Abstract
Alteration in the timing of particular developmental events can lead to major morphological changes that have profound effects on the life history of an organism. Insights into developmental timing mechanisms have been revealed in the model organism C. elegans, in which a regulatory network of heterochronic genes times events during larval development, ensuring that stage-specific programs occur in the appropriate sequence and on schedule. Developmental timing studies in C. elegans led to the landmark discovery of miRNAs and continue to enhance our understanding of the regulation and activity of these small regulatory molecules. Current views of the heterochronic gene pathway are summarized here, with a focus on the ways in which miRNAs contribute to temporal control and how miRNAs themselves are regulated. Finally, the conservation of heterochronic genes and their functions in timing, as well as their related roles in stem cells and cancer are highlighted.
Introduction
In the development of an organism from a single cell, an intricate sequence of events is regulated over developmental time to ensure proper formation of the adult animal. Temporal cues act together with spatial and sexual signals to coordinate development throughout the body plan. Variations in relative timing of developmental events are thought to contribute to morphological diversification and speciation events (Gould, 1992). In addition, disruption of temporal programs can cause organism-wide changes in development that can result in catastrophic birth defects (Wilson, 1988). One example of a developmentally timed event in humans is the onset of puberty, the time at which an individual undergoes a semi-synchronous set of physiological changes to become a reproductive adult. Several new studies identify genetic variations that affect the age of puberty onset (He et al., 2009; Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009). One of the genes characterized in these reports is Lin28B, the homolog of a known regulator of timing in the nematode C. elegans, suggesting that regulation of developmental time may be highly conserved evolutionarily.
C. elegans provides a powerful system for dissecting developmental timing mechanisms because of the robust genetic tools available, the ability to visualize tissues in the context of a living animal, and its essentially invariant cell lineage, which allows analysis of development at the level of individual cellular events (Sulston and Horvitz, 1977). Upon completion of embryogenesis, worms hatch and develop through four larval stages (L1-L4), each ending with a molt, before becoming a fully-differentiated and reproductively-competent adult. A number of stage-specific developmental events in the worm facilitate isolation of timing mutants, in which these events occur in the right place (i.e., the correct cell lineage), but at the wrong developmental time. For example, the hypodermal seam cells, which run laterally along the left and right sides of the worm and contribute to formation of the cuticle, execute two hallmark stage-specific events. In the early L2 stage a subset of seam cells undergo a proliferative division that increases their number (Fig. 1). Later, at the L4-to-adult molt, as seam cells exit the cell cycle and differentiate into their adult form, adjacent seam cells fuse and they generate a specialized cuticle with lateral ridges (alae), providing a convenient morphological marker of the adult state (Fig. 1). Two types of heterochronic mutants have been described: precocious mutants, in which the developmental events of a particular larval stage are skipped causing subsequent events to occur too early; and retarded mutants, in which the program from a particular larval stage is reiterated causing subsequent patterns to be delayed (Ambros and Horvitz, 1984). Heterochronic mutations often cause timing defects in other tissues in addition to the seam, including the vulva, the nervous system, and the gut.
Figure 1. Heterochronic mutations alter the timing of events in the seam cell lineage.
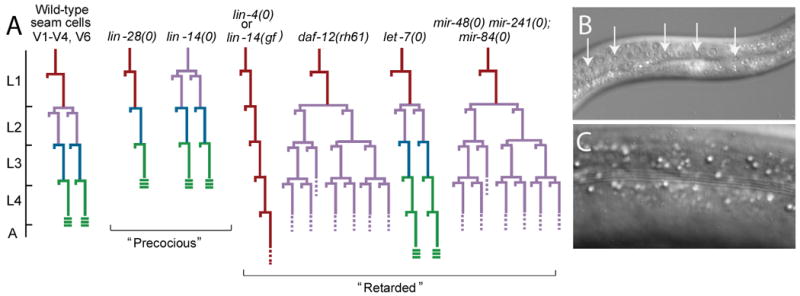
A. Seam cell lineages of some heterochronic mutants. Wild type is shown on the left, followed by the lineages of selected precocious and retarded mutants. The developmental time-scale is indicated at the left with hatch marks representing the molts. In the cell lineage diagrams, each cell division is denoted by a horizontal line, with vertical lines indicating cells. The larval stages are color coded. The L2 stage is characterized by a proliferative seam cell division and the L4-to-adult transition is marked by the formation of alae. L3 and L4 stages lack hallmark patterns and are harder to resolve, therefore color designations of these stages represent the most probable temporal identity.
B. Nomarski DIC image of L3 larval seam cells. Arrows indicate seam cell nuclei.
C. Nomarski DIC image of an adult cuticle, exhibiting characteristic ridges (alae)
Study of timing in the nematode has had a major impact on understanding of gene regulation and developmental biology through the discovery of microRNAs (miRNAs), a class of non-coding regulatory RNAs of approximately 22 nucleotides (Lee et al., 1993; Reinhart et al., 2000). These first-identified C. elegans miRNAs paved the way for discovery of hundreds more miRNAs involved in diverse biological processes. Several miRNAs and miRNA-regulatory genes were first characterized in C. elegans timing research and subsequently found to be conserved in mammalian systems where they are important in human development and disease. Prominent among these, the let-7 miRNA plays key roles in tumor progression and lin-28 regulates stem cell identity through its role in miRNA processing (Bussing et al., 2008; Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). Ongoing research into the roles of miRNAs in the nematode heterochronic gene pathway continues to shed light on fundamental biological questions of developmental timing and also provides new insights into complex mechanisms of miRNA-based gene regulatory networks. This review examines our understanding of developmental timing in C. elegans, with an emphasis on the roles of miRNAs.
Discovery of miRNAs, in timing and beyond
Two C. elegans heterochronic genes, lin-4 and let-7, provided separate but similar puzzles at the time of their characterization (Lee et al., 1993; Reinhart et al., 2000). In many ways they were stereotypical regulators of developmental timing; loss-of-function mutations in each resulted in retarded phenotypes (Chalfie et al., 1981; Ambros and Horvitz, 1984). The mystery arose when these genes were cloned and mutant alleles did not alter protein-coding genes. Subsequent analyses revealed that these genes were unlike any that had been previously identified: lin-4 and let-7 express short non-coding RNAs, the founding members of the miRNA family (Lee et al., 1993; Reinhart et al., 2000). That the first two miRNAs were discovered not only in the same organism, but also in the same genetic pathway was striking and raised the possibility that tiny RNAs were a phenomenon specific to C. elegans and perhaps to temporal control mechanisms. Far from being limited to nematodes, miRNAs are now understood to be nearly ubiquitous (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001), found in diverse plant and animal species, and to play an integral role in repression of mRNA targets (Filipowicz et al., 2008; Ghildiyal and Zamore, 2009). miRNAs are key regulators in myriad biological programs including neuronal development, stem cell regeneration, apoptosis, and others; an understanding of the roles of miRNAs is expanding rapidly.
