Abstract
Previous studies indicated that these genetic elements could be involved in the regulation of lysogenization and prophage induction processes. The effects were dramatic in Shiga toxin-converting phage Φ24B after treatment with oxidative stress-inducing agent, hydrogen peroxide, while they were less pronounced in bacteriophage λ and in both phages irradiated with UV. The hydrogen peroxide-caused prophage induction was found to be RecA-dependent. Importantly, in hydrogen peroxide-treated E. coli cells lysogenic for either λ or Φ24B, deletion of the exo-xis region resulted in a significant decrease in the levels of expression of the S.O.S. regulon genes. Moreover, under these conditions, a dramatic decrease in the levels of expression of phage genes crucial for lytic development (particularly xis, exo, N, cro, O, Q, and R) could be observed in Φ24B-, but not in λ-bearing cells. We conclude that genes located in the exo-xis region are necessary for efficient expression of both host S.O.S regulon in lysogenic bacteria and regulatory genes of Shiga toxin-converting bacteriophage Φ24B.
1. Introduction
Infection of humans by enterohemorrhagic Escherichia coli (EHEC) strains causes hemorrhagic colitis, and in some patients it may result in various complications, including, the most severe of them, the hemolytic-uremic syndrome and neurological dysfunctions [1–3]. The main causes of EHEC-mediated complications are Shiga toxins, produced by the infecting bacteria [4]. The severity of EHEC infections and significance of the medical problem related to them are exemplified by local outbreaks, occurring in various geographical regions around the world. One of the most famous of them took place in 2011 in Germany, where over 4,000 patients developed severe symptoms, and 54 died [5–10].
In EHEC strains, Shiga toxins are encoded by genes (called stx genes) located in genomes of prophages [11, 12]. The phages bearing stx genes are referred to as Shiga toxin-converting bacteriophages, and all of them belong to the family of lambdoid phages (with phage λ serving as a paradigm) [12]. stx genes are present between Q antiterminator gene and the genes coding for proteins causing cell lysis; thus, in the lysogenic state, these genes are not transcribed [13–15] and Shiga toxins are not produced. Their expression is possible only after prophage induction [11, 12] which usually requires activation of the bacterial S.O.S. response, mediated by RecA protein, though RecA-independent induction of Shiga toxin-converting prophages by EDTA has also been reported [16].
During infection of human intestine by EHEC, the oxidative stress appears to be the most likely condition causing the bacterial S.O.S. response and subsequent induction of Shiga toxin-converting prophages [17]. In fact, it was demonstrated that hydrogen peroxide (which is produced by neutrophils as a response to infection) enhanced production of Shiga toxins by EHEC [18] due to oxidative stress-mediated induction of Shiga toxin-converting prophages [19, 20].
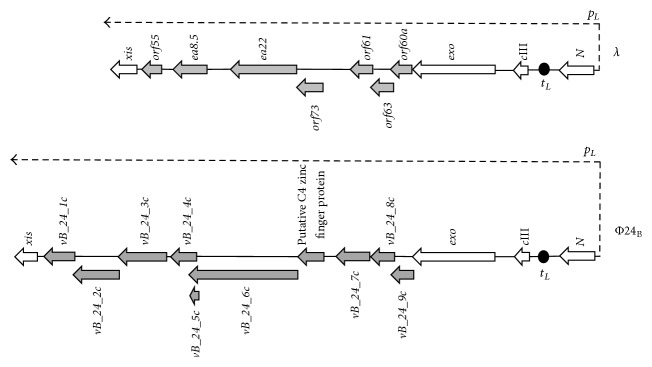
Since many antibiotics not only kill bacteria and inhibit their growth but also induce prophage lytic development, their use is not recommended when EHEC infection is confirmed or even suspected (reviewed in [12]). Therefore, there are serious problems with treatment of patients, indicating that searching for new targets of potential therapies against Shiga toxin-producing bacteria is important. One might consider that such therapies should be focused on inhibition of Shiga toxin-converting prophage induction which would impair production of the EHEC virulence factor. All lambdoid phages, including Shiga toxin-converting bacteriophages, contain the b region in their genomes which is dispensable for the development under standard laboratory conditions [14, 15]. Inside this part of the phage genome, there is an evolutionarily conserved fragment, located between exo and xis genes and transcribed from p L promoter, called the exo-xis region (Figure 1). This region encompasses several genes and open reading frames whose functions in phage development are largely unknown, and only a few articles are available in the literature that focused on them. Nevertheless, some interesting observations have been reported. Namely, induction of expression of genes from the exo-xis region resulted in synchronization of the host cell cycle [21] and inhibition of host DNA replication [22]. Moreover, overexpression of these genes impaired lysogenization of E. coli by bacteriophage λ [23] and enhanced induction of prophages λ and Φ24B (one of Shiga toxin-converting phages) [24]. Ea8.5 protein, encoded by a gene located in the exo-xis region, contains a fused homeodomain/zinc finger fold [25] which suggests a regulatory role for this protein. Interestingly, prophage induction with mitomycin C or hydrogen peroxide caused different expression patterns of genes from the exo-xis region; such differences were observed in both phages, λ and Φ24B [26]. In this work, we used the deletion mutants to investigate the role of the exo-xis region in induction of λ and Φ24B prophages under oxidative stress conditions.
Figure 1.
Schematic maps of the exo-xis regions of bacteriophages λ and Φ24B. Genes from the exo-xis region are marked as thick grey arrows, and other genes are shown as thick open arrows. Directionality of transcription from p L promoter is indicated as thin dashed arrow. t L1 terminator is marked as black oval.
2. Materials and Methods
2.1. Bacteria and Bacteriophages
E. coli MG1655 strain [27] and its derivatives, used in this work, are listed in Table 1. Bacteria were routinely cultured in the Luria-Bertani (LB) medium at 30°C (most experiments) or 37°C (lysogenization and recombination procedures during construction of strains and SOS ChromoTest, according to the instructions of kits' manufacturers), under aerobic conditions. Where appropriate, the following antibiotics were added: chloramphenicol up to 20 μg/mL, kanamycin up to 50 μg/mL, and/or tetracycline up to 12.5 μg/mL.
Table 1.
Escherichia coli strains.
