Abstract
With extended lifespans in modern humans, menopause has become a significant risk factor for depression, anxiety, loss of cognitive functions, weight gain, metabolic disease, osteoporosis, cardiovascular disease and neurodegenerative diseases. Clinical studies have found beneficial neural effects of ovarian steroid hormone therapy (HT) during the menopausal transition and data are emerging that it can be continued long-term. To further understand molecular underpinnings of the clinical studies, we used qRT-PCR to examine gene expression in the serotonergic dorsal raphe of old (>18 yrs) rhesus macaques, focusing on genes related to depression, cellular resilience and neurodegenerative diseases. The animals were ovariectomized (Ovx, surgically menopausal) and subjected to either estradiol or estradiol plus progesterone HT, or to placebo, starting 2 months after Ovx and continuing for ~4 years. Significant changes were observed in 36 out of 48 genes examined that encode proteins supporting serotonin neurotransmission, synapse assembly, glutamate neurotransmission, DNA repair, chaperones, ubiquinases and transport motors, as well as genes encoding proteins that have potential to delay the onset of neuropathologies. The data reveal important gene targets for chronic HT that contribute to neural health. Alternatively, the loss of ovarian steroids may lead to loss of functions at the gene level that contribute to many of the observable neural deficits following menopause.
Keywords: serotonin, estrogen, progesterone, dorsal raphe, macaques, glutamate, synapse, DNA repair, chaperones, ubiquinases, transport, neurodegeneration
Introduction
Menopause in women and female rhesus macaques is associated with a marked decline in the production and secretion of 17β-estradiol (E) and progesterone (P) from the ovaries (Burger, et al., 2002; Downs and Urbanski, 2006; McKinlay, et al., 2008). This attenuation of circulating concentrations of E and P adversely impacts many physiological processes, and is thought to play a major role in the etiology of age-related pathologies, such as hot flushes and disrupted sleep-wake cycles (Mittelman-Smith, et al., 2012; Sarti, et al., 2005). Depression, anxiety and cognitive loss also accompany the onset of menopause in a significant number of women (Epperson, et al., 2012; Gordon, et al., 2015; Maki, et al., 2010). Bio-identical hormone therapy (HT) during peri-menopause, or the menopausal transition, alleviates or attenuates many of the neural dysfunctions that occur during menopause (Heikkinen et al, 2006; Schmidt and Rubinow, 2009; Epperson et al, 2012), but it is not possible to probe the underlying neural mechanisms in living humans.
Ovariectomy of nonhuman primates has provided a reasonable model of abrupt human menopause. Studies have shown that E replacement in surgically menopausal macaques improves or maintains cognitive function over placebo controls, with corresponding improvement in synapses, dendritic spines and mitochondria in prefrontal cortex (Tang et al., 2004; Tinkler et al., 2005; Hao et al., 2006; Hao et al., 2007; Voytko et al., 2009; Hara et al., 2014). However, clinically relevant, continuous HT protocols failed to improve cognitive function or spinogenesis in prefrontal cortex of old female macaques, raising questions regarding the best method of delivery (Ohm et al., 2012; Baxter et al., 2013).
Many of the neural symptoms of menopause have been linked to serotonin in both human and animal studies (Albert, et al., 2014; Amin, et al., 2006; Araragi and Lesch, 2013; Diaconescu, et al., 2011; Stollstorff, et al., 2013). Serotonin neurons in adult ovariectomized (Ovx) female macaques express nuclear estrogen receptor beta (ERβ) at the same level, with or without HT (Gundlah, et al., 2000; Gundlah, et al., 2001). E, acting through ERβ increased expression of progesterone (P) receptors (PR) in serotonin neurons (Bethea, 1994). In adult short-term Ovx monkeys (age 6–12 years) with 1 month of E with or without supplemental P, significantly altered gene expression in serotonin neurons in a manner that would lead to increased serotonin neurotransmission (Bethea, et al., 2002), increased proliferation of dendritic spines (Bethea and Reddy, 2010; Rivera and Bethea, 2012), increased glutamate receptors (Bethea and Reddy, 2012b), increased synaptogenesis (Bethea and Reddy, 2012a), and increased neuronal resilience (Bethea and Reddy, 2015). These functions have largely been confirmed at the transmitter or protein levels (Sanchez et al., 2013; Rivera and Bethea, 2012; Rivera and Bethea, 2013; Lima and Bethea, 2009).