A brief summary of the miRNA biogenesis pathway is provided here to establish context for understanding miRNA regulation and function with respect to control of developmental time. For recent reviews of miRNA biogenesis, see Filipowicz et al. (2008) and Ding et al. (2009). miRNAs are transcribed as long primary transcripts (pri-miRNA) that are cleaved to generate a precursor form (pre-miRNA) of approximately 70 nucleotides that folds into a hairpin structure (Fig. 2). The pre-miRNA then undergoes a second round of processing to release a ∼22 nucleotide duplex, which is loaded into the pre-miRISC (miRNA-induced silencing complex). The duplex is passively unwound, and the miRNA strand is retained, while its complement is released and degraded (Kawamata et al., 2009). The mature miRNA then directs the miRISC to partially complementary sites in the 3′ UTR of target transcripts, which are negatively regulated through translational repression or mRNA destabilization. Many steps in miRNA biogenesis are differentially regulated to allow modulation of miRNA levels, including transcription, processing, and degradation of the miRNA itself, particularly when it is present in excess of its target mRNAs (Chatterjee and Grosshans, 2009; Ding et al., 2009).
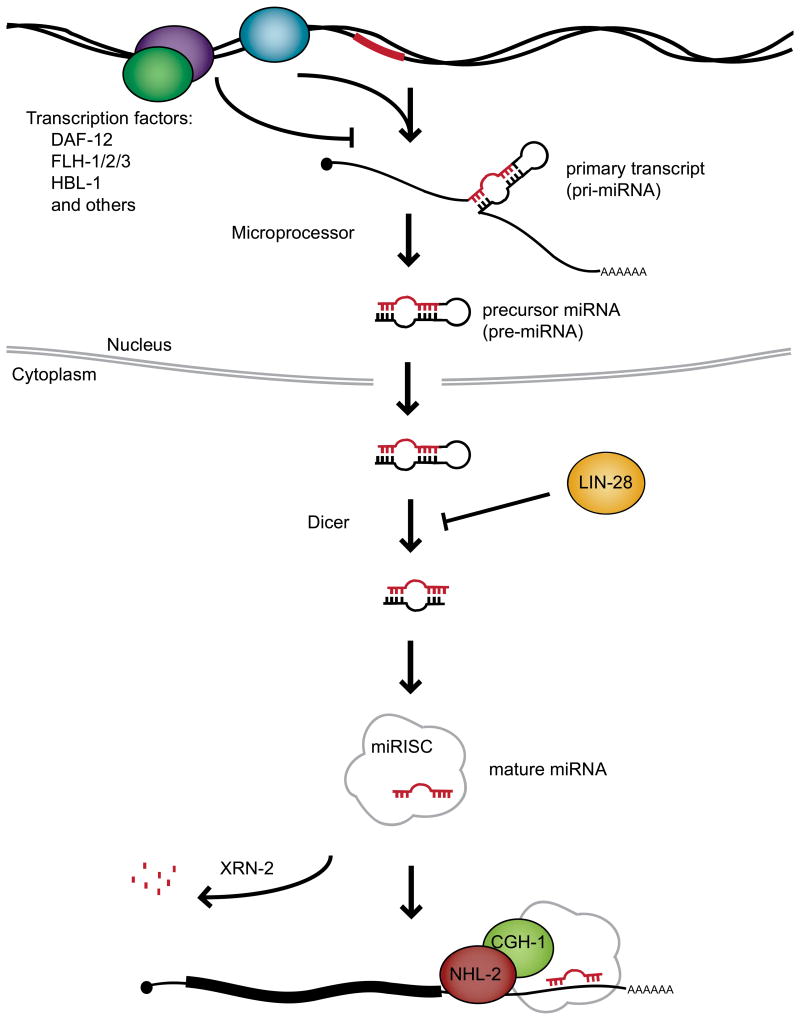
Figure 2. Overview of miRNA biogenesis.
miRNAs are transcribed most commonly by RNA polymerase II as long primary transcripts that are capped and polyadenylated. The Microprocessor, which includes the endoribonuclease Drosha/DRSH-1 and Pasha/PASH-1/DGCR8, cleaves the pri-miRNA to generate the ∼70 nucleotide pre-miRNA. This intermediate form is exported from the nucleus and cleaved again by the endoribonuclease Dicer to generate a ∼22 nucleotide duplex, which is assembled into the pre-miRISC. The duplex is passively unwound and the miRNA-complementary strand is released. The resulting miRISC, contains the mature miRNA and proteins including ALG-1/2 (Argonaute) and AIN-1/2 (GW182). The miRISC binds mRNAs at sites of imperfect pairing with the miRNA and represses targets through translational inhibition or message destabilization. In some cases the seed region is sufficient for targeting, though complementarity along the rest of the miRNA can also contribute. miRNA biogenesis and activity is regulated at many steps; some important regulators that play a role in developmental timing are illustrated and described in the text. XRN-2 is a 5′-3′ exoribonuclease that degrades mature miRNAs. Target mRNAs stabilize the mature miRNAs, thus XRN-2 has been suggested to regulate the relative levels of miRNAs and their targets.
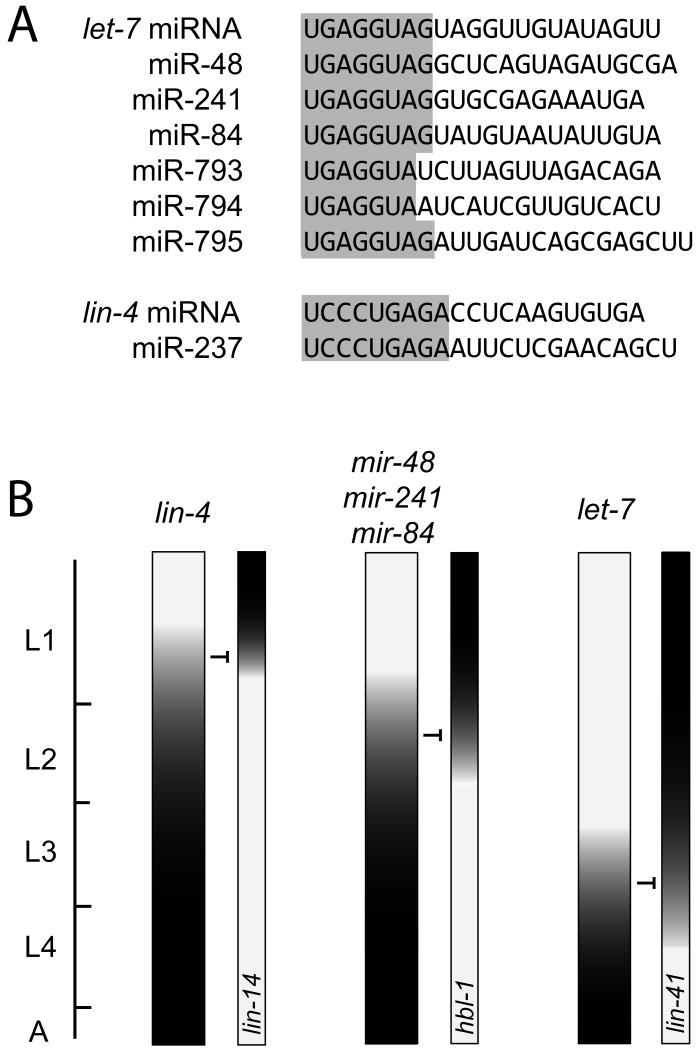
Nucleotides 2-8 at the 5′ end of the miRNA, termed the seed region, are particularly important in recognition of binding sites on target mRNAs. Many miRNAs comprise families in which related miRNAs share identity in the 5′ seed region and may target an overlapping set of mRNAs. For example, there are six miRNAs that share the same 5′ seed sequence with the let-7 miRNA: miR-48, miR-241, and miR-84, whose roles in developmental timing are described below, and the less characterized miR-793, miR-794, and miR-795 (Abbott et al., 2005; Ruby et al., 2006). There is one miRNA, miR-237, that shares a seed sequence with the lin-4 miRNA, but its deletion alone does not cause major morphological defects (Miska et al., 2007) and a heterochronic phenotype has not been reported.
