Abstract
Germ cells are the special cells in the body that undergo meiosis to generate gametes and subsequently entire new organisms after fertilization, a process that continues generation after generation. Recent studies have expanded our understanding of the factors and mechanisms that specify germ cell fate, including the partitioning of maternally supplied ‘germ plasm’, inheritance of epigenetic memory and expression of transcription factors crucial for primordial germ cell (PGC) development. Even after PGCs are specified, germline fate is labile and thus requires protective mechanisms, such as global transcriptional repression, chromatin state alteration and translation of only germline-appropriate transcripts. Findings from diverse species continue to provide insights into the shared and divergent needs of these special reproductive cells.
Germ cells have a crucial role in development. They are uniquely capable of undergoing meiosis, the specialized cell cycle necessary to generate oocytes and sperm. The fusion of these gametes at fertilization launches the development of a new organism, which will have all of the diverse somatic cell types and a fresh set of germ cells. To serve this crucial purpose of making gametes and subsequently generating entire new organisms, generation after generation, germ cells must be properly specified early in development and then protected from deviating from their germline-differentiation path during later stages of development.
There are two general modes of germ cell specification (BOX 1). In some animal species, germline identity is continuous and is passed via the oocyte to the primordial germ cells (PGCs), which are formed during early embryogenesis. In such animals, specification of germ cell fate involves segregated cytoplasmic ‘determinants’. In other animal species, including mammals, the germline is discontinuous, as PGCs must be newly induced from a subset of embryonic cells later in development. These two modes of germ cell specification are often called preformation and induction, respectively1.
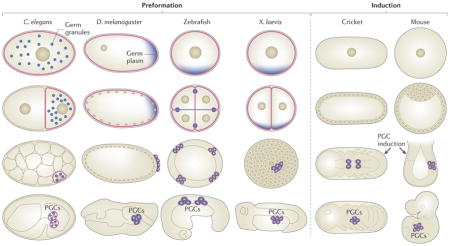
Box 1. Formation of PGCs in diverse animal embryos.
Common animal models used to investigate germ cell specification and maintenance include the nematode Caenorhabditis elegans, the fruitfly Drosophila melanogaster, the zebrafish (Danio rerio), the frog Xenopus laevis, the cricket Gryllus bimaculatus and the mouse Mus musculus.
In C. elegans, D. melanogaster, zebrafish and X. laevis, germ cell specification occurs by preformation: germline identity is continuous and is passed via the oocyte to the primordial germ cells (PGCs) formed during early embryogenesis.
In crickets and mice, specification of germ cell fate occurs by induction: the germline is discontinuous, as PGCs must be newly induced from a subset of embryonic cells later in development (see the figure).
Embryogenesis starts at the one-cell zygote stage, followed by cleavage and the initiation of gastrulation, then by organogenesis (see the figure; top row, middle two rows and bottom row, respectively). In preformation embryos, such as those of C. elegans, D. melanogaster, zebrafish and X. laevis, the germline is present at all stages (see the figure; germline cells are outlined in pink), and the PGCs inherit maternally supplied germ plasm (see the figure; shown in blue). In induction embryos, such as those of crickets and mice, the PGCs do not inherit germ plasm and instead are induced by cell signalling.
Protection of germ cell fate is accomplished by several mechanisms. For many animals, a first level of protection from extrinsic somatic signals is accomplished by the separation of PGCs from somatic cells during early embryogenesis. As somatic cells turn on their transcriptional programmes in response to maternal and zygotic cues, newly formed PGCs are protected by their transcriptional quiescence, which is regulated at the level of RNA polymerase II (Pol II) activity and chromatin state. Later in development, after PGCs initiate their transcriptional programme, germ cell identity is protected by preventing expression of transcripts that are inappropriate for germ cells. Disruption of these protective mechanisms in the germline induces germ cells to lose their germ cell fate.
Recent discoveries have expanded and challenged traditional models of germ cell specification and protection. This Review revisits previously known factors and mechanisms and introduces new players in germ cell specification and protection. The first half of the article discusses the mechanisms of germ cell specification in diverse animal types. The second half discusses common and unique strategies different animals use to protect germ cell fate.
Specifying germ cell fate
The search for germ cell determinants in preformation animals has yielded conflicting views of the roles of unique cytoplasmic organelles generically termed ‘germ granules’ and has focused our attention on the ‘germ plasm’ in which germ granules reside. The search for factors that are critical for de novo induction of germ cells in mammalian embryos has identified a small set of signalling molecules and transcription factors. In both types of animals, chromatin-level epigenetic regulation plays a key part.
Specification of PGCs by germ plasm and germ granules
Germ granules are a well-known feature of germ cells and have been suggested to function as determinants of germ cell fate ever since their segregation to germ cells was first observed2. By electron microscopy, they are seen as amorphous, non-membrane-bound, electron-dense aggregates in the cytoplasm of germ cells in numerous phyla (reviewed in REF. 3). The discovery of antibodies to germ granules in Caenorhabditis elegans and Drosophila melanogaster paved the way for analysis of germ-granule behaviour, composition and function4,5. In both organisms, germ granules are maternally loaded into embryos and segregated to the PGCs (BOX 1). In D. melanogaster, segregation occurs during oogenesis, positioning germ granules at the end of the oocyte where PGCs will bud off during embryogenesis5–7. In C. elegans, segregation occurs progressively during the four asymmetric embryonic divisions that ultimately generate a germline blastomere and several somatic founder cells4. Segregation of maternally loaded germ granules to the PGCs has also been documented in Xenopus laevis and zebrafish (BOX 1). In these vertebrates, the transmission of maternal germ granules evolved independently from the ancestral condition of absence of maternal germ granules in early embryos, as displayed by mammals8. In mammals, PGCs are induced at the post-implantation epiblast stage of embryogenesis. After they migrate to the genital ridge, mammalian PGCs synthesize new germ-granule components and assemble them into diverse granule types as development proceeds9–11. Thus, germline-specific granules are observed in diverse animals, but segregation of maternally supplied germ granules to PGCs is not a universal rule.
Conflicting evidence that germ granules are determinants
The notion that germ granules have a role in germline specification was initially suggested by cytoplasm-leak and cytoplasm-transfer experiments in early beetle, amphibian and fruitfly embryos2,12,13 (FIG. 1). Illmensee and Mahowald’s classic cytoplasm-transplantation experiments in D. melanogaster showed that the cytoplasm that contains germ granules, when transferred to an ectopic location, is sufficient to induce the formation of PGCs in that new location14. Similarly, transplantation of germ-granule-containing cytoplasm to ectopic sites in X. laevis embryos leads to the formation of ectopic PGCs15. In both systems, the ectopic PGCs were shown to be functional, as evidenced by their ability to generate progeny after the PGCs were transferred to positions that allowed them to migrate to the gonad. Those experiments have been highly influential but do not directly demonstrate that germ granules are crucial germline determinants. The isolation of D. melanogaster mutants defective in the assembly of germ granules, and the molecular identification of germ-granule components, enabled more refined analyses. Notably, mutants that fail to assemble germ granules fail to form PGCs and as a result develop into sterile adults16,17; additionally, mislocalization or overexpression of Oskar, a key component for germ-granule assembly, leads to the formation of ectopic PGCs18,19. These findings led to the view that germ granules are necessary and sufficient to specify germline fate, apparently validating the textbook claims that germ granules are germline determinants.