The goal of this study was to determine if old animals responded as well as young animals to HT, albeit in a chronic, but clinically relevant model. Therefore, genes were preselected that responded to E or E+P in serotonin neuron-enriched preparations from young adult animals treated for 1 month with hormone therapy (Bethea and Reddy, 2008; 2010; 2012a; 2012b; 2015; Rivera and Bethea, 2012). The serotonin related genes were selected from our knowledge base and earlier experiments demonstrating their regulation by ovarian steroids at gene and protein levels. Dendritic spines are the elementary structural units of neuronal plasticity and their proliferation and stabilization involve components of synapse assembly and glutamate neurotransmission. Therefore, genes were selected that related to dendritic spine proliferation, glutamate transmission and synapse assembly, and that had shown regulation by ovarian steroids in young adult females. We showed that serotonin neurons in OVX animals had more DNA fragmentation (TUNEL positive) than in animals with hormone therapy for 1 month. DNA fragmentation and other neurodegenerative mechanisms are controlled by DNA repair enzymes, chaperones (protein folding), ubiquinases, transportation motors and mutations within specific genes. Hence, we selected genes related to these categories and that showed regulation by ovarian hormones in young adult females. This paradigm is also similar to the “Econtinuous + Pcyclic” treatment group in NHP studies by Ohm et al. (2012) and Baxter et al. (2013).
Materials and Methods
This experiment was approved by the IACUC of the Oregon National Primate Research Center and conducted in accordance with the 2011 Eighth Edition of the National Institute of Health Guide for the Care and Use of Laboratory Animals. All animals were born at ONPRC, were aged between 18–22 years, weighed between 5 and 8 kg, and were in good health. The animals were housed indoors under controlled environmental conditions: 24 C temperature; 12-h light, 12-h dark photoperiods (lights on at 0700 h); regular meals at 0800 and 1500 h (Purina High Protein Monkey Chow; Purina Mills, Inc., St. Louis, MO) supplemented with fresh fruit and vegetables, and fresh drinking water available ad libitum.
Animals and Treatments
Twelve ovarian intact, old female rhesus monkeys (Macaca mulatta) were dedicated to this project from a larger cohort of study animals. The ovarian intact, aged animals were trained in a Delayed Response (DR) task (Voytko, 2000; Rapp et al., 2003) and a Spatial Foodport Maze task (Voytko, 2002; Haley, et al., 2009).
Once delayed response training was achieved, the animals were Ovx by ONPRC surgical personnel according to accepted veterinary surgical protocol. After two months of Ovx, subgroups of animals received placebo, E alone or E+P. They were tested for 2 years. The results of the cognitive testing during the first 12 months after Ovx have been published in abstract form (Renner et al., 2010) and are in preparation for publication in a separate article.
The E-treated monkeys were implanted with one 4-cm E-filled Silastic capsules (i.d. 0.132 in.; o.d. 0.183 in.; Dow Corning, Midland, MI, USA). The capsule was filled with crystalline estradiol (1,3,5(10)-estratrien-3, 17-b-diol; Steraloids, Wilton, NH, USA). The E + P-treated animals received an E-filled capsule and were administered micronized progesterone orally for 11 days out of every month to model the menstrual cycle rather than various clinical prescriptions. The capsules were placed in the periscapular area under ketamine anesthesia (ketamine HCl, 10 mg/kg, Fort Dodge Laboratories, Fort Dodge, IA, USA). The treatments were maintained for ~4 years. Placebo controls received one empty Silastic capsule.
The E capsules were designed to last up to 1 year and to maintain serum E concentrations between 100–200 pg/mL, which is similar to concentrations during the mid-to-late follicular phase of the menstrual cycle. They were replaced annually, to ensure sustained long-term delivery of the steroid for the entire duration of the study; the untreated Ovx animals maintained the same empty capsules throughout. Serum E and P concentrations were measured at various times across the 4-year experiment to confirm that the target hormone concentrations were being maintained in the old Ovx animals.
At regular intervals during the treatment period, circadian activity was measured with accelerometers and included day activity, dark:light activity ratio, sleep latency and wake bouts. At regular intervals, the animals were vaccinated and the T cell response was determined. MRIs on brain morphology were regularly performed.
