide sequences are explicitly read out directly from fragment ion spectra as well as hybrid computational approaches, such as those based on the extraction of short sequence tags of three to five residues in length, followed by “error-tolerant” database searching (27). For the majority of signal transduction laboratories, database searching remains the most frequently used peptide identification method. The use of MS-based techniques to identify quantitative protein profiles from animals/tissues has been excellently reviewed elsewhere (28–30) and therefore the focus of the rest of this overview is the predictive pathway analysis of mass datasets either from MS- or array-based experiments to appreciate factor expression at a network level.
While the accurate and unbiased collection of factor data is paramount, one extremely important caveat with respect to data retrieval and metabolic pathway analysis, is the need to physically retain both significant and nonsignificant factor data. The nature of the “nonsignificantly regulated” data may yet yield significance when the co-existence of related factors is analyzed using functional annotation-based bioinformatic strategies. Often subtle differences between experimental conditions may be missed as no individually dramatically modulated factors may present themselves. If, however, we consider our posit that metabolic and signaling functions are indeed composed of multiple interlaced network activities, the appreciation and functionally relevant correlation of these small changes with each other may illuminate a more realistic view of cellular physiology.
3. Bioinformatic Analysis of Quantitative Mass Analytical Datasets
With application of an initial data-filtering statistical analysis to each factor individually (compared to background), it is frequently the case that a large (100–1,000s) dataset of significantly regulated factors remains. In the first decade of mass biological data analysis only the highest and lowest regulated factors were often considered for further analyses. This approach, despite yielding some actionable data to describe the signaling function or physiological state under study, is often criticized for ignoring the correlated biological relevance of the multiple factors arranged in the large dataset that do not individually demonstrate significant differential regulation. Hence, we assume that genes and proteins function together and interact with each other in relevant groups and in specific microdomains but the analysis of the datasets often does not include this biologically vital information. However, if we consider that functional signaling responses or physiological states are the functional composite of multiple linked networks then an appreciation of the entire set in a mechanism analogous to signaling networks is needed. Gene-class, or pathway-level testing, integrates factor annotation and significance signaling pathway population tests (with geneset enrichment analysis) for coordinated changes at the system level. These approaches can both increase power for detecting differential factor expression and allow for a better understanding of the underlying biological processes associated with variations in signal transduction outcome. One of the earliest developed processes that allowed facile classification of factor function was Gene Ontology (http://www.geneontology.org/index.shtml) analysis.
3.1. Gene Ontology Classification
To create a rational and physiological/pharmacologically relevant appreciation of large datasets the first most reasonable goal is to look for methods in which to cluster the factors that are related to each other either by function, linkage in a metabolic process, or by subcellular localization. The number of these associations and the strength of observing multiple factors possessing the same associations within a large dataset provides the first level of “contextual” relevance of the mass dataset. An exemplar of the importance of elucidating common functional attributes for factors would be a protein such as actin, which conceivably may be directly involved in approximately 90% of all cellular processes either directly or distant by just one level from nearly all the factors in the dataset. To begin to appreciate what particular functional relevance the presence of actin has in one's dataset, the ability to look for functional groups in which to assign actin would start to narrow down the number of functional effects that the experimental changes in actin may be inducing. One of the primary levels of analysis of mass datasets to yield functional metabolic insights into its nature is the use of functional Gene Ontology (GO) analysis.
After many of the genomes of the major experimental eukaryotic organisms were fully sequenced, it became clear that a large majority of the genes controlling the fundamental biological processes and signaling pathways were common across multiple species. Therefore, an analytical method to allow inference and analogy of data between the diverse experimental organisms was required to potentially identify conserved signaling mechanisms. The GO project is an ongoing academic effort to address the need for consistent descriptions of gene products in different databases. The project began in 1998 as a collaboration between three model organism databases, FlyBase (Drosophila: http://flybase.org/), the Saccharomyces Genome Database (SGD: http://www.yeastgenome.org/) and the Mouse Genome Database (MGD: http://www.informatics.jax.org/). Since inception, the GO Consortium has grown to include many databases, including several of the world's major repositories for plant, animal, and microbial genomes. Functional biological knowledge is inherently complex and so cannot readily be integrated into existing databases of molecular (for example, sequence) data. An ontology is a formal way of representing knowledge in which concepts are described both by their meaning and their relationship to each other. Unique identifiers that are associated with each concept in biological ontologies (bio-ontologies) can be used for linking to and querying molecular databases.
The Gene Ontology Consortium (http://www.geneontology.org/GO.doc.shtml) was developed to provide a dynamic and controllable functional terminology syntax that can be used to accommodate the exponential increase in knowledge of factor connectivity in functional metabolic pathways. To initiate a mechanism by which factors (genes initially) could be associated with an expanding list of signaling functions, three major ontological databases were created, freely available on the internet (http://www.geneontology.org). These three databases would assist in assigning biologically relevant information to identified factors so that associations between functions and factors in a dataset can be ascertained and the relative significance of these within the dataset can be assessed. Biological Gene Ontology has two fundamental components: the ontologies themselves, which are the defined terms and the structured relationships between them (GO ontology), and the associations between gene products and the terms (GO annotations). GO provides both ontologies and annotations for three distinct areas of cell biology: molecular function, biological process, and cellular component or location.
