Abstract
Breast cancer is the most common cancer in women worldwide. Breast cancer in one third of all patients will go on to metastasis, which is the main cause of mortality in cancer cases. Tumor cells detach from primary tumor and enter into the circulation as circulating tumor cells (CTCs) which can form metastatic lesions. In this study, the expression of CK-19 gene in blood samples of metastatic breast cancer women was investigated and compared to control group. Twenty one patients with metastatic breast cancer and 20 healthy female volunteers enrolled in this study. For every patient and healthy donor 10 ml peripheral blood was collected. Peripheral blood mononuclear cells were isolated by gradient density centrifugation using Ficoll Hypaque. CK-19 gene expression was evaluated using SYBR green-based real-time quantitative polymerase chain reaction assays. The relative expression level of CK-19 was calculated using the 2−ΔΔCt analysis method. The mean of CK-19 expression was increased in metastatic breast cancer when compared to those of normal women (1.50 fold). 38.1% of the metastatic breast cancer patients showed CK-19 mRNA-detectable CTCs in their blood samples. There was no statistically significant difference between the relative expression level of CK-19 and the patient's clinicopathological characteristics. According to our knowledge, no study for determining CTC biomarkers in Iranian breast cancer women patients has yet been established. Our results suggest that the CK-19 mRNA expression investigation may be useful for monitoring CTCs in the blood of metastatic breast cancer patients, predicting early metastatic relapse or monitoring of anti-metastasis treatments.
Keywords: Circulating tumor cells, Breast cancer, CK-19, Metastasis
INTRODUCTION
Breast cancer accounts for 25% of invasive cancers and is the most common malignancy of women in females worldwide (1). Although it is the fifth cause of death from cancer overall, but in females it is the most frequent cause of cancer death in less developed regions (14.3% of total) and the second cause of cancer death in more developed regions (15.4% of total) after lung cancer. In Iran, incidence, mortality and 5-year prevalence of breast cancer in 2012 was estimated 24.5%, 14.2% and 37.7% respectively (2). Breast cancer in one third of all patients lead to metastasis, which is the main cause of mortality in cancer cases (3). The forming of metastasis is a complex process including many steps and is not still understood completely. The first step in metastasis is tumor's micrometastasis. Tumor cells detach from primary tumor and enter into the circulation, via a blood or a lymphatic vessel and if they remain alive in the circulation, come to the secondary organ and then proliferate which can form metastatic lesions. So circulating tumor cells (CTCs) are the vector of metastasis (4). These cells also named disseminated tumor cells (DTCs) when they enter bone marrow and cause tumor metastasis (5). The first publication on CTCs described by Ashworth in 1869 when he found that a few cells in peripheral blood and primary tumor of a dead cancer case were similar (6).
CTCs capture and analysis can be used as a surrogate unique method for understanding how the primary tumor proceeds to the metastasis. The number of CTCs reflects prognosis in metastatic patients. CTCs detection can be a liquid biopsy and a marker of response to anti-tumor therapy (4). Liquid biopsy is a new diagnostic method based on CTC analysis, whiles liquid phase of tumor progression mentions a specific stage of disease that is characterized by CTCs (7). Enumeration and analysis of CTCs can be used for finding early metastatic relapse, prognosis, treatment monitoring and decreasing side effects (8). Also CTC analysis may correlate with clinical parameters such as disease-free survival (DFS), progression-free survival (PFS) and overall survival (OS) (9). CTCs have been studied in the most cancers such as breast, colorectal, urothelial, prostate, small cell lung, non-small cell lung, pancreatic and gastric cancers (10,11,12,13,14,15,16,17). Since gene expression analysis of CTCs is important, researchers have studied many different markers in this field (18). An ideal CTC marker is expressed only on all CTCs but no other blood cells. In real situation it may express at higher levels in cancer compared to normal cells and they are specific to certain tumor types (7). Several markers have been used to detect breast cancer CTCs in nucleic-acid based techniques such as cytokeratin-19 (CK-19), CK-8, CK-18 (19), CK-20, epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER2) (20), Mammaglobin, Maspin (21), epithelial cell adhesion molecule (EpCAM) (22), and MUC1 (23). The most current marker for the detection of CTCs in breast cancer is CK-19 (20). CK-19, a general marker of epithelial cells, is strongly expressed in epithelial but not in mesenchymal cells and has been extensively studied as a potential marker for the detection of CTCs in breast cancer (24). It is also a cytoskeletal component present in normal and cancerous epithelial cells, which is expressed in 90% of invasive breast cancer tissue and has been extensively used for the detection of breast cancer cells in mesenchymal tissues and seems to be the most sensitive and reliable tumor marker in both patients with operable and metastatic breast cancer (20,25). In this study, we investigated the expression of CK-19 gene in CTCs of blood samples of metastatic breast cancer women and compared to control group. The results of this study may be applicable clinically for improvement in treatment and chemotherapy, diagnosis and predicting recurrence in cancer patients.
MATERIALS AND METHODS
Patients and samples
Twenty one patients with metastatic breast cancer and 20 female healthy volunteers were recruited to this study in Seyed Al-Shohada hospital in Isfahan, Iran. For all patients, a complete diagnostic evaluation was done, including of chest X-rays, abdominopelvic M.D.C.T scan, brain C.T scan, liver ultrasonography, and whole-body bone scan to confirm distant metastasis. Healthy volunteers serve as negative controls and had no history of previous cancer. All donors and patients have their written informed consent. Blood samples were collected before the starting a new treatment or after the last cycle of chemotherapy. All the patients were followed up during the period of performing this study.
For every patient and healthy donor, peripheral blood was collected in 10 ml ethylenediaminetetraacetic acid-containing tubes. To avoid contamination with epithelial skin cells, all blood samples were obtained at the middle of vein puncture after the first several milliliters of blood was discarded as previously described (25).