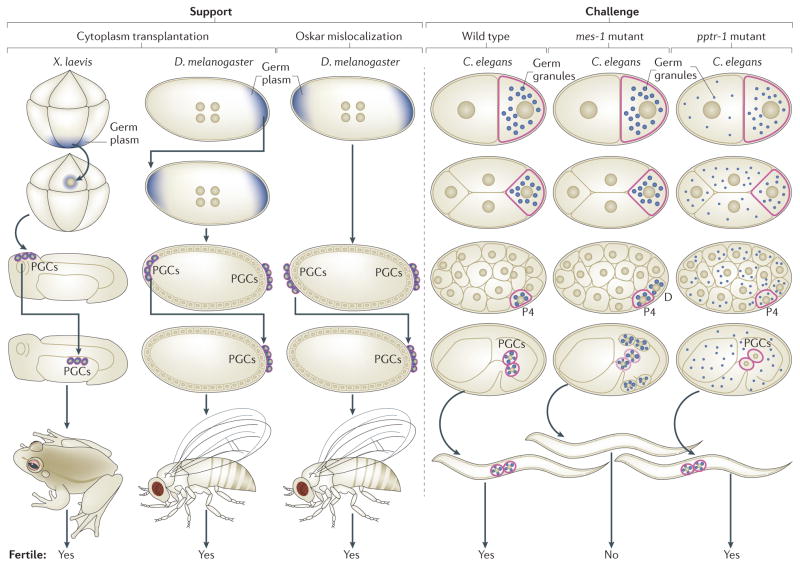
Figure 1. Findings that support or challenge the notion that germ granules are ‘determinants’ of PGC fate.
Germline cells are outlined in red. Support: cytoplasm-transfer experiments in Xenopus laevis and Drosophila melanogaster demonstrated that the cytoplasm containing germ granules (germ plasm; shown in blue) can cause formation of primordial gem cells (PGCs) at ectopic sites. Those ectopic PGCs were functional and able to generate a fertile germline after they were transferred to the normal PGC location in embryos. Mislocalization of the germ-granule component Oskar to the wrong (anterior) end of D. melanogaster embryos caused induction of anterior PGCs that, after transfer to the normal PGC location, were functional and capable of generating a fertile germline. Challenges: in Caenorhabditis elegans maternal effect sterile 1 (mes 1)-mutant embryos, germ granules (shown in blue) are mis-segregated to both the PGC (P4) and its somatic sister cell (D). Despite containing germ granules, both cells develop as muscle (the normal fate of D) instead of germline (the normal fate of the PGC). In C. elegans protein phosphatase 2A regulatory subunit 1 (pptr 1)-mutant embryos, maternal germ granules do not become enriched in embryonic PGCs. The PGCs nevertheless turn on a germline programme, including zygotic synthesis of germ granules, and can develop into a fertile germline.
Findings from diverse organisms have challenged the view that germ granules are germline determinants. When X. laevis PGCs were transplanted to ectopic sites, they were observed to differentiate into diverse somatic cell types and join the somatic tissues surrounding them20. This revealed that germ-granule-containing frog PGCs are not irreversibly determined to follow a germline fate and that PGCs require protection from somatic influences. C. elegans maternal effect sterile 1 (mes-1)-mutant embryos fail to segregate germ granules to the final germline blastomere P4 cell and instead deliver granules to both P4 and its sister cell, D, which in normal embryos generates muscle (FIG. 1). The observation that mis-segregated granules do not drive the D cell towards a germline fate suggests that germ granules are not sufficient for germline fate in worms21. An alternative interpretation is that, in mes-1-mutant embryos, neither the P4 nor the D cell inherits a sufficient amount of germ-granule material to specify germline fate. Indeed, in D. melanogaster embryos, cells with reduced levels of germ-granule material do not form PGCs22.
Evidence that segregation of maternal germ granules may not be necessary for specification of germline fate in worms comes from analysis of C. elegans protein phosphatase 2A regulatory subunit 1 (pptr-1) and maternal effect germ-cell defective (meg) mutants. These mutants fail to partition germ granules to the germ lineage during the asymmetric divisions of the early embryo but nevertheless develop into fertile adults23,24. It should be noted that pptr-1- and meg-mutant animals are not devoid of germ granules; the PGCs in mutant embryos inherit a reduced level of maternally provided germ-granule factors, perhaps in the form of small granules, but then synthesize and assemble new germ granules as they develop, so that larval and adult animals may have a full complement of germ granules.
Taken together, the above findings challenge the simple view that segregated germ granules are necessary and sufficient to instruct cells to develop as germ cells, and they raise important questions about the roles of individual germ-granule components and of the cytoplasm in which germ granules reside.
RNA-binding and other properties of germ plasm and granules
Germ granules reside in specialized cytoplasm called germ plasm. The relationship between and the relative functions of germ granules and germ plasm are not well understood. In D. melanogaster and X. laevis, germ plasm appears as a zone of cytoplasm in which distinct germ granules can only be observed by high-resolution immunofluorescence imaging or electron microscopy25,26. In C. elegans, germ granules are readily seen even by low-resolution immunofluorescence imaging, and some germ-granule proteins (for example, the P-granule abnormality (PGL) proteins) seem to be almost quantitatively associated with granules27. Fluorescence recovery after photo-bleaching (FRAP) experiments revealed that PGL-1 dynamically associates with granules, so there is a dynamic equilibrium between PGL-1 in granules and PGL-1 diffuse in the cytoplasm28,29. In fact, C. elegans germ granules are thought to be segregated to the germline cytoplasm by condensation of granules in germline-destined cytoplasm and dissolution of granules in non-germline-destined cytoplasm29. C. elegans pharynx intestine in excess 1 (PIE-1) exemplifies a different pattern. Some PIE-1 associates with germ granules, but much of it is diffusely distributed in the germ plasm. Importantly, the granule association of PIE-1 is not necessary for its segregation to the germline blastomeres during the embryonic divisions, and PIE-1 is still segregated to the PGCs in pptr-1-mutant embryos that fail to segregate large germ granules23. The latter finding underscores the importance of germ plasm and the dynamic relationship between germ granules and the cytoplasm in which they reside.
The vast majority of germ-granule and germ-plasm components are known or suspected to bind RNA. Among the components conserved across species are the VASA-related RNA helicases, the Tudor-domain proteins, NANOS, the Arg methyltransferase PRMT5 and certain Argonaute proteins30–32. These conserved RNA-binding proteins are each essential for determining and maintaining germ cell fate, which may or may not be dependent on their association with germ granules or germ plasm. Teasing apart the specific functions of individual germ-granule components from their roles in germ-granule assembly is challenging, as mutations in these conserved components also compromise germ-granule assembly. Interestingly, Arg methylation of germline Argonaute proteins by PRMT5 facilitates interactions with proteins containing multiple Tudor domains, which in turn may nucleate germ-granule aggregate formation33,34. Some components that seem to be species-specific are the PGL proteins in C. elegans, Oskar in D. melanogaster and bucky ball in zebrafish6,27,35,36.