3.2. Gene Ontology Categorization
The three main GO categories commonly used to cluster factors into related and biologically relevant groups are as follows: biological process (GObp), molecular function (GOmf), and cellular component (GOcc). Biological process, molecular function, and cellular component are all attributes of genes, gene products, or gene-product groups. Each of these may be assigned independently to factors in a dataset. The relationships between a given factor and biological process, molecular function, and cellular component are one-to-many, reflecting the biological reality that a particular protein may function in several processes, contain domains that carry out diverse molecular functions, and participate in multiple alternative interactions with other proteins, organelles, or locations in the cell. Within all of these three subgroups, there are hierarchies of GO terms ranging from extremely broad categories that can encompass hundreds of factors to GO terms that may only be associated with a handful of factors. An ontology comprises a set of well-defined terms with well-defined relationships. The ontological structure itself reflects the current representation of biological knowledge and therefore should be considered highly plastic and can act as a guide for organizing new data. Data can be annotated to varying levels depending on the amount and completeness of the available information. This flexibility also allows users to narrow or widen the focus of queries (31). The Gene Ontologies are formalized representations of current molecular and cellular biology knowledge. The GO ontology functional classification structure can be represented as a directed acyclic graph (DAG) in which the terms are nodes and the relationships among them are edges. Key characteristics of a DAG in the context of GO are that: parent–progeny relationships are defined, with parent terms representing more general biochemical functions than their progeny terms; and, unlike a simple tree (Fig. 3), a term in a DAG can have multiple parents. These characteristics of the GO structure facilitate facile grouping, searching, and analysis of multiple relevant factors.
Fig. 3.
Representation of ontological structures. Ontology of biologically relevant factors can be represented in a simple graphical structure in which parent Gene Ontology terms give rise to progeny terms (a). Parent terms are typically of a broad nature with their successive progeny possessing increasingly specific annotation (level 1 to 4). This simple graphical ontology representation though can be governed by both directed and nondirected rules. Directed ontological relationships imply a classical hierarchical parent–progeny linking between the terms, that is, parent–progeny relationships are directed downward from less complex terms to more complex terms (black arrows, panel A). However, as broad-level parent terms may lead to multiple more specific ontological terms the simple one-parent one-progeny relationship may be less likely to reflect physiological systems than the one-parent multiple-progeny ontology (b). Undirected ontological representations, however, may allow nondirected progeny to parent relationships (c). Undirected representations may lead to cyclic closed relationship loops. If, however, all of the ontological relationships are directed then it is possible to represent biological linkages into a directed acyclic graph (DAG). (d) An example of an actual DAG from input signaling data. The three major classes of ontology (GObp, GOmf, GOcc) are shown. GO term specificity increases with descent into progeny branches of the DAG. Therefore, the most statistically significantly populated ontology terms are found in the lowest areas of the DAG diagram (e.g., circled GO term groups).
GObp terms refer to biological objectives to which the factor contributes. The process is accomplished via one or more ordered assemblies of molecular functions. The specific functional processes often involve a chemical or physical transformation of a protein or a gene, for example, broad (high level) GObp terms are “cell communication” or “negative regulation of cellular process.” Examples of more specific (lower level) process terms include, “pyrimidine metabolism” or “cAMP biosynthesis” and the most specific GObp terms include items such as “cytoplasmic sequestering of transcription factor” or “protein import into mitochondrial matrix.” GOmf terms are defined as a biochemical activity (including specific binding to ligands or structures) of an individual factor. This definition also applies to the capability that a factor carries as a potential. GOmf terms describe only what the factor can carry out without specifying where or when the biochemical event actually occurs. Examples of broad functional terms are “enzyme,” “transporter,” or “ligand.” Examples of narrower functional terms are “Insulysin activity” or “Peptide YY receptor activity.” GOcc terms refer to the subcellular localization in the cell where the given factor is active. GOcc terms includes such terms as “ribosome” or “proteasome,” “nuclear membrane” or “Golgi apparatus” specifying where multiple factors would be found. An important note, however, with respect to the usage of GO terms is the fact that due to the multispecies nature of their inception, GO terms may often not be fully transferable across species boundaries. Therefore, not all GO terms are applicable to all organisms; however, the full gamut of GO terminology is meant to be as inclusive as possible.
3.3. Application of Gene Ontology Annotation
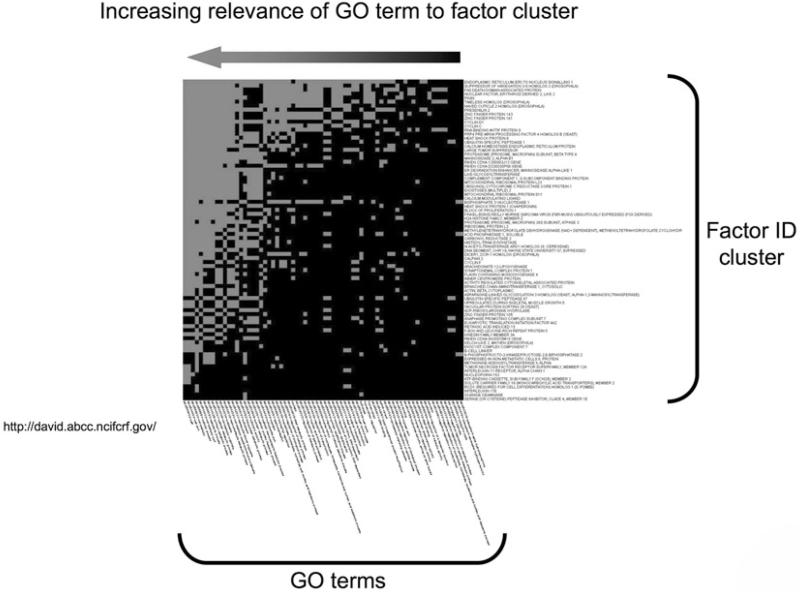
The GO project is currently one of the most widely used biological annotation databases for bioinformatic computational analyses. Upon interrogation of NCBI-Pubmed (http://www.ncbi.nlm.nih.gov/sites/entrez) there are currently over 2605 publications citing gene ontology as a crucial technique in functional signaling annotation, despite the first citation only occurring in 1997. GO annotation of datasets has been demonstrated to be vital for a variety of applications, for example, genome sequencing (32), network modeling (33), text data mining (34, 35), and for applied clinical situations (36). One of the first large-scale applications of GO term analysis of mass datasets was the creation of gene-GO term matrices, generating heatmap structures, to annotate sections of the Drosophila Melanogaster genome (37, 38). The ability to show increases in relevance (demonstrated by heatmap clusters) of certain GO terms ascribed to a subfamily of factors often represents the first level of revelation of the potential functional outputs of the experimental dataset (Fig. 4). The application of the appropriate GO terms to a dataset of significant factors is the first step in the process by which the statistical elucidation of the most likely clustering of the factors to a certain set of GO terms that can predict biologically relevant actions. There are now a plethora of excellent computational devices to achieve this first level of dataset functional analysis (Table 1). For the majority of the analytical tools indicated in Table 1, GO term annotation is used to analyze results from mass analytical techniques, primarily gene arrays but also more recently from quantitative proteomic studies. For these datasets, GO annotations are applied to greatly simplify and to determine which biological processes, functions and/or cellular locations are significantly over- or under-represented in the whole group of factors. This classification facilitates the determination of what new functions can be inferred on the basis of the data and how the given factors are distributed across a predefined set of biological GO term categories. As the primary goal of analysis of mass datasets is the revelation of physiologically/biologically relevant predictive functions that are distinct between the control and experimental scenarios, a quantitative assessment of the presence or absence of certain GO term groups is vital. The relative over- or under-representation of certain GO term groups can then be statistically assessed using various techniques.