| Strain |
Genotype or relevant characteristics | Reference |
|---|---|---|
| E. coli MG1655 | F− λ − ilvG rfb-50 rph-1 | [27] |
| E. coli MG1655 (λ) | MG1655 bearing λ prophage | [24] |
| E. coli MG1655 (λΔexo-xis) | MG1655 bearing λ prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by recombination |
| E. coli MG1655 (λΔorfs) | MG1655 bearing λ prophage with deletion of orf60a, orf63, orf61, and orf73 | This study, by recombination |
| E. coli MG1655 (λΔorf60a) | MG1655 bearing λ prophage with deletion of orf60a | This study, by recombination |
| E. coli MG1655 (λΔorf63) | MG1655 bearing λ prophage with deletion of orf63 | This study, by recombination |
| E. coli MG1655 (λΔorf61) | MG1655 bearing λ prophage with deletion of orf61 | This study, by recombination |
| E. coli MG1655 (λΔorf73) | MG1655 bearing λ prophage with deletion of orf73 | This study, by recombination |
| E. coli MG1655 (λΔea22) | MG1655 bearing λ prophage with deletion of ea22 | This study, by recombination |
| E. coli MG1655 (λΔea8.5) | MG1655 bearing λ prophage with deletion of ea8.5 | This study, by recombination |
| E. coli MG1655 (Φ24B) | MG1655 bearing Φ24B prophage | [24] |
| E. coli MG1655 (Φ24BΔexo-xis) | MG1655 bearing Φ24B prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by recombination |
| E. coli MG1655 (Φ24BΔorfs) | MG1655 bearing Φ24B prophage with deletion of 4 orfs being homologues of λorf60a, λorf63, λorf61, and λorf73 | This study, by recombination |
| E. coli MG1655 (Φ24BΔorf60a) | MG1655 bearing Φ24B prophage with deletion of vb_24B_9c, the homologue of λorf60a | This study, by recombination |
| E. coli MG1655 (Φ24BΔorf63) | MG1655 bearing Φ24B prophage with deletion of vb_24B_8c, the homologue of λorf63 | This study, by recombination |
| E. coli MG1655 (Φ24BΔorf61) | MG1655 bearing Φ24B prophage with deletion of vb_24B_7c, the homologue of λorf61 | This study, by recombination |
| E. coli MG1655 (Φ24BΔorf73) | MG1655 bearing Φ24B prophage with deletion of the sequence of putative C4 zinc finger protein, the homologue of λorf73 | This study, by recombination |
| E. coli MG1655 (Φ24BΔea22) | MG1655 bearing Φ24B prophage with deletion of vb_24B_6c, the analogue of λea22 | This study, by recombination |
| E. coli MG1655recA13 | MG1655 but recA13 | [33] |
| E. coli MG1655recA13 (λ) | MG1655recA13 bearing λ prophage | This study, by lysogenization |
| E. coli MG1655recA13 (λΔexo-xis) | MG1655recA13 bearing λ prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by lysogenization |
| E. coli MG1655recA13 (Φ24B) | MG1655recA13 bearing Φ24B prophage | This study, by lysogenization |
| E. coli MG1655recA13 (Φ24BΔexo-xis) | MG1655recA13 bearing Φ24B prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by lysogenization |
| E. coli PQ37 | sfiA::Mud(Ap lac) cts, lacΔU169, mal +, galE, galY, PhoC, rfa, F−, thr, leu, his, pyrD, thi, trp::MUC +, srl300::Tn10, rpoB, uvrA + | [31] |
| E. coli PQ37 (λ) | PQ37 bearing λ prophage | This study, by lysogenization |
| E. coli PQ37 (λΔexo-xis) | PQ37 bearing λ prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by lysogenization |
| E. coli PQ37 (Φ24B) | PQ37 bearing Φ24B prophage | This study, by lysogenization |
| E. coli PQ37 (Φ24BΔexo-xis) | PQ37 bearing Φ24B prophage with deletion of all orfs and genes localized between genes exo and xis | This study, by lysogenization |
Bacteriophages λ papa (from our collection) [26] and Φ24B (Δstx2::cat) [28] were employed in this study. Phage suspensions were stored in the TM buffer (10 mM Tris-HCl, 10 mM MgSO4, pH 7.2) at 4°C.
The deletion mutants were constructed as described previously [29], by using the Quick and Easy E. coli Gene Deletion Kit (from Gene Bridges). The deletion of the indicated region was performed according to the manufacturer's protocol using primers listed in Table 2. In the first step, the targeted sequence has been replaced with the FRT-flanked kanamycin resistance cassette, and the selection marker was subsequently removed in the FLP-recombinase step, leaving only 87 nucleotides of the cassette in the place of the original sequence. Each deletion was confirmed by DNA sequencing.
Table 2.
Primers used for construction of E. coli strains.
| Primer name | Sequence (5′→3′) |
|---|---|
| pF_λ_exo-xis = pF_λ_orf60a pR_λ_exo-xis |
ATATCCGGGTAGGCGCAATCACTTTCGTCTACTCCGTTACAAAGCGAGGAATTAACCCTCACTAAAGGGCG AGGCGGCTTGGTGTTCTTTCAGTTCTTCAATTCGAATATTGGTTACGTCTTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_orfs = pF_λ_orf60a pR_λ_orfs = pR_λ_orf73 |
ATATCCGGGTAGGCGCAATCACTTTCGTCTACTCCGTTACAAAGCGAGGAATTAACCCTCACTAAAGGGCG ACCTCTCTGTTTACTGATAAGTTCCAGATCCTCCTGGCAACTTGCACAAGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_orf60a pR_λ_orf60a |
ATATCCGGGTAGGCGCAATCACTTTCGTCTACTCCGTTACAAAGCGAGGAATTAACCCTCACTAAAGGGCG AGATGCTTTGTGCATACAGCCCCTCGTTTATTATTTATCTCCTCAGCCAGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_orf63 pR_λ_orf63 |
CACAAAGCATCTTCTGTTGAGTTAAGAACGAGTATCGAGATGGCACATAGAATTAACCCTCACTAAAGGGCG TCATACCTGGTTTCTCTCATCTGCTTCTGCTTTCGCCACCATCATTTCCATAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_orf61 pR_λ_orf61 |
AGAGAAACCAGGTATGACAACCACGGAATGCATTTTTCTGGCAGCGGGCTAATTAACCCTCACTAAAGGGCG TTATCCGGAAACTGCTGTCTGGCTTTTTTTGATTTCAGAATTAGCCTGACTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_orf73 pR_λ_orf73 |
ACATCATTGATTCAGCATCAGAAATAGAAGAATTACAGCGCAACACAGCAAATTAACCCTCACTAAAGGGCG ACCTCTCTGTTTACTGATAAGTTCCAGATCCTCCTGGCAACTTGCACAAGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_ea22 pR_λ_ea22 |
GAAATTAACTCTCAGGCACTGCGTGAAGCGGCAGAGCAGGCAATGCATGAAATTAACCCTCACTAAAGGGCG GTCAGACATCATATGCAGATACTCACCTGCATCCTGAACCCATTGACCTCCTAATACGACTCACTATAGGGCTC |
|
| |
| pF_λ_ea8.5 pR_λ_ea8.5 |
ATGAGTATCAATGAGTTAGAGTCTGAGCAAAAAGATTGGGCGTTATCAATAATTAACCCTCACTAAAGGGCG TAATCATCTATATGTTTTGTACAGAGAGGGCAAGTATCGTTTCCACCGTATAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_exo-xis = pF_24B_orf60a pR_Φ24B_exo-xis |
TGGTAATGAAGCATCCTCACGATAATATCCGGGTAGGCACGATCACTTTCAATTACCTCACTAAAGGGCG TGGCAATATGCTTTTCCTTCTCAATTTCCGCTTTAATCATATGCAGTTCGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_orfs = pF_Φ24B_orf60a pR_Φ24B_orfs = pR_Φ24B_orf73 |
TGGTAATGAAGCATCCTCACGATAATATCCGGGTAGGCACGATCACTTTCAATTACCTCACTAAAGGGCG CTTCGAACCTCTCTGTTTACTGATAAGCTCCAGATCCTCCTGGCAACTTGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_orf60a pR_Φ24B_orf60a |
TGGTAATGAAGCATCCTCACGATAATATCCGGGTAGGCACGATCACTTTCAATTACCTCACTAAAGGGCG GGAGATGCTTTGTGCATACAGCCCCTCGTTATTATTTATCTCTTCAGCCTAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_orf63 pR_Φ24B_orf63 |
TGCACAAAGCATCTCCTGTTGAATTAAGAACGAGTATCGGGATGGCACATAATTAACCCTCACTAAAGGGCG CATCATTTCCAGCTTTTGTGAAAGGGATGTGGCTAACGTATGAAATTCTTTAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_orf61 pR_Φ24B_orf61 |
TTCTGGCAGCAGGCTTCATATTCTGTGTGCTTATGCTTGCCGACATGGGAAATTAACCCTCACTAAAGGGCG CACCGTTCCTTAAAGACGCCGTTTAACATGCCGATCGCCAGACTTAAATGTACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_orf73 pR_Φ24B_orf73 |
TGGCAGACCTCATTGATTCAGCATCAGAAATTGAAGAATTACAGCGCAACAATTAACCCTCACTAAAGGGCGG CTTCGAACCTCTCTGTTTACTGATAAGCTCCAGATCCTCCTGGCAACTTGTAATACGACTCACTATAGGGCTC |
|
| |
| pF_Φ24B_ea22 pR_Φ24B_ea22 |
ATCAGGCACTGCGTGAAAAGGCAGAGAAAGCAACTAAAGGAAGCTACATCAATTAACCCTCACTAAAGGGCG ATCCAGTGTGACGGTTTCCACGACGCACCAGGAATTATCCACCCATCATTATACGACTCACTATAGGGCTC |
Lysogenic strains were constructed according to the procedure described previously [24], with slight modifications. Briefly, host bacteria were cultured to A600 of 0.5 in LB medium supplemented with MgSO4 and CaCl2 (to final concentrations of 10 mM each) at 37°C with shaking. At this point, one milliliter of the culture was withdrawn and centrifuged (10 min, 2000 ×g). Pellet was washed twice with TCM buffer (10 mM Tris-HCl, pH 7.2, 10 mM MgSO4, 10 mM CaCl2) and then suspended in 1 mL of the same buffer. Next, bacteria were incubated for 30 min at 30°C and mixed with phage suspensions at multiplicity of infection (m.o.i.) = 5. Mixtures of bacteria and phages were incubated in TMC buffer for 30 min at 30°C; then serial dilutions were prepared in TM buffer (10 mM Tris-HCl, 10 mM MgSO4; pH 7.2), and the mixture was plated onto LB agar. Plates were incubated at 37°C overnight. Lysogens were verified by sensitivity to UV irradiation and confirmed by PCR with primers designed to amplify phage sequence (Table 3).