A complete blood count (CBC) was performed on all samples. Peripheral blood samples were diluted with sterile Phosphate-Buffered Saline (PBS, pH 7.0) (v/v). Peripheral blood mononuclear cells (PBMC) were isolated by gradient density centrifugation using Ficoll Hypaque (Innotrain, Kronberg, Germany) at 2000 rpm for 20 min at room temperature. The interface cells were removed, washed twice with 30 ml of sterile PBS at 2000 rpm for 5 min and resuspended in 1 ml of PBS. The cells were pelleted and kept at –80 °C until RNA extraction.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from the cells using the Total RNA Purification Kit (Jena Bioscience, Germany) according to the manufacturer's instructions. All RNA preparation and handling steps were done under RNAse-free conditions. Isolated RNA was stored at –80 °C until use. RNA concentration was quantified spectrophotometrically at 260 nm and the purity and integrity were determined using the A260/A280 ratio. The quality and concentration of the RNA samples were further confirmed by electrophoresis on denaturated 1% agarose gel. First-strand cDNA was synthesized from the total RNA using the AccuPower® CycleScript RT PreMix (dN6) (Bioneer, Inc. USA) according to the manufacturer's protocol. The cycling protocol consisted of annealing step at 20 °C for 30 s, cDNA synthesis at 44 °C for 4 min, melting secondary structure and cDNA synthesis at 55 °C for 30 s, and repeated 12 times. The normalization of cDNA was performed with the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Real-time quantitative PCR
SYBR green-based real-time quantitative polymerase chain reaction assays were used for the gene expression analysis of CK-19 and the housekeeping gene GAPDH. The reaction mixtures (20 µl) consisted of 10 µl SYBR Green qPCR Master Mix (Yektatajhiz, Iran), 0.1 µl of the CK-19 forward primer, 0.1 µl of the CK-19 reverse primer, and 2 µl cDNA. Amplification was performed using the Rotor-Gene 6000 (Corbett Research, Australia) at the following cycling conditions: 1 cycle of 2 min at 95 °C for the initial denaturation step and 40 cycles of 15 s at 95 °C for the denaturation step, 40 s at 60 °C for the annealing and extension step. The generation of a specific PCR product was tested using the ΔΔCT method and GAPDH as endogenous control.
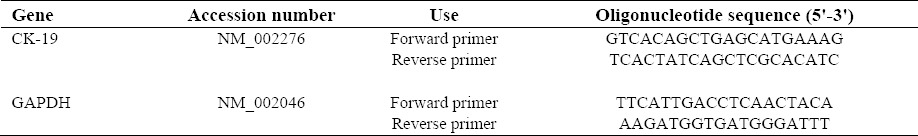
The primers for CK-19 (NM_002276) and GAPDH (NM_002046) were designed using Gene Runner Software (Version 3.05). These primers were exon-exon junction, so a probable DNA contamination would not cause a significant problem. The corresponding sizes of PCR products for CK-19 and GAPDH were 138 and 122 base pairs, respectively. The primer sequences are presented in Table 1.
Table 1.
Sequences of primers used in this study.
Statistical analysis
The association between CK-19 relative expression value and clinicopathological factors, such as menopausal status, tumor size, histology grade, lymph nodes involvement, ER and PR status were analyzed using Mann-Whitney Test. The Kaplan–Meier method was used to estimate DFS and OS curves, and the log-rank test was used to compare the different groups. The results were analyzed using SPSS 18.00 statistical software. All statistical tests were performed at the 5% level of significance and P value < 0.05 was considered statistically significant.
RESULTS
Patient's characteristics
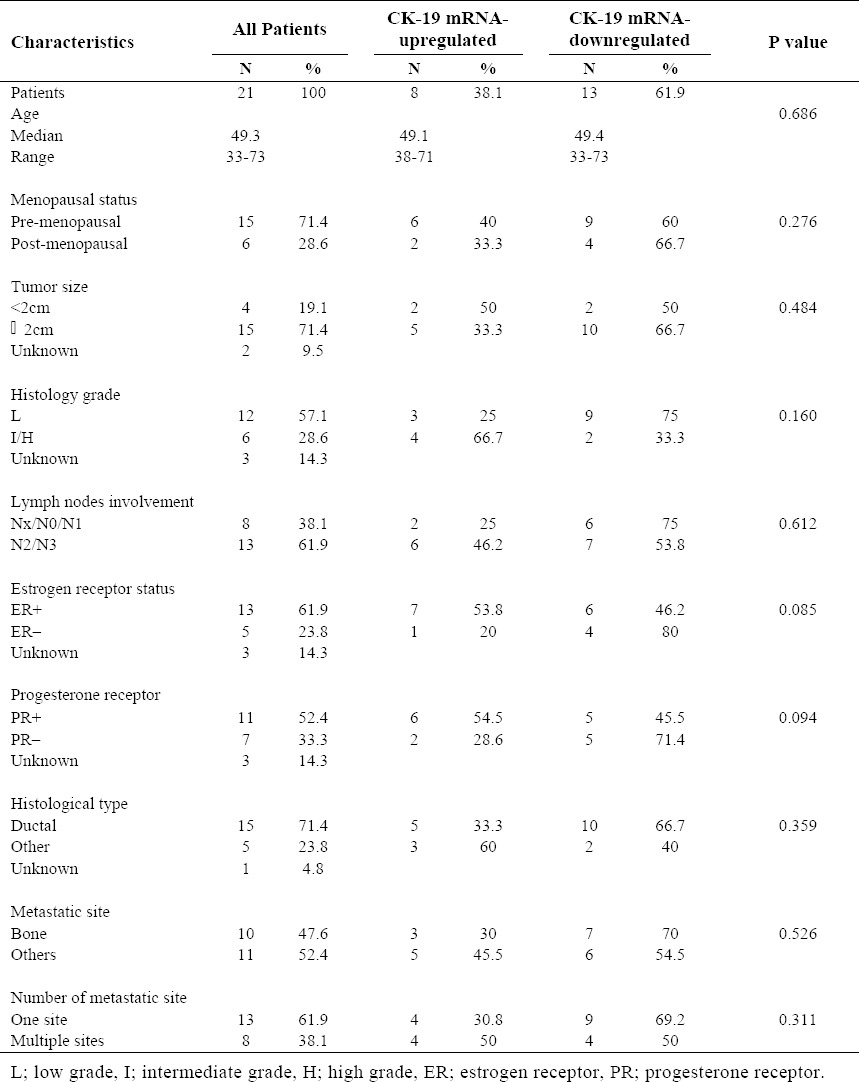
The clinical, pathological and histological characteristics of the patients are summarized in Table 2 and 3. Median patient age was 49.3 years (range, 33-73) and most of them were premenopausal (71.4%). 71.4% of the patients had tumors measuring > 2 cm, 57.1% had low-grade disease and 61.9% had > 4 involved lymph nodes. ER-positive (61.9%) and PR-positive (52.4%) tumors were predominant. Most patients had ductal breast cancer (71.4%). Bone was the major metastatic site (47.6%), and 38.1% of the patients had two or more metastatic sites.