miRNAs as regulators of developmental timing
Control of developmental timing is achieved through a complex regulatory network (Fig. 4). The basic framework of this regulatory system in the seam cell lineage relies on downstream transcription factors that direct stage-specific programs. For example, LIN-14 is required for execution of L1 cell division patterns, HBL-1 for L2 patterns, and LIN-29 for triggering terminal differentiation in the adult. Other heterochronic genes then act to ensure proper temporal control of these transcription factors, and among these, miRNAs play key roles. Strikingly, each of these heterochronic transcription factors is regulated by stage-specifically activated miRNAs (Fig. 3). lin-4 miRNA directly represses lin-14, while miR-48, miR-241, and miR-84 together repress hbl-1 expression (Arasu et al., 1991; Lee et al., 1993; Wightman et al., 1993; Abrahante et al., 2003; Lin et al., 2003; Abbott et al., 2005). Direct miRNA-mediated regulation of lin-29 mRNA is also likely as it physically associates with proteins of the miRISC and reporters fused to the lin-29 3′ UTR show rapid down-regulation after the L4-to-adult molt (Zhang et al., 2007), a pattern consistent with miRNA-mediated repression after specification of adult patterns. These miRNA-transcription factor interactions, together with the additional players in the heterochronic gene network, direct precise and reliable transitions between developmental stages. This temporal regulatory system also provides an excellent model for analysis of miRNA action, the relationships between miRNAs and their targets, as well as the regulation of the miRNAs themselves.
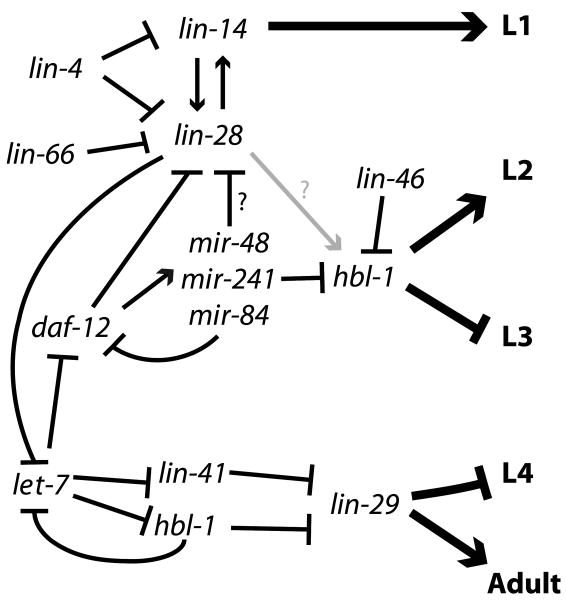
Figure 4. The heterochronic gene pathway of C. elegans.
The heterochronic pathway regulating larval development is shown. The arrows indicate activation; bars indicate repression. Regulatory relationships shown here are supported by genetic data, but not all are direct. lin-42 is not shown, as its complex expression pattern and genetic interactions suggest that it acts at multiple steps in the pathways with different genes, and its precise placement is uncertain.
Figure 3. Alignments and expression patterns of lin-4 and let-7-family miRNAs.
A. Alignments of the lin-4-family and let-7-family miRNAs.
B. Developmental expression patterns of lin-4 and let-7-family miRNA genes, shown in conjunction with expression profiles of representative targets.
lin-4 and initiation of the heterochronic gene pathway
Transcriptional activation of lin-4 in the mid-L1 is a trigger of temporal progression. The lin-4 miRNA accumulates and down-regulates two core developmental timing genes, lin-14 and lin-28, which are expressed highly at hatching and persist through the L1 and L2 stages, respectively (Fig. 4). As mentioned above, lin-14, which encodes a novel nuclear protein, directs L1 patterns and when mutated causes the L2 pattern to occur in the first larval stage and subsequent events each occur one stage too early (Chalfie et al., 1981; Ambros and Horvitz, 1984). lin-28 encodes a predominantly cytoplasmic protein with conserved RNA-binding motifs (Moss et al., 1997), which has been recently shown to play a role in regulation of miRNA biogenesis, as discussed in more detail below (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008; Lehrbach et al., 2009). When lin-28 is disrupted, the L2 proliferative division is skipped, resulting in decreased seam cell number and precocious adult cuticle synthesis. Timely down-regulation of lin-14 and lin-28 requires lin-4, which targets these genes directly through their 3′ UTRs (Lee et al., 1993; Wightman et al., 1993; Moss et al., 1997). If lin-4 activation does not occur in mid-L1 or if their lin-4 binding sites are deleted, lin-14 and lin-28 expression levels remain high. This causes L1 cell division patterns to be reiterated in subsequent larval stages; seam cells do not undergo L2 proliferative divisions and extra larval molts are executed (Chalfie et al., 1981; Ambros and Horvitz, 1984). Additional layers of regulation are required for proper expression of these genes, including a 3′ UTR-mediated positive feedback loop between lin-14 and lin-28, in which each promotes expression of the other in a lin-4-independent manner (Arasu et al., 1991; Moss et al., 1997; Seggerson et al., 2002).
The let-7 family and specification of L2 patterns
One difficulty in the study of miRNA families is the potential for redundancy, such that single loss-of-function mutations do not result in robust phenotypes. Indeed, most C. elegans miRNAs appear individually dispensable (Miska et al., 2007). Available genetic tools facilitate analysis of redundant gene families through construction of multiply mutant loss-of-function strains and also through identification of rare gain-of-function alleles and analysis of over-expression in transgenic animals. Together these studies implicate mir-48, mir-241, and mir-84 as important regulators of the L2-to-L3 transition. When mir-48 or mir-84 is over-expressed from multi-copy arrays precocious phenotypes are observed, including skipping of the L2 proliferative seam cell division and premature adult alae synthesis (Johnson et al., 2005; Li et al., 2005). In contrast, although deletion of mir-48, mir-241, or mir-84 individually does not cause pronounced heterochronic defects, loss of all three genes together results in a strong phenotype opposite to that caused by over-expression, reiteration of L2 patterns (Abbott et al., 2005; Esquela-Kerscher et al., 2005; Hayes et al., 2006). Thus, these miRNAs function redundantly to promote progression to L3 identity. The let-7 homologs may also have some specialized individual roles, as suggested by variations in transcriptional regulation within the family (Abbott et al., 2005; Esquela-Kerscher et al., 2005; Li et al., 2005; Bethke et al., 2009). The extent to which the let-7 family members overlap functionally, the ways in which they may act independently, and how much function they share with let-7 itself are interesting questions for further study.