Table 3.
Primers used for PCR.
| Primers for bacterial sequences | Primers for phage sequences | ||
|---|---|---|---|
| Name | Sequence (5′→3′) | Name | Sequence (5′→3′) |
| pF_recA | AGATTTCGACGATACGGCCC | pF_λ_int | TTTGATTTCAATTTTGTCCCACT |
| pR_recA | AACCATCTCTACCGGTTCGC | pR_ λ_int | ACCATGGCATCACAGTATCG |
| pF_lexA | ATGGATGGTGACTTGCTGGC | pF_λ_xis | TACCGCTGATTCGTGGAACA |
| pR_lexA | TTCGTCATCAATACGTGCGAC | pR_ λ_xis | GGGTTCGGGAATGCAGGATA |
| pF_ssb | ATCGAAGGTCAGCTGCGTAC | pF_λ_exo | TGCCGTCACTGCATAAACC |
| pR_ssb | CGACTTCTGTGGTGTAGCGA | pR_ λ_exo | TCTATCGCGACGAAAGTATGC |
| pF_recF | CGATACCGGCGCTATACTCC | pF_λ_cIII | ATTCTTTGGGACTCCTGGCTG |
| pR_recF | TTACGAACAGCTACGCCCG | pR_ λ_cIII | GTAAATTACGTGACGGATGGAAAC |
| pF_rpoD | GAATCTGAAATCGGGCGCAC | pF_λ_N | CTCGTGATTTCGGTTTGCGA |
| pR_rpoD | GTCAACAGTTCAACGGTGCC | pR_ λ_N | AAGCAGCAAATCCCCTGTTG |
| pF_rpoH | GCTTTGGTGGTCGCAACTTT | pF_λ_cI | ACCTCAAGCCAGAATGCAGA |
| pR_rpoH | TCGCCGTTCACTGGATCAAA | pR_ λ_cI | CCAAAGGTGATGCGGAGAGA |
| pF_rpoS | TTGCTCTGCGATCTCTTCCG | pF_λ_cro | ATGCGGAAGAGGTAAAGCCC |
| pR_rpoS | GAACGTTTACCTGCGAACCG | pR_ λ_cro | TGGAATGTGTAAGAGCGGGG |
| pF_uvrA | GTCCATATCCGCCACTACCG | pF_λ_cII | TCGCAATGCTTGGAACTGAGA |
| pR_uvrA | TTACCCAACGTCTTGCCGAG | pR_ λ_cII | CCCTCTTCCACCTGCTGATC |
| pF_ftsK | ACAAACCGTTTATCTGCGCG | pF_λ_O | AATTCTGGCGAATCCTCTGA |
| pR_ftsK | ATCTTTACCCAGCACCACGG | pR_ λ_O | GAATTGCATCCGGTTT |
| pF_16SrRNA | CCTTACGACCAGGGCTACAC | pF_λ_Q | TTCTGCGGTAAGCACGAAC |
| pR_16SrRNA | TTATGAGGTCCGCTTGCTCT | pR_ λ_Q | TGCATCAGATAGTTGATAGCCTTT |
| pF_λ_R | ATCGACCGTTGCAGCAATA | ||
| pR_ λ_R | GCTCGAACTGACCATAACCAG | ||
| pF_Φ24B_int | CAGTTGCCGGTATCCCTGT | ||
| pR_ Φ24B_int | TGAGGCTTTCTTGCTTGTCA | ||
| pF_Φ24B_xis | TATCGCGCCGGATGAGTAAG | ||
| pR_ Φ24B_xis | CGCACAGCTTTGTATAATTTGCG | ||
| pF_Φ24B_exo | TGCCGTCACTGCATAAACC | ||
| pR_ Φ24B_exo | TCTATCGCGACGAAAGTATGC | ||
| pF_Φ24B_cIII | ATTCTTTGGGACTCCTGGCTG | ||
| pR_ Φ24B_cIII | GTAAATTACGTGACGGATGGAAAC | ||
| pF_Φ24B_N | AGGCGTTTCGTGAGTACCTT | ||
| pR_ Φ24B_N | TTACACCGCCCTACTCTAAGC | ||
| pF_Φ24B_cI | TGCTGTCTCCTTTCACACGA | ||
| pR_ Φ24B_cI | GCGATGGGTGGCTCAAAATT | ||
| pF_Φ24B_cro | CGAAGGCTTGTGGAGTTAGC | ||
| pR_ Φ24B_cro | GTCTTAGGGAGGAAGCCGTT | ||
| pF_Φ24B_cII | TGATCGCGCAGAAACTGATTTAC | ||
| pR_ Φ24B_cII | GACAGCCAATCATCTTTGCCA | ||
| pF_Φ24B_O | AAGCGAGTTTGCCACGAT | ||
| pR_ Φ24B_O | GAACCCGAACTGCTTACCG | ||
| pF_Φ24B_Q | GGGAGTGAGGCTTGAGATGG | ||
| pR_ Φ24B_Q | TACAGAGGTTCTCCCTCCCG | ||
| pF_Φ24B_R | GGGTGGATGGTAAGCCTGT | ||
| pR_ Φ24B_R | TAACCCGGTCGCATTTTTC | ||
2.2. Phage Lytic Development after Prophage Induction
Bacteria lysogenic with tested phages were cultured in LB medium at 30°C to A600 of 0.1. Two induction agents were tested: H2O2 (1 mM) and UV irradiation (50 J/m2; this dose was achieved by 20 sec incubation of the bacterial suspensions in Petri dishes under UV lamp hanged 17 cm above the laboratory table). At indicated times after induction, samples of bacterial cultures were harvested, and 30 μL of chloroform was added to 0.5 mL of each sample. The mixture was vortexed and centrifuged for 5 min in a microcentrifuge. Then, serial dilutions were prepared in TM buffer, and phage titer (number of phages per mL) was determined by spotting 2.5 μL of each dilution of the phage lysate on a freshly prepared LB agar (1.5%) or LB agar with 2.5 μg/mL chloramphenicol (according to a procedure described previously [30]), with a poured mixture of 1 mL of the indicator E. coli MG1655 strain culture and 2 mL of 0.7% nutrient agar (prewarmed to 45°C), supplemented with MgSO4 and CaCl2 (to a final concentration of 10 mM each). When full-plate titration was used, 0.1 mL of phage lysate dilutions was plated onto LB agar. Plates were incubated at 37°C overnight. Analogous experiment but without induction agents (control experiments), which allows estimation of effects of spontaneous prophage induction, was performed with each lysogenic strain. The relative phage titer, expressed as plaque forming units (pfu)/mL, was calculated by subtracting the values obtained in the control experiment from the values determined in the main experiment, and as a consequence it represents the ratio of phage titers in induced and noninduced cultures. Each experiment was repeated three times.
