Abstract
While sexual reproduction is universal in eukaryotes, and shares conserved core features, the specific aspects of sexual reproduction can differ dramatically from species to species. This is also true in Fungi. Among fungal species, mating determination can vary from tetrapolar with more than a thousand different mating types, to bipolar with only two opposite mating types, and finally to unipolar without the need of a compatible mating partner for sexual reproduction. Cryptococcus neoformans is a human pathogenic fungus that belongs to the Basidiomycota. While C. neoformans has a well-defined bipolar mating system with two opposite mating types, MATa and MATα, it can also undergo homothallic unisexual reproduction from one single cell or between two cells of the same mating type. Recently, it was shown that, as in a-α bisexual reproduction, meiosis is also involved in α-α unisexual reproduction in C. neoformans. Briefly, recombination frequencies, the number of crossovers along chromosomes, as well as frequencies at which aneuploid and diploid progeny are produced, are all comparable between a-α bisexual and α-α unisexual reproduction. The plasticity observed in C. neoformans sexual reproduction highlights the extensive diversity in mating type determination, mating recognition, as well as modes of sexual reproduction across fungal species.
1. Introduction
Sexual reproduction, the process of producing progeny by combining genetic material from the two parents, is pervasive and present in all of the major groups of the eukaryotic tree of life. However, while sexual reproduction is ubiquitous and conserved in core features (e.g. ploidy changes, zygote formation, mixing and segregation of parental genetic material), the detailed aspects of sexual reproduction, including how sex is determined, as well as how sexual reproduction is accomplished, are diverse (Dellaporta and Calderon-Urrea, 1993; Goodfellow and Lovell-Badge, 1993; Heitman et al., 2013; Korpelainen, 1990; McNair Senior et al., 2015). For example, while for most animal species the sex is genetically determined by sex chromosomes, for some reptile species, sex is decided by environmental factors, such as the temperature at which the egg is hatched (Bull, 1980). Additionally, sex can change in some fish species due to changes in the social dynamics within the group of fish (Lorenzi et al., 2006), and in the ciliate Paramecium tetraurelia, it has been shown that a small RNA-mediated genome defense mechanism is involved in the maternal inheritance of mating types in this species (Singh et al., 2014). Also, while most animals and plants have only two sexes, this is far from a universal rule in many other species. In the social amoeba Dictyostelium discoideum, there are three mating types that are decided by different gene compositions at one mat locus (Bloomfield et al., 2010). More strikingly, in the ciliate protozoa Tetrahymena thermophila there are seven different mating types, and each mating type can undergo successful mating with individuals of any of the other six mating types, but not with individuals of its own type. The mating type of T. thermophila is determined by a specific chromosomal region called the mating type locus in the active somatic nuclear genome, and the presence of one of the seven different alleles at this mating type locus decides the mating type of the individual (Cervantes et al., 2013). Interestingly, it has recently been shown that the mating type locus in the T. thermophila germline nucleus that is dormant during vegetative growth actually contains all seven elements that are required to define the seven different mating types, and that these seven elements form a cluster at the mating type locus that is interspersed with repetitive elements. During the development of the somatic nucleus from the germline nucleus, the mating type locus undergoes repeated homologous recombination events mediated by these repetitive elements, which results in successive elimination of mating type elements until only one of the original seven elements is left, with this remaining element defining the mating type of that individual (Cervantes et al., 2013).
In fungi, it is thus not surprising that the processes of sexual reproduction also show extensive plasticity in mating type determination and the number of sexes, as well as in mating recognition, leading to different modes of sexual reproduction.
2. Mating type loci and mating recognition in fungi
In fungi, sexual identity is defined by the mating type, which is typically established by a specialized region of the genome that is termed the mating type locus (MAT) (Ni et al., 2011). Some fungi (e.g. many of the Ascomycetes) have only one MAT locus in their genome that encodes mating-type-specific transcription factors. In these species, the MAT locus is usually biallelic and defines two mating types, and mating can only occur between individuals with opposite mating types. Thus, these fungi have a bipolar mating system, because a maximum of two different mating types can be produced from one meiotic event. In some other fungal species (e.g. many of the Basidiomycetes), the mating type is established by the allelic combination of two unlinked MAT loci, where one encodes the pheromones and pheromone receptors (P/R locus) and the other encodes homeodomain transcription factors (HD locus) that activate mating specific pathways upon successful mating recognition by the P/R locus. In these fungi, successful mating can only occur between isolates with different alleles at both of the two MAT loci. Thus, these species have a tetrapolar mating system because a maximum of four different mating types can be produced through one round of meiosis (Casselton and Olesnicky, 1998).