Fig. 4.
Heatmap clustering for Gene Ontology annotation. Functional annotation of factor datasets using analytical tools such as DAVID (Database for Annotation, Visualization and Integrated Discovery: http://david.abcc.ncifcrf.gov/) allow the creation of visual factor heatmap clusters according to their most commonly descriptive GO terms. A large input dataset is broken down into smaller clusters that demonstrate commonality of related GO terms. The degree of correlation intensity between the input factors and the GO terms that most closely link the majority of the factors is demonstrated by the increased presence of correlating blocks (grey). Hence, in the figure depicted the GO terms (arranged horizontally) on the far left (end of arrow) are more likely to describe the functional output of the vertically arranged factor list.
Table 1.
Computational programs for Gene Ontology term analysis of large datasets
3.4. Functional GO Term Enrichment and Categorization
Clustering of functionally correlated factors into common GO term groups can be used to infer which specific signaling functions the genes/proteins may be creating. The co-expression of these factors and the most common similarities in their functional common GO term annotation can demonstrate a potential predictive output of the dataset. The goal of mass analytical experimentation is the generation of differential datasets that, with variable isolation, can be linked to a biochemical function, physiological response, or even an organismal phenotype. This generation of a functional signaling “profile” of the dataset will allow correlation of factor expression to resultant function, with the most profoundly enriched factor clusters in the dataset being more reliably linked to the resultant output. Practically the “profile” of the dataset is often conducted by determining which GO terms are represented differently, in a significant fashion more or less often than expected by chance within the factor set compared to say their expression in a reference set (39–42). The most commonly applied approach for this is the calculation of “enrichment” for each GO term (i.e., a higher proportion of factors with certain common annotations among the differentially expressed factors than among all of the background factors in the study). The main problem here is that any enrichment value can occur just by chance. Therefore, enrichment alone should not be interpreted as unequivocal evidence implicating the GO term in the phenomenon studied without application of an appropriate statistical test. More sophisticated approaches calculate the probability of observing a particular enrichment value just by chance using a binomial model (43). This is a good approximation for large reference sets (e.g., whole-genome microarrays). However, it has been demonstrated that in many practical examples, better-suited models include the hypergeometric distribution or the Chi-squared (44) distribution, both of which take into consideration how the probabilities change when a factor is picked. More recent approaches perform the analysis while considering information about the relative position of the GO terms in the hierarchical tree (Fig. 3, 45–47).
Two types of questions can be addressed when performing functional GO term profiling: hypothesis-generating queries, for example, “which GO terms are significant in a particular set of factors?” or hypothesis-driven queries. An unbiased search for significant GO term associations can be performed with a standard “bottom-up” approach: for every progeny GO term, p-values for the factors are directly associated with it. If any term is significant, then analysis is not propagated to factors above it in the hierarchy. This would provide the most specific node that is significant in that particular DAG branch. If a term is not significant, the annotations are propagated to its parent and are recalculated with the parent term. The factor analysis will then propagate upward until a significant node is found or until the root is reached. To minimize false discovery rates, it may be more prudent in the future to precollapse many of the possible DAG branches to prevent “overtesting” of the dataset. To do this, a specific section of the tree organization may be reduced before any p-values are calculated, on the basis of the biological hypotheses tested. Unfortunately, most tools that are currently available are limited to performing analysis either at a fixed depth or with all nodes, thus preventing the customized collapsing of the GO that could improve significance in most circumstances. However, one of the more recently developed GO term analytical tools, QuickGO, was created to specifically facilitate this form of flexible analysis (31). QuickGO (http://www.ebi.ac.uk/QuickGO) allows users to individually tailor annotation sets using multiple filtering options as well as to construct specific and targeted subsets of the GO terms, called “GO slims” to “map-up” annotations allowing a general overview of the attributes of a set of factors. Collections of initial enriched GO terms primary dataset analysis can then be employed to construct a desired GO slim analytical subset. Broad “first pass” analysis annotations can then be “mapped up” or “slimmed” to these selected GO terms. Predetermined GO slims created by groups in the GO Consortium can also be used. However, it is likely for anything other than primary discovery analysis that the majority of users in the future will be primarily interested in using their personal GO slims based on empirical data from other experimental sources.
Another common application of GO is to categorize genes on the basis of a relatively small set of heavily factor-populated high-level GO terms. Results of the functional categorization are frequently shown as pie charts or bar charts (48) based on the number or p-value of the factors present in that GO term group from the primary dataset. This involves the mapping of a set of annotations for the factors of interest to a specified subset of high-level GO terms. This is a typical way of providing an overview of the broad biology encoded by a differential expression patterns (49).