One important target of the let-7 family of miRNAs is hbl-1, the homolog of the Drosophila hunchback transcription factor (Fay et al., 1999). Depletion of hbl-1 expression by RNAi results in developmental arrest late in embryogenesis (Fay et al., 1999; Abrahante et al., 2003). However, if RNAi is performed post-embryonically, depletion of hbl-1 expression leads to skipping of L2 patterns, resulting in a decrease in seam cell number and precocious alae formation (Abrahante et al., 2003; Lin et al., 2003), indicating that hbl-1 is required to promote L2 developmental programs. Consistent with this role, hbl-1 is highly expressed during L1, and begins to disappear during the L2 stage. By the early L3 stage, hbl-1 reporter expression is absent from the seam and hypodermis, though it remains expressed in neuronal cells. Disruption of the let-7-family miRNAs causes mis-expression of hbl-1 into the L3 stage in the hypodermis, resulting in the L2-reiteration phenotype of these mutants (Abbott et al., 2005). A similar repeat of L2 patterns is observed in lin-46 mutants (Pepper et al., 2004). lin-46 acts upstream of hbl-1, and lin-46 loss-of-function (lf) enhances the retarded phenotypes of let-7-family mutants (Abbott et al., 2005), suggesting that lin-46 acts in parallel with mir-48, mir-241, and mir-84 to regulate L2 programs. LIN-46 is a putative scaffolding protein, suggesting that it may act as part of a complex of heterochronic factors to mediate hbl-1 activity. Also recently placed upstream of hbl-1 in the regulation of L2 patterns is sel-7, a transcription factor previously identified as a positive regulator of Notch signaling (Xia et al., 2009).
In addition to regulating hbl-1, members of the let-7 family may also target lin-28. The lin-28 3′ UTR contains a predicted let-7-family binding site, and LacZ-reporter fusions are temporally mis-regulated if this site is deleted (Morita and Han, 2006). However, the expression of a rescuing lin-28 fluorescent reporter gene was not disrupted in animals mutant for let-7-family miRNAs, mir-48, mir-241, and mir-84 (Abbott et al., 2005), leaving open the question of whether endogenous lin-28 is a target. The lin-28 3′ UTR also appears to be required for regulation by LIN-66, a novel cytoplasmic protein suggested to act in parallel with the let-7 family to inhibit lin-28 post-transcriptionally (Morita and Han, 2006). Indeed, mutation of lin-66 gives rise to retarded phenotypes including reiteration of the L2 proliferative seam cell division, a phenotype that resembles lin-28 gain-of-function (gf) and loss of the let-7-related miRNAs.
Finally, specification of L2 patterns may also involve lin-42, an atypical member of the heterochronic pathway. Unlike most heterochronic genes, which are expressed for contiguous blocks of developmental time (e.g. see Fig. 3), lin-42 is expressed in a cyclical pattern with mRNA and protein present at high levels in the intermolts and disappearing at the molts (Jeon et al., 1999; Tennessen et al., 2006). This pattern suggests that lin-42 may have a reiterative role or multiple functions during larval development. Genetic analyses also imply that lin-42 functions at multiple developmental times. lin-42(lf) mutants execute a wild-type seam cell lineage until the L3 molt when the cells precociously exit the cell cycle and synthesize an adult-type cuticle (Abrahante et al., 1998). However, loss of lin-42 function in combination with a weak lin-14 allele causes skipping of the L2 proliferative division, a phenotype not observed in either single mutant, suggesting an early redundant role for lin-42 in L2 specification (Liu, 1990). Furthermore, lin-42 interacts genetically with both early- and late-acting miRNA genes, lin-4 and let-7 (Abrahante et al., 1998; Reinhart et al., 2000; Tennessen et al., 2006). Although the mechanism of lin-42 action remains unknown, its cyclical expression pattern is particularly striking because lin-42 is the C. elegans homolog of the core Drosophila circadian clock gene period, the expression of which oscillates over a 24-hour cycle (Hardin, 2005). Indeed, lin-42 has recently been implicated in regulating circadian locomotory behavior in C. elegans (Simonetta et al., 2009), although how its expression pattern relates to circadian behavior remains unclear.
In summary, temporally-specified activation of miRNAs, together with inputs from less-understood components, decreases LIN-14 and HBL-1 levels in early development and thereby directs progression through L1- and L2-stage programs, respectively, to the L3 stage. A crucial question then becomes, what genes are regulated by these transcription factors? Recent studies have made strides in this direction with the identification of potential direct targets of the HBL-1 (Niwa et al., 2009; Roush and Slack, 2009), as described below, and of LIN-14 (Hristova et al., 2005). The role for miRNAs in temporal programming continues in late larval development, when activation of let-7 regulates the transition to the adult state.
let-7 and the larval-to-adult transition
let-7 expression is strongly up-regulated in the L3 stage, significantly later in development than are its homologs or lin-4, and it plays important roles in timely differentiation of the adult tissues (Reinhart et al., 2000). let-7 mutants develop normally until the L4 molt, at which time they undergo one extra larval stage before seam cells terminally differentiate. let-7 directly represses lin-41, a particularly intriguing member of the heterochronic pathway (Slack et al., 2000; Vella et al., 2004). lin-41 encodes a member of the TRIM-NHL family of RNA-binding proteins (containing the tripartite motif, which includes RING, B-box, and coiled-coil domains; and the NHL domain, named for its inclusion in NCL-1, HT2A, and LIN-41) and is highly conserved evolutionarily. Moreover, several recent reports show that its let-7-mediated regulation is also maintained in diverse organisms including Drosophila, mice, and humans (Lancman et al., 2005; Schulman et al., 2005; Kanamoto et al., 2006; Lin et al., 2007; Maller Schulman et al., 2008; O'Farrell et al., 2008).
In addition to being a prominent miRNA target, lin-41 is also emerging as a regulator of miRNA function. LIN-41 binds to Dicer, the endonuclease that cleaves pre-miRNAs, in worms (Duchaine et al., 2006), and this interaction is conserved in mice (Rybak et al., 2009). The mouse homolog, mLin41, also binds Argonaute proteins, conserved core members of the miRISC; and it localizes to P-bodies, cytoplasmic locales specializing in mRNA repression and turnover. Many TRIM proteins function as E3 ubiquitin ligases (Meroni and Diez-Roux, 2005) and this activity may also be relevant to the role of LIN-41 in miRNA function. mLin41 displays E3 ubiquitin ligase activity, and specifically ubiquitylates the Argonaute protein Ago2 (Rybak et al., 2009), suggesting a model in which mLin41 inhibits miRNA activity through destabilization of Ago2 and possibly other members of the miRISC.
In worms, lin-41 is required for specification of late larval stages, and lin-41 mutants execute the larval-to-adult transition precociously at the L3 molt (Slack et al., 2000). LIN-41 prevents early accumulation of the zinc finger transcription factor LIN-29, which triggers the cell cycle exit and cell fusion events associated with seam cell terminal differentiation (Rougvie and Ambros, 1995). Insight into whether lin-41-mediated regulation of lin-29 is through modulation of miRNA activity, as might be suggested by the recent mammalian results, is eagerly awaited. Genetic experiments imply that hbl-1 also contributes to late larval pattern specification (Abrahante et al., 2003; Lin et al., 2003) and that it is a let-7 target in parallel to lin-41. Thus, terminal differentiation of adult tissues is directed by lin-29 only after let-7 relieves repression by lin-41 and hbl-1.