Clinical Observations
Biannual physical exams were conducted by a clinical veterinarian throughout the study. At the beginning of the study, the uterus was noted to be small and difficult to palpate; the cervix was small in all animals. The uterus and cervix remained small throughout the study in the placebo- and E+P-treated animals. In the E-treated animals in years 2–4, the uterus was noted to be palpable, enlarged, of firm consistency and no cysts were observed with ultrasound. One uterus was irregularly shaped. At the same time points, the cervix was noted to be enlarged. Menses were irregular and up to 10 days long in one animal, but not heavy. The pathology report indicated uterine enlargement with endometrial hyperplasia. One animal had leiomyoma.
Steroid Hormone Assays
The E and P assays were performed by the ONPRC Endocrine Technology and Support Core using a chemilluminescence-based automatic clinical platform (Immulite 2000; Siemens Healthcare Diagnostics, Deerfield, IL, USA). The sensitivity limit of the E assay was 20 pg/mL, and the sensitivity limit of the P assay was 0.2 ng/mL. The intra-assay and inter-assay coefficients of variation were all less than 15%. Before these analyses, measurements of E and P on this platform were compared to traditional RIAs as previously reported (Bethea et al., 2005). The E + P treatment regimen has been shown to cause proliferation and differentiation of the uterine endometrium in a manner similar to a normal 28-day menstrual cycle (Brenner and Slayden, 1994).
Euthanasia
The monkeys were euthanized at the end of the treatment periods according to procedures recommended by the 2013 Edition of the American Veterinary Medical Association Guidelines for the Euthanasia of Animals. Each animal was sedated with ketamine, administered pentobarbital (30 mg/kg, i.v.), and exsanguinated by severance of the descending aorta. The E+P group was euthanized at the end of the P treatment period.
Tissue preparation
The left ventricle of the heart was cannulated, and the head of each animal was flushed with 1 L of 0.9% saline (made with DEPCtreated water [0.1% diethyl pyrocarbonate] to minimize RNase contamination) via a vascular catheter. The brain was removed from the cranium and dissected into blocks. The midbrain block was bisected at the midline and the dorsal raphe was micro dissected from one half. The half raphe was then frozen in liquid nitrogen and stored at −80 °C. The decussation of the cerebellar peduncles and the central canal were used as dorsal-ventral borders and the edges of the periaqueductal gray were aligned as lateral borders. The block extended from the appearance of the dorsal raphe nucleus to the loss of the decussation.
qRT-PCR assays
RNA was obtained from the microdissected raphe blocks using TriReagent and further cleaned with a Qiagen RNAeasy column (Velencia, CA). The quality of the RNA from the Qiagen column was examined on an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) and found acceptable equivalent within all individuals. Reverse transcription and complementary DNA (cDNA) synthesis was performed using Oligo-dT 15 and Random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA) plus Superscript III reverse transcriptase (200 U/μg of RNA, Invitrogen Life Technologies) at 42°C for 1 hr. cDNA was treated with RNAse H to digest dsDNA. Total RNA from the individual animals and from a standard pool of rhesus tissues was transcribed and stored at −80°C as cDNA at a concentration of 250 ng/μl.
Custom primers from the ABI Rhesus Monkey Library (Applied Biosystems, Foster City, CA) were used in individual assays conducted in 96-well plates, or preloaded on custom 384-well Taqman cards for qPCR. The primers utilize a 5′ fluorescent reporter, FAM (Fluorescein amidite; Molecular Probes, Eugene, OR) and a 3′ quencher, TAMRA (tetramethylrhodamine), which improves sensitivity. The exact primer sequences are proprietary, but the gene names, symbols, AB assay IDs and NCBI gene reference information are shown in Table 1.
Table 1.
The gene name, symbol, ABI Taqman Assay Identifier (ID) and NCBI reference sequence of the primers used in this study.