The RNA-binding theme suggests that, across species, germ granules and germ plasm regulate RNA stability and translation in germ cells, and/or that RNAs have structural roles. Interestingly, germ granules may be the germline version of the RNA–protein hydrogels that were recently shown to result from the aggregation of low-complexity, or intrinsically disordered, sequence domains of some RNA-binding proteins, which can recruit other RNA-binding proteins via RNA linkages37–41. As an example, C. elegans PGL-1 and PGL-3 are both capable of aggregating into granules when expressed in non-germline cells (such as the worm intestine or mammalian tissue culture cells)42,43. Most other germ-granule proteins tested did not aggregate into granules on their own but were recruited to PGL granules, perhaps via RNA. Movies of the behaviour of C. elegans germ granules in vivo revealed them to have properties of liquid droplets29. Analysis of CAR-1 (cytokinesis, apoptosis, RNA-associated 1) granules in the C. elegans germline exemplifies how RNA granules can undergo ‘phase transitions’ from diffuse to liquid granules to crystals and how such transitions may be regulated44. Such phase transitions may contribute to a dynamic equilibrium between important germline components diffusely residing in germ plasm and being concentrated in liquid-droplet-like germ granules.
In summary, although germ granules are obvious and unique occupants of germline cytoplasm, the assembly of large granules and their segregation to the PGCs seems not to be required for germ cell specification. Other components of germ plasm probably have key roles; in fact, the distinction between germ granules and germ plasm is not well understood and may be artificial. It remains to be tested whether the phase transition of germ-plasm components into large aggregates is crucial for their function. Moreover, certain species specify their germ cells in the absence of maternally provided germ plasm and germ granules, as discussed in the next section.
De novo induction of PGCs
Mammalian embryos do not transmit distinctive germ plasm to a subset of early embryonic cells and do not set PGCs apart during the early embryonic divisions. Instead, PGCs are induced in post-implantation epiblasts. In mouse embryos at embryonic day 5.75 (E5.75), WNT3-expressing cells in the posterior corner of the proximal epiblast are subject to bone morphogenetic protein 4 (BMP4) signalling from neighbouring extra-embryonic tissue (FIG. 2). WNT3 signalling induces expression of the mesodermal transcription factor T (also known as BRACHYURY), but BMP4 signalling blocks T from binding the promoters of mesodermal genes, allowing T to instead turn on expression of transcription factors needed for germ cell development45,46. Recent reviews nicely present the details of PGC specification in mice47–51. Here we summarize a few of the key concepts.
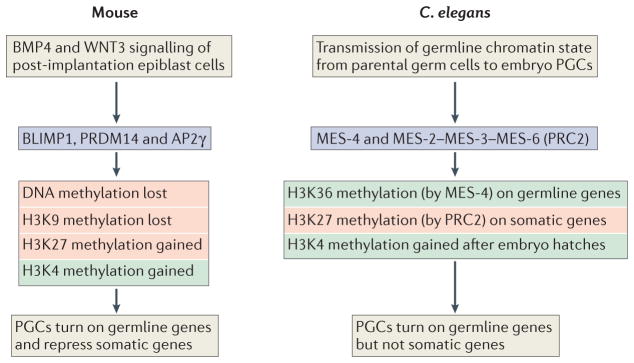
Figure 2. Chromatin regulation in the PGCs of mice and C. elegans.
In mice, bone morphogenetic protein 4 (BMP4) signalling from extra-embryonic tissue causes a small cluster of WNT3-primed post-implantation epiblast cells to express the transcription factors BLIMP1 (also known as PRDM1), PR domain zinc-finger protein 14 (PRDM14) and activating enhancer-binding protein 2γ (AP2γ). This set of three transcription factors is necessary and sufficient to cause the indicated chromatin changes, turn on germline genes and repress somatic genes in the newly induced primordial germ cells (PGCs). In Caenorhabditis elegans, the memories of germline gene expression and repression are transmitted from parent germ cells to progeny germ cells by maternal effect sterile 4 (MES-4) and by MES-2 MES-3 MES-6, the worm version of polycomb repressive complex 2 (PRC2), respectively. MES-4 achieves this via methylation of Lys36 on histone H3 (H3K36), and MES-2 MES-3 MES-6 operates via methylation of H3K27 PGCs are kept in a relatively quiescent chromatin state, lacking H3K4 methylation, until after the embryo hatches and starts feeding. Chromatin marks associated with gene expression are in green, and those associated with repression are in red.
Expression of three transcription factors, BLIMP1 (also known as PRDM1), PR domain zinc-finger protein 14 (PRDM14) and activating enhancer-binding protein 2γ (AP2γ), induced by WNT3 and BMP4 signalling in a small set of cells in post-implantation epiblasts, is necessary to specify PGC fate. Interestingly, it is thought that all epiblast cells are competent to be induced to PGCs. These three transcription factors, with PRDM14 playing a central part, can even programme cells in vitro to become PGC-like cells, which can produce gametes and offspring in vivo, revealing that the transcription factor trio is sufficient for PGC fate45,52,53. Among the gene-expression changes that the transcription factor trio causes are repression of somatic genes and activation of germ cell genes, including pluripotency genes. At least part of the mode of action of BLIMP1, PRDM14 and AP2γ is resetting the ‘epigenetic landscape’ in post-implantation epiblasts to germline (reviewed in REF. 51). In culture, epiblast-like cells seem to be poised for expression of somatic genes and for somatic differentiation, as those cells contain ‘bivalent’ chromatin marks (a mixture of activating and repressive histone modifications) on a large number of developmental regulatory genes54. On PGC induction, bivalent chromatin marks are lost from many regions, global DNA demethylation proceeds to what is considered a ground state, and the repressive chromatin mark of dimethylation of Lys9 on histone H3 (H3K9me2) is replaced by that of trimethylation of Lys 27 on H3 (H3K27me3) (FIG. 2). H3K9me2 is associated with heterochromatin and H3K27me3 with more dynamically regulated repressed chromatin. Loss of any of the three key transcription factors from mice causes the mutant PGCs to be lost or to develop more like their somatic neighbours45,55,56.
PGC induction in mice illustrates an alternative mode of specification to the segregation of germ plasm used in the preformation organisms discussed in the previous section. Although germ plasm is not segregated to mouse PGCs, components of germ plasm, including the mouse VASA homologue, MVH (also known as DDX4), are expressed after PGCs are specified10. Eventually, these components can be observed to assemble into distinct granules (reviewed in REF. 57). In embryos and during gametogenesis, germ granules include VASA, Argonaute proteins, PRDM5 and Tudor-domain proteins. Although germ granules and germ plasm do not have a specification role in mouse, they are a feature of mouse germ cells at various stages of development.
A recent hallmark paper investigated PGC specification in human embryos58. Human embryonic stem cells were induced by BMP4 and other factors to develop into human PGC-like cells (PGCLCs). Unexpectedly, SOX17, a transcription factor needed for endoderm, was identified as being expressed early in human PGCLC derivation and shown to be essential for germ cell fate. BLIMP1 was shown to act downstream of SOX17 to repress endoderm and other somatic genes. This study highlights important similarities and differences between human and mouse PGC specification. Both require BLIMP1 and a transcription factor that functions in somatic differentiation. In mouse embryos, the mesodermal transcription factor T is diverted to a germ cell-promoting function by BMP4 signalling, whereas in humans the endodermal transcription factor SOX17 promotes germ cell development through collaboration with BLIMP1. Other differences underscore that mammalian embryos do not necessarily use the same set of factors and mechanisms to drive critical developmental events such as PGC specification.