Regulators of miRNAs in developmental timing
Proper temporal progression through larval stages relies upon strict control of the biogenesis, function, and stability of heterochronic miRNAs. Identification of additional protein factors required to regulate miRNA expression and activity is an important goal to expand our understanding of developmental timing and of miRNA-mediated gene regulation in general. The well-characterized requirement for miRNAs in the heterochronic pathway makes this system uniquely well-suited for identification and evaluation of such factors. Indeed, disruption of core components of the miRISC, which interact broadly with mature miRNAs to target and regulate mRNAs post-transcriptionally, results in heterochronic phenotypes (Grishok et al., 2001). Depletion of alg-1/2 (C. elegans Argonaute genes) expression leads to reiteration of the L2 proliferative seam cell division and retarded alae synthesis, which can be explained by impaired activity of let-7-family miRNAs. Similarly, other regulators of miRNA function or expression are expected to impact the timing mechanism. Recent findings from the developmental timing field that inform our understanding of the proteins involved in the activity of miRNAs, the transcriptional mechanisms of miRNA expression, and the post-transcriptional regulation of miRNA biogenesis are discussed below.
Co-factors regulating miRNA function
Efficient repression of mRNA targets by miRNAs relies on the function of protein cofactors; the roles of two such proteins, NHL-2 and CGH-1, have recently been described (Hammell et al., 2009b). Genetic analyses demonstrate that nhl-2 and cgh-1 are regulators of developmental timing; mutation of either enhances the weak retarded phenotypes caused by mutation of certain let-7-family members. Depletion of hbl-1, a principal target of these miRNAs, suppresses the timing defects, suggesting that nhl-2 and cgh-1 act together with the let-7 family in its established role in the heterochronic pathway, targeting of hbl-1.
NHL-2 is a member of the conserved TRIM-NHL family of proteins that includes LIN-41. nhl-2 interacts genetically with cgh-1, which encodes a DEAD-box RNA helicase known to modulate miRNA activity in other systems (Chu and Rana, 2006; Eulalio et al., 2007) and to regulate mRNA stability and interact with P-bodies in worms (Navarro et al., 2001; Boag et al., 2008; Rajyaguru and Parker, 2009).
Mutation of nhl-2 or cgh-1 does not alter mature miRNA levels, demonstrating that these genes do not affect miRNA biogenesis and suggesting that they are required for miRNA function in a manner similar to RISC components (Hammell et al., 2009b). Indeed, NHL-2 and CGH-1 physically interact, and they are present in a complex that includes core members of the miRISC, including ALG-1/2, in an RNA-dependent manner. CGH-1 and NHL-2 co-localize to cytoplasmic foci in a pattern that partially overlaps with a P-body marker. Thus genetic, molecular, and cell-biological approaches all support a model in which these two proteins interact with core miRISC components to promote robust miRNA-mediated inhibition of mRNAs.
The precise mechanism by which nhl-2 and cgh-1 are involved in miRNA function is not yet clearly defined. Neither of these factors on its own is essential for miRNA activity; individually, nhl-2 and cgh-1 mutants have only a very mild timing defect, but the double mutant displays a completely penetrant retarded phenotype. The shared domain structure of LIN-41 and NHL-2 raises the possibility that these proteins might functionally overlap. Mutation of nhl-2 does not, however, enhance lin-41 mutant phenotypes; instead it suppresses these phenotypes, suggesting that the two genes function in opposition to each other. Whether NHL-2 functions as a E3 ubiquitin ligase as do mLin-41 and other TRIM proteins (Meroni and Diez-Roux, 2005; Rybak et al., 2009), and if so, whether it targets regulators of miRNA function, are exciting areas for further research.
One important question about new regulators of miRNA activity is whether they are required for all or just a subset of miRNA-mRNA pairs. In the case of NHL-2, its role in miRNA function is neither limited to the heterochronic pathway nor to the let-7 family. Mutation of nhl-2 enhances a hypomorphic allele of lsy-6, a miRNA involved in neuronal fate specification (Johnston and Hobert, 2003). In addition, nhl-2 cgh-1 double mutants display partially penetrant lethality, suggesting an effect on other miRNAs beyond the let-7 family and lsy-6. However, Hammell et al. (2009b) indicate that not all C. elegans miRNA mutants show genetic interactions with nhl-2, suggesting there is some specificity to nhl-2 action. Defining the characteristics that are important for these protein-miRNA interactions will provide new insights into miRNA-mediated gene regulation.
Transcriptional regulation of miRNA expression
Although much is known about miRNA function, only recently have studies begun to focus on their transcriptional regulation (Martinez and Walhout, 2009; Turner and Slack, 2009). This holds true for the heterochronic gene pathway as well, where stage-specific accumulation of miRNAs has long been acknowledged as a key regulatory step that guides temporal progression. Factors that regulate the transcription of heterochronic miRNA genes are just now being identified; some of these transcriptional regulators are previously identified members of the pathway, while others are novel factors.
One known heterochronic gene recently linked to miRNA transcriptional control is the nuclear hormone receptor (NR) DAF-12, which acts as a molecular switch by mediating the choice between reproductive development and the stress response. daf-12 regulates both developmental timing and the decision to enter a stress-resistant alternative L3 stage, the dauer diapause (Antebi et al., 1998; Antebi et al., 2000). In this state, the larva is developmentally arrested, long lived, and optimized for dispersal and survival (Cassada and Russell, 1975). Recent work reveals that DAF-12 can both repress and activate the transcription of let-7 family members, some directly, and that these miRNAs also target daf-12 in a regulatory feedback mechanism (Bethke et al., 2009; Hammell et al., 2009a).
Similar to other NRs, DAF-12 contains an N-terminal DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD), and interactions at the LBD regulate DAF-12 activity (Antebi et al., 2000; Magner and Antebi, 2008). The role of daf-12 in the stress response has been studied extensively (Hu, 2007; Fielenbach and Antebi, 2008). In favorable environments, insulin and TGFβ signals promote the synthesis of steroid hormones that bind to the LBD of DAF-12, and ligand-bound DAF-12 promotes reproductive development (McElwee et al., 2003; Murphy et al., 2003; Liu et al., 2004; Motola et al., 2006). In conditions of environmental stress, such as lack of food, insulin and TGFβ signaling decrease and hormone levels fall. When ligand levels are low, DAF-12 binds its co-repressor, DIN-1, and directs formation of a dauer larva (Ludewig et al., 2004).
The precise role of daf-12 in developmental timing has long provided an enigma for the field: genetic studies revealed that the daf-12(null) heterochronic phenotype is much less severe than the timing defects caused by mutations that affect the LBD alone (Antebi et al., 1998; Antebi et al., 2000). For example, the daf-12(rh61) allele contains a point mutation in the LBD that disrupts ligand binding (Motola et al., 2006), and unlike the null, causes severe retarded heterochronic defects with an L2 reiteration (Antebi et al., 1998; Antebi et al., 2000).