The MAT locus/loci are evolutionarily dynamic, and undergo expansions and contractions, as well as chromosomal rearrangements including translocations and inversions, even between closely related species (James, 2007). This is likely due to the fact that these regions are normally repressed for meiotic recombination and are typically enriched with transposable elements and repetitive sequences. Additionally, transitions between tetrapolar and bipolar mating systems have also been shown to occur in several fungal species (Fraser and Heitman, 2004; James et al., 2006). For example, in the mushroom Coprinellus disseminatus, it was shown that the extant bipolar mating system has likely evolved from the ancestral tetrapolar mating system through the loss of mating-type-specific pheromone receptor function, rendering the P/R locus no longer involved in mating recognition. Also, the human fungal pathogen Cryptococcus neoformans has a well-defined bipolar mating system with an unusually large MAT locus (>100 kb in size with more than 20 genes) (Fraser et al., 2004; Lengeler et al., 2002). When compared to closely related species, it is clear that this large MAT locus has evolved through some sort of chromosomal rearrangement that resulted in the physical linkage between the ancestrally separated P/R and HD locus (Findley et al., 2012). However, the detailed mechanisms by which this linkage was established have yet to be elucidated.
3. Heterothallic vs. homothallic
We mentioned that in species with bipolar mating systems, mating can only occur between individuals with different mating types; and for species with tetrapolar mating systems, only individuals that differ at both P/R and HD loci can undergo successful sexual reproduction (Raper, 1966). For these species, we say they undergo heterothallic sexual reproduction, because the two individuals involved in mating have different mating types and are most likely not genetically identical by descent. However, there are also some fungal species that do not require a mating partner to initiate and complete sexual reproduction, and can initiate and complete the sexual cycle from a single cell. We call these species homothallic, and there are several mechanisms that could enable a species to undergo homothallic sexual reproduction.
First, homothallic sexual reproduction can be achieved by mating type switching, such as in the model yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, which independently evolved one active mating type locus and two silenced mating type cassettes that enable gene conversion mating type switching evoked in response to recombinogenic lesions (Dalgaard and Klar, 1999; Egel, 2005; Haber, 2012; Klar, 2007; Klar, 2010). In the genome of S. cerevisiae, there is one active MAT locus that defines a bipolar mating system with two mating types, MATa and MATα. In addition, there are also two silent mating cassettes, HMR and HML, which carry epigenetically silenced copies of MATa and MATα alleles, respectively. So, an individual with a MATa allele at the active MAT locus behaves as a mating type even though it carries a silenced MATα allele at the HML locus, and vice versa. Now, S. cerevisiae can undergo a unique process called mating type switching, where during the G1 phase of the cell cycle in haploid cells, the allele at the active MAT locus can be excised and eliminated through the activities of a specialized DNA endonuclease called HO and exonucleases. This gap will be subsequently filled in through gene conversion using genetic information present at either the HML or the HMR locus. Typically, MATa cells tend to repair the gap at the active MAT locus using the MATα allele from the HML locus, and vise versa, and thus, undergo mating type switching. While mating in S. cerevisiae still requires the two partners to have opposite mating types, because of mating type switching, the two partners could be identical by descent except at the active MAT locus, that is, they could be the mitotic products of the same cell. Thus, mating between these individuals can be considered functionally homothallic (Haber, 2012).
Recently, another novel mechanism for mating type switching has been discovered in Hansenula polymorpha (=Ogataea angusta), an Ascomycetous species that does not possess the HO gene (Hanson et al., 2014; Maekawa and Kaneko, 2014). It turned out that H. polymorpha has two MAT loci: MATa and MATα, which are approximately 18 kb apart on the same chromosome. Additionally, the chromosomal region encompassing the MATa and MATα lies beside a centromere. As a consequence, depending on the orientation of this chromosomal region, one of the two MAT loci will be silenced by the centromeric chromatin, leaving only one locus maintained in an active state to define the mating type of the cell. Moreover, inversion of the MAT intervening region was found to be induced by nutrition limitation, resulting in the switching of the locations of the two MAT loci on the chromosome, their respective activity status, and consequently, the mating type of the cell. Thus, similar to S. cerevisiae, individuals of different mating types can be produced from one single parental isolate, which can then undergo homothallic sexual reproduction. A very similar two loci switching system has also been found in Pichia pastoris (=Komagataella pastoris) a species that is closely related to H. polymorpha (Hanson et al., 2014). In P. pastoris, the two MAT loci are approximately 138 kb apart on the same chromosome, and depending on the orientation, one of the two MAT loci will be located close to the end of the chromosome, and consequently silenced due to its close proximity to the telomere (Hanson et al., 2014). It has been proposed that this switching by inversion mechanism identified in H. polymorpha and P. pastoris represents the ancestral state of the mating type switching and silencing mechanism from which the three loci system observed in S. cerevisiae has evolved (Hanson et al., 2014).