Along the same lines, recent studies of diverse insects have revealed that segregation of maternally provided germ plasm is not shared by all insects and that several use the mammalian mode of PGC induction. The cricket Gryllus bimaculatus has been particularly illustrative. First, it was discovered that G. bimaculatus has an oskar homologue and that, in contrast to D. melanogaster Oskar, G. bimaculatus OSKAR is not required for PGC specification59. Instead, G. bimaculatus OSKAR functions in neural development. The ancestral role of OSKAR is thought to be neural development, with a co-opted role in PGC specification in higher insects like D. melanogaster. Subsequently, analysis of G. bimaculatus VASA and PIWI demonstrated that G. bimaculatus lacks maternally inherited germ plasm60. How does G. bimaculatus specify its PGCs? Similarly to mouse PGCs, G. bimaculatus PGCs are induced later in development60 (BOX 1). G. bimaculatus PGCs arise from abdominal mesoderm; notably, embryos that lack mesoderm fail to form PGCs. The important implications of these findings are that the ancestral mechanism of PGC specification in insects is induction in later-stage embryos, and that reliance on inherited germ plasm by D. melanogaster and other higher insects is a derived mechanism.
Transmission of an ‘epigenetic memory of germline’
The epigenetic landscape plays a key part in PGC specification (as described above for mice), even in preformation embryos, which inherit and transmit germ plasm. Studies in C. elegans illustrate how a ‘memory of germline’ is passed from the parental germline to the PGCs and is critical for PGCs to correctly launch their germline developmental programme. The major players in this transmission of germline memory are the MES histone modifiers (FIG. 2) (reviewed in REF. 61). MES-2, MES-3 and MES-6 form the worm version of polycomb repressive complex 2 (PRC2) and generate the repressive modification H3K27me3. MES-4 generates modifications associated with active gene expression: H3K36me2 and H3K36me3. Loss of any of the MES proteins from the mother worm’s germline causes her progeny to be sterile, as a result of death of the PGCs after a few divisions. Thus, maternal MES product is needed to ensure survival and proper development of PGCs in her progeny. Immunostaining, chromatin immunoprecipitation (ChIP) experiments and transcript profiling have led to the following model of MES regulation62–64. The MES-2–MES-3–MES-6 complex concentrates repressive H3K27me3 on the X chromosomes and on somatic genes located on the autosomes; both categories of genes need to be kept repressed in germ cells. MES-4 concentrates H3K36 methylation on germline-expressed genes, to maintain them in an open chromatin state. H3K36 methylation prevents H3K27 methylation on the same histone tails65,66, providing a mechanism for MES-4 to keep germline genes open, by preventing H3K27 methylation of those genes by MES-2–MES-3–MES-6. An attractive scenario is that MES-4-catalysed H3K36 methylation of germline-expressed genes in the parental germline and during early embryo development protects those genes from being shut down by H3K27 methylation during the window between fertilization and formation of PGCs, thus poising those genes for expression when the PGCs initiate their gene expression programme.
Any transgenerational epigenetic memory model, like that described above, must address several key questions, including: are epigenetic marks transmitted from the parental germline to the embryo? If so, by the oocyte, the sperm or both, and in what form? How are epigenetic marks transmitted through embryonic cell divisions? Recent analysis of H3K27me3 transmission provides answers to these questions67. H3K27me3 is transmitted to the one-cell embryo on the chromosomes from both the sperm and the oocyte. The MES-2–MES-3–MES-6 enzyme complex is supplied to the one-cell embryo via the oocyte, not the sperm. Strategic crosses were used to generate two informative types of embryos. Embryos that lacked MES-2–MES-3–MES-6 and had H3K27me3 on the sperm chromosomes but not the oocyte chromosomes revealed that the H3K27me3 mark is passed through at least four rounds of DNA replication and cell division. Remarkably, H3K27me3 remained restricted to sperm-derived chromosomes. Thus, histone marks themselves can provide short-term memory. Embryos that possessed MES-2–MES-3–MES-6 and had H3K27me3 on the oocyte chromosomes but not the sperm chromosomes revealed that H3K27me3 is passed through many divisions, in this case remaining restricted to oocyte-derived chromosomes. Thus, the histone-modifying enzyme can provide long-term memory. These findings illustrate how epigenetic memory can be passed from parents to progeny to influence developmental decisions, in this case how PGCs initiate their developmental programme.
Protecting germ cell fate
Germ cell fate is labile and can be lost when germ cells experience inappropriate conditions or signals. This was elegantly demonstrated by transplanting X. laevis PGCs from their normal migratory route (towards the gonadal ridge) to an ectopic site (the blastocoel cavity) of host embryos and observing that the transplanted PGCs contributed to somatic tissues in all three germ layers20, as mentioned in an earlier section. Germ cell fate is protected by separating PGCs from their somatic neighbours and then by protective mechanisms that operate in both the nucleus and cytoplasm of germ cells.
Protection by transcriptional silencing in PGCs
Newly formed PGCs are protected from somatic differentiation by inhibition of transcription. In both C. elegans and D. melanogaster, transcription is blocked via inhibition of positive transcription elongation factor b (P-TEFb)68. P-TEFb is a cyclin-dependent kinase that phosphorylates the carboxy-terminal domain (CTD) of RNA Pol II to activate transcription elongation69. Although both C. elegans and D. melanogaster silence transcription by inhibiting P-TEFb, they use different proteins and mechanisms to accomplish this inhibition (FIG. 3). In C. elegans, the germline protein PIE-1 contains CTD-like domains that bind and sequester P-TEFb and prevent it from interacting with Pol II70. PIE-1 has also been shown to block Pol II transcription initiation in a P-TEFb-independent manner71. When PIE-1 is absent, germline blastomeres adopt somatic cell fates72. D. melanogaster PGCs express a protein called polar granule component, which physically interacts with P-TEFb and inhibits the recruitment of this complex to chromatin73. In the absence of polar granule component function, somatic transcripts are expressed and nascent germ cells degenerate74,75. Later in D. melanogaster development, during PGC migration to the embryonic gonads, the inhibition of CTD phosphorylation is maintained independently of P-TEFb, requiring the germ-granule component Nanos76. nanos-mutant PGCs express numerous somatic transcripts77,78. NANOS is also required to repress CTD phosphorylation and somatic gene expression in the germline blastomeres and PGCs of X. laevis and the sea urchin Strongylocentrotus purpuratus79–81. Because NANOS is a cytoplasmic RNA-binding protein, its effect on transcription is likely to be indirect1,80,82. The sea squirt Ciona intestinalis represses CTD phosphorylation and zygotic gene expression in germline blastomeres through an unknown mechanism involving the ascidian-specific protein posterior end mark 1 (PEM1)83. A variation on the transcription repression theme is found in mouse embryos, in which PGCs become specified from proximal post-implantation epiblast cells by expression of BLIMP1. During early PGC specification in mice (E6.25–E8), transcription is not globally repressed. Instead, BLIMP1 specifically represses transcription of genes required for mesodermal development84–86. In blimp1 mutants, a small cluster of aberrant PGCs continue to express somatic markers and are eventually lost55,87. However, after PGC specification (E8–E9), the Pol II CTD becomes dephosphorylated, suggesting that transcription is globally repressed as PGCs migrate to the somatic gonad86. Thus, blocking transcription in PGCs, primarily by targeting the CTD of Pol II, is a conserved mechanism to maintain germ cell fate in early development.