| Gene Name | Gene Symbol | Assay ID | NCBI Gene Reference |
|---|---|---|---|
| Serotonin regulation | |||
| Fifth ewing variant (PET1 in rodent) | FEV | Rh02872593 | XM_001095962.2 |
| Tryptophan hydroxylase 2 | TPH2 | Rh02788839 | NM_001039946.1 |
| Serotonin reuptake transporter | SLC6A4 | Rh02787892 | NM_001032823.1 |
| Serotonin receptor 1A | 5HT1A | Rh02902683 | NM_001198700.1 |
| Corticotropin releasing hormone receptor type 1 | CRHR1 | Rh02787591 | NM_001032803.1 |
| Corticotropin releasing hormone receptor type 2 | CRHR2 | Rh01120857 | XM_001085987.2 |
| Urocortin 1 (stresscopin) | UCN1 | Rh03986716 | NM_001265661.1 |
| Synapse assembly | |||
| Neuroligin 3 | NLGN3 | Rh03986723 | XM_001111843.1 |
| Neurotrophic tyrosine kinase, receptor, type 2, trk-B | NTRK | Rh02831788 | NM_001261297.1 |
| Synaptosomal-associated protein, 25kDa | SNAP25 | Rh03043048 | NM_001032864.1 |
| Neural cell adhesion molecule 1 | NCAM1 | Rh00354742 | XM_001083366.2 |
| Glutamate Receptors | |||
| Glutamate [NMDA2A] receptor subunit epsilon 1 | GRIN2A | Rh03986722 | XM_001105525.1 |
| Glutamate receptor, ionotropic, AMPA 2 | GRIA2 | Rh02790124 | NM_001039946.1 |
| Glutamate receptor, metabotropic 1 | GRM1 | Rh01068380 | XM_001085942.1 |
| DNA Repair | |||
| Nibrin, part of double strand break repair complex | NBS1 | Rh01039845 | XM_001085033.1 |
| Nth endonuclease III-like 1, DNA N-glycosylas | NTHL | Rh00959765 | XM_001082772.2 |
| Ligase IV, DNA double strand break repair | LIG4 | Rh04269856 | XM_001084107 |
| General transcription factor IIH, polypeptide 5, subunit of TFIIH | GTF2H5 | Rh02792678 | NM_001190969.1 |
| RAD23 homolog A, involved in nucleotide excision repair (NER) | RAD23A | Rh00908422 | XM_001110103.2 |
| Apurinic/apyrimidinic endonuclease 1 | APEX1 | Rh02793202 | XM_001090240.2 |
| Proliferating cell nuclear antigen, BER gap filling | PCNA | Rh02806147 | XM_001115746.2 |
| Split hand/foot malformation (ectrodactyly) type 1 | SHFM1 | Rh02860294 | XM_001091523.2 |
| Chaperones | |||
| Heat shock protein 90kD | HSP90B1 | Rh02790147 | NM_001195524.1 |
| Heat shock protein 60kD protein1 | HSPD1 | Hs01036753 | NM_002156.4 |
| Heat shock protein 27kD protein 1 | HSPB1 | Rh02980144 | XM_001109274.2 |
| Ubiquinases | |||
| Ubiquitin-like modifier activating enzyme 1 | UBE1 | Hs01566989 | NM_024818.3 |
| Ubiquitin-conjugating enzyme E2D 3 | UBE2D3 | Hs01518517 | NM_181892.2 |
| ubiquitin protein ligase E3A | UBE3A | Rh00963674 | XM_002804686.1 |
| Transport | |||
| Kinesin family member 5B | KIF5B | Rh01037194 | XM_002805607.1 |
| Dynactin 4 (p62) | DCTN4 | Rh01092757 | XM_001109230.2 |
| Dynein, light chain, LC8-type 1 | DYNCL1 | Hs00867659 | NM_001037495.1 |
| Microtubule-associated protein tau | MAPT | Rh04269822 | XM_001115803.2 |
| Disease specific | |||
| α-secretase cleavage of APP precludes amyloid beta formation | ADAM10 | Rh01109565 | XM_001097016.2 |
| α-synuclein, presynaptic signaling, membrane trafficking | SNCA | Rh1103386 | XM_001095402.2 |
| Amyloid beta (A4) precursor | APP | Rh01552279 | XM_002803170.1 |
| Presenilin (γ-secretase component)produces toxic α-beta 40–42 | PSEN1 | Rh01552279 | XM_002803170.1 |
Four concentrations of the standard pool and 7.5 ng of cDNA from the experimental samples were loaded into each well. There was a log linear increase in fluorescence detected as the concentration of amplified double-stranded product cDNA increased during the reaction. The fluorescence was detected as cycle threshold (Ct) with an ABI 7900 thermal cycler (Applied Biosystems Inc.) during 40 cycles. The slope of the standard curve was used to calculate the relative picograms of each transcript in the RNA extracted from the laser-captured pools. Then, the ratio of each transcript to GAPDH was calculated for each sample.