Two recent studies have clarified our understanding of how daf-12 regulates developmental timing and have helped to explain the genetic complexities of the daf-12 locus. In daf-12(null) mutants the levels of several let-7-family miRNAs are somewhat reduced (Bethke et al., 2009; Hammell et al., 2009a). In contrast, these miRNAs are strongly down-regulated in daf-12(rh61) mutants. DAF-12-mediated regulation of at least two family members, mir-84 and mir-241, is direct, through response elements in their promoters (Bethke et al., 2009). These DAF-12-mediated changes in miRNA expression impact the expression of the let-7-family target gene hbl-1; the expression of a hbl-1 reporter persists later in development in daf-12(rh61) mutants compared to wild-type animals, and importantly, this effect requires the hbl-1 3′UTR. Taken together, these experiments demonstrate that ligand-bound DAF-12 contributes to the up-regulation of let-7-family miRNAs, which results in down-regulation of hbl-1 and developmental progression. In the absence of ligand, the expression of these miRNAs is strongly repressed by the DIN-1/DAF-12 complex, leading to developmental arrest. These results help explain why the daf-12(rh61) mutation results in a stronger heterochronic phenotype than does the null allele. The strong repression of the let-7-family genes from a functional DIN-1/DAF-12 complex in daf-12(rh61) mutants results in persistence of hbl-1 expression later in development, whereas a daf-12(null) mutation eliminates both activating and repressing functions, causing a much milder effect.
The regulatory relationship between daf-12 and let-7-family miRNAs is not a simple linear pathway. Not only does daf-12 regulate expression of the let-7 family, these miRNAs also appear to affect daf-12 expression via a regulatory feedback loop. Animals triply mutant for mir-241, mir-48, and mir-84 exhibit enhanced expression of a functional daf-12 fluorescent reporter and increased penetrance of some daf-12-dependent phenotypes, indicating that these miRNAs may modulate daf-12 expression (Hammell et al., 2009a). let-7 itself also targets daf-12 in later larval stages (Grosshans et al., 2005). This cell-autonomous regulatory circuit can also be modulated by environmental signaling. Mildly stressful environmental conditions, which are not dauer inducing, also affect miRNA levels. let-7-family miRNA levels in animals grown in crowded conditions are lower than those in animals from uncrowded environments, showing that external cues regulate these miRNAs (Hammell et al., 2009a).
These data support a model whereby daf-12 and the let-7 family are part of a negative feedback loop in which organism-wide life-history decisions in C. elegans are integrated with cell autonomous regulators of developmental progression (Fig. 5; Hammell et al., 2009a; Bethke et al., 2009). In favorable environments, ligand-bound DAF-12 contributes to the activation of miRNAs that promote L3 patterns by down-regulating hbl-1. As let-7-family miRNAs begin to accumulate, they also contribute to the repression of daf-12, locking in commitment to reproductive development. In dauer-inducing conditions, ligand is limiting, and the DAF-12 repressor complex then down-regulates the let-7-family miRNAs and the animal enters the dauer diapause. The discovery of this regulatory circuit between daf-12 and the let-7-family miRNAs places daf-12 more firmly in the heterochronic pathway and clarifies its role in mediating the choice between continuous reproductive development and dauer formation.
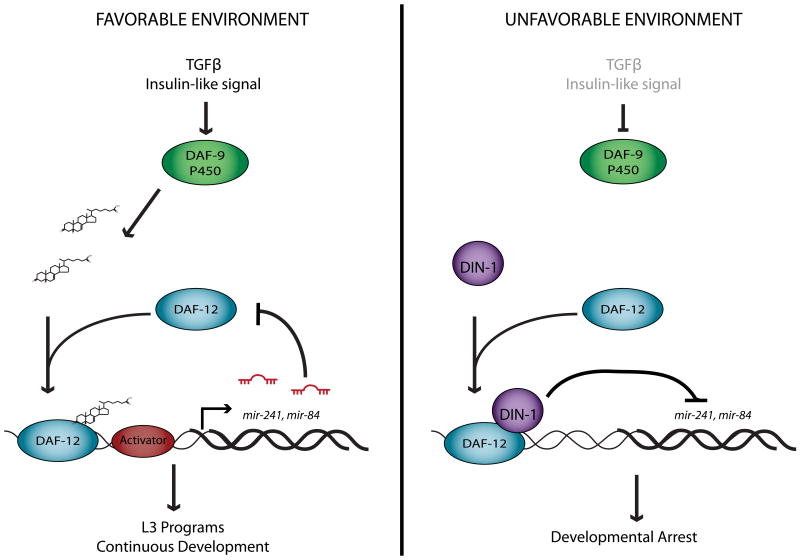
Figure 5. daf-12 and let-7-family miRNAs regulate each other through a negative feedback loop.
A simplified model for daf-12 regulation in the response to environmental cues modified from Hammell et al., (2009a) and Bethke et al., (2009). In favorable environmental conditions, TGFβ and insulin signaling are up-regulated, resulting in the synthesis of dafachronic acids, which bind to DAF-12. Ligand-bound DAF-12 up-regulates the expression of let-7-family miRNAs. An as yet unidentified transcriptional activator is independently required for expression of these miRNAs, because mature let-7-family miRNAs are present in daf-12(null) mutants. As these miRNAs accumulate, they contribute to the repression of daf-12, locking in the decision to progress through development.
In unfavorable environments, enzymes required for dafachronic acid synthesis are down-regulated. In the absence of ligand, DAF-12 binds to DIN-1, a co-repressor, and this repressor complex strongly inhibits the expression of let-7-family miRNAs, leading to developmental arrest.
The HBL-1 transcription factor also appears to regulate expression of a developmental timing miRNA gene, let-7 (Roush and Slack, 2009). Deletion of a predicted HBL-1 binding site from a let-7 reporter fusion results in precocious GFP expression in the seam, and let-7 levels are elevated in hbl-1(lf) animals compared to wild type. However, let-7 is expressed in the seam primarily during later larval stages, times when reporter fusions do not reveal hbl-1 expression, so some details of this regulation remain unsolved (Abrahante et al., 2003; Johnson et al., 2003; Lin et al., 2003).
In addition to previously identified pathway members, other factors have been discovered as transcriptional regulators of the heterochronic miRNA genes. Included among these are the C. elegans homologs of the FLYWCH family of transcription factors (Ow et al., 2008), which contain a C2H2-type zinc finger domain first described in Drosophila, where the family member mod4 was identified through its effect on position-effect variegation (Dorn and Krauss, 2003). Of the four described C. elegans FLYWCH genes flh-1, flh-2, and flh-3 function redundantly to repress embryonic expression of several heterochronic miRNAs including lin-4, mir-48, and mir-241 (Ow et al., 2008). Although absence of FLH proteins results in precocious miRNA expression during embryogenesis and the early L1 stage, this increase surprisingly does not result in heterochronic phenotypes (Ow et al., 2008). This result suggests that the repression of miRNA transcription by the flh genes may not be essential to regulation of the developmental clock.
Stage-specific accumulation of miRNAs is one key feature that guides temporal development, therefore an important goal is to identify activators that act in concert with the repressing activities of the ligand-free DAF-12, HBL-1 and the FLH proteins to instruct these expression profiles. One step toward this goal comes from Martinez et al. (Martinez et al., 2008) who used a yeast one-hybrid assay to identify more than 300 transcription-factor interactions with nematode miRNA promoters, including factors that bind to the promoters of the heterochronic miRNA genes. Furthermore, integration of these data with miRNA target predictions has begun to reveal the breadth of transcription factor-miRNA regulatory networks in C. elegans through prediction of 23 miRNA-transcription factor feedback loops. The combination of high-throughput genome-wide analyses with the classical genetic strengths of C. elegans provides excellent tools with which to validate these predictions and illuminate how miRNA expression is regulated in the heterochronic pathway.