2.3. The S.O.S. Assay
The S.O.S. assay was performed using the SOS-ChromoTest Kit (Environmental Bio-Detection Products Inc.), following the manufacturer's protocol and using provided 4-nitro-quinoline oxide (4-NQO) as a positive reference standard, and 1 mM H2O2 and UV irradiation (50 J/m2) as tested inducers of the S.O.S. response [31, 32]. In the case of UV light irradiation, the production of β-galactosidase was evaluated immediately after the exposure, without 2 h incubation at 37°C (recommended by the manufacturer), as, without this modification, the visual detection of the blue color was not possible due to rapid S.O.S. response after UV irradiation. Before use, the SOS-ChromoTest bacterial strain (E. coli PQ37, provided with the kit) was lysogenized by following phages: λ, λΔexo-xis, Φ24B, or Φ24BΔexo-xis, according to procedure described above.
2.4. Preparation of RNA and cDNA from Bacteria
For the isolation of total RNA, the previously described [26] procedure was employed. Briefly, the prophage induction was performed with 1 mM H2O2 or UV irradiation (at the dose of 50 J/m2). Following induction, the samples were withdrawn at indicated times and the growth of bacteria was inhibited by the addition of NaN3 (Sigma-Aldrich) to a final concentration of 10 mM. Total RNA was isolated from 109 bacterial cells by using the High Pure RNA Isolation Kit (Roche Applied Science). Bacterial genomic DNA carryover was removed by incubation with TURBO DNase from TURBO DNA-free Kit (Life Technologies) for 60 min at 37°C, according to the manufacturer's guidelines. To evaluate the quality and quantity of the isolated RNA, a NanoDrop spectrophotometer was employed, considering the absorbance ratio (which should be 1.8 ≤ A260/A280 ≤ 2.0). Moreover, band patterns of total RNA were visualized by electrophoresis. The absence of DNA from RNA samples was controlled by PCR amplification and by real-time PCR amplification (all analyzed genes were tested). RNA preparations were stored at −80°C. cDNA was obtained with Transcriptor Reverse Transcriptase and random hexamer primers (Roche Applied Science), using total RNA samples (1.25 μg) as templates. cDNA reaction mixtures were diluted 10-fold for use in real-time PCR.
2.5. Real-Time PCR Assay and Data Analysis
The patterns of genes' expression were determined by quantitative real-time reverse transcription-PCR (qRT-PCR), using the LightCycler 480 Real-Time PCR System (Roche Applied Science) and cDNA samples from lysogenic bacteria. Transcripts of tested phage and bacterial genes were compared in parallel to 16S rRNA housekeeping gene (according to a procedure described previously [34]), whose expression was found to be constant. Primers were developed by Primer3web version 4.0.0 and produced by Sigma-Aldrich or GENOMED. The transcriptional analysis of phage and bacterial genes from lysogenic strains was performed with primers presented in Table 3. Real-time PCR amplifications were carried out for 55 cycles in 20 μL reaction volume, using LightCycler 480 SYBR Green I Master (Roche Applied Science) as a fluorescent detection dye. Reactions were performed in Roche 96-well plates containing 10 μL 2x SYBR Green I Master Mix, 6.25 ng/μL cDNA, and 200 nM of each gene-specific primer (Table 3). Relative quantification assays were performed with cDNA of 16S rRNA and phage/bacterial genes multiplex assay. All templates were amplified using the following program: incubation at 95°C for 5 min, followed by 55 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. No template control was included with each run. The specificity of amplified products was examined by melting curve analysis immediately after the final PCR cycle and confirmed by gel electrophoresis. Each experiment was conducted in triplicate.
The relative changes in gene expression revealed by quantitative real-time PCR experiments were analyzed using the calibrator, normalizing relative quantification method with efficiency correction (called the E-Method). This method has been used and described in detail previously [26, 29, 35]. The values obtained at time 0 (representing the conditions of spontaneous prophage induction) were used as calibrators. Thus, the following equation was employed to calculate the final results:
| (1) |
where t is target and r is reference.
2.6. In Silico Analyses
The multiple sequence alignment was performed using the ClustalW algorithm available at the website http://www.genome.jp/tools/clustalw/. The Pfam protein families database [36], available at the website http://pfam.xfam.org/, was used to identify protein domains.
3. Results and Discussion
3.1. Deletion of the Exo-Xis Region Impairs Φ24B but Not λ Prophage Induction after Treatment with Hydrogen Peroxide
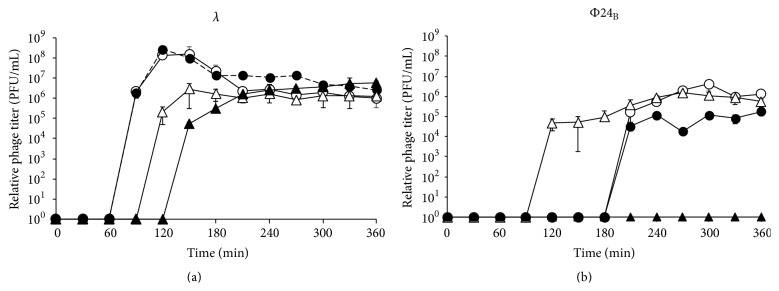
Until now, all in vivo studies on effects of the exo-xis region on host or phage development were performed with the use of strains overexpressing genes from this region [21–24, 26]. In this work, we have constructed a series of bacteriophage λ and Φ24B mutants with deletions of either the whole exo-xis region or individual genes or open reading frames (Table 1). When wild-type λ and Φ24B prophages were induced by UV irradiation (employed in this work as positive control conditions causing effective prophage induction) or hydrogen peroxide treatment of the lysogenic cells, efficiencies of induction and further phage lytic development were comparable in both phages, though some differences were observed in the duration of the lag phase of the phage development (Figure 2). Induction of λΔexo-xis mutant with UV irradiation was similar to that observed for the wild-type λ, and treatment with hydrogen peroxide caused only a slight delay in the mutant phage development. The decrease in the phage titer at later times of the experiments is characteristic for λ and most probably arises from adsorption of the progeny virions on fragments of disrupted cell envelopes [15, 24]. However, induction of Φ24BΔexo-xis prophage by UV irradiation was less efficient than that of the wild-type Φ24B, and induction of the mutant by hydrogen peroxide was severely impaired (Figure 2). More detailed analyses, based on the full-plate phage titration method, allowing detection of 10 pfu/mL (see Section 2 for details), indicated that the number of pfu per mL of Φ24BΔexo-xis phage after induction with hydrogen peroxide was at the same range (103/mL) as that measured without specific induction (i.e., representing efficiency of spontaneous prophage induction). Nevertheless, the titer of Φ24BΔexo-xis measured at 240 and 360 min after induction was 9.0 ± 0.2 × 103 and 7.9 ± 0.9 × 103, respectively, that is, still 3-4 times higher than that without induction, which was 2.1 ± 0.3 × 103 and 2.6 ± 0.4 × 103, respectively (note that the titer of the wild-type Φ24B after prophage induction was several orders of magnitude higher than that without induction, Figure 2).