4. Geneset Enrichment and Pathway Analysis
While GO-based annotation techniques provide an excellent appreciation of the biologically relevant biases in a dataset there are additional, more in-depth, formats that can be applied to mass datasets. For example, analysis can be focused upon individual chemical molecular activity, promoter and regulatory network analysis, or by employing the vast-accumulated knowledge from the literature to carry out metabolic signaling pathway analysis. Signaling pathway analysis focuses on physical and functional interactions between factors within a preset signal transduction framework rather than taking the factor-centered view of GO-based database analyses (50). The simplest forms of pathway analysis analyze the distribution of factors within the dataset into precompiled functional signaling pathways in order to elucidate the most likely functional signaling relationships between the individual factors in the dataset. This is typically conducted using a process termed geneset enrichment analysis (GSEA). As this was primarily developed for genomics, the term GSEA has remained although this can be directly applied to proteomic data as well. GSEA typically employs predefined factor sets to identify significant biological changes in microarray/proteomic datasets. The EcoCyc database was perhaps one of the first computational attempts to methodically apply pathway analysis (51, 52). There are various efforts aimed toward the establishment of an accepted standard or ontology to represent functional pathway data. Defined signaling pathways usually include three major classes, (1) the molecules involved in the pathways, (2) the chemical reactions in which these molecules are involved, and (3) the location of the reactions. A pathway ontology should not only represent all these three classes of data, but also capture the intricate relationships among them. For example, a molecule can be related to a reaction as a reactant or a product. The transition from a reactant to a product can be affected by another molecule called a modifier. The modifier can exert various effects to the transition, such as catalysis, stimulation, inhibition, or modulation. Furthermore, the relationship between reactions and cellular components describes the location of these reactions. Such a higher level of functional correlation cannot be adequately captured using GObp as it does not capture all the dynamic inter-relationships in the pathways.
4.1. Statistical Analysis of Pathway Enrichment
Pathway enrichment analysis is a statistical approach used to discover a statistically significant representation of a functional pathway class within a selection of factors from a heterogeneous factor population. Enrichment analysis can be applied in any situation where important physiological/pharmacological activity is suspected in the choice of a subset of members from a reference dataset. Enrichment analysis requires calculations on thousands of sets against thousands of candidate classifiers, generating often large output datasets containing both significant and nonsignificant data. There are multiple freely available pathway databases and facile calculation programs now able to facilitate these computational issues for molecular biologists (Table 2). As with GO term analysis, there are several important issues to consider with respect to the enrichment analysis. The appropriate choice of the reference dataset with which the experimental dataset is compared is vital. Unlike many simple statistical algorithms for accurate enrichment analysis, the accommodation of nonindependent association of factors is required. This allows empirically known physiological interactions to be included into the enrichment inference. In addition, as with GO term analysis, multiple-testing errors need to be accounted for as lack of independence among factor classifiers (seen in many datasets), for example, the hierarchical organization of multiple ontologies, often complicates estimation of false discovery. A simple paradigm for the statistical elucidation of enrichment analysis for a given signaling pathway is depicted in Fig. 5. As with all technological applications subsequent iterations and developments can quickly surpass previous techniques. For example, in recent years the use of simple GSEA has been largely replaced by a parametric version of this process (PAGE, parametric geneset enrichment analysis; 53). GSEA employs a distribution-free, nonparametric approach to the analysis of the significance of population (normally at least two factors in each pathway are required for effective “population” of that pathway) of signaling pathways by the input dataset. PAGE and other parametric GSEA tools use a Central Limit Theorem, which states that “when the sampling size is large enough, distribution of an average of sampled observations is normal regardless of the nature of parent distribution.” Statistical PAGE analysis intentionally directs the analysis of predefined signaling pathways in datasets rather than of individual factors. To generate easy to appreciate data with respect to differential metabolic/physiological states, PAGE uses the fold change between the control and experimental groups to calculate Z-scores of the predefined gene sets (various database sources can be used) and normal distribution to assign statistical significance to the gene sets (53). The list of all of the factors used in the dataset and their Z-scores are put into the analysis and Z-scores are assigned to the functional signaling sets within each experimental group. Traditional large dataset analysis requires that individual genes have significantly different expression levels in order for them to be considered differentially regulated. PAGE specifically takes into account that factors are both co-regulated and co-present, to help populate discrete signaling pathways. Therefore, it is possible that factors individually may not be significantly regulated above or below baseline, but significant regulation of pathways can be generated by such factors by grouping them significantly into the predefined signaling sets. By looking at groups of factors involved in a specific function, significant differences between their relative population may represent a biologically meaningful result. The polarity (up or downregulated) of the respective PAGE signaling pathway is determined by the sum of the Z-scores of the factors present in the experimental dataset that then fall into the set of factors used to describe the predetermined signaling pathway.
Table 2.