Mating type switching has also been observed in basidiomycetes, as reported in one species, Agrocybe aegerita (Labarère and Noël, 1992), further indicating that this form of homothallism has evolved repeatedly and independently.
Homothallic sexual reproduction can also be the result of special configurations at the MAT locus, including pseudohomothallism where meiotic spores contain separate nuclei with opposite mating types and can germinate into heterokaryotic mycelia (e.g. Neurospora tetrasperma (Merino et al., 1996), Agrocybe semiorbicularis, Conocybe tenera for. bispora, and Coprinellus ephemerus (Raper, 1966), Agaricus bisporus (Li et al., 2004)), the presence of opposite MAT loci in one genome, either unlinked (e.g. Aspergillus nidulans (Galagan et al., 2005)) or linked (e.g. Neurospora pannonica and Neurospora terricola (Pöggeler, 1999)), and sometimes even from cells with only one and the same mating type (e.g. Cryptococcus neoformans (Lin et al., 2005), Candida albicans (Alby et al., 2009), Neurospora spp. (Pöggeler, 1999), and Stemphylium spp. (Inderbitzin et al., 2005)).
Detailed discussions about these types of homothallic sexual reproduction have been provided in several recent reviews (Fu et al., 2014; Ni et al., 2011). In the following sections we will focus on recent developments in the study of the homothallic sexual reproduction in a major human fungal pathogen, C. neoformans.
4. Unisexual reproduction in C. neoformans
C. neoformans is a basidiomycete yeast that has been shown to be associated with soil, trees and pigeon guano in the natural environment (Heitman et al., 2011). Together with its sister species Cryptococcus gattii, this fungus has been estimated to cause more than one million new infections and over 600,000 deaths each year (Park et al., 2009). C. neoformans has two varieties, var. grubii (serotype A) and var. neoformans (serotype D). The infection starts with inhalation of the infectious propagules by the host. These propagules lodge in the lung alveoli and then disseminate to the central nervous system, resulting in meningoencephalitis (Heitman et al., 2011). As mentioned earlier, C. neoformans has a well-characterized bipolar mating system with two mating types, MATa and MATα, which are determined by one unusually large MAT locus (Kwon-Chung, 1975; Kwon-Chung, 1976; Lengeler et al., 2002). Cells of opposite mating type can undergo heterothallic sexual reproduction that produces basidiospores. Because of the small sizes of these basidiospores, they have been long suspected to be primary infectious propagules. While C. neoformans does not produce asexual spores, it should be noted that another type of cell, called blastospores, are also frequently produced through budding off of the clamp connection along the hyphae during sexual reproduction. However, while called blastospores, these cells are in fact simply budding yeast cells based on analyses by light and electron microscopy (Lin and Heitman, 2005). Additionally, blastospores are produced prior to meiosis and thus are asexual, and there is no testing if blastospores can serve as primary infectious propagules. For a long time, researchers had been puzzled by a very unique characteristic of the C. neoformans natural populations, that is, among all of the isolates collected from natural environment or clinic, the vast majority (>99%) are of α mating type, with the exception of a few geographical regions (e.g. Africa) where the ratio between the two mating types is more balanced (Heitman et al., 2011). Given such a biased mating type composition, how often does C. neoformans undergo sexual reproduction in nature, and produce infectious spores? Additionally, what is the selection pressure that helps C. neoformans maintain the ability to undergo sexual reproduction, if there is little chance for mating (at least for the MATα individuals) in nature anyway?