Figure 2.
Lytic development of bacteriophages λ (a) and Φ24B (b), either wild-type (open symbols) or Δexo-xis (closed symbols), after induction of lysogenic E. coli MG1655 with UV irradiation (50 J/m2, circles) or hydrogen peroxide (1 mM, triangles). The presented results are mean values from 3 independent experiments (biological samples), with error bars indicating SD. Statistically significant differences (p < 0.05 in t-test) between wild-type and Δexo-xis phages were found at times 270, 300, and 360 min of experiments with hydrogen peroxide and at 270, 300, 330, and 360 min of experiments with UV for λ and at times 120, 240, 270, 300, 330, and 360 min of experiments with hydrogen peroxide and at 270, 300, 330, and 360 min of experiments with UV for Φ24B.
Deletions of individual genes and open reading frames from the exo-xis region in Φ24B did not affect significantly the phage titer. However, such deletions resulted in delays in prophage induction by hydrogen peroxide (Table 4). Interestingly, when prophage induction was stimulated by UV irradiation, such effect was not observed, and in some cases even more rapid induction of the mutant prophages occurred. In bacteriophage λ, only slight effects of deletions of individual genes and open reading frames were detected (Table 4).
Table 4.
Duration of the lag phase of the phage lytic development after prophage induction with either hydrogen peroxide (1 mM) or UV irradiation (50 J/m2).
| Strain | The time range of the switch from lag to log phase | |
|---|---|---|
| H2O2 (1 mM) | UV (50 J/m2) | |
| MG1655 (λ) | 60–90 min | 60 min |
| MG1655 (λΔexo-xis) | 90–120 min | 30–60 min |
| MG1655 (λΔorfs) | 60–90 min | 30–60 min |
| MG1655 (λΔorf60a) | 60–90 min | 30–60 min |
| MG1655 (λΔorf63) | 60–90 min | 30–90 min |
| MG1655 (λΔorf61) | 60–90 min | 30–60 min |
| MG1655 (λΔorf73) | 30–60 min | 0–30 min |
| MG1655 (λΔea22) | 60–90 min | 30–60 min |
| MG1655 (λΔea8.5) | 60–90 min | 30–60 min |
| MG1655 (Φ24B) | 60–90 min | 150–180 min |
| MG1655 (Φ24BΔexo-xis) | a | 150–180 min |
| MG1655 (Φ24BΔorfs) | 150–180 min | 150–180 min |
| MG1655 (Φ24BΔorf60a) | 120–150 min | 90–120 min |
| MG1655 (Φ24BΔorf63) | 120–150 min | 30–60 min |
| MG1655 (Φ24BΔorf61) | 90–120 min | 30–60 min |
| MG1655 (Φ24BΔorf73) | 90–120 min | 120–150 min |
| MG1655 (Φ24BΔea22) | 120–150 min | 30–60 min |
aThe value was not determined due to a very low efficiency of prophage induction under these conditions (as shown in Figure 2).
We conclude that the genes and open reading frames from the exo-xis region play important roles in the regulation of lambdoid prophage induction, as deletions of the whole region or single loci caused significant changes in efficiency and timing of this process. The effects of mutations are more pronounced in Shiga toxin-converting phage Φ24B than in λ and in lysogenic E. coli cells treated with hydrogen peroxide than in UV-irradiated ones. Thus, the exo-xis region seems to be particularly important for Φ24B phage under conditions of the oxidative stress, the most likely conditions causing Shiga toxin-converting prophage induction during infection with EHEC.
3.2. Hydrogen Peroxide-Mediated Prophage Induction Is a RecA-Dependent Process
Efficient induction of lambdoid prophages is a RecA-dependent process due to a molecular mimicry between the phage cI repressor and the host-encoded LexA repressor which is self-cleaved after stimulation by the activated form of RecA protein under the S.O.S. response conditions [12–15]. Such a mimicry is well known for bacteriophage λ cI protein and LexA [12, 13], and we found that both domain structure and amino acid residues crucial for the self-cleavage are also conserved in cI repressor of phage 24B (Figure 3) (note that cI sequence of 24B is identical to that of another Shiga toxin-converting bacteriophage, 933 W [37]). Nevertheless, since RecA-independent induction of Shiga toxin-converting prophages has also been reported [16], we asked whether hydrogen peroxide-caused prophage induction depends on the activation of the S.O.S. response.
Figure 3.

Alignment of amino acid sequences of E. coli LexA protein and cI repressors of bacteriophages λ and Φ24B. Specific protein domains are indicated by grey background. Self-cleavage sites are underlined (two amino acid residues between which the cleavage occurs). The active sites of the peptidase domains are framed. Symbols under the sequence alignment indicate conserved sequence (∗), conservative mutations (:), semiconservative mutations (.), and nonconservative mutations ().
When testing H2O2- or UV-dependent induction of prophages λ, λΔexo-xis, Φ24B, and Φ24BΔexo-xis in recA13 mutant host, in the assays analogous to those presented in Figure 2, pfu/mL values were at the levels of those estimated for the uninduced controls (1.8 ± 0.1 × 102, 4.1 ± 1.4 × 102, 1.3 ± 0.1 × 103, and 2.8 ± 0.3 × 103 pfu/mL for λ, λΔexo-xis, Φ24B, and Φ24BΔexo-xis, resp.). Therefore, we conclude that induction of the investigated prophages under conditions of the oxidative stress (treatment with hydrogen peroxide) strongly depends on RecA function. Indeed, in cells lysogenic for λ or Φ24B and treated with UV light or hydrogen peroxide, efficient induction of the S.O.S. response was evident, as estimated with the SOS ChromoTest (Figure 4). Intriguingly, while induction of the S.O.S. response by hydrogen peroxide in λΔexo-xis lysogen was comparable to that in λ lysogen, the signal in the SOS ChromoTest in Φ24BΔexo-xis lysogen was considerably weaker than in the analogous experiment with Φ24B lysogen (Figure 4). No such difference could be observed in UV-irradiated bacteria (Figure 4).
Figure 4.