Table 2.
Patient's clinical, pathological and histological characteristics and the relative expression level of CK-19.
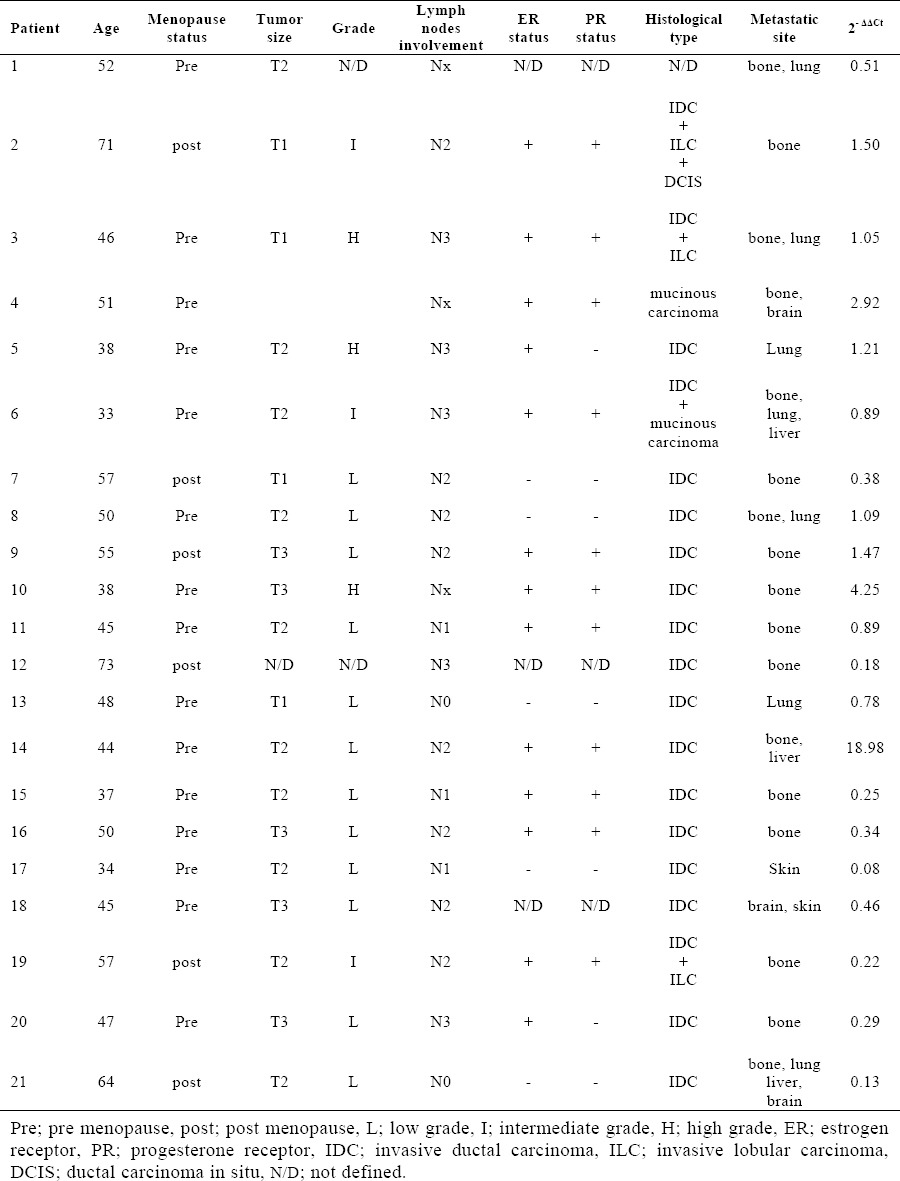
Table 3.
Patient's characteristics and correlation between CK-19 relative expression value and clinicopathologic factors.