Post-transcriptional regulation of miRNAs
In addition to control of miRNA transcription, recent studies are revealing new layers of post-transcriptional regulation of miRNA biogenesis. The conserved heterochronic gene lin-28 has been linked to regulation of the let-7 family of miRNAs. In mammalian tissue culture systems, Lin28 inhibits processing of the let-7 transcript into the mature miRNA form, by preventing cleavage of pri-let-7 and/or pre-let-7 through direct binding to the let-7 RNA hairpin (Newman et al., 2008; Piskounova et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). Lin28 targets pre-let-7 for terminal uridylation, which prevents cleavage by Dicer and promotes degradation of the intermediate form (Heo et al., 2008). The enzyme responsible for the uridylation is TUT4/Zcchc11, which binds pre-let-7 in a Lin28-dependent manner. These interactions have important implications, as depletion of Lin28 and TUT4 from embryonic stem cells increases the amount of mature let-7 miRNA present and hastens the disappearance of pluripotency markers (Hagan et al., 2009; Heo et al., 2009).
A role for lin-28 in processing of the let-7 miRNA is a conserved function (Lehrbach et al., 2009). C. elegans let-7 is also regulated post-transcriptionally and levels of the mature form are elevated in lin-28 mutants (Fig. 2). However, the loop in the pre-let-7 hairpin is not well conserved between worms and mammals, and the loop sequence appears to be dispensable for regulation of C. elegans let-7 miRNA by LIN-28, in contrast to the mammalian findings (Heo et al., 2008; Newman et al., 2008; Piskounova et al., 2008; Rybak et al., 2008; Lehrbach et al., 2009). Interestingly, the let-7 family members, mir-48, mir-241, and mir-84, appear not to be regulated by LIN-28, despite the fact that they share homology in the seed sequence. If, indeed, neither the seed region nor the loop of the pre-miRNA is the essential element responsible for recognition by LIN-28, then the mechanism by which LIN-28 targets specific miRNAs remains an important topic for further study. The observation that LIN-28 does not appear to broadly regulate all miRNAs also raises the question of what proteins influence processing of other miRNAs and whether they do so by a mechanism similar to that of LIN-28.
C. elegans LIN-28 also likely targets pre-let-7 for uridylation. A poly(U) polymerase, PUP-2, interacts physically with LIN-28 and depletion of pup-2 expression results in accumulation of pre-let-7 miRNA (Lehrbach et al., 2009). Additional layers of regulation must also contribute to proper temporal expression of let-7, as LIN-28 disappears during the L2 stage but mature let-7 miRNA does not accumulate until the L3, and let-7-mediated activation of lin-29 does not occur until the L4 (Reinhart et al., 2000). These recent findings underscore, once again, the strong conservation of these temporal control mechanisms and the importance that findings made in this robust model system can have for some of the most pressing questions in mammalian development.
miRNAs and Timing – Beyond Worms
Once thought to be a peculiarity of C. elegans, miRNAs are now the focus of intense study across the evolutionary spectrum. Conservation of these and other C. elegans heterochronic genes has led to important new insights into the development of diverse species. Some genes have conserved roles in timing mechanisms and others have been adapted to regulate related developmental events. lin-28 and let-7 are both widely conserved evolutionarily and, as described above, recent reports demonstrate that even their regulatory relationship is maintained from worms to mammals (Pasquinelli et al., 2000; Moss and Tang, 2003; Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008; Lehrbach et al., 2009). Expression patterns of both of these genes also show similarities between nematodes and mammalian systems. LIN-28 is highly expressed early and then decreases as development progresses in both worms and mammals, whereas let-7 displays the opposite pattern, with low levels of let-7 early in development and increasing expression as cells differentiate. This reciprocal expression pattern of lin-28 and let-7 is not a coincidence. Mammalian Lin28 is instrumental in maintaining pluripotency of embryonic stem cells and decreasing Lin28 expression appears to be important for relieving the inhibition on let-7 miRNA processing, thereby allowing tissues to differentiate. This role for let-7 in differentiation parallels the function of C. elegans let-7 in directing differentiation of adult tissues, albeit in an earlier developmental window. A paradigm shift in the understanding of stem cells occurred in 2006 when somatic cells were first induced to behave like embryonic stem cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). Lin28 is among the genes that can contribute to restoration of pluripotent stem cell characteristics (Yu et al., 2007). The therapeutic potential of induced pluripotent stem cells is vast and it is the focus of fervent ongoing research.
In addition to its roles in early embryonic differentiation, let-7 mis-expression is an important contributor to tumorigenesis in mammals (Jerome et al., 2007; Bussing et al., 2008). The let-7 miRNA plays a role in regulation of oncogenes, including direct targeting and down-regulation of Ras. This connection was first demonstrated in C. elegans and subsequently shown in mammalian tissue culture cells (Johnson et al., 2005). Microarrays from patient tumors show a correlated decrease in let-7 and increase in Ras expression. In addition, let-7 miRNA levels are especially low in cancer cells that display strong tumor-formation potential and resistance to chemotherapeutic agents. Impressively, the net result of these findings is that let-7, a miRNA identified in the nematode, is now being explored as a diagnostic and therapeutic tool for certain forms of human cancer (Bussing et al., 2008).
The general mechanism of regulating the transition from early to late programs through miRNAs appears to be a common theme in development, and studies in Drosophila have further emphasized that miRNAs are conserved regulators of developmental timing. The Drosophila genome contains many homologs of the miRNAs initially characterized in worms, and several are developmentally expressed (Pasquinelli et al., 2000; Lagos-Quintana et al., 2001; Sempere et al., 2002; Bashirullah et al., 2003; Sempere et al., 2003). Recent work has shown that Drosophila let-7 also guides temporal progression. Cells of the wing imaginal disc fail to terminally differentiate and exit the cell cycle in let-7 mutants, thus retaining juvenile characteristics that result in increased wing cell number and decreased cell size (Caygill and Johnston, 2008). Flies mutant for let-7 also fail to complete juvenile-to-adult remodeling of the neuromusculature, resulting in locomotion defects (Sokol et al., 2008). These were the first studies to report developmental timing functions of miRNAs in a system other than C. elegans.
Finally, regulatory relationships between NRs and microRNAs as exemplified by the recent daf-12-let-7-family feedback loop are also emerging as a common theme in diverse systems (Varghese and Cohen, 2007; McCafferty et al., 2009). For example in flies, the ecdysone receptor and the miRNA miR-14 participate in a feedback loop that ensures proper cell fate transitions during development. With the recent findings in worms, regulatory feedback between NRs and miRNAs may provide a conserved mechanism for coordinating developmental timing networks with environmentally responsive endocrine signals.
Perspectives
The study of developmental timing programs in C. elegans has not only contributed to our knowledge of the molecular and genetic basis of temporal progression in animals, but has also had impacts in many areas beyond timing, including the critical role of miRNAs in development, stem cell biology, and cancer. Many genes that were first isolated as members of the C. elegans heterochronic pathway are highly conserved, and some appear to regulate developmental timing in these other systems as well. Understanding of miRNAs and their roles in developmental biology has expanded rapidly since lin-4 and let-7 were first discovered, and the study of the heterochronic pathway in C. elegans has continued to yield key insights into miRNA regulation and function. However, many important issues in miRNA-mediated gene regulation and developmental timing remain unresolved.