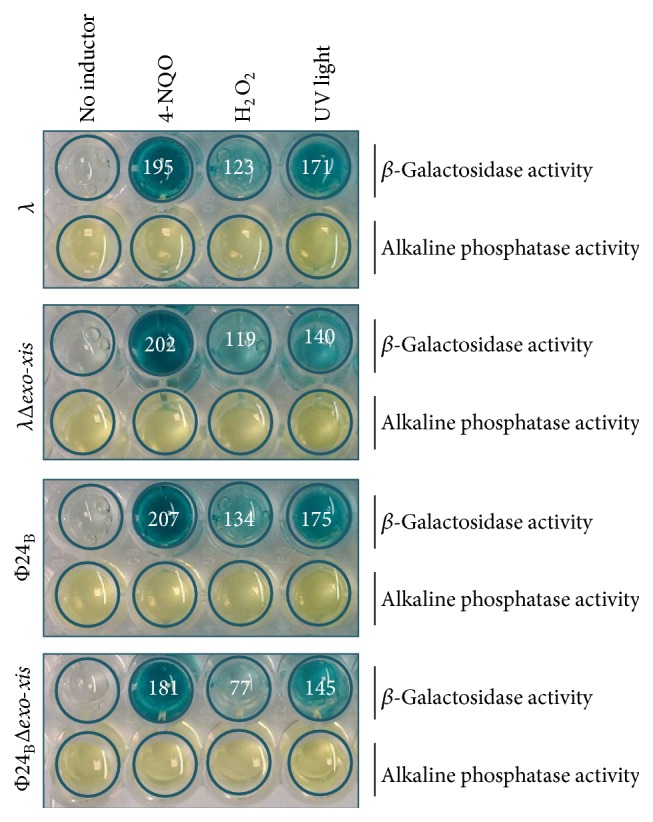
Induction of the S.O.S. response in E. coli PQ37 lysogenic with λ, λΔexo-xis, Φ24B, or Φ24BΔexo-xis, treated with 4-NQO (4-nitro-quinoline oxide, positive control), H2O2 (1 mM), or UV (50 J/m2), using the SOS ChromoTest. β-Galactosidase activity (identified by the blue spots) represents induction of the S.O.S. regulon. Alkaline phosphatase activity (identified by yellow spots) evaluates viability of tested bacteria. Quantification of β-galactosidase activity was performed by densitometry, using the ImageJ software (available at http://imagej.nih.gov/ij/index.html). The results (in arbitrary units reflecting value = 1 ascribed to samples with no inductor), presented as numbers inside the corresponding spots, are mean values from three measurements (with SD < 10% in each case). All these values were significantly (p < 0.001 in t-test) higher than that in the control experiments with no inductor. When Δexo-xis mutants were compared to wild-type phages, the only significant difference (p < 0.05 in t-test) occurred between Φ24B and Φ24BΔexo-xis lysogens induced with hydrogen peroxide.
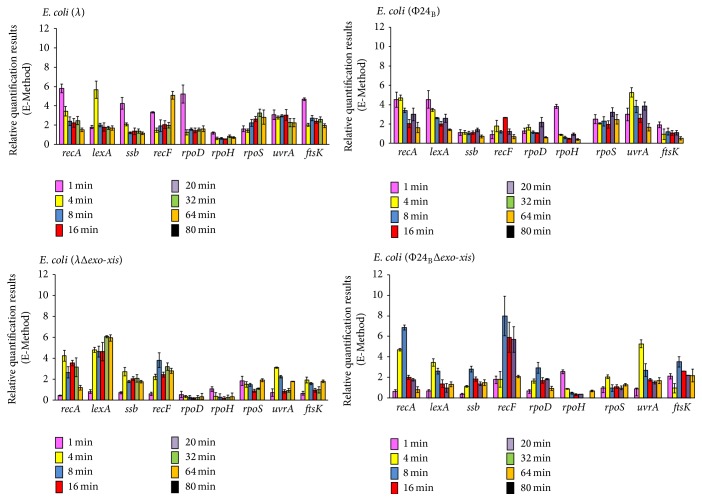
3.3. Deletion of the Exo-Xis Region Negatively Influences Expression of Genes from the S.O.S. Regulon in Hydrogen Peroxide-Treated Lysogenic Bacteria
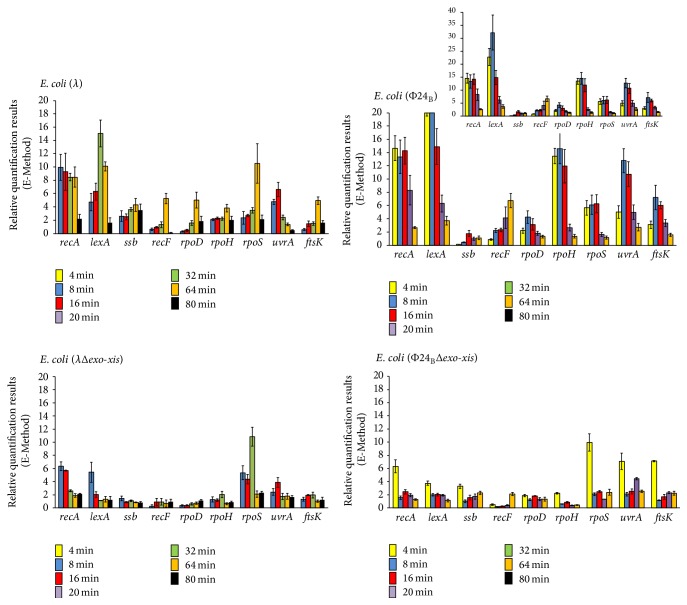
Since unexpected results were obtained in experiments with hydrogen peroxide-treated Φ24BΔexo-xis lysogenic cells (Figure 4), we aimed to investigate the phenomenon of a less efficient induction of the S.O.S. response in more detail. Thus, expression of genes from the S.O.S. regulon was tested by reverse transcription quantitative real-time PCR in E. coli cells lysogenic for λ, λΔexo-xis, Φ24B, and Φ24BΔexo-xis and treated with hydrogen peroxide. In both λ and Φ24B, deletion of the exo-xis region caused a significant reduction in the mRNA levels of most of the S.O.S. regulon genes relative to wild-type prophages, with exceptions of rpoS gene in both phages and ssb, uvrA, and ftsK genes in Φ24B, especially at later times after the treatment (Figure 5). Interestingly, in the case of wild-type Φ24B lysogenic cells, the enhanced expression of particular genes from the S.O.S. regulon persisted longer, in most cases until 16 min after induction, whereas in the deletion mutant it decreases after 4 min (Figure 5). The impairment in expression of genes from the S.O.S. regulon (in particular recA and lexA genes, encoding the main regulators of the S.O.S. response) in the absence of the exo-xis region was more pronounced in Φ24B than in λ. Moreover, induction of the S.O.S. regulon occurred significantly earlier in Φ24B and Φ24BΔexo-xis lysogens than in cells bearing λ and λΔexo-xis prophages (Figure 5). These results might explain, at least partially, effects of deletions of exo-xis genes on prophage induction, demonstrated in Figure 2 and Table 4, particularly delayed induction of Φ24B prophage devoid of certain genes and open reading frames, and less pronounced effects of their lack in λ than in Φ24B.
Figure 5.
Expression of genes from the S.O.S. regulon in E. coli MG1655 lysogenic with λ, λΔexo-xis, Φ24B, or Φ24BΔexo-xis, at indicated times after treatment with 1 mM H2O2, as estimated by reverse transcription quantitative real-time PCR. The values obtained with untreated cells were used as calibrators and were subtracted from the values determined at particular time points; thus, the presented values indicate the induction of expression of tested genes. The presented results are mean values from 3 independent experiments (biological samples), with error bars indicating SD. The additional panel for E. coli (24B) represents the results with different scale.
Indications that overexpression of some genes from the exo-xis region of λ can influence host cell cycle and DNA replication have been reported previously [21, 22]. Suggestions that some genes of Φ24B prophage may affect host growth were also published [38]. However, the results described in this subsection demonstrate for the first time that the exo-xis region can significantly modulate one of global cellular responses, the S.O.S. response, after treatment with hydrogen peroxide.