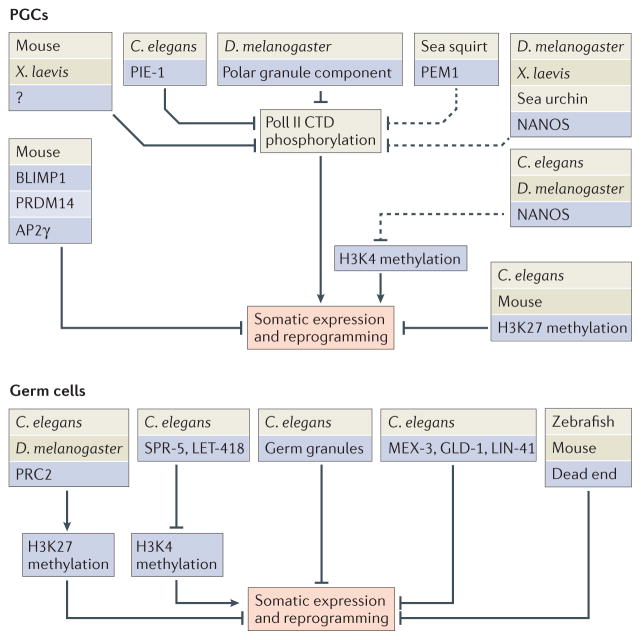
Figure 3. Diverse factors and mechanisms that repress expression of somatic genes and protect germ cells from reprogramming towards somatic cells.
In embryos, a common target of regulation in the primordial germ cells (PGCs) is the carboxy-terminal domain (CTD) of RNA Polymerase II (Pol II). Phosphorylation of the Pol II CTD is required for transcription elongation, and organisms use diverse methods to prevent CTD phosphorylation in PGCs. Inhibition of Pol II keeps early PGCs transcriptionally silent. At later stages, repression is more selective, to keep somatic genes switched off while germline genes are being expressed. In adults, this selective repression is achieved at the level of chromatin regulation by histone modifiers and remodellers (for example, polycomb repressive complex 2 (PRC2) promotes methylation of Lys27 on histone H3 (H3K27) and gene repression, and suppressor of presenilin defect 5 (SPR-5) and lethal 418 (LET-418) remove H3K4 methylation, which is associated with gene activation), as well as at the level of translational regulation by germ granules, translational regulators (for example, muscle excess 3 (MEX-3), defective in germline development 1 (GLD-1) and abnormal cell lineage 41 (LIN-41)) and proteins that stabilize germline-specific RNA (such as dead end). C. elegans, Caenorhabditis elegans; D. melanogaster, Drosophila melanogaster; PEM1, posterior end mark 1; PIE-1, pharynx intestine in excess 1; PRDM14, PR domain zinc-finger protein 14; X. laevis, Xenopus laevis.
Later in development, PGCs switch from Pol II CTD-based transcriptional repression to chromatin-based repression. In C. elegans embryos, the disappearance of PIE-1 from PGCs at the ~100-cell stage is coincident with loss of H3K4 methylation and H4K8 acetylation88,89, marks of transcriptionally competent or active chromatin. This suggests that, when PIE-1 repression of Pol II is lifted, chromatin regulation maintains a degree of transcriptional repression in PGCs. In D. melanogaster embryos, transcription and H3K4 methylation are upregulated in somatic nuclei at the syncytial blastoderm stage but are not upregulated in the PGCs until after their migration through the midgut during gastrulation75. In both D. melanogaster and C. elegans, NANOS participates, probably indirectly, in clearing or repressing H3K4 methylation, and nanos mutations result in loss of PGCs. The loss of H3K4 methylation is thought to be due to histone replacement instead of demethylase activity90. D. melanogaster PGCs increase transcription as they migrate to the gonad. C. elegans PGCs transition from limited transcription during late embryogenesis to robust transcription after embryos hatch and begin to feed88,91. In mice, repressive H3K27 methylation is acquired after PGC specification, but as PGCs enter the genital ridge, this repressive histone mark is lost and H3K4 methylation is acquired92,93. These observations suggest that, in addition to the conservation of Pol II CTD-based transcriptional repression, a transition to chromatin-based repression is conserved across species.
Protection by the chromatin state of germ cells
Even after germ cells initiate transcription, their chromatin state continues to have a protective role: it inhibits their reprogramming towards somatic fates. In the soma, master regulatory proteins promote differentiation into specific cell types. For example, in C. elegans, abnormal chemotaxis 1 (CHE-1) specifies neural fate and helix–loop–helix 1 (HLH-1), the worm homologue of myoblast determination 1 (MYOD), specifies muscle fate. Forced expression of those somatic master regulators in the germ cells of C. elegans larvae and adults does not drive germ cells towards somatic fates, revealing that germ cells are protected against reprogramming94. This protection is conferred by a repressed chromatin state mediated by MES-2–MES-3–MES-6 (worm PRC2), which generates repressive H3K27me3, as described above. A histone chaperone called abnormal cell lineage 53 (LIN-53) probably functions with MES-2–MES-3–MES-6, and the distribution of MES-2–MES-3–MES-6 activity is influenced by the H3K36 methyltransferase MES-4. Loss of any of these components renders germ cells reprogrammable by experimentally driven expression of somatic master regulatory proteins94,95. For example, in germ cells lacking MES-2–MES-3–MES-6, expression of CHE-1 causes germ cells to express neuronal markers and to extend neurite-like projections, and expression of HLH-1 causes germ cells to express muscle markers (FIG. 4). Therefore, the MES-2–MES-3–MES-6 complex creates a chromatin state that can buffer germ cells against the effects of forced expression or stochastic mis-expression of somatic cell fate factors. A recent study in D. melanogaster suggests that, in flies, germ cell pluripotency is also maintained by PRC2 but in a non-cell autonomous and probably indirect manner: compromising PRC2 in the somatic gonad is sufficient to induce the expression of somatic cell markers in germ cells96. Whether MES-2–MES-3–MES-6-mediated repression of somatic fates in the C. elegans germline is cell-autonomous (that is, in germ cells) or non-cell-autonomous (that is, in the surrounding somatic gonad) remains to be tested.
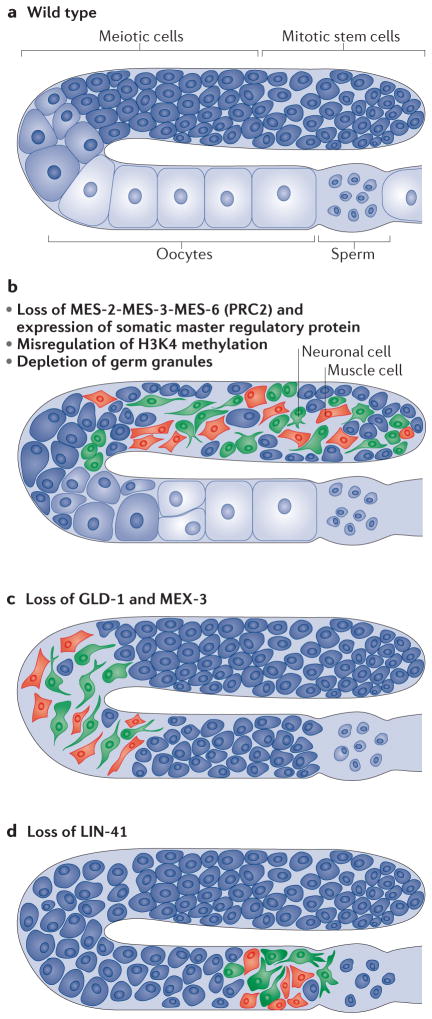
Figure 4. Conditions that cause adult germ cells to reprogramme towards somatic cells in C. elegans.
a | Wild-type Caenorhabditis elegans germlines contain a progression of mitotic stem cells, meiotic cells, oocytes and sperm. b | Somatic cell types most commonly observed after germ soma reprogramming are neuronal (shown in green) and muscle (shown in red). Loss of the complex of maternal effect sterile 2 (MES-2), MES-3 and MES-6 (the worm version of polycomb repressive complex 2 (PRC2)), mis-regulation of methylation of Lys4 on histone H3 (H3K4), and depletion of germ granules cause mitotic stem cells to reprogramme towards somatic lineages. c | Loss of defective in germline development 1 (GLD-1) and muscle excess 3 (MEX-3) causes meiotic cells to reprogramme towards soma. d | Loss of abnormal cell lineage 41 (LIN-41) causes oocytes to reprogramme towards somatic cells, owing to the embryonic programme being turned on prematurely.