We chose to examine a technically reasonable number of genes in triplicate that would be representative of complex functions. The genes selected were regulated by hormone therapy in laser captured serotonin neurons from adult Ovx females ± hormone therapy for 1 month (Bethea and Reddy, 2010; 2012a; 2012b; 2015; Rivera and Bethea, 2012).
Genes examined and reported:
Serotonin regulation- FEV (Fifth Ewing variant, determines serotonin phenotype), TPH2 (tryptophan hydroxylase, rate limiting enzyme in serotonin synthesis, SERT (serotonin reuptake transporter), 5HT1A (serotonin 1A receptor), CRHR1 (corticotropin releasing hormone receptor type 1), CRHR2 (corticotropin releasing hormone receptor type 2) and UNC1 (urocortin 1, stresscopin).
Synapse assembly- NLGN3 (neuroligin 3), NTRK2 (TrkB, neurotrophic tyrosine kinase), SNAP25 (synaptosomal-associated protein, 25kd), and NCAM (neural cell adhesion molecule).
Glutamate receptors- GRIN2A (NMDA receptor type 2A), GRIA2 (AMPA receptor type 2), and GRM1 (metabotropic glutamate receptor type 1).
DNA repair- related - NSB1 (nibrin, double strand break repair complex), NTHL1 (endonuclease family III), LIG4 (DNA double-strand break repair, nonhomologous end joining), GTF2H5 (subunit of transcripton/repair factor TFIIH), RAD23A (double strand break repair; DNA damage recognition), APEX1 (multifunctional DNA repair), PCNA (single base repair) and SHFM1- (homologous recombination double strand break).
Chaperones – heat shock proteins HSP90B1 (HSP90), HSPD1 (HSP60), HSPB1 (HSP27).
Ubiquinases - UBEA5 (type E1 activating), UBE2D3 (type E2 conjugating), UBE3A (type E3 ligase).
Transport motors - KIF5B (kinesin), DYNCL1 (dynein), DCTN4 (dynactin), MAPT (tau).
Disease specific genes - ADAM10 (α secretase), SCNA (α synuclein), APP (amyloid beta precursor protein), PSEN1 (presenilin1).
Genes examined and not reported due to no regulation were: NRXN1 (neurexin 1), ITGA8 (integrin A8), SCD2 (syndycan 2), GADD45 (DNA-damage inducible gamma), PARP2 (poly (ADP-ribosyl) transferase-like 2), POLG2 (mitochondrial DNA polymerase gamma subunit), SHPRH (motifs of DNA repair and helicases), XRCC1 (single strand break repair), ERCC1 (DNA nucleotide excision repair), HSPA8 (HSP70 chaperone), APOE (apolipoprotein E).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), GUSB, and PIAA were measured for reference. GAPDH was not significantly altered by treatment enabling its use for normalization of the expression results and comparison to previous articles.
Statistical analysis
The expression level of each gene was normalized against the level of GAPDH independently of the other triplicates. The mean of the normalized result of the triplicates was considered the individual animal’s result. The average of the results of individual animals in a group, or group mean, was used in statistical analyses. Thus, the standard error of the mean represents within group variance between animals.
The average relative expressions of the 3 groups from the qRT-PCR assays were compared with ANOVA followed by Newman-Keuls posthoc pairwise comparisons. Variance between animals is not unusual for this type of preparation, and more animals would reduce the chance of making a type 2 error. Therefore, negative results need further confirmation. Comparisons were considered significantly different when the chance of making a type 1 error was less than 5% (p<0.05). The expression of each gene was correlated to all the other genes. Those that exhibited a significant difference between groups, or a strong trend, with ANOVA and shown in the figures (n=36 genes) were further subjected to a multiple comparison False Discovery Rate (FDR) procedure. Prism 5.0 from Graph Pad (San Diego, CA) was used for statistical tests; Excel was used to obtain the correlation matrix and the Benjamini-Hochberg test for False Discovery Rate was applied using a freeware FDR calculator obtained from the Internet (http://www.sdmproject.com/utilities/?show=FDR). Four genes that demonstrated non-lines significant trends were further examined with Cohen’s d test for effect size.