CK-19 relative expression
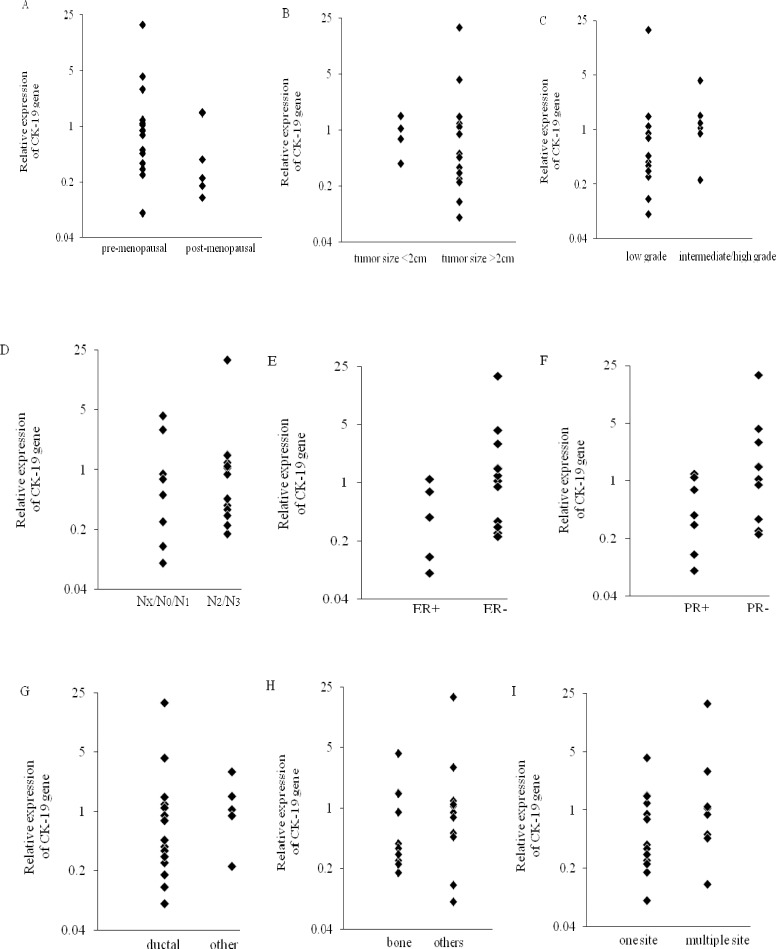
The relative expression level of CK-19 was calculated using the 2−ΔΔCt analysis method. Relative expression values > 1 were considered upregulated for the respective gene in comparison to the negative control samples (26). The peripheral blood mononuclear cells isolated from metastatic breast cancer samples and healthy individuals were analyzed for the mRNA expression patterns of CK-19. We found that all samples express CK-19 mRNA. Fig. 1 shows a higher mean value of CK-19 Fig. 1. Relative expression of CK-19 expression in metastatic breast cancer patients compare to healthy control group (1.50 fold). CK-19 is found upregulated in 8 out of 21 patients. Thus 38.1% of the metastatic breast cancer patients showed CK-19 mRNA-detectable CTCs in their blood samples. Furthermore, 14.3% of the patients showed relative expression level > 2. Patients 4 and 10 revealed the relative expression value 2.92-fold, 4.25-fold upregulated expression level, respectively. It is noteworthy that the relative expression value of patient 14 was 18.89-fold upregulated expression level. The relative expression values of patients are listed in Table 2.
Fig. 1.
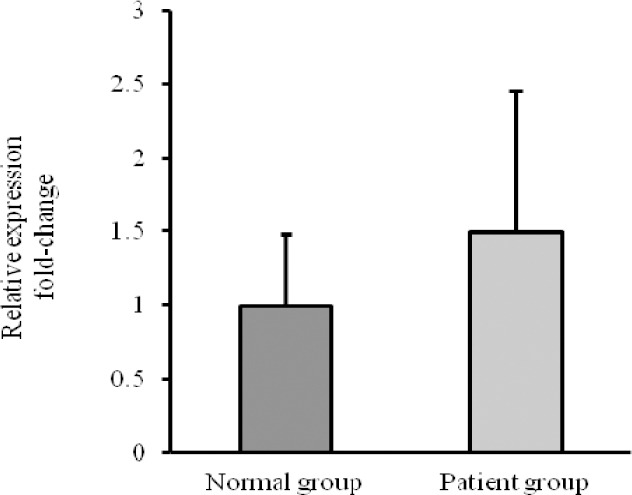
Relative expression of CK-19 in metastatic breast cancer compared to normal group.
There was no statistically significant difference between the patient's clinicopathological characteristics and the relative expression level of CK-19 (P>0.05). The correlations are shown in Table 3 and Fig. 2.
Fig. 2.
Relative expression of cytokeratin-19 (CK-19) in patients with metastatic breast cancer according to A; menopausal status: pre-menopausal [2.27 ± 4.75], post-menopausal [0.65 ± 0.55], B; tumor size: tumor size <2cm [2.07 ± 4.79], tumor size >2cm [0.93 ± 0.47], C; histology grade: low grade [2.1 ± 5.33], intermediate/high grade [1.52 ± 1.4], D; lymph node involvement: Nx/N0/N1 [1.23 ± 1.52], N2/N3 [2.16 ± 5.07], E; estrogen receptor status: ER + [2.64 ± 5.04], ER – [0.5 ± 0.43], F; progesterone receptor status: PR + [2.98 ± 5.44], PR – [0.57 ± 0.46], G; histological type: ductal [2.1 ± 4.79], other [1.3 ± 1], H; metastatic site: bone [0.98 ± 1.25], others [2.55 ± 5.5] and I; number of metastatic site: one site [0.91 ± 1.11], multiple sites [3.25 ± 6.41] (P>0.05).