Computational programs for signaling and metabolic pathway analysis of large datasets
| Applications | URL |
|---|---|
| Signaling pathway databases | |
| BBID | http://bbid.grc.nia.nih.gov/ |
| BioCarta | http://www.biocarta.com/genes/index.asp |
| BioModels - biomodels database | http://www.ebi.ac.uk/biomodels-main/ |
| DOQCS - database of quantitative cellular signaling | http://doqcs.ncbs.res.in/ |
| DSM - dynamic signaling maps | http://www.hippron.com/hippron/index.html |
| eMIM - electronic molecular interaction map | http://discover.nci.nih.gov/mim/index.jsp |
| GeneNet - genetic networks | http://wwwmgs.bionet.nsc.ru/mgs/gnw/genenet/ |
| GenMAPP - gene microarray pathway profiler | http://www.genmapp.org/ |
| GON - genomic object net | http://genome.ib.sci.yamaguchi-u.ac.jp/~gon/index.html |
| HCPIN - human cancer protein interaction network | http://nesg.org:9090/HCPIN/ |
| INOH - integrating network objects with hierarchies | http://www.inoh.org/ |
| JWS online - online cellular systems modeling | http://jjj.biochem.sun.ac.za/ |
| KEGG | http://www.genome.jp/kegg/pathway.html |
| Millipore pathways | http://www.millipore.com/pathways/pw/pathways |
| NetPath | http://www.netpath.org/ |
| PANTHER - protein analysis through evolutionary relationships | http://www.pantherdb.org/ |
| PC - pathway commons | http://www.pathwaycommons.org/pc/ |
| PDS - pathways database system | http://nashua.case.edu/pathwaysweb/ |
| PID - NCI-nature pathway interaction database | http://pid.nci.nih.gov/ |
| pSTIING | http://pstiing.licr.org/ |
| Reactome - reactome knowledgebase | http://www.reactome.org/ |
| RGD - rat genome database pathway resource | http://rgd.mcw.edu/wg/pathway |
| ROSPath - reactive oxygen species related signaling pathway | http://rospath.ewha.ac.kr/ |
| Signaling gateway - UCSD-nature signaling gateway | http://www.signaling-gateway.org/ |
| SigPath - signaling pathway information system | http://icb.med.cornell.edu/crt/SigPath/index.xml |
| SMPDB - small molecule pathway database | http://www.smpdb.ca/ |
| SPIKE - signaling pathway integrated knowledge engine | http://www.cs.tau.ac.il/~spike/ |
| STCDB - signal transduction classification database | http://bibiserv.techfak.uni-bielefeld.de/stcdb/ |
| TRMP - therapeutically relevant multiple pathways database | http://bidd.nus.edu.sg/group/trmp/trmp_ns.asp |
| TRRD - transcription regulatory regions database | http://wwwmgs.bionet.nsc.ru/mgs/gnw/trrd/ |
| WikiPathways - WikiPathways | http://wikipathways.org/index.php/WikiPathways |
| Metabolic pathway databases | |
| aMAZE - protein function and biochemical pathways project | http://www.amaze.ulb.ac.be/ |
| BioCyc - biocyc knowledge library | http://biocyc.org/ |
| BioModels - biomodels database | http://www.ebi.ac.uk/biomodels-main/ |
| Biopath - biochemical pathways database | http://www.molecular-networks.com/databases/biopath |
| BRENDA - braunschweig enzyme database | http://www.brenda-enzymes.info/ |
| CellML repository - CellML model repository | http://models.cellml.org/ |
| CPDB - ConsensusPathDB | http://cpdb.molgen.mpg.de/ |
| ERGO - ERGO genome analysis and discovery system | http://www.ergo-light.com/ |
| ExPASy biochemical pathways | http://www.expasy.org/tools/pathways/ |
| GeneNet - genetic networks | http://wwwmgs.bionet.nsc.ru/mgs/gnw/genenet/ |
| HMDB - human metabolome database | http://www.hmdb.ca/ |
| HumanCyc - encyclopedia of homo sapiens genes and metabolism | http://humancyc.org/ |
| IntEnz - integrated relational enzyme database | http://www.ebi.ac.uk/intenz/index.jsp |
| LIGAND - database of chemical compounds and reactions | http://www.genome.jp/ligand/ |
| MetaCyc - metabolic pathway database | http://metacyc.org/ |
| MetNetDB - metabolic network exchange | http://www.metnetdb.org/MetNet_db.htm |
| MouseCyc - mouse pathway database | http://mousecyc.jax.org/ |
| NetBiochem - medical biochemistry resource | http://library.med.utah.edu/NetBiochem/NetWelco.htm |
| PathCase - CASE pathways database system | http://nashua.cwru.edu/PathwaysWeb/ |
| PATRIC - PathoSystems resource integration center | http://patric.vbi.vt.edu/ |
| PharmGKB | http://www.pharmgkb.org/ |
| Pathway analytical applications | |
| Ariadne genomics: pathway studio | http://www.ariadnegenomics.com/pathway-studio/ |
| ArrayXPath | http://www.snubi.org/software/ArrayXPath/ |
| Biochip core laboratory - CRSD | http://140.120.213.10:8080/crsd/ |
| Cpath | http://cbio.mskcc.org/software/cpath/ |
| D-GEM (disease-to-gene expression mapper) | http://dgem.cs.iupui.edu/ |
| ErmineJ | http://www.bioinformatics.ubc.ca/ermineJ/ |
| Gene set enrichment analysis - molecular signatures database | http://www.broadinstitute.org/gsea/ |
| GeneTrial | http://genetrail.bioinf.uni-sb.de/ |
| GenMAPP | http://www.genmapp.org/ |
| Genome expression pathway analysis tool | http://gepat.sourceforge.net/ |
| Ingenuity pathway analysis | www.ingenuity.com/ |
| KEGG pathway database | http://www.genome.jp/kegg/pathway.html |
| KOBAS: KO-based annotation system | http://kobas.cbi.pku.edu.cn/ |
| Onto-express - intelligent systems and bioinformatics laboratory | http://vortex.cs.wayne.edu/ontoexpress/ |
| PathExpress | http://bioinfoserver.rsbs.anu.edu.au/utils/PathExpress/ |
| PathJam - biological pathway integration tool | http://www.pathjam.org/ |
| Pathway miner - genes and their pathways | http://www.biorag.org/index.php |
| PROPA: probabilistic pathway annotation | http://www.stat.duke.edu/research/software/west/propa/ |
| VisANT: an integrative platform for network/pathway analysis | http://visant.bu.edu/ |
| WebGestalt: Web-based gene set analysis toolkit | http://bioinfo.vanderbilt.edu/webgestalt |
Fig. 5.