One area in which many questions remain is understanding of how miRNAs identify and regulate their target mRNAs in vivo. Tools for prediction of miRNA targets through sequence analysis have improved significantly in recent years, with better understanding of the roles of the seed region and downstream sequence complementarity, and with analysis of evolutionary conservation of target sites. Yet the manner in which miRNAs interact with particular targets and direct their precise temporal control in vivo is more intricate, and understanding the specifics of this regulation would be an important step forward in the field. One example can be seen in differential regulation of mRNAs by lin-4, which strongly down-regulates lin-14 in the L1 stage and not until later, in the L2 stage, does it repress lin-28. Determination of whether the number of target sites, the exact pattern of base pairing between miRNA and target site, other features of the target's 3′ UTR, or additional proteins of the RISC contribute to this specificity will shed light on the complex role of miRNAs in their biological context. Another question awaiting further analysis in vivo surrounds the manner in which a particular miRNA chooses whether to regulate an individual target through translation inhibition, active turnover, or some other mechanism. Identification of the sequences and trans-acting factors that mediate the decision between these modes of repression is an important and on-going undertaking. This decision is likely a complex one, influenced by a variety of factors. For example, a recent report suggests that environmental conditions may affect the mechanism of repression (Holtz and Pasquinelli, 2009).
Important goals toward fully understanding the heterochronic pathway remain as well. The number of factors that act to time the L2 proliferative division is strikingly large when compared to the number of genes identified that control later stages. The reason for this difference is unclear; perhaps it is an experimental bias influenced by the ease of scoring the double division. Alternatively, it may be a biological consequence of the importance of the L2 stage to integrating environmental cues into life-history decisions. Identification of markers that clearly distinguish L3 and L4 programs should aid in the discovery of additional late-acting factors, potentially including a transcription factor that specifies the L3-L4 transition. Furthermore, identification and analysis of additional target genes for the transcription factors that regulate stage-specific programs should reveal how changes in transcription factor activity result in temporal patterning of the hypodermis. Analyses of the roles of individual targets in developmental timing will be of great interest.
Finally, the heterochronic regulatory pathway drawn in Fig. 1 contains (at least) one glaring omission relative to timing. If lin-4 is key to triggering the pathway, we need to understand how it becomes transcriptionally activated in a stage-specific manner. The FLH transcription factors play a role in repressing lin-4 in the embryo (Ow et al., 2008), but their absence does not result in timing defects, so some other cue must be required for activation of lin-4 in larval development. At least part of the answer seems to reside in nutritional cues, as lin-4 is not activated in larvae hatched in the absence of a food source and hypodermal programs do not progress (Arasu et al., 1991; Abbott et al., 2005). An interesting piece of the puzzle may reside in insulin signaling pathways. daf-16/FOXO is a downstream transcriptional effector of insulin signaling in worms, and a lin-4 reporter fusion is inappropriately activated in daf-16 mutant animals hatched in the absence of food, conditions in which the reporter gene is inactive in wild-type animals (Baugh and Sternberg, 2006). Further investigation into the environmental and genetic regulation of lin-4, and therefore the activation of the heterochronic pathway, will be critical in broadening our understanding of miRNAs and their role in developmental timing. C. elegans provides a beautifully simple system in which to study these regulatory networks and reveal how environmental signals regulate miRNAs and ultimately developmental timing programs.
Acknowledgments
We thank members of the Twin Cities worm community for helpful discussions. The work of T.D.R. was supported by a College of Biological Sciences Developmental Biology Fellowship and the National Institutes of Health (NIH) NRSA (F32 GM087845-01). Work in the laboratory of A.E.R. was supported by grants from the NIH (R01 GM-050227) and the National Science Foundation (IOB -0515682).
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Miller EA, Rougvie AE. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen∷green fluorescent protein fusion gene. Genetics. 1998;149:1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008;182:543–557. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27:27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Dorn R, Krauss V. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica. 2003;117:165–177. doi: 10.1023/a:1022983810016. [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, 3rd, Mello CC. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. Ontogeny and phylogeny--revisited and reunited. Bioessays. 1992;14:275–279. doi: 10.1002/bies.950140413. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009a;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009b;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009 doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Holtz J, Pasquinelli AE. Uncoupling of lin-14 mRNA and protein repression by nutrient deprivation in Caenorhabditis elegans. RNA. 2009;15:400–405. doi: 10.1261/rna.1258309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PJ. WormBook. 2007. Dauer; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- Jerome T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8:229–233. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kanamoto T, Terada K, Yoshikawa H, Furukawa T. Cloning and regulation of the vertebrate homologue of lin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev Dyn. 2006;235:1142–1149. doi: 10.1002/dvdy.20712. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, Fallon JF. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn. 2005;234:948–960. doi: 10.1002/dvdy.20591. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- Liu T, Zimmerman KK, Patterson GI. Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev Biol. 2004;4:11. doi: 10.1186/1471-213X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. thesis. Harvard University; 1990. [Google Scholar]
- Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, Gerisch B, Hutter H, Antebi A. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magner DB, Antebi A. Caenorhabditis elegans nuclear receptors: insights into life traits. Trends Endocrinol Metab. 2008;19:153–160. doi: 10.1016/j.tem.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–445. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty MP, McNeill RE, Miller N, Kerin MJ. Interactions between the estrogen receptor, its cofactors and microRNAs in breast cancer. Breast Cancer Res Treat. 2009;116:425–432. doi: 10.1007/s10549-009-0429-7. [DOI] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, Mangelsdorf DJ. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 2001;128:3221–3232. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Hada K, Moliyama K, Ohniwa RL, Tan YM, Olsson-Carter K, Chi W, Reinke V, Slack FJ. C. elegans sym-1 is a downstream target of the hunchback-like-1 developmental timing transcription factor. Cell Cycle. 2009;8 doi: 10.4161/cc.8.24.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009 doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow MC, Martinez NJ, Olsen PH, Silverman HS, Barrasa MI, Conradt B, Walhout AJ, Ambros V. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev. 2008;22:2520–2534. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pepper AS, McCane JE, Kemper K, Yeung DA, Lee RC, Ambros V, Moss EG. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 2004;131:2049–2059. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van Meurs J, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009 doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of MicroRNA Processing Inhibition by the Developmentally Regulated RNA-binding Protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Rajyaguru P, Parker R. CGH-1 and the control of maternal mRNAs. Trends Cell Biol. 2009;19:24–28. doi: 10.1016/j.tcb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- Roush SF, Slack FJ. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev Biol. 2009;334:523–534. doi: 10.1016/j.ydbio.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009 doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Simonetta SH, Migliori ML, Romanowski A, Golombek DA. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS One. 2009;4:e7571. doi: 10.1371/journal.pone.0007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009 doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol. 2006;289:30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Turner MJ, Slack FJ. Transcriptional control of microRNA expression in C. elegans: promoting better understanding. RNA Biol. 2009;6:49–53. doi: 10.4161/rna.6.1.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilson GN. Heterochrony and human malformation. Am J Med Genet. 1988;29:311–321. doi: 10.1002/ajmg.1320290210. [DOI] [PubMed] [Google Scholar]
- Xia D, Huang X, Zhang H. The temporally regulated transcription factor sel-7 controls developmental timing in C. elegans. Dev Biol. 2009;332:246–257. doi: 10.1016/j.ydbio.2009.05.574. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]