It has been known for a long time that C. neoformans strains, particularly those of α mating type, can undergo monokaryotic fruiting, during which cells form filaments and produce spores through sporulation (Wickes et al., 1996). While monokaryotic fruiting was originally thought to be asexual, later studies (Lin et al., 2005) showed that fruiting α cells undergo cell fusion or endoreplication to form α/α diploid or dikaryon zygotes, and then undergo a morphological transition to filamentous growth, and eventually form basidia and produce recombinant haploid basidiospores. Therefore, monokaryotic fruiting is a unisexual reproduction process that involves two cells of the same mating type. This is also supported by subsequent studies where α/α diploids (fusion/mating products of two MATα isolates of the same serotype), as well as αADα hybrids (fusion/mating products of two MATα isolates of different serotypes, A and D) were found to be among isolates collected from natural environments, indicating unisexual reproduction occurs in nature (Lin et al., 2007; Lin et al., 2009).
Unisexual reproduction and bisexual reproduction share similarities. First, studies have shown that the two modes of sexual reproduction respond to similar environmental cues. For example, both modes of sexual reproduction are stimulated by room temperature (22–24 °C), nitrogen limitation, dehydrated substrates, absence of light, as well as the presence of mating pheromones, inositol, the plant hormone auxin, and copper ions (Fu et al., 2014; Kozubowski and Heitman, 2012). Second, the C. neoformans Cpk1 mitogen activated protein kinase (MAPK) pheromone response pathway, as well as several transcription factors, including Mat2, Znf2 and Znf3, have been shown to regulate both unisexual and bisexual reproduction (Feretzaki and Heitman, 2013; Fu et al., 2014; Kozubowski and Heitman, 2012; Lin et al., 2010). It should be noted that compared to the dikaryotic hyphae with fused clamp connections that are produced during bisexual reproduction, the hyphae produced during unisexual reproduction typically are monokaryotic with unfused clamp cells, and the diploid nuclei could be the result of either endoduplication of MATα cells, or nuclear fusion after zygote formation by two MATα cells (Lin et al., 2005).
The next key question then is whether unisexual reproduction in C. neoformans is meiotic, or a parasexual process that bypasses meiosis? It has been shown that C. neoformans monokaryotic fruiting requires Spo11 and Dmc1, two conserved proteins that are not located within the MAT locus but are essential for the formation and repair of DNA double-strand breaks during meiosis in diverse eukaryotes (Feretzaki and Heitman, 2013; Lin et al., 2005), suggesting meiosis is involved. However, it should be noted that Spo11 is also required during the parasexual cycle in C. albicans even though conventional meiosis is not involved (Forche et al., 2008). Additionally, in the original study that described the α-α unisexual reproduction in C. neoformans, while clear evidence of independent chromosomal segregation as well as recombination was found, the analyses was done on 8 progeny using 20 genetic markers located on 5 different chromosomes. Thus, the relatively sparse distribution of the markers, as well as the relatively small size of the progeny population, prevented a more quantitative assessment of recombination frequencies during α-α unisexual reproduction, as well as comparison to those observed in a-α bisexual reproduction.
In a recent study by Sun et al. (Sun et al., 2014), the authors showed that meiotic recombination in a-α bisexual and α-α unisexual reproduction occurs in a very similar fashion. Specifically, by analyzing large numbers of meiotic progeny from both a-α bisexual and α-α unisexual reproduction, they found that: 1) the numbers of crossovers along chromosomes have comparable distributions during the two modes of sexual reproduction; 2) the chromosomal average recombination frequencies (physical distance/genetic distance, kb/cM) are similar between unisexual and bisexual reproduction (average ~7 kb/cM), and are comparable to those that have been previously reported in bisexual reproduction in C. neoformans (average ~13 kb/cM) (Marra et al., 2004), and much higher than those observed during hybridization between serotypes A and D (average ~85 kb/cM) (Sun and Xu, 2007); and 3) recombination hotspots, such as those that flank the MAT locus during a-α bisexual reproduction (Hsueh et al., 2006), also operate during α-α unisexual reproduction. They also found that recombination within the MAT locus is, like during a-α bisexual reproduction, highly repressed during α-α unisexual reproduction, even though in this case the two MAT alleles are co-linear and there are no physical barriers (e.g. translocations or inversions within the MAT locus between MATa and MATα alleles) to prevent recombination from occurring during meiosis. Specifically, they analyzed 156 meiotic progeny recovered from α-α unisexual reproduction and found only one crossover event within the MAT locus. This recombination frequency is significantly lower than expected given the size of the MAT locus (~100 kb) and the average recombination frequency during α-α unisexual reproduction (~7 kb/cM). Interestingly, this crossover breakpoint within the MAT locus during α-α unisexual reproduction coincides with the region that contains a previously identified gene conversion hot spot during a-α bisexual reproduction (Sun et al., 2012). Taken together, their study provides definitive evidence that α-α unisexual reproduction in C. neoformans involves meiosis.