Survival
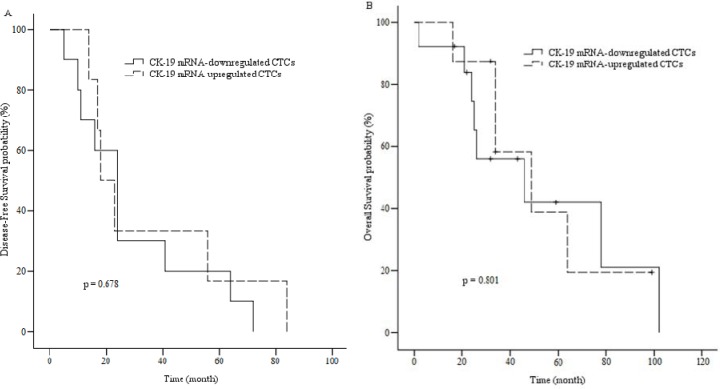
DFS and OS were defined as the time from primary surgery until the day of the first evidence of disease relapse, and death from breast cancer, respectively. 13 of 21 patients (61.9%) died during follow-up as a result of disease progression which 5 of them had CK-19 mRNA-detectable CTCs in their bloods (38.5%). For all patients, the median DFS was approximately 31.4 months (5–84) and the median OS was approximately 40.9 months (2–102). The median OS during follow-up period for upregulated patients was approximately 45.2 months (16–99). Kaplan-Meier curves indicated no statistically significant association (P>0.05) between DFS/OS and clinicopathological characteristics of the patients (Table 4). Kaplan-Meier plots comparing DFS and OS of patients with CK-19 mRNA changes are shown in Fig. 3.
Table 4.
Associations between DFS/OS and Patient's characteristics.
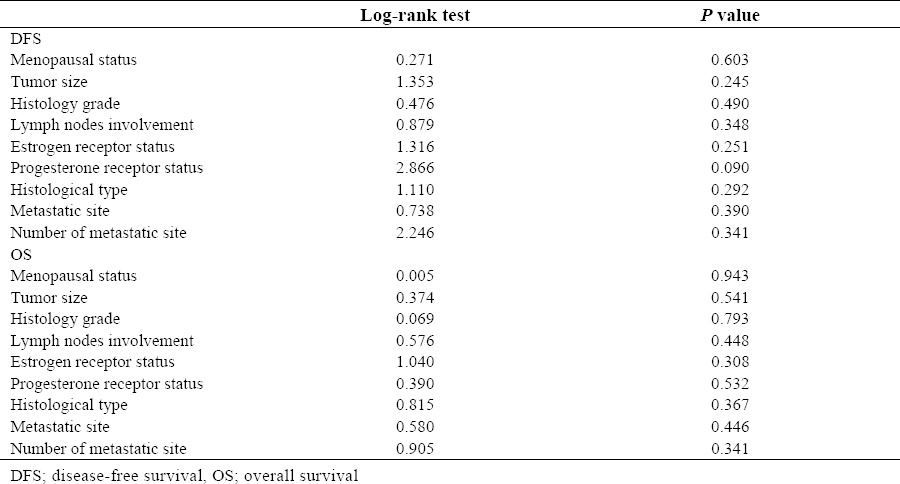
Fig. 3.
A; Disease-free survival of patients with CK-19 mRNA-up or downregulated CTCs. B; Overall survival of patients with CK-19 mRNA-up or downregulated CTCs.
DISCUSSION
Mortality from breast cancer is mainly a result of distant metastasis. Clinical practice shows that current metastasis detection imaging tools are not able to detect early spreading of tumor cells (27). To the best of our knowledge, no study for determining CTC biomarkers in Iranian women with breast cancer has yet been established. Therefore, in this study, the expression of CK-19 gene in blood samples of metastatic breast cancer women was investigated and compared to control group. The results of this study may be applicable clinically for improvement in the treatment and chemotherapy, diagnosis and predicting recurrence of the disease in cancer patients.
Although CTCs as a prognostic factor can lead to disease progression and metastasis, their detection and analysis is very difficult. This is because CTC does not have remarkable specificity which can be used to separate them from other blood cells (28). The number of CTCs in peripheral blood is about only one in 106–107 leukocytes and CTC count of ≥5 CTCs per 7.5 ml was significantly associated with shortened OS and PFS (24). CTCs have very different types, molecular phenotypes and markers (28). Also, CTCs are heterogeneous. Some of them are single cells, some are clumps, several related with blood cells especially platelets and many cannot be viable cells in circulation, but only one in 104 CTCs can cause metastatic disease (4). Thus, investigators have been using different detection techniques such as immunomagnetic isolation, flow cytometry, immunofluorescent microscopy, fluorescence in situ hybridization, PCR, and reverse transcriptase-PCR (RT-PCR) to find special CTC markers (28). In this study we first showed that all samples express CK-19 mRNA and the mean of CK-19 expression was increased in metastatic breast cancer when compared to those of normal women. Moreover, our data demonstrated that 38.1% of the metastatic breast cancer patients showed CK-19 mRNA-detectable CTCs in their blood samples.
The positive rate of CK-19 has been observed at various ratios (from 0-55%) in different studies (29). This may be because CK-19 as epithelial marker is neither breast nor cancer specific and studies performed in peripheral blood, bone marrow, and lymph nodes show a relatively high rate of false positives, both by immunohistochemistry and RT-PCR (30). The normal mammary gland is chiefly composed of luminal epithelial cells expressing different cytokeratins. Brotherick and coworkers examined cytokeratin protein and mRNA expression in malignant and benign breast tissues and found that CK-8, 18, and 19 revealed a consistent pattern of expression with respect to tumor grade. The significant correlation with increasing tumor size only has been shown for CK-19 (27). Brown and colleagues also showed that mRNA from women without evidence of breast cancer including 13 normal lymph nodes and 10 buffy coat samples were positive for CK-19 (60% of all samples). In addition, they also showed that normal tissues including prostate, testis, pancreas, liver, stomach, colon, kidney, muscle and spleen had detectable levels of CK-19 (30). Also, immunohistochemical studies revealed that CK-19 expression is heterogeneous basically on normal cells and with minority on cancerous cells (31).