Several important new studies provide further evidence that the chromatin state of germ cells protects their germline fate. Even without driving the expression of somatic master regulators in germ cells, simultaneous loss of two chromatin factors, suppressor of presenilin defect 5 (SPR-5) and lethal 418 (LET-418), the worm homologues of LSD1 (also known as KDM1A) and Mi2, respectively, causes C. elegans germ cells to express neuronal markers and extend neurite-like processes or to express muscle markers97 (FIG. 4). SPR-5 is an H3K4 demethylase that interacts with LET-418 within two complexes, the nucleosome remodelling and deacetylase (NuRD) complex and the MEC complex. Compromising the H3K4 methyltransferase SET domain-containing 2 (SET-2) or its cofactor, WD-repeat 5.1 (WDR-5.1), also leads to expression of somatic markers in the germline and causes soma-like differentiation of germ cells98. Thus, compromising the ability to either methylate or demethylate H3K4 causes germ–soma reprogramming. This suggests that H3K4 methylation levels must be carefully regulated. One crucial question is whether germ cells that express somatic markers are germ cells ‘gone wrong’ (in other words, rerouted to express an inappropriate fate) or instead are germ cells undergoing ‘precocious differentiation’ (in other words, expressing some of the somatic markers they will later express after fertilization and during early embryogenesis). Notably, in mice, PGCs must first dedifferentiate before developing into somatic cell types99,100. Loss of the NuRD complex component methyl-CpG-binding domain 3 (MBD3) promotes dedifferentiation of PGCs into pluripotent embryonic germ cells, which can then respond to somatic differentiation cues101. Perhaps altering the chromatin state in C. elegans germ cells promotes their dedifferentiation, after which they can respond to somatic cues and initiate somatic development.
Protection by cytoplasmic factors in germ cells
Chromatin-level repression is not the only mechanism used by germ cells to prevent somatic reprogramming. Germ cells also rely on translational regulation. For example, two cytoplasmic RNA-binding translational regulators, muscle excess 3 (MEX-3) and defective in germline development 1 (GLD-1), function redundantly in C. elegans to prevent somatic reprogramming and maintain germ cell totipotency. The germlines of worms with mutations in both mex-3 and gld-1 are tumorous and contain differentiated somatic cell types from all three germ layers102 (FIG. 4). Interestingly, somatic reprogramming in mex-3 gld-1 mutants does not occur until after germ cells have entered meiosis, and requires overexpression of the GLD-1 target cyclin E, a factor that promotes mitosis and somatic gene expression in fertilized embryos103. Similarly, precocious activation of cyclin A in D. melanogaster oocytes causes them to re-enter mitosis and produce a tumorous germline phenotype104. The tripartite motif (TRIM)-NHL protein LIN-41 is another cytoplasmic component that is required in C. elegans oocytes to prevent them from re-entering mitosis and expressing somatic genes105 (FIG. 4). In D. melanogaster, TRIM-NHL functions similarly to prevent mitosis in neuroblasts and ovarian stem cells106–108. In humans, LIN-41 inhibits the translation of prodifferentiation genes, and elevated LIN-41 can promote the reprogramming of differentiated cells into induced pluripotent stem cells109. The appearance of differentiated somatic cell types from all three germ layers in lin-41 and mex-3 gld-1 C. elegans mutants is reminiscent of human germ cell teratomas (GCTs). As the name suggests, GCTs are derived from germ cells, and they are the most common cancer in men between the ages of 20 and 39 (REF. 110). The high rate of occurrence of GCTs suggests that the phenotype can be caused by the loss of any of several genes111. Indeed, studies in mice have identified numerous factors whose loss increases the incidence of GCTs, the most heavily studied being a germ-granule component called dead end, which post-transcriptionally regulates the accumulation and translation of several mRNA targets in the germline112,113. By determining how germ cells maintain pluripotent potential at the level of translation, we will gain a better understanding of what is required to repress the formation of GCTs.
A common theme in the C. elegans germ–soma reprogramming events discussed above is the disappearance of germ granules from reprogrammed germ cells. To examine whether the loss of germ granules is a cause or a consequence of germ cell reprogramming, a recent study tested whether depletion of germ-granule components by RNAi can cause germ cell reprogramming. Germ cells depleted of germ-granule factors were observed to express muscle and pan-neuronal markers and to send out neurite-like projections114 (FIG. 4). Reprogrammed germ cells did not seem to terminally differentiate, but they could be induced to do so by forced expression of a master regulatory protein, such as CHE-1. Taken together, these findings support a model in which germ granules selectively destabilize and/or impair the translation of mRNAs encoding proteins that promote somatic differentiation. Germ granules may buffer germ cells from the effects of stochastic mis-transcription of inappropriate mRNAs, thereby maintaining pluripotent potential by preventing the reprogramming of germ cells towards somatic cell types. Whether this role of germ granules is conserved in other species has yet to be examined. Interestingly, the converse scenario is frequently observed: germ-granule components expressed in somatic cancers may promote cell proliferation, pluripotency and tumorigenesis115–117.
How might germ granules antagonize somatic fate? In C. elegans, germ granules overlie the cytoplasmic face of nuclear pores during most stages of germline development, and several tests suggest that worm germ granules extend the nuclear pore complex environment28,42,118. First, the association of germ granules with the nuclear periphery requires intact nuclear pores. Second, germ granules exhibit size-exclusion properties similar to those of nuclear pores. Third, chemicals that disrupt the integrity of nuclear pores also disrupt the integrity of germ granules. In adult germ cells, it is estimated that 75% of nuclear pores are covered by germ granules119. Perinuclear granules may serve as a germline-specific compartment through which proteins and RNAs pass on their way into or out of germ nuclei. In that location, germ granules could survey and silence transcripts not appropriate for germline development. This possibility raises the question of how germ granules could selectively silence the translation of somatic transcripts while allowing the translation of germline-appropriate transcripts. Several recent studies suggest a possible mechanism involving the Argonaute proteins chromosome-segregation and RNAi-deficient 1 (CSR-1) and PIWI-related gene 1 (PRG-1) functioning together in C. elegans germ granules to distinguish between self (that is, germline) transcripts and non-self (that is, transposon, foreign transgene and perhaps somatic) transcripts120,121. PRG-1 is the C. elegans PIWI orthologue, and it binds over 30,000 different piRNAs produced from the genome122,123. If mismatches are allowed, piRNAs can target almost all mRNA substrates to inhibit their translation124. CSR-1 binds to over 4,000 specific small RNAs, which collectively are complementary to nearly all germline transcripts. The current model is that these small RNAs guide CSR-1 to its mRNA targets, protecting them from PRG-1 silencing and effectively licensing their germline expression125. It remains to be determined whether piRNA-mediated repression of non-self targets extends to somatic transcripts produced in the germline, whether loss or impairment of CSR-1 and PRG-1 function underlies the somatic reprogramming of germ cells when C. elegans germ granules are depleted, and whether other organisms use germ-granule-associated Argonaute proteins to prevent germ–soma reprogramming.