5. Generation of genetic diversity through sexual reproduction in C. neoformans
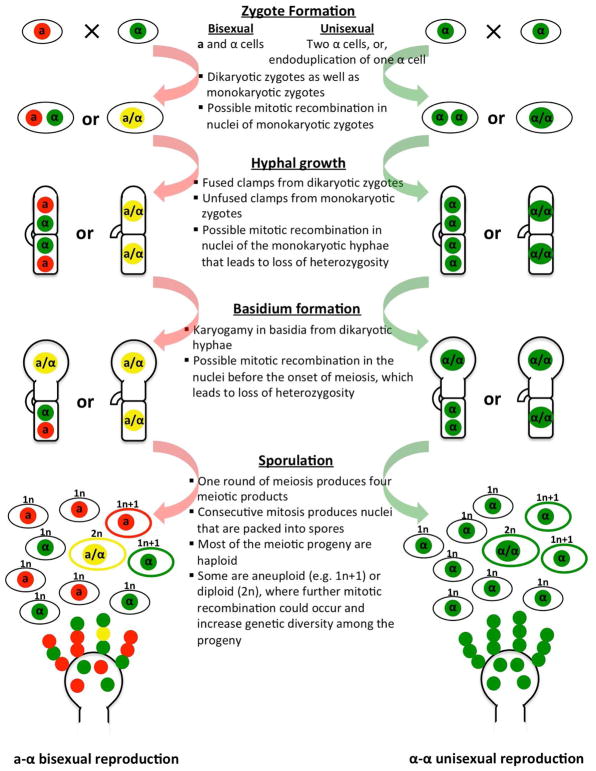
It is generally accepted that when linkage disequilibrium (LD) is present in the population, sexual reproduction, through meiotic recombination, typically increases genetic diversity by breaking down the preexisting linkage disequilibrium. However, more and more studies have shown that during sexual reproduction genetic diversity can also be generated de novo through many different mechanisms other than meiosis (Forche et al., 2008; Ni et al., 2013). This is also true for C. neoformans, and for both a-α bisexual and α-α unisexual reproduction (Figure 1).
Figure 1. Illustration of major developmental stages of mating during a-α bisexual (left) and α-α unisexual (right) reproduction in C. neoformans.
Listed in the middle are the differences, as well as common features between the two modes of sexual reproduction, and the possible origins of genetic diversity that can be generated at each stage
First, while homologous recombination can generate novel allele combinations through crossovers, it can also introduce novel nucleotide substitutions, as well as short insertions and deletions, during the induction, and subsequently, the repair of double strand breaks (DSBs). For example, in the yeast S. cerevisiae, it was shown that recombinational repair of DSBs elevated the local mutation rate by 100-fold when compared to those repaired during S-phase replication (Strathern et al., 1995). Because meiotic crossovers are mostly mediated through homologous recombination, meiosis thus could be a mutagenic process.
Second, recombination can occur not only during sexual reproduction in the form of meiotic recombination, but also during vegetative growth in the form of mitotic recombination. In a recent study of hybridization between serotypes A and D in C. neoformans, the authors found that more than four haplotypes could be identified among progeny dissected from one single basidium (Vogan et al., 2013). This contradicts the paradigm that only one meiosis occurs within the basidium, which produces a maximum number of four different meiotic progeny (Idnurm, 2010). The authors proposed that the observed excessive haplotypes from a single basidium are the result of post-meiotic mitotic recombination within aneuploid/diploid progeny that are frequently produced during A-D hybridization (Vogan et al., 2013). Evidence of mitotic recombination has also been found in study of intra-serotype a-α bisexual and α-α unisexual reproduction in serotype D C. neoformans (Sun et al., 2014). Specifically, the authors found that among progeny from α-α unisexual reproduction, certain chromosomal regions exhibited highly skewed allele inheritance, while among progeny from a-α bisexual reproduction, alleles from one of the two parents at certain chromosomal regions were absent in the progeny recovered from a single basidium. Because these markers cluster together and encompass large chromosomal regions, the most plausible explanation for this skewed allele inheritance is mitotic recombination occurring after karyogamy and prior to meiosis that resulted in loss of heterozygosity in these chromosomal regions. Indeed, in both a-α bisexual and α-α unisexual reproduction, nuclear fusion can occur immediately following cell fusion, resulting in heterozygous diploid cells. These diploid cells produce monokaryotic hyphae with unfused clamp cells, prior to the formation of basidium and the onset of meiosis. Thus, ample opportunities are present for mitotic recombination to occur during both modes of sexual reproduction in C. neoformans (Heitman et al., 2011).