Results
General
Gene expression was determined with qRT-PCR in a micro-dissected block of midbrain containing the serotoninergic dorsal raphe nucleus from old Ovx macaques treated with placebo (n=4), or E (n=3) or E+P (n=4) for ~4 years. The data was collated to obtain the mean ratio of gene/GAPDH for individual animals and then averages of multiple animals in each treatment group were statistically compared. A total of 48 mRNAs were examined and 36 mRNAs showed significant differences between the groups or trends that warranted reporting. The ANOVA significance is stated with posthoc differences defined from Newman-Keuls test (p<0.05) in the text below.
One-way ANOVA was applied across all groups to obtain the F and p-values. In the figures, letters on the histogram columns are derived from the post hoc Neuman-Keuls pairwise comparison. Groups labeled with different letters were significantly different in the post hoc test with p < 0.05. Columns labeled with crosses indicate the 2 groups were significantly different in a Student t test, whereas carrots indicate there was a large effect size that did not reach a level of significance.
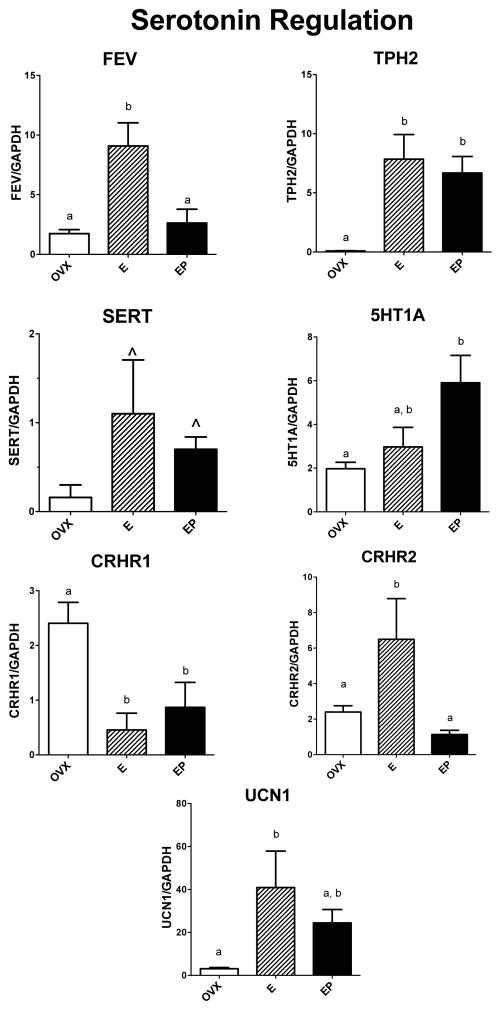
Serotonin Regulation
The relative mRNA expression of 7 examined genes known to have pivotal roles in the regulation of serotonin neurotransmission is illustrated in Figure 1. TPH2 mRNA was significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=10.6; p=0.006). FEV mRNA and CRHR2 (anxiolytic) mRNAs were significantly different across groups with E treatment increased over placebo and E+P treatments (F [2,10]=10.5; p=0.006 and F [2,10]=6.177; p=0.024 respectively). 5HT1A mRNA was different across groups with E+P treatment elevated over placebo (F [2,10]=5.36; p=0.033). UCN1 mRNA (stresscopin) was significantly different across groups with E treatment increased over placebo (F [2,10]=4.57; p=0.047). SERT mRNA trended toward elevation with E treatment over placebo (F [2,10] = 2.36; p =0.156). CRHR1 mRNA (anxiogenic) was significantly different across groups with E and E+P treatments decreased from placebo (F [2,10]=6.46; p=0.021).
Figure 1.