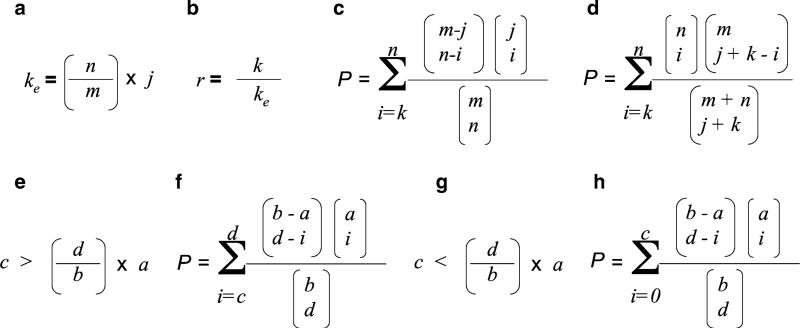
Functional factor enrichment. To identify functional categories with significantly enriched factor numbers within the input experimental dataset comparison is needed between the input dataset with a reference dataset. The input dataset needs in this case to be a subset of the reference dataset. For a theoretical scenario we may have n factors in the experimental dataset (a) and m factors in the reference dataset (b). For a given functional category of interest (e.g., a KEGG signaling pathway, c) there may be k number of factors from A and j number of factors from B. Based on the reference dataset (b) the expected value of k (ke) is depicted in panel A. If k exceeds ke then the specific category C is said to be enriched. Derivation of the index of the degree of pathway C enrichment (r) in the experimental dataset A is depicted in panel B. Analysis of the significance of the enrichment of pathway C in dataset B compared to dataset A, using a hypergeometric test is demonstrated in panel C. If, however, datasets A and B are independent, a Fisher's exact test may be more appropriate (d). Advanced pathway analysis software such as WebGestalt also allow the user to reduce their scope of pathway analysis in a similar manner to GO slims, for example, inspecting tissue-specific enrichment. For another factor, there may be d examples of a selected factor in all tissues and b examples for all factors in all tissues. In addition, if there are c number of a selected factor in a selected tissue and a number of all factors in that tissue, the over-representation of the specific factor in that tissue can be calculated as depicted (e). Calculation of the significance of over-representation in the specific tissue is depicted in panel (f). Mathematical under-representation of the specific factor in the selected tissue is described by the equation in panel (g) with the significance of the under-representation denoted in panel (h).
4.2. Pathway Analysis Applications
GSEA is especially powerful for the largest datasets that will have an increased likelihood of retrieved factor identity variation between experiments (especially the case for MS-based proteomics) or when there are subtle differences between control and experimental paradigms. With respect to the latter issue, a specific example of the power of GSEA techniques was the successful demonstration of prediction of significant metabolic pathway activation (oxidative phosphorylation) from a human dataset in which no one single gene out of 20,000 tested yielded an individually significant perturbation between control and diabetic patient muscle tissue (54). Thus the ability to apply significance of predicted functional output no longer rests upon individual factors but on co-expression and coherent regulation of these factors, reflecting the coordinated, interconnected nature of metabolic pathways themselves. Therefore, across diverse samples the signaling functionality can be correlated even if the identity of the regulated factors are not identical but still fall within the same functional preset pathway. Such flexibility is crucial for the analysis of MS-based quantitative proteomic data as the detection of exactly the same stream of proteins is highly unlikely over what can be long term experiments (10–20 h of run time).
In complex biological systems, coordinated metabolic functions are created by the summation of multiple interconnected pathways forming networks of varying sizes and relative importance. Using statistical processes to specifically search for these may greatly expand our understanding of the subtleties of disease processes or drug responses. Not only can these techniques be used for the investigation of dynamic experimental responses but may also illuminate how cells/tissues/animals react in response to spontaneous disease or genetically implied pathophysiology (48). Hence, not only may “disease-causing” networks of factors exist; “disease-management” factor networks are also likely as flexible and reactive biological systems attempt to ameliorate perturbations and achieve homeostasis.
With respect to the practical implementation of pathway analysis for large datasets there are multiple excellent databases of precompiled pathways available for pathway analysis as well as freely accessible software applications to perform the analysis (Table 2). However, not all signaling pathways are equally suitable for various experimental paradigms. For example, metabolic signaling pathways are controlled to a large extent by protein-based events that are not observable on microarrays as only steady-state levels of mRNAs are monitored. Kinase-based signaling cascades also do not necessarily involve changes in mRNA levels. The best case for microarray-based pathway analysis is transcriptional-signaling pathways that are directly coupled to de novo transcription. One of earliest developed tools for pathway analysis is the GenMAPP tool (55) that allots factors to preset pathways, as well as allowing user-based pathway generation. There are many excellent Web-based Pathway analysis tools such as Pathway Miner that provides ranking of the gene/pathway groups via a Fisher's exact test on top of the gene–pathway association analysis (56) and WebGestalt, that can generate GO DAG diagrams as well as KEGG and BioCarta pathway enrichment analysis (57). An example of the practical workflow and functioning of pathway analysis tools (e.g., WebGestalt) is depicted in Fig. 6. An extensive list of available programs is listed in Table 2. These tools often share similar lists of signaling pathways consisting of the relative factors allotted to them based on meta-literature searches. Again, as with the analytical tools themselves there are multiple sources of rationally created signaling and metabolic pathways. Some of the most commonly employed are the KEGG database (http://www.genome.jp/kegg/pathway.html) of metabolic and signaling pathways (58), the BioCarta database (http://www.biocarta.com/genes/index.asp) and the excellent and authoritative MIT/Harvard Broad Institute Molecular Signatures Database (MsigDB: http://www.broadinstitute.org/gsea/msigdb/). All of these databases provide easy open access to the pathways and associated diagrams for use with geneset enrichment software. In addition to these excellent resources for metabolic pathway analysis, correlated investigational technologies employing similar methodologies of functional inference are now widely used for transcription promoter analysis, protein–protein interaction and resultant mammalian phenotype prediction (Table 3). These analysis modules can often be used to supplement and support findings derived from GO and signaling pathway analysis.