Third, while the majority of the meiotic products of C. neoformans are euploid and haploid, studies have shown that aneuploid, as well as diploid progeny can be frequently produced during both a-α bisexual and α-α unisexual reproduction (Ni et al., 2013; Sun et al., 2014). Mitotic recombination occurring on the non-monosomic chromosomes could result in loss of heterozygosity (LOH), and consequently phenotypic variation. Recently, it was found that in C. neoformans, diploid progeny from both a-α bisexual and α-α unisexual reproduction could undergo whole chromosome loss and reduplication, resulting in LOH affecting a whole chromosome (Sun et al., 2014). So the production of these aneuploid and diploid progeny not only increases the phenotypic diversity among the progeny by itself, it also provides the raw material for further phenotypic and genotypic variation to accumulate. Interestingly, it has been found that in C. neoformans mitotic recombination breakpoints are enriched in certain chromosomal regions (Sun et al., 2014), suggesting the presence of fragile sites in the C. neoformans genome that are vulnerable to mitotic recombination, similar to those that have been recently reported in the yeast S. cerevisiae (Song et al., 2014). Additionally, some of these potential fragile sites overlap with previously identified meiotic recombination hot spots, suggesting certain characteristics of these chromosomal regions, such as high GC content (Hsueh et al., 2006), might render them more prone to induction of both meiotic and mitotic recombination.
6. Evolutionary consequences of unisexual reproduction in C. neoformans
As mentioned earlier, the natural population (environmental and clinical) of C. neoformans has a biased mating type distribution, with MATα individuals comprising the majority of the individuals isolated from natural environment, with the exception of certain geographic regions, such as in sub-Saharan Africa where the natural population of C. neoformans (molecular group VNB) appears to have a relatively balanced mating type composition (Litvintseva et al., 2003; Litvintseva et al., 2006). Thus, unisexual reproduction increases the chances that C. neoformans can undergo successful sexual reproduction, and consequently enjoy the benefits conferred by meiosis, e.g. breaking up existing regions of LD, thus increasing genetic diversity and the efficiency of natural selection (Hiremath et al., 2008). Additionally, several studies have shown that MATα strains are at least equally, and in some cases more virulent when compared to their MATa isogenic counterparts. Thus, α-α unisexual reproduction, and the production of MATα basidiospores, may also have consequences on the epidemiology of this fungal pathogen (Kwon-Chung et al., 1992; Nielsen et al., 2005a; Nielsen et al., 2003; Nielsen et al., 2005b). It should be noted that from the standpoint of pathogens such as C. neoformans, asexual reproduction could sometimes also be beneficial, such as when a particular genotype emerges that confers advantages to the individual when surviving in the environment or within the host. However, clonal expansion will produce linkage disequilibrium in the population, which will then require sexual reproduction to break up and generate novel allele combinations at the population level.
Another evolutionary aspect of unisexual reproduction that has not been fully investigated yet is its effect on the inheritance and evolutionary dynamics of mitochondria. In C. neoformans, mitochondrial inheritance is uniparental from the MATa parent during a-α bisexual reproduction (Yan et al., 2004; Yan et al., 2007; Yan and Xu, 2003). However, this mitochondrial uniparental inheritance is compromised during α-α unisexual reproduction, and consequently, the chances of mitochondrial mixing and recombination increases. How this would affect the population structure, as well as the evolutionary trajectory of the mitochondria in C. neoformans, remains to be elucidated.