Histograms illustrating the expression of 5 genes that are involved in serotonin neurotransmission. FEV mRNA was significantly increased by E over placebo and E+P treatments. TPH2 mRNA was significantly increased by E and EP over placebo treatment. SERT mRNA was not different between the groups. 5HT1A mRNA was significantly increased by E+P over placebo and E treatments. CRHR1 mRNA (anxiogenic) was significantly decreased by E and E+P treatments compared to placebo. CRHR2 mRNA was significantly increased by E over placebo and E+P treatments. UNC1 mRNA (stresscopin) was significantly increased by E over placebo and there was no difference between E and E+P treatment.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
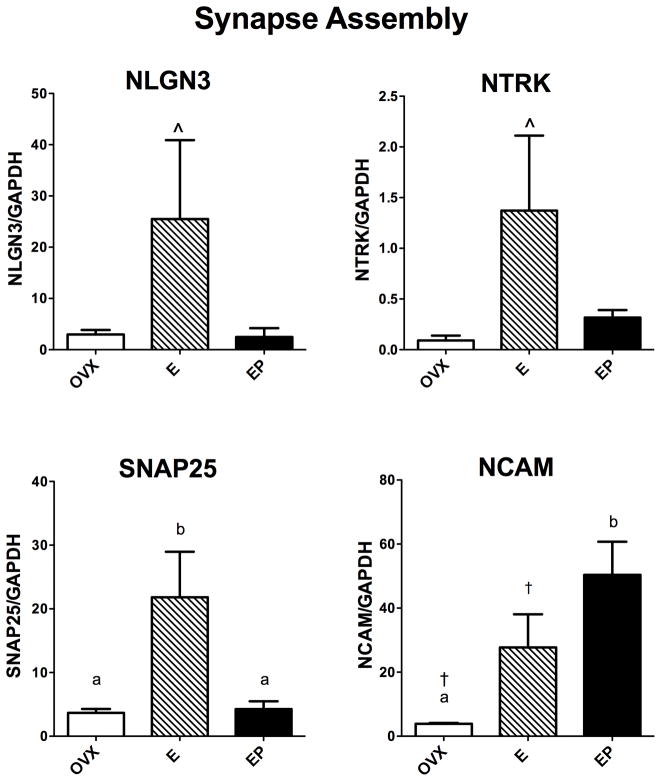
Synapse Assembly
The relative mRNA expression of 4 out of 7 genes examined that are known to have pivotal roles in synapse assembly and maintenance is illustrated in Figure 2. SNAP25 mRNA was significantly different across groups with E treatment increased over placebo and E+P treatments (F [2,10]=8.45; p=0.011). NCAM mRNA was significantly different across groups with E+P treatment increased over placebo (F [2,10]=8.97; p=0.009). NLGN3 and NTRK mRNAs trended toward elevation with E treatment over placebo (F [2,10]=3.08; p=0.102 and F [2,10]=3.66; p=0.074, respectively).
Figure 2.
Histograms illustrating the expression of 4 genes that are involved in synapse assembly. NGLN3 and NRK mRNAs showed a marked elevation with E treatment over placebo and E+P treatments, but neither increase reached statistical significance. SNAP25 mRNA was significantly increased by E over placebo or E+P treatments. NCAM mRNA was significantly increased by E with a t-test, and significantly increased by E+P treatment with ANOVA and the posthoc test.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
† - increased over placebo in Student’s t-test, p<0.05
^ - non-significant trend
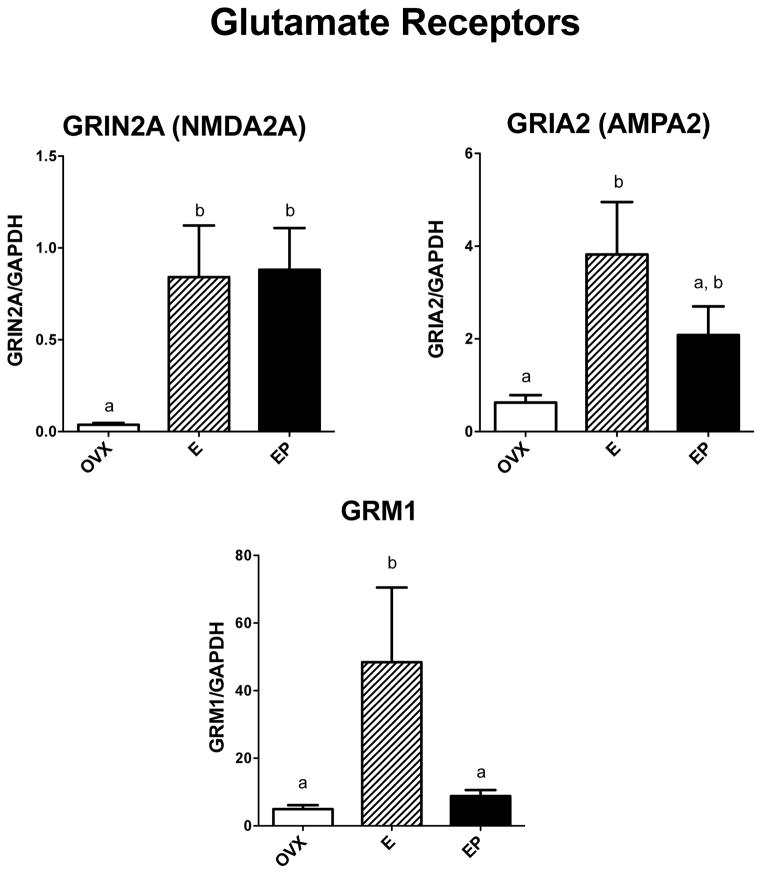
Glutamate (excitatory) receptors
Figure 3 illustrates the relative expression of 3 examined genes that represent three types of glutamate receptors, NMDA, AMPA and metabotropic receptors. NMDA2A (GRIN2A) mRNA was significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=6.36; p=0.022); AMPA2 (GRIA2) mRNA was significantly different across groups with E treatment increased over placebo (F [2,10]=5.57; p=0.030); and the metabotropic receptor (GRM1) mRNA was significantly different across groups with E treatment increased over placebo and E+P treatment (F [2,10]=5.08; p=0.038).