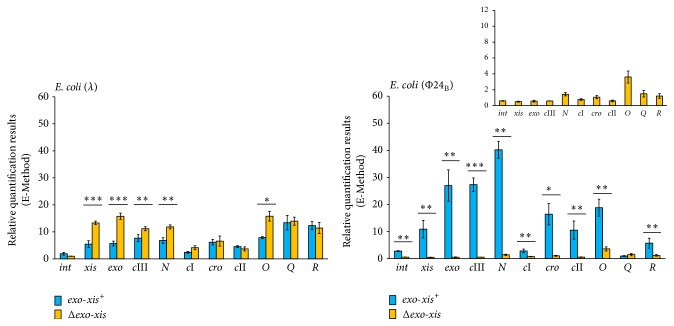
3.4. Expression of Crucial Phage Genes Is Dramatically Decreased after Treatment of Lysogenic Cells with Hydrogen Peroxide in the Absence of the Exo-Xis Region in Φ24B Prophage
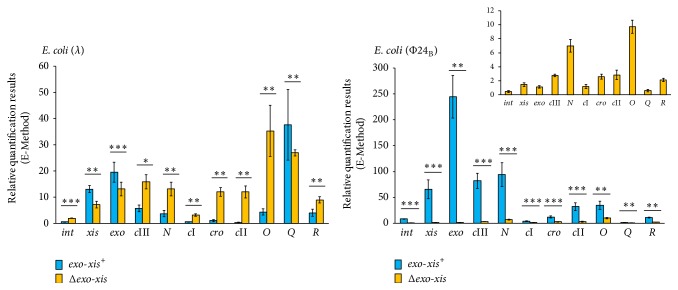
Expression of phage genes, crucial for the regulatory processes and lytic development, has been tested under the same conditions as described in the preceding subsection. The specific conditions and time after addition of hydrogen peroxide into the cell culture at which samples were withdrawn were chosen on the basis of similar experiments reported previously [26]. Interestingly, different effects of the deletion of the exo-xis region were observed for phages λ and 24B. In λ, deletion of genes and open reading frames located between exo and xis genes did not cause considerable effects on mRNA levels for xis, exo, and Q, whereas expression of int, cIII, N, cI, cro, cII, O, and R was enhanced upon treatment with hydrogen peroxide (Figure 6). Completely different results were obtained when Φ24B and Φ24BΔexo-xis lysogens were studied; namely, expression of all tested genes was drastically impaired in hydrogen peroxide-treated bacteria in the absence of the exo-xis region on the prophage (Figure 6).
Figure 6.
Expression of selected bacteriophage genes in E. coli MG1655 lysogenic with λ or Φ24B, either wild-type (blue columns) or Δexo-xis (yellow columns) at 160 min after treatment with 1 mM H2O2, as estimated by reverse transcription quantitative real-time PCR. The values obtained with untreated cells were used as calibrators and were subtracted from the values determined at particular time points; thus, the presented values indicate the induction of expression of tested genes. The presented results are mean values from 3 independent experiments (biological samples), with error bars indicating SD. The additional panel for E. coli (24B) represents the results of Δexo-xis variant with different scale, due to very small values measured. Statistically significant differences (in t-test) are marked as follows: ∗ p < 0.05, ∗∗ p < 0.01, and ∗∗∗ p < 0.001.
While negative regulation of transcription from cII-dependent promoters by overexpression of the exo-xis region has been reported previously in phage λ [23], this study demonstrated for the first time significant effects of this region on expression of a battery of phage genes under conditions of the oxidative stress. The results presented in Figure 6 for phage λ are compatible with those published previously (though obtained with different methods) [23], as overexpression of the exo-xis region had opposite effects to those observed in its absence. On the other hand, severely impaired expression of all tested phage genes in Φ24BΔexo-xis was unexpected. However, these results (Figure 6) can explain a strong defect in the induction of Φ24BΔexo-xis prophage (and perhaps further lytic development) by hydrogen peroxide (Figure 2). Similarly, drastic differences between effects of Δexo-xis mutations on hydrogen peroxide-mediated prophage induction between λ and 24B (Figure 2) can be ascribed to opposite regulation of expression of phage genes in the absence of the exo-xis region.
3.5. Effects of the Exo-Xis Region on Expression of Host and Phage Gene in UV-Irradiated Lysogenic Cells
Experiments analogous to those described in two preceding subsections were performed with lysogenic cells irradiated with UV. Interestingly, in both λ and 24B phages, deletion of the exo-xis region caused only moderate effects on expression of most genes from the S.O.S. regulon (Figure 7), contrary to hydrogen peroxide-treated bacteria where the differences were significantly higher (compare Figures 5 and 7). The exceptions in UV-irradiated cells were rpoD, rpoH, and rpoS genes in λ and rpoH and rpoS in 24B, whose expressions were at considerably lower level in the absence of the exo-xis region (Figure 7). One should also note that the induction of the S.O.S. regulon with UV irradiation was quicker than that with hydrogen peroxide. These results indicate that the influence of the exo-xis region on the S.O.S. response is particularly well pronounced under conditions of the oxidative stress.
Figure 7.
Expression of genes from the S.O.S. regulon in E. coli MG1655 lysogenic with λ, λΔexo-xis, Φ24B, or Φ24BΔexo-xis, at indicated times after UV irradiation (50 J/m2), as estimated by reverse transcription quantitative real-time PCR. The values obtained with untreated cells were used as calibrators and were subtracted from the values determined at particular time points; thus, the presented values indicate the induction of expression of tested genes. The presented results are mean values from 3 independent experiments (biological samples), with error bars indicating SD.
Unlike the S.O.S. regulon expression, levels of mRNAs of bacteriophage genes in UV-irradiated cells were affected similarly to those in hydrogen peroxide-treated lysogenic bacteria by the absence of the exo-xis region (Figure 8). Again, although some differences were observed between λ and λΔexo-xis, the differences between Φ24B and Φ24BΔexo-xis were dramatic. This indicates that the influence of the exo-xis region on expression of phage genes after prophage induction does not depend on the induction agent.
Figure 8.
Expression of selected bacteriophage genes in E. coli MG1655 lysogenic with λ or Φ24B, either wild-type (blue columns) or Δexo-xis (yellow columns) at 160 min after UV irradiation (50 J/m2), as estimated by reverse transcription quantitative real-time PCR. The values obtained with untreated cells were used as calibrators and were subtracted from the values determined at particular time points; thus, the presented values indicate the induction of expression of tested genes. The presented results are mean values from 3 independent experiments (biological samples), with error bars indicating SD. The additional panel for E. coli (24B) represents the results of Δexo-xis variant with different scale, due to very small values measured. Statistically significant differences (in t-test) are marked as follows: ∗ p < 0.05, ∗∗ p < 0.01, and ∗∗∗ p < 0.001.
4. Conclusions
The exo-xis region is necessary for effective, RecA-dependent induction of Shiga toxin-converting bacteriophage Φ24B under conditions of the oxidative stress. In hydrogen peroxide-treated E. coli, this region positively influences expression of the S.O.S. regulon in both Φ24B and λ lysogens and expression of phage genes crucial for lytic development (particularly xis, exo, N, cro, O, Q, and R) in Φ24B, but not in λ. Since the oxidative stress appears to be the major cause of induction of Shiga toxin-converting prophages during infections of human intestine by enterohemorrhagic E. coli (EHEC), the exo-xis region and/or products of its expression might be considered as potential targets for anti-EHEC drugs.
Acknowledgment
This work was supported by the National Science Center (Poland) Grant no. 2013/09/B/NZ2/02366 to Alicja Węgrzyn.
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Katarzyna Licznerska and Aleksandra Dydecka contributed equally to this work.