7. Conclusion
It is now clear that α-α unisexual reproduction in C. neoformans is a canonical meiotic process, with recombination frequency, presence, and distribution of recombination hot spots and cold spots, as well as the frequency at which aneuploid and diploid progeny are generated all comparable to those observed during a-α bisexual reproduction (Sun et al., 2014). By comparison to the modes of sexual reproduction in closely related species, it appears that the α-α unisexual reproduction in C. neoformans is a derived state from bisexual reproduction, as the MATa and MATα alleles of the mating type determining genes (SXI1, SXI2, and STE3, etc.) in C. neoformans all show mating type specific clustering together with their corresponding alleles from the closely related heterothallic sister species. It is yet to be elucidated why and how some of the MATα isolates obtained the ability to undergo both unisexual and bisexual reproduction. It could still be that this ability to undergo both unisexual and bisexual reproduction is an intrinsic characteristic of MATα isolates, and the initiation of unisexual reproduction is due to some yet to be identified environmental cues. Or, it could be due to genetic changes in the MATα allele that resulted in relaxed mating partner recognition, thus, enabling the individual to undergo both homothallic and heterothallic sexual reproduction. Such isolates will inevitably have an advantage over the original MATα isolates in finding suitable mating partners, given the highly skewed mating type composition towards the α mating type in the natural population of C. neoformans.
Being capable of undergoing either homothallic or heterothallic sexual reproduction allows the organism to alleviate costs associated with mating, and to better survive and expand in hostile or changing environments by generating novel genotypes, increasing genetic diversity to facilitate natural selection, producing spores that are more resistant to harsh environments, as well as by transitioning into hyphal growth forms that are better at acquiring nutrients from the environment (Fu et al., 2014; Phadke et al., 2013). The frequent transitions between vegetative growth and mating, or between homothallic and heterothallic sexual reproduction, illustrate the balance between inbreeding and outcrossing, and are likely a response to specific environmental cues that the organism has evolved to recognize and respond to by undergoing one mode of reproduction or the other.
Acknowledgments
We thank Dr. Ursula Kües and Blake Billmyre for comments and suggestions during the preparation of the manuscript. This study was supported by R37 MERIT award AI39115-17 and R01 grant AI50113-10 from the NIH/NIAID to J. H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield G, Skelton J, Ivens A, Tanaka Y, Kay RR. Sex determination in the social amoeba Dictyostelium discoideum. Science. 2010;330:1533–1536. doi: 10.1126/science.1197423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Sex determination in reptiles. Quart Rev Biol. 1980;55:3–21. [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in Basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, Miao W, Orias E. Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol. 2013;11:e1001518. doi: 10.1371/journal.pbio.1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJS. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature. 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Calderon-Urrea A. Sex determination in flowering plants. The Plant Cell Online. 1993;5:1241–1251. doi: 10.1105/tpc.5.10.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. Fission yeast mating-type switching: programmed damage and repair. DNA Repair. 2005;4:525–536. doi: 10.1016/j.dnarep.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet. 2013;9:e1003688. doi: 10.1371/journal.pgen.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Sun S, Fraser JA, Hsueh Y-P, Averette AF, Li W, Dietrich FS, Heitman J. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genet. 2012;8:e1002528. doi: 10.1371/journal.pgen.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Evolution of fungal sex chromosomes. Mol Microbiol. 2004;51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- Fu C, Sun S, Billmyre RB, Roach KC, Heitman J. Unisexual versus bisexual mating in Cryptococcus neoformans: consequences and biological impacts. Fungal Genet Biol. 2014 doi: 10.1016/j.fgb.2014.08.008. pii: S1087–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Goodfellow PN, Lovell-Badge R. SRY and sex determination in mammals. Annu Rev Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SJ, Byrne KP, Wolfe KH. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. PNAS. 2014;111:E4851–E4858. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Kozel TR, Kwon-Chung JK, Perfect JR, Casadevall A. Cryptococcus: from human pathogen to model yeast. ASM Press; Washington, D. C: 2011. [Google Scholar]
- Heitman J, Sun S, James TY. Evolution of fungal sexual reproduction. Mycologia. 2013;105:1–27. doi: 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, Xu J. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology. 2008;154:1513–1524. doi: 10.1099/mic.0.2007/015594-0. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Idnurm A, Heitman J. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2006;2:e184. doi: 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2010;185:153–163. doi: 10.1534/genetics.109.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. Lateral transfer of mating system in Stemphylium. PNAS. 2005;102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY. Analysis of mating-type locus organization and synteny in mushroom fungi: Beyond model species. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in fungi. ASM Press; Washington, DC: 2007. pp. 317–331. [Google Scholar]