Several studies investigated CTCs status in breast cancer patients. Stathopoulou and coworkers studied 47 patients with verified metastases of whom 19 (40.4%) and 20 (42.6%) had detectable CK-19-mRNA cells before and after chemotherapy (32). They also detected CK-19-mRNA positive cells in 33 out of 160 (20.6%) patients with early breast cancer (33). Zhao and colleagues investigated the expressions of EpCAM, CK-19, and hMAM in metastatic breast cancer patients that were detected in 50 (51.0%), 43 (43.9%), and 68 (69.4%) of the 98 patients, respectively (24). Reinholz and coworkers determined the expressions of CK-19 and mammaglobin in metastatic breast cancer patients. CK-19 and mammaglobin were detectable in 56%-75% and 23%-38% of 86 patients at baseline (29). Tunca and colleagues evaluated CK-20, EGFR, CK-19, and HER2 mRNA expression in primary invasive ductal breast cancer patients. The positive rates of which were 28.57% (24/84), 20.23% (17/84), 5.95% (5/84) and 2.17% (1/46), respectively (20). Daskalaki and coworkers detected CK-19 mRNA-positive CTCs and DTCs in 55.2% and 57.6% of patients before chemotherapy, but in 44 (52.4%) and 43 (51.2%) of 84 patients after chemotherapy (34). Tokudome and colleagues determined the presence of CTCs with the CellSearch System in 28 metastatic breast cancers. Nine of 28 patients (32%) had ≥ 5 CTCs before starting a new treatment and 2–3 months after starting chemotherapy, 5 of 23 patients (22%) had ≥ 5 CTCs per 7.5 ml of blood (35). Trastuzumab could decrease the frequency of CK-19 positive cells to undetectable levels in 28 out of 30 treated patients or after surgery (36). These findings are in contrast with Daskalaki and coworkers regarding the high CK-19 positive patients after surgery (34).
Many studies in the clinical evaluation and standardization of quantitative RT-PCR methodology for transcripts of CK-19 mRNA are too complex and not easily adaptable and applicable. Because CTCs are highly heterogeneous and may express different levels of the indicated marker, it is difficult to correlate the number of CTCs to the corresponding mRNA levels. Another major limitation is the fact that normalization to the expression of a reference gene makes variability between different patients (32).
The relationship between CTCs and clinicopathological features is controversial. Several studies have determined that the presence of CTCs in breast cancer is associated with a poor clinical outcome (37,38,39,40,41), but further studies of the same research groups did not confirm previous findings and failed to demonstrate any association between CTCs status and prognosis (34,42,43,44,45). In our study, there was no statistically significant association between clinicopathological features of the patients and CK-19 mRNA-detectable CTCs or DFS/OS.
Several studies have demonstrated the relationship between the CTCs status and DFS/OS. Daskalaki and coworkers showed that the detection of CK-19 mRNA-positive CTCs before chemotherapy correlated with reduced OS but did not associate with patient's clinical or pathological characteristics (34). Stathopoulou and colleagues found that the detection of CK-19 mRNA-positive cells in the peripheral blood of patients with stages I and II breast cancer associated with decreased OS but did not correlated with the patient's characteristics (46). Also, Reinholz and coworkers observed that the presence of baseline CK-19 mRNA-positive was associated with shorter OS, hormone receptor positivity, > 3 metastatic sites, and non-ductal histology (29). Xenidis and colleagues found that the detection of CK-19 mRNA-positive CTCs before and after chemotherapy was an independent factor correlated with decreased DFS and OS. They also reported that the detection of CK-19 mRNA-positive CTCs postchemotherapy was associated with a more than three axillary lymph nodes involvement (40).
Saloustros and coworkers observed that the group of CK-19 mRNA-persistently positive patients compared to persistently negative had a higher risk of disease relapse and shorter OS, but the persistent detection of CTCs did not associate with the patient's characteristics (25). In the current study, due to the small number of patients in the metastatic group a significant association between clinicopathological factors and CK-19 mRNA-detectable CTCs was not observed. Although this important aspect should be addressed in future studies, but it is rather difficult to recruit a large number of patients with metastatic cancers. Also for improvement of the detection of CTCs by real-time PCR, a combination of different promising marker genes needs to be tested (19).
CONCLUSION
In conclusion, the RT-PCR detection of the mRNA of CK-19 biomarker in the peripheral blood of metastatic breast cancer patients may be used to monitor CTCs in the blood circulation and to detect the occurrence of metastasis, recurrence and therapeutic outcome.
ACKNOWLEDGEMENTS
The content of this paper is extracted from the Master thesis NO. 391417 submitted by S. Soltani which was financially supported by the Research Department of Isfahan University of Medical Sciences, Isfahan, I.R. Iran. We gratefully acknowledge all the patients who participated in this study. We also appreciate the great assistance of Seyed Al-Shohada hospital personnels.