Fig. 6.
Archetypical Pathway Analysis workflow. A typical flow of information processing to create a metabolic signaling output pathway using the WebGestalt analytic process is demonstrated in a series of logical steps. After data retrieval from mass analytical techniques primary statistical analysis can be employed using empirically derived cutoffs or whole-dataset data may be used instead. After uploading, the data can be converted to various identifiers, for example, Locus Links, Uniprot, or Unigene symbols. The software allows simple dataset Boolean operations as well before the two major forms of dataset analysis, that is, molecular or non-molecular-based. Non-molecular-based analyses include the investigation of enriched tissue or chromosome-specific expression of factors in the dataset. In addition, Pubmed (Gene-Association publication database) or GRIF (Gene expression into Function: http://generifs_basic.gz) Tables demonstrate co-expression of various factors in the dataset within the same publications. Multiple forms of biological signaling information can also be generated in parallel to these outputs. With selection of appropriate comparative base datasets (built-in) statistical enrichment of factors in the primary dataset into protein domain tables (Pfam: http://pfam.sanger.ac.uk/), directed acyclic gene ontologies (DAG) or discrete KEGG/BioCarta signaling pathways is determined.
Table 3.
Databases and computational tools for mass analysis of promoter activity, protein-protein interaction and mammalian phenotype annotation
5. Future Aspects for Signaling Pathway Analysis
The combined employment of mass data collection and signaling pathway analytical tools is likely to revolutionize signal transduction research in the next several decades. The ability to accurately appreciate and perhaps predict a global cellular impact of physiological or pharmacological perturbations may facilitate an understanding of disease etiology and eventual drug control of disease at the level of the factor network rather than the linear signaling pathway level. The appreciation of a network hypothesis for biological activity presents many important new avenues for signal transduction and pharmacological research. For example, the ability to identify “keystone” factors within a network that exert the most profound actions upon the state of a given pathological network may facilitate the creation of indirect pharmacological strategies. Such agents may be able to ensure a profound regulation of the keystone factors via modulation of multiple parts of the signaling network that have subsequent synergistic actions upon the keystones. These agents may be therefore more efficacious in smaller doses as their effects are amplified greatly by the reinforced network before hitting the keystone itself. In addition, as they may be inducing regulation of the network keystone through multiple mechanisms, such therapeutics may be more resistant to the development of desensitization, tolerance, or resistance. Hence these agents may present a polypharmacological network profile, but through careful knowledge-based design may effectively result in a more discrete resultant phenotypic action.
One important consideration of signaling pathway analysis that is often overlooked is the huge potential for temporal plasticity in signaling networks. The majority of mass analytical datasets are usually “snapshots” in time, as the expense of gaining multiple, temporally distinct, datasets is currently prohibitive. However, as the cost of mass analysis is likely to be reduced, our conversion of signaling pathways from rigid to plastic will undoubtedly assist in the greater appreciation of how signaling systems are integrated to form the basis of complicated physiological states and also drug responses. An understanding of the therapeutic at effective temporal windows may increase the potentiation of drug efficacy, again allowing a potential reduction in applied dose, thus minimizing side-effects or contra-indications. At a very crude level we are already demonstrating such a temporal drug response concept by the use of “chronotherapeutics” for anti-cancer drugs (59).
In conclusion, it is clear that the relentless increase in the intricacy of our understanding of molecular signaling has presented many challenges both in technological methodology and in computational analysis. Our ability to combine these two approaches for diagnostic and predictive capacities will only serve to improve our appreciation of disease pathophysiology and the mechanism of action of pharmacological agents. Appreciating these two coordinated factors at a systemic network level may allow the generation of far more efficacious and better-tolerated drug treatments for a wide variety of diseases and pathophysiological states.
Acknowledgments
This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.Luttrell LM. “Location, location, location”: activation and targeting of MAP kinases by G protein-coupled receptors. J Mol Endocrinol. 2003;30:117–26. doi: 10.1677/jme.0.0300117. [DOI] [PubMed] [Google Scholar]
- 2.Maudsley S, Martin B, Luttrell LM. The origins of diversity and specificity in G protein-coupled receptor signaling. J Pharmacol Exp Ther. 2005;314:485–494. doi: 10.1124/jpet.105.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–7. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;45:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilsenbeck SG, Friedrichs WE, Schiff R, O'Connell P, Hansen RK, Osborne CK, Fuqua SA. Statistical analysis of array expression data as applied to the problem of tamoxifen resistance. J Natl Cancer Inst. 1999;91:453–9. doi: 10.1093/jnci/91.5.453. [DOI] [PubMed] [Google Scholar]
- 6.Martin B, Pearson M, Brenneman R, Golden E, Wood W, Prabhu V, Becker KG, Mattson MP, Maudsley S. Gonadal transcriptome alterations in response to dietary energy intake: sensing the reproductive environment. PLoS One. 2009;4:e4146. doi: 10.1371/journal.pone.0004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin B, Brenneman R, Golden E, Walent T, Becker KG, Prabhu VV, Wood W, 3 rd, Ladenheim B, Cadet JL, Maudsley S. Growth factor signals in neural cells: coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J Biol Chem. 2009;284:2493–511. doi: 10.1074/jbc.M804545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Li MC, Simon R. An adaptive method for cDNA microarray normalization. BMC Bioinformatics. 2005;6:28. doi: 10.1186/1471-2105-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kepler TB, Crosby L, Morgan KT. Normalization and analysis of DNA microarray data by self-consistency and local regression. Genome Biol. 2002;3:RESEARCH0037. doi: 10.1186/gb-2002-3-7-research0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucl Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zien A, Aigner T, Zimmer R, Lengauer T. Centralization: a new method for the normalization of gene expression data. Bioinformatics. 2001;17:S323–31. doi: 10.1093/bioinformatics/17.suppl_1.s323. [DOI] [PubMed] [Google Scholar]