- James TY, Srivilai P, Kües U, Vilgalys R. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics. 2006;172:1877–1891. doi: 10.1534/genetics.105.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJS. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- Klar AJS. The yeast mating-type switching mechanism: A memoir. Genetics. 2010;186:443–449. doi: 10.1534/genetics.110.122531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen H. Sex ratios and conditions required for environmental sex determination in animals. Biological Reviews. 1990;65:147–184. doi: 10.1111/j.1469-185x.1990.tb01187.x. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Heitman J. Profiling a killer, the development of Cryptococcus neoformans. FEMS Microbiol Rev. 2012;36:78–94. doi: 10.1111/j.1574-6976.2011.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarère J, Nöel T. Mating type switching in the tetrapolar basidiomycete Agrocybe aegerita. Genetics. 1992;131:307–319. doi: 10.1093/genetics/131.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Challen M, Elliott T, Casselton L. Molecular analysis of breeding behavior in Agaricus species Mushroom. Science. 2004;16:103–109. [Google Scholar]
- Lin X, Heitman J. Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot. Cell. 2005;4:1746–1754. doi: 10.1128/EC.4.10.1746-1754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell. 2003;2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi V, Earley R, Grober M. Preventing behavioural interactions with a male facilitates sex change in female bluebanded gobies, Lythrypnus dalli. Behav Ecol Sociobiol. 2006;59:715–722. [Google Scholar]
- Maekawa H, Kaneko Y. Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet. 2014;10:e1004796. doi: 10.1371/journal.pgen.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra RE, Huang JC, Fung E, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans) Genetics. 2004;167:619–631. doi: 10.1534/genetics.103.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNair A, Senior, Lokman PM, Closs GP, Nakagawa S. Ecological and evolutionary applications for environmental sex reversal of fish. The Quarterly Review of Biology. 2015;90:23–44. doi: 10.1086/679762. [DOI] [PubMed] [Google Scholar]
- Merino ST, Nelson MA, Jacobson DJ, Natvig DO. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics. 1996;143:789–799. doi: 10.1093/genetics/143.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 2013;11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Sun S, Wang X, Heitman J. Sex in fungi. Annu Rev Genet. 2011;45:405–430. doi: 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK, Jr, Giles SS, Mitchell TG, Casadevall A, Perfect JR, Heitman J. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect Immun. 2005a;73:4922–4933. doi: 10.1128/IAI.73.8.4922-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, Heitman J. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005b;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Phadke SS, Feretzaki M, Heitman J. Unisexual reproduction enhances fungal competitiveness by promoting habitat exploration via hyphal growth and sporulation. Eukaryot Cell. 2013;12:1155–1159. doi: 10.1128/EC.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S. Phylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes. Curr Genet. 1999;36:222–231. doi: 10.1007/s002940050494. [DOI] [PubMed] [Google Scholar]
- Raper JR. Genetics of sexuality in higher fungi. Ronald Press Co; New York: 1966. [Google Scholar]
- Singh DP, Saudemont B, Guglielmi G, Arnaiz O, Gout JF, Prajer M, Potekhin A, Przybos E, Aubusson-Fleury A, Bhullar S, Bouhouche K, Lhuillier-Akakpo M, Tanty V, Blugeon C, Alberti A, Labadie K, Aury JM, Sperling L, Duharcourt S, Meyer E. Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014;509:447–452. doi: 10.1038/nature13318. [DOI] [PubMed] [Google Scholar]
- Song W, Dominska M, Greenwell PW, Petes TD. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. PNAS. 2014;111:E2210–E2218. doi: 10.1073/pnas.1406847111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Billmyre RB, Mieczkowski P, Heitman J. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet. 2014;10:e1004849. doi: 10.1371/journal.pgen.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hsueh YP, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012;8:e1002810. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Xu J. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics. 2007;177:1475–1486. doi: 10.1534/genetics.107.078923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan AA, Khankhet J, Xu J. Evidence for mitotic recombination within the basidia of a hybrid cross of Cryptococcus neoformans. PLoS ONE. 2013;8:e62790. doi: 10.1371/journal.pone.0062790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. PNAS. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hull CM, Heitman J, Sun S, Xu J. SXI1α controls uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Biol. 2004;14:R743–744. doi: 10.1016/j.cub.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hull CM, Sun S, Heitman J, Xu JP. The mating-type specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Genet. 2007;51:187–195. doi: 10.1007/s00294-006-0115-9. [DOI] [PubMed] [Google Scholar]
- Yan Z, Xu J. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2003;163:1315–1325. doi: 10.1093/genetics/163.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]