REFERENCES
- 1.Maliheh A, Noghabaei G. Comparison of age-standard incidence rate trends of gyneco¬ logic and breast cancer in iran and other countries. Iran J Public Health. 2014;43:1372–1379. [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Molloy TJ, Roepman P, Naume B, van’t Veer LJ. A prognostic gene expression profile that predicts circulating tumor cell presence in breast cancer patients. PloS one. 2012;7:e32426. doi: 10.1371/journal.pone.0032426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Flaherty JD, Gray S, Richard D, Fennell D, O’Leary JJ, Blackhall FH, et al. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer. 2012;76:19–25. doi: 10.1016/j.lungcan.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Andergassen U, Kölbl AC, Hutter S, Friese K, Jeschke U. Detection of circulating tumour cells from blood of breast cancer patients via RT-qPCR. Cancers. 2013;5:1212–1220. doi: 10.3390/cancers5041212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghossein R, Bhattacharya S. Molecular detection and characterisation of circulating tumour cells and micrometastases in solid tumours. Eur J Cancer. 2000;36:1681–1694. doi: 10.1016/s0959-8049(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 7.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 8.Lianidou E, Mavroudis D, Georgoulias V. Clinical challenges in the molecular characterization of circulating tumour cells in breast cancer. Br J Cancer. 2013;108:2426–2432. doi: 10.1038/bjc.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinert G, Schölch S, Koch M, Weitz J. Biology and significance of circulating and disseminated tumour cells in colorectal cancer. Langenbecks Arch Surg. 2012;397:535–542. doi: 10.1007/s00423-012-0917-9. [DOI] [PubMed] [Google Scholar]
- 10.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 11.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 12.Hou J-M, Greystoke A, Lancashire L, Cummings J, Ward T, Board R, et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am J Pathol. 2009;175:808–816. doi: 10.2353/ajpath.2009.090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher D, Milowsky M, Ishill N, Trout A, Boyle M, Riches J, et al. Detection of circulating tumor cells in patients with urothelial cancer. Ann Oncol. 2009;20:305–308. doi: 10.1093/annonc/mdn627. [DOI] [PubMed] [Google Scholar]
- 14.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 15.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara T, Itoi T, Sofuni A, Itokawa F, Tsuchiya T, Tsuji S, et al. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J Hepatobiliary Pancreat Surg. 2008;15:189–195. doi: 10.1007/s00534-007-1250-5. [DOI] [PubMed] [Google Scholar]
- 17.Mimori K, Fukagawa T, Kosaka Y, Kita Y, Ishikawa K, Etoh T, et al. Hematogenous metastasis in gastric cancer requires isolated tumor cells and expression of vascular endothelial growth factor receptor-1. Clin Cancer Res. 2008;14:2609–2616. doi: 10.1158/1078-0432.CCR-07-4354. [DOI] [PubMed] [Google Scholar]
- 18.Mostert B, Sleijfer S, Foekens JA, Gratama JW. Circulating tumor cells (CTCs): detection methods and their clinical relevance in breast cancer. Cancer Treat Rev. 2009;35:463–474. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Andergassen U, Hofmann S, Kölbl AC, Schindlbeck C, Neugebauer J, Hutter S, et al. Detection of tumor cell-specific mrna in the peripheral blood of patients with breast cancer—evaluation of several markers with real-time reverse transcription-PCR. Int J Mol Sci. 2013;14:1093–1104. doi: 10.3390/ijms14011093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tunca B, Egeli U, Cecener G, Tezcan G, Gökgöz S, Tasdelen I, et al. CK19, CK20, EGFR and HER2 status of circulating tumor cells in patients with breast cancer. Tumori. 2012;98:243–251. doi: 10.1177/030089161209800211. [DOI] [PubMed] [Google Scholar]
- 21.Mercatali L, Valenti V, Calistri D, Calpona S, Rosti G, Folli S, et al. RT-PCR determination of maspin and mammaglobin B in peripheral blood of healthy donors and breast cancer patients. Ann Oncol. 2006;17:424–428. doi: 10.1093/annonc/mdj109. [DOI] [PubMed] [Google Scholar]
- 22.Taback B, Chan AD, Kuo CT, Bostick PJ, Wang H-J, Giuliano AE, et al. Detection of occult metastatic breast cancer cells in blood by a multimolecular marker assay correlation with clinical stage of disease. Cancer Res. 2001;61:8845–8850. [PubMed] [Google Scholar]
- 23.Uen YH, Lin SR, Wu CH, Hsieh JS, Lu CY, Yu FJ, et al. Clinical significance of MUC1 and c-Met RT-PCR detection of circulating tumor cells in patients with gastric carcinoma. Clin Chim Acta. 2006;367:55–61. doi: 10.1016/j.cca.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Zhao S, Yang H, Zhang M, Zhang D, Liu Y, Liu Y, et al. Circulating tumor cells (CTCs) detected by triple-marker EpCAM, CK19, and hMAM RT-PCR and their relation to clinical outcome in metastatic breast cancer patients. Cell Biochem Biophys. 2013;65:263–273. doi: 10.1007/s12013-012-9426-2. [DOI] [PubMed] [Google Scholar]
- 25.Saloustros E, Perraki M, Apostolaki S, Kallergi G, Xyrafas A, Kalbakis K, et al. Cytokeratin-19 mRNA-positive circulating tumor cells during follow-up of patients with operable breast cancer: prognostic relevance for late relapse. Breast Cancer Res. 2011;13:R60. doi: 10.1186/bcr2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Brotherick I, Robson C, Browell D, Shenfine J, White M, Cunliffe W, et al. Cytokeratin expression in breast cancer: phenotypic changes associated with disease progression. Cytometry. 1998;32:301–308. [PubMed] [Google Scholar]