Concluding remarks
Early in development, multicellular animals must specify which of their cells will serve as the seeds, or ‘germs’ to produce gametes and subsequent generations, and throughout development the immortal and totipotent potential of those germ cells must be protected by repressing somatic differentiation. This Review focuses on the diverse mechanisms used to specify and protect germline fate.
Both specification and protection are critical to ensure reproductive capacity, and often the two processes are interwoven. For example, C. elegans and D. melanogaster embryos exemplify physical separation of PGCs from developing somatic cells early in development, with segregation of germ plasm containing germline determinants to the PGCs, and with maintenance in PGCs of transcriptional repression to prevent expression of somatic genes. Later in development, chromatin barriers and germ granules prevent the expression of somatic genes in germ cells. Mouse embryos do not set germ cells apart early on. Instead, they exemplify a different and probably ancestral mode of germ cell specification, signalling a small set of WNT-primed post-implantation epiblast cells to express a trio of transcription factors that specify PGCs. As in worms and flies, protection of germline fate is likely to involve chromatin regulation and RNA granules.
Transmission of an epigenetic memory of the germline programme from parent to progeny germ cells is a newly recognized contributor in worms and a new frontier for exploration in other systems. Although the broad themes and some of the important players are known in a few organisms, much remains to be elucidated. Recent advances in technology, such as high-throughput genome sequencing and protein analysis and CRISPR-based genome editing, are expanding our understanding of germ cell specification and protection. Soon, this understanding will extend to more than just a handful of well-defined model organisms, providing a broader picture of how this process has evolved and diversified life.
Acknowledgments
The authors thank Amander Clark and anonymous reviewers for helpful discussion and comments on this review.
Glossary
- Primordial germ cells
(PGCs) The nascent germ cells formed during embryogenesis, which ultimately generate oocytes and/or sperm in adults
- Blastomere
A type of cell that is produced by cleavage divisions during early embryogenesis
- Genital ridge
A ridge of embryonic mesoblast cells that envelopes primordial germ cells in vertebrates and develops into the sex organs
- Tudor-domain proteins
Proteins containing a conserved structural motif that binds symmetrically dimethylated Arg. This domain was originally characterized in the Drosophila melanogaster protein Tudor
- Argonaute proteins
Proteins that contain a PAZ and a PIWI domain and that bind different classes of small RNAs (for example, small interfering RNAs, micro RNAs and PIWI-interacting RNAs), which guide the Argonaute proteins to their specific target mRNAs
- Ground state
A cellular condition that is liberated from epigenetic and developmental constraints, like that found in pluripotent epiblasts
- Open chromatin
Non-compacted euchromatin that is accessible to transcriptional machinery and for gene expression
- CRISPR-based genome editing
A gene-editing technique that is derived from the bacterial immune system and is widely used to create deletions, insertions and modifications of targeted genome sequences. Since 2013, this technique has been used for genome editing in a wide range of different organisms
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Hegner RW. Effects of removing the germ-cell determinants from the eggs of some chrysomelid beetles. Biol Bull. 1908;16:19–26. [Google Scholar]
- 3.Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 4.Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 5.Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- 6.Ephrussi A, Dickinson LK, Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- 8.Evans T, Wade CM, Chapman FA, Johnson AD, Loose M. Acquisition of germ plasm accelerates vertebrate evolution. Science. 2014;344:200–203. doi: 10.1126/science.1249325. [DOI] [PubMed] [Google Scholar]
- 9.Spiegelman M, Bennett D. A light- and electron-microscopic study of primordial germ cells in the early mouse embryo. J Embryol Exp Morphol. 1973;30:97–118. [PubMed] [Google Scholar]
- 10.Toyooka Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 11.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith LD. The role of a ‘germinal plasm’ in the formation of primordial germ cells in Rana pipiens. Dev Biol. 1966;14:330–347. doi: 10.1016/0012-1606(66)90019-4. [DOI] [PubMed] [Google Scholar]
- 13.Okada M, Kleinman IA, Schneiderman HA. Restoration of fertility in sterilized Drosophila eggs by transplantation of polar cytoplasm. Dev Biol. 1974;37:43–54. doi: 10.1016/0012-1606(74)90168-7. [DOI] [PubMed] [Google Scholar]
- 14.Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tada H, Mochii M, Orii H, Watanabe K. Ectopic formation of primordial germ cells by transplantation of the germ plasm: direct evidence for germ cell determinant in Xenopus. Dev Biol. 2012;371:86–93. doi: 10.1016/j.ydbio.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Nüsslein-Volhard C, Frohnhöfer HG, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 17.St Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 18.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. This paper demonstrates that mislocalization of germ-plasm components to an ectopic site, the anterior pole of the D. melanogaster embryo, is sufficient to induce formation of functional PGCs at that site. [DOI] [PubMed] [Google Scholar]
- 19.Smith JL, Wilson JE, Macdonald PM. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell. 1992;70:849–859. doi: 10.1016/0092-8674(92)90318-7. [DOI] [PubMed] [Google Scholar]
- 20.Wylie CC, Holwill S, O’Driscoll M, Snape A, Heasman J. Germ plasm and germ cell determination in Xenopus laevis as studied by cell transplantation analysis. Cold Spring Harb Symp Quant Biol. 1985;50:37–43. doi: 10.1101/sqb.1985.050.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Strome S, Martin P, Schierenberg E, Paulsen J. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121:2961–2972. doi: 10.1242/dev.121.9.2961. [DOI] [PubMed] [Google Scholar]
- 22.Lerit DA, Gavis ER. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol. 2011;21:439–448. doi: 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. This paper describes a C. elegans mutant that fails to partition maternally provided germ granules to the germline blastomeres but nevertheless forms and correctly specifies PGCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JT, et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep. 2013;5:1169–1177. doi: 10.1016/j.celrep.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy EM, Ito S. Fine structural and radioautographic observations on dense perinuclear cytoplasmic material in tadpole oocytes. J Cell Biol. 1971;49:90–108. doi: 10.1083/jcb.49.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawasaki I, et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- 28.Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 30.Gao M, Arkov AL. Next generation organelles: structure and role of germ granules in the germline. Mol Reprod Dev. 2012;80:610–623. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seervai RNH, Wessel GM. Lessons for inductive germline determination. Mol Reprod Dev. 2013;80:590–609. doi: 10.1002/mrd.22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J Androl. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arkov AL, Wang JYS, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki I, et al. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167:645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bontems F, et al. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 37.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han TW, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt HB, Görlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife. 2015;4:1–30. doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589:15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Updike DL, Hachey SJ, Kreher J, Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanazawa M, Yonetani M, Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol. 2011;192:929–937. doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell. 2013;27:161–173. doi: 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnúsdóttir E, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aramaki S, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. This paper documents how mouse PGCs are induced by WNT3 and BMP4 signalling: WNT3 promotes expression of the mesodermal transcription factor T, and BMP4 permits T to directly activate two key transcription factors (BLIMP1 and PRDM14) for PGC fate. [DOI] [PubMed] [Google Scholar]