References
- 1.Gyles C. L. Shiga toxin-producing Escherichia coli: an overview. Journal of Animal Science. 2007;85(13, supplement):E45–E62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 2.Hunt J. M. Shiga toxin-producing Escherichia coli (STEC) Clinics in Laboratory Medicine. 2010;30(1):21–45. doi: 10.1016/j.cll.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razzaq S. Hemolytic uremic syndrome: an emerging health risk. American Family Physician. 2006;74(6):991–998. [PubMed] [Google Scholar]
- 4.Law D. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli . Journal of Applied Microbiology. 2000;88(5):729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 5.Beutin L., Martin A. Outbreak of shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. Journal of Food Protection. 2012;75(2):408–418. doi: 10.4315/0362-028x.jfp-11-452. [DOI] [PubMed] [Google Scholar]
- 6.Bloch S. K., Felczykowska A., Nejman-Falenczyk B. Escherichia coli O104:H4 outbreak—have we learnt a lesson from it? Acta Biochimica Polonica. 2012;59(4):483–488. [PubMed] [Google Scholar]
- 7.Karch H., Denamur E., Dobrindt U., et al. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Molecular Medicine. 2012;4(9):841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laing C. R., Zhang Y., Gilmour M. W., et al. A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0037362.e37362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellmann A., Harmsen D., Cummings C. A., et al. Prospective genomic characterization of the german enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0022751.e22751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werber D., Krause G., Frank C., et al. Outbreaks of virulent diarrheagenic Escherichia coli—are we in control? BMC Medicine. 2012;10, article 11 doi: 10.1186/1741-7015-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison H. E. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiology. 2007;2(2):165–174. doi: 10.2217/17460913.2.2.165. [DOI] [PubMed] [Google Scholar]
- 12.Loś J. M., Loś M., Wegrzyn G. Bacteriophages carrying Shiga toxin genes: genomic variations, detection and potential treatment of pathogenic bacteria. Future microbiology. 2011;6(8):909–924. doi: 10.2217/fmb.11.70. [DOI] [PubMed] [Google Scholar]
- 13.Ptashne M. A Genetic Switch: Phage Lambda Revisited. 3rd. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 14.Wegrzyn G., Wegrzyn A. Genetic switches during bacteriophage λ development. Progress in Nucleic Acid Research and Molecular Biology. 2005;79:1–48. doi: 10.1016/s0079-6603(04)79001-7. [DOI] [PubMed] [Google Scholar]
- 15.Węgrzyn G., Licznerska K., Węgrzyn A. Phage λ-new insights into regulatory circuits. Advances in Virus Research. 2012;82:155–178. doi: 10.1016/b978-0-12-394621-8.00016-9. [DOI] [PubMed] [Google Scholar]
- 16.Imamovic L., Muniesa M. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0032393.e32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Łoś J. M., Łoś M., Węgrzyn A., Węgrzyn G. Altruism of Shiga toxin-producing Escherichia coli: recent hypothesis versus experimental results. Frontiers in Cellular and Infection Microbiology. 2013;2, article 166 doi: 10.3389/fcimb.2012.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner P. L., Acheson D. W. K., Waldor M. K. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli . Infection and Immunity. 2001;69(3):1934–1937. doi: 10.1128/iai.69.3.1934-1937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Łoś J. M., Łoś M., Węgrzyn G., Węgrzyn A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microbial Pathogenesis. 2009;47(6):289–298. doi: 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Łoś J. M., Łoś M., Wȩgrzyn A., Wȩgrzyn G. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunology and Medical Microbiology. 2010;58(3):322–329. doi: 10.1111/j.1574-695x.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- 21.Kourilsky P., Knapp A. Lysogenization by bacteriophage lambda. III.—multiplicity dependent phenomena occuring upon infection by lambda. Biochimie. 1975;56(11-12):1517–1523. doi: 10.1016/s0300-9084(75)80275-6. [DOI] [PubMed] [Google Scholar]
- 22.Sergueev K., Court D., Reaves L., Austin S. E. coli cell-cycle regulation by bacteriophage λ . Journal of Molecular Biology. 2002;324(2):297–307. doi: 10.1016/s0022-2836(02)01037-9. [DOI] [PubMed] [Google Scholar]
- 23.Łoś J. M., Łoś M., Węgrzyn A., Węgrzyn G. Role of the bacteriophage λexo-xis region in the virus development. Folia Microbiologica. 2008;53(5):443–450. doi: 10.1007/s12223-008-0068-0. [DOI] [PubMed] [Google Scholar]
- 24.Bloch S., Nejman-Faleńczyk B., Łoå J. M., et al. Genes from the exo-xis region of λ and Shiga toxin-converting bacteriophages influence lysogenization and prophage induction. Archives of Microbiology. 2013;195(10-11):693–703. doi: 10.1007/s00203-013-0920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan J. J., Smirnova E., Khazai S., Evanics F., Maxwell K. L., Donaldson L. W. The solution structures of two prophage homologues of the bacteriophage λ Ea8.5 protein reveal a newly discovered hybrid homeodomain/zinc-finger fold. Biochemistry. 2013;52(21):3612–3614. doi: 10.1021/bi400543w. [DOI] [PubMed] [Google Scholar]
- 26.Bloch S., Nejman-Faleńczyk B., Dydecka A., et al. Different expression patterns of genes from the Exo-Xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Φ24B following infection or prophage induction in Escherichia coli . PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0108233.e108233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen K. F. The Escherichia coli K-12 ‘wild types’ W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. Journal of Bacteriology. 1993;175(11):3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allison H. E., Sergeant M. J., James C. E., et al. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infection and Immunity. 2003;71(6):3409–3418. doi: 10.1128/iai.71.6.3409-3418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nejman-Faleńczyk B., Bloch S., Licznerska K., et al. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24B . Scientific Reports. 2015;5 doi: 10.1038/srep10080.10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Łoś J. M., Golec P., Wȩgrzyn G., Wȩgrzyn A., Łoś M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Applied and Environmental Microbiology. 2008;74(16):5113–5120. doi: 10.1128/aem.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quillardet P., Huisman O., D'Ari R., Hofnung M. SOS chromotest, a direct assay of induction of an SOS function in Escherichia coli K-12 to measure genotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(19):5971–5975. doi: 10.1073/pnas.79.19.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fish F., Lampert I., Halachmi A. The SOS chromotest kit: a rapid method for the detection of genotoxicity. Toxicity Assessment. 1987;2(2):135–147. [Google Scholar]
- 33.Herman-Antosiewicz A., Wȩgrzyn G. Regulation of copy number and stability of phage λ derived pTCλ1 plasmid in the light of the dimer/multimer catastrophe hypothesis. FEMS Microbiology Letters. 1999;176(2):489–493. doi: 10.1016/s0378-1097(99)00278-5. [DOI] [PubMed] [Google Scholar]
- 34.Strauch E., Hammerl J. A., Konietzny A., et al. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infection and Immunity. 2008;76(12):5466–5477. doi: 10.1128/iai.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowicki D., Bloch S., Nejman-Faleńczyk B., Szalewska-Pałasz A., Węgrzyn A., Węgrzyn G. Defects in RNA polyadenylation impair both lysogenization by and lytic development of Shiga toxin-converting bacteriophages. Journal of General Virology. 2015;96(7):1957–1968. doi: 10.1099/vir.0.000102. [DOI] [PubMed] [Google Scholar]
- 36.Finn R. D., Bateman A., Clements J., et al. Pfam: the protein families database. Nucleic Acids Research. 2014;42(1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koudelka A. P., Hufnagel L. A., Koudelka G. B. Purification and characterization of the repressor of the shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. Journal of Bacteriology. 2004;186(22):7659–7669. doi: 10.1128/JB.186.22.7659-7669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley L. M., Veses-Garcia M., Hillman J. D., Handfield M., McCarthy A. J., Allison H. E. Identification of genes expressed in cultures of E. coli lysogens carrying the Shiga toxin-encoding prophage Φ24B. BMC Microbiology. 2012;12, article 42 doi: 10.1186/1471-2180-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]