- 28.Jiang W, Zhang H. Enrichment and detection of circulating tumor cells in peripheral blood. Chin Ger J Clin Oncol. 2011;10:240–244. [Google Scholar]
- 29.Reinholz MM, Kitzmann KA, Tenner K, Hillman D, Dueck AC, Hobday TJ, et al. Cytokeratin-19 and mammaglobin gene expression in circulating tumor cells from metastatic breast cancer patients enrolled in North Central Cancer Treatment Group trials, N0234/336/436/437. Clin Cancer Res. 2011;17:7183–7193. doi: 10.1158/1078-0432.CCR-11-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown NM, Stenzel TT, Friedman PN, Henslee J, Huper G, Marks JR. Evaluation of expression based markers for the detection of breast cancer cells. Breast Cancer Res Treat. 2006;97:41–47. doi: 10.1007/s10549-005-9085-8. [DOI] [PubMed] [Google Scholar]
- 31.Peehl DM, Sellers RG, McNeal JE. Keratin 19 in the adult human prostate: tissue and cell culture studies. Cell Tissue Res. 1996;285:171–176. doi: 10.1007/s004410050633. [DOI] [PubMed] [Google Scholar]
- 32.Stathopoulou A, Gizi A, Perraki M, Apostolaki S, Malamos N, Mavroudis D, et al. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin Cancer Res. 2003;9:5145–5151. [PubMed] [Google Scholar]
- 33.Stathopoulou A, Ntoulia M, Perraki M, Apostolaki S, Mavroudis D, Malamos N, et al. A highly specific real-time RT-PCR method for the quantitative determination of CK-19 mRNA positive cells in peripheral blood of patients with operable breast cancer. Int J Cancer. 2006;119:1654–1659. doi: 10.1002/ijc.22017. [DOI] [PubMed] [Google Scholar]
- 34.Daskalaki A, Agelaki S, Perraki M, Apostolaki S, Xenidis N, Stathopoulos E, et al. Detection of cytokeratin-19 mRNA-positive cells in the peripheral blood and bone marrow of patients with operable breast cancer. Br J Cancer. 2009;101:589–597. doi: 10.1038/sj.bjc.6605183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokudome N, Ito Y, Takahashi S, Kobayashi K, Taira S, Tsutsumi C, et al. Detection of circulating tumor cells in peripheral blood of heavily treated metastatic breast cancer patients. Breast cancer. 2011;18:195–202. doi: 10.1007/s12282-011-0259-4. [DOI] [PubMed] [Google Scholar]
- 36.Bozionellou V, Mavroudis D, Perraki M, Papadopoulos S, Apostolaki S, Stathopoulos E, et al. Trastuzumab administration can effectively target chemotherapy-resistant Cytokeratin-19 Messenger RNA-Positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin Cancer Res. 2004;10:8185–8194. doi: 10.1158/1078-0432.CCR-03-0094. [DOI] [PubMed] [Google Scholar]
- 37.Ignatiadis M, Perraki M, Apostolaki S, Politaki E, Xenidis N, Kafousi M, et al. Molecular detection and prognostic value of circulating Cytokeratin-19 messenger RNA–positive and HER2 messenger RNA–Positive cells in the peripheral blood of women with early-stage breast cancer. Clin Breast Cancer. 2007;7:883–889. doi: 10.3816/CBC.2007.n.054. [DOI] [PubMed] [Google Scholar]
- 38.Ignatiadis M, Kallergi G, Ntoulia M, Perraki M, Apostolaki S, Kafousi M, et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin Cancer Res. 2008;14:2593–2600. doi: 10.1158/1078-0432.CCR-07-4758. [DOI] [PubMed] [Google Scholar]
- 39.Apostolaki S, Perraki M, Kallergi G, Kafousi M, Papadopoulos S, Kotsakis A, et al. Detection of occult HER2 mRNA-positive tumor cells in the peripheral blood of patients with operable breast cancer: evaluation of their prognostic relevance. Breast Cancer Res Treat. 2009;117:525–534. doi: 10.1007/s10549-008-0239-3. [DOI] [PubMed] [Google Scholar]
- 40.Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S, et al. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27:2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Zou TN, Wu ZP, Zhou YC, Gu YL, Liu X, et al. Detection of cytokeratin 19, human mammaglobin, and carcinoembryonic antigen-positive circulating tumor cells by three-marker reverse transcription-PCR assay and its relation to clinical outcome in early breast cancer. Int J Biol Markers. 2010;25:59–68. doi: 10.1177/172460081002500201. [DOI] [PubMed] [Google Scholar]
- 42.Suchy B, Austrup F, Driesel G, Eder C, Kusiak I, Uciechowski P, et al. Detection of mammaglobin expressing cells in blood of breast cancer patients. Cancer lett. 2000;158:171–178. doi: 10.1016/s0304-3835(00)00520-6. [DOI] [PubMed] [Google Scholar]
- 43.Lin YC, Chen SC, Hsueh S, Lo YF, Chow-Wu YH, Liaw I, et al. Lack of correlation between expression of human mammaglobin mRNA in peripheral blood and known prognostic factors for breast cancer patients. Cancer Sci. 2003;94:99–102. doi: 10.1111/j.1349-7006.2003.tb01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benoy I, Elst H, Philips M, Wuyts H, Van Dam P, Scharpe S, et al. Real-time RT–PCR detection of disseminated tumour cells in bone marrow has superior prognostic significance in comparison with circulating tumour cells in patients with breast cancer. Br J Cancer. 2006;94:672–680. doi: 10.1038/sj.bjc.6602985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marques AR, Teixeira E, Diamond J, Correia H, Santos S, Neto L, et al. Detection of human mammaglobin mRNA in serial peripheral blood samples from patients with non-metastatic breast cancer is not predictive of disease recurrence. Breast Cancer Res Treat. 2009;114:223–232. doi: 10.1007/s10549-008-0002-9. [DOI] [PubMed] [Google Scholar]
- 46.Stathopoulou A, Vlachonikolis I, Mavroudis D, Perraki M, Kouroussis C, Apostolaki S, et al. Molecular detection of cytokeratin-19–positive cells in the peripheral blood of patients with operable breast cancer: Evaluation of their prognostic significance. J Clin Oncol. 2002;20:3404–3412. doi: 10.1200/JCO.2002.08.135. [DOI] [PubMed] [Google Scholar]