- 47.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4:a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development. 2014;141:245–252. doi: 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- 49.Cantú AV, Laird DJ. Wnt and Bmp fit germ cells to a T. Dev Cell. 2013;27:485–487. doi: 10.1016/j.devcel.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leitch HG, Tang WWC, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol. 2013;104:149–187. doi: 10.1016/B978-0-12-416027-9.00005-X. [DOI] [PubMed] [Google Scholar]
- 51.Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 53.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 54.Kurimoto K, et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell. 2015;16:1–16. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 56.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 57.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 58.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. This paper investigate the specification of human PGCs and show that SOX17, an endoderm-required transcription factor, is the key regulator and that the role of BLIMP1 is to repress somatic fates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ewen-Campen B, Srouji JR, Schwager EE, Extavour CG. oskar predates the evolution of germ plasm in insects. Curr Biol. 2012;22:2278–2283. doi: 10.1016/j.cub.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 60.Ewen-Campen B, Donoughe S, Clarke DN, Extavour CG. Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Curr Biol. 2013;23:835–842. doi: 10.1016/j.cub.2013.03.063. This paper shows that, in G. bimaculatus (cricket) embryos, PGCs arise from abdominal mesoderm and are not specified by cytoplasmic inheritance of maternal determinants. [DOI] [PubMed] [Google Scholar]
- 61.Strome S, Kelly WG, Ercan S, Lieb JD. Regulation of the X chromosomes in Caenorhabditis elegans. Cold Spring Harb Perspect Biol. 2014;6:a018366. doi: 10.1101/cshperspect.a018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bender LB, et al. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 67.Gaydos LJ, Wang W, Strome S. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345:1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–3827. doi: 10.1242/dev.022434. This paper provides an excellent overview of transcriptional-silencing mechanisms during PGC specification in C. elegans, D. melanogaster and mice. [DOI] [PubMed] [Google Scholar]
- 69.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seydoux G, et al. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh D, Seydoux G. Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics. 2008;178:235–243. doi: 10.1534/genetics.107.083212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- 73.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 75.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 76.Deshpande G, Calhoun G, Jinks TM, Polydorides AD, Schedl P. Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mech Dev. 2005;122:645–657. doi: 10.1016/j.mod.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 77.Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 1999;99:271–281. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- 78.Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 79.Palancade B, Bellier S, Almouzni G, Bensaude O. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J Cell Sci. 2001;114:2483–2489. doi: 10.1242/jcs.114.13.2483. [DOI] [PubMed] [Google Scholar]
- 80.Swartz SZ, et al. Deadenylase depletion protects inherited mRNAs in primordial germ cells. Development. 2014;141:3134–3142. doi: 10.1242/dev.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;1486:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ewen-Campen B, Schwager EE, Extavour CGM. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- 83.Shirae-Kurabayashi M, Matsuda K, Nakamura A. Ci-Pem-1 localizes to the nucleus and represses somatic gene transcription in the germline of Ciona intestinalis embryos. Development. 2011;138:2871–2881. doi: 10.1242/dev.058131. [DOI] [PubMed] [Google Scholar]
- 84.Kurimoto K, et al. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 86.Seki Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 87.Vincent SD, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 88.Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Checchi PM, Kelly WG. emb-4 is a conserved gene required for efficient germline-specific chromatin remodeling during Caenorhabditis elegans embryogenesis. Genetics. 2006;174:1895–1906. doi: 10.1534/genetics.106.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katz DJ, Edwards TM, Reinke V, Kelly WGA. C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 93.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Patel T, Tursun B, Rahe DP, Hobert O. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2012;2:1178–1186. doi: 10.1016/j.celrep.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eun SH, Shi Z, Cui K, Zhao K, Chen X. A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker. Science. 2014;343:1513–1516. doi: 10.1126/science.1246514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Käser-Pébernard S, Müller F, Wicky C. LET-418/Mi2 and SPR-5/LSD1 cooperatively prevent somatic reprogramming of C. elegans germline stem cells. Stem Cell Rep. 2014;2:547–559. doi: 10.1016/j.stemcr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robert VJ, et al. The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Rep. 2014;9:443–450. doi: 10.1016/j.celrep.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 99.Leitch HG, et al. On the fate of primordial germ cells injected into early mouse embryos. Dev Biol. 2014;385:155–159. doi: 10.1016/j.ydbio.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leitch HG, et al. Rebuilding pluripotency from primordial germ cells. Stem Cell Rep. 2013;1:66–78. doi: 10.1016/j.stemcr.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rais Y, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. This paper describes how depletion of MBD3, a component of the NuRD chromatin-repressor complex, allows nearly complete reprogramming of mouse and human somatic cells to induced pluripotent stem cells. [DOI] [PubMed] [Google Scholar]
- 102.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 103.Biedermann B, et al. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell. 2009;17:355–364. doi: 10.1016/j.devcel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 104.Sugimura I, Lilly MA. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev Cell. 2006;10:127–135. doi: 10.1016/j.devcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 105.Tocchini C, et al. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 2014;10:e1004533. doi: 10.1371/journal.pgen.1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor Brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 107.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 108.Neumüller RA, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Worringer KA, et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Richardson LC, Wingo PA, Zack MM, Zahran HS, King JB. Health-related quality of life in cancer survivors between ages 20 and 64 years: population-based estimates from the Behavioral Risk Factor Surveillance System. Cancer. 2008;112:1380–1389. doi: 10.1002/cncr.23291. [DOI] [PubMed] [Google Scholar]
- 111.Kanetsky PA, et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet. 2011;20:3109–3117. doi: 10.1093/hmg/ddr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Youngren KK, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bustamante-Marín X, Garness JA, Capel B. Testicular teratomas: an intersection of pluripotency, differentiation and cancer biology. Int J Dev Biol. 2013;57:201–210. doi: 10.1387/ijdb.130136bc. [DOI] [PubMed] [Google Scholar]
- 114.Updike DL, Knutson AK, Egelhofer TA, Campbell AC, Strome S. Germ-granule components prevent somatic development in the C. elegans germline. Curr Biol. 2014;24:970–975. doi: 10.1016/j.cub.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- 116.Yuan L, et al. Proteomic analysis reveals that MAEL, a component of nuage, interacts with stress granule proteins in cancer cells. Oncol Rep. 2014;31:342–350. doi: 10.3892/or.2013.2836. [DOI] [PubMed] [Google Scholar]
- 117.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. This paper shows that brain tumours in lethal (3) malignant brain tumour-mutant D. melanogaster display expression of key genes for germline development and that knockdown of those genes is sufficient to suppress tumour growth. [DOI] [PubMed] [Google Scholar]
- 118.Voronina E, Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 2010;137:1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- 120.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gu W, et al. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang G, Reinke VA. C elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bagijn MP, et al. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wedeles CJ, Wu MZ, Claycomb JM. Silent no more: endogenous small RNA pathways promote gene expression. Worm. 2014;3:e28641. doi: 10.4161/worm.28641. [DOI] [PMC free article] [PubMed] [Google Scholar]