Figure 3.
Histograms illustrating the expression of 3 representative receptor-encoding genes involved in glutamate transmission. NMDA2a (GRIN2A) mRNA was significantly increased by E and E+P treatments over placebo treatment. AMPA2 (GRIA2) mRNA was significantly increased by E treatment compared to placebo, and E+P treatment was not different from E alone. GRM1 mRNA was significantly increased by E treatment compared to placebo and E+P treatment.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
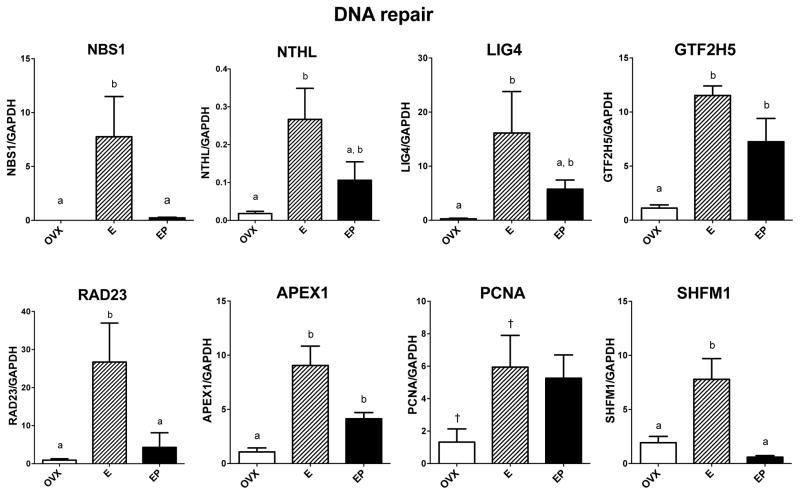
DNA repair
Eight of the 16 examined genes that relate to DNA repair showed elevated expression with one or both steroid treatments over placebo, and their responses are illustrated in Figure 4. GTF2H5 and APEX1 mRNAs were significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=5.32; p=0.034 and F [2,10]=17.8; p=0.001, respectively). NBS1, RAD23 and SHFM1 mRNAs were significantly different between groups with E treatment increased over placebo and E+P treatments (F [2,10]=6.10; p=0.025, F [2,10]=6.40; p=0.022, and F [2,10]=14.7; p=0.002, respectively). NTHL and LIG4 mRNAs were significantly different across groups with E treatment increased over placebo (F [2,10]=6.16; p=0.024 and F [2,10]=4.51; p=0.048, respectively). PCNA mRNA increased with E in a Student’s t-test. Both E and E+P treatments exhibited an elevated trend over placebo, with 91% confidence by ANOVA (F [2,10]=3.34; p=0.088).
Figure 4.
Histograms illustrating the expression of 8 genes encoding proteins that are involved in DNA repair. GTF2H5 and APEX1 mRNAs were significantly increased by E and E+P treatments over placebo. NBS1, NTHL, LIG4, RAD23, and SHFM1 mRNAs were significantly increased with E treatment and decreased by E+P to a degree that was significantly different from E alone (NBS1, RAD23, SHFM1) or not different from E alone (NTHL, LIG4). PCNA mRNA was significantly increased by E treatment over placebo in a Student’s t-test, but ANOVA did not disclose a difference between the groups.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
† - increased over placebo in Student’s t-test, p<0.05
Neuroendangerment