- 13.Sasik R, Calvo E, Corbeil J. Statistical analysis of high-density oligonucleotide arrays: a multiplicative noise model. Bioinformatics. 2002;18:1633–40. doi: 10.1093/bioinformatics/18.12.1633. [DOI] [PubMed] [Google Scholar]
- 14.Troyanskaya OG, Garber ME, Brown PO, Botstein D, Altman RB. Nonparametric methods for identifying differentially expressed genes in microarray data. Bioinformatics. 2002;18:1454–61. doi: 10.1093/bioinformatics/18.11.1454. [DOI] [PubMed] [Google Scholar]
- 15.Lee ML, Whitmore GA, Björkbacka H, Freeman MW. Nonparametric methods for microarray data based on exchangeability and borrowed power. J Biopharm Stat. 2005;15:783–97. doi: 10.1081/BIP-200067778. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Wood CL, Getchell TV, Getchell ML, Stromberg AJ. Analysis of oligonucleotide array experiments with repeated measures using mixed models. BMC Bioinformatics. 2004;5:209. doi: 10.1186/1471-2105-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meuwissen TH, Goddard ME. Bootstrapping of gene-expression data improves and controls the false discovery rate of differentially expressed genes. Genet Sel Evol. 2004;36:191–205. doi: 10.1186/1297-9686-36-2-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 19.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics. Mol Systems Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin TJ, Xie H, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J Proteome Res. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dayon L, Pasquarello C, Hoogland C, Sanchez JC, Scherl A. Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J Proteomics. 2010;73:769–77. doi: 10.1016/j.jprot.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem massspectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 26.Zhang N, Aebersold R, Schwilkowski B. ProbID: a probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectral data. Proteomics. 2002;2:1406–12. doi: 10.1002/1615-9861(200210)2:10<1406::AID-PROT1406>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 27.Creasy DM, Cottrell JS. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2:1426–34. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787–97. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 29.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–27. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 30.Mueller LN, Brusniak M-Y, Mani DR, Aebersol R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomic data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 31.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–6. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Liberzon A, Kong SW, Lai WR, Park PJ, Kohane IS, Kasif S. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genetics. 2007;3:e96. doi: 10.1371/journal.pgen.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6:S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camon EB, Barrell DG, Dimmer EC, Lee V, Magrane M, Maslen J, Binns D, Apweiler R. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA. BMC Bioinformatics. 2005;6:S1–S17. doi: 10.1186/1471-2105-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, Nevins JR, Chute JP. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Medicine. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisen M, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faustino RS, Behfar A, Perez-Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008;9:R6. doi: 10.1186/gb-2008-9-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin B, Pearson M, Brenneman R, Golden E, Keselman A, Iyun T, Carlson OD, Egan JM, Becker KG, Wood W, 3 rd, Prabhu V, de Cabo R, Maudsley S, Mattson MP. Conserved and differential effects of dietary energy intake on the hippocampal transcriptomes of females and males. PLoS One. 2008;3:e2398. doi: 10.1371/journal.pone.0002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stranahan AM, Lee K, Becker KG, Zhang Y, Maudsley S, Martin B, Cutler RG, Mattson MP. Hippocampal gene expression patterns underlying the enhancement of memory by running in aged mice. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.10.016. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 43.Draghici S, Kulaeva O, Hoff B, Petrov A, Shams S, Tainsky MA. Noise sampling method: an ANOVA approach allowing robust selection of differentially regulated genes measured by DNA microarrays. Bioinformatics. 2003;19:1348–59. doi: 10.1093/bioinformatics/btg165. [DOI] [PubMed] [Google Scholar]
- 44.Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16:953–9. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
- 45.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 46.Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene Ontology annotations with parent child analysis. Bioinformatics. 2007;23:3024–31. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- 47.Schlicker A, Rahnenfuhrer J, Albrecht M, Lengauer T, Domingues FS. GOTax: investigating biological processes and biochemical activities along the taxonomic tree. Genome Biol. 2007;8:R33. doi: 10.1186/gb-2007-8-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer's mice: understanding the interface between physiology and disease. PLoS One. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin X, Ahn S, Speed TP, Rubin GM. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8:R63. doi: 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas PD, Mi H, Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Chem Biol. 2007;11:4–11. doi: 10.1016/j.cbpa.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 51.Karp PD, Riley M, Paley SM, Pelligrini-Toole A. EcoCyc: an encyclopedia of Escherichia coli genes and metabolism. Nucl Acids Res. 1996;24:32–9. doi: 10.1093/nar/24.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucl Acids Res. 2005;33:D334–7. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S-Y, Volsky DJ. PAGE: Parametric analysis of geneset enrichment. BMC Bioinformatics. 2005;6:144–156. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 55.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 56.Pandey R, Guru RK, Mount DW. Pathway Miner: extracting gene association networks from molecular pathways for predicting the biological significance of gene expression microarray data. Bioinformatics. 2004;20:2156–8. doi: 10.1093/bioinformatics/bth215. [DOI] [PubMed] [Google Scholar]
- 57.Zhang B, Kirrov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucl Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucl Acids Res. 2002;30:42–6. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouchahda M, Adam R, Giacchetti S, Castaing D, Brezault-Bonnet C, Hauteville D, Innominato PF, Focan C, Machover D, Lévi F. Rescue chemotherapy using multidrug chronomodulated hepatic arterial infusion for patients with heavily pretreated metastatic colorectal cancer. Cancer. 2009;115:4990–9. doi: 10.1002/cncr.24549. [DOI] [PubMed] [Google Scholar]
- 60.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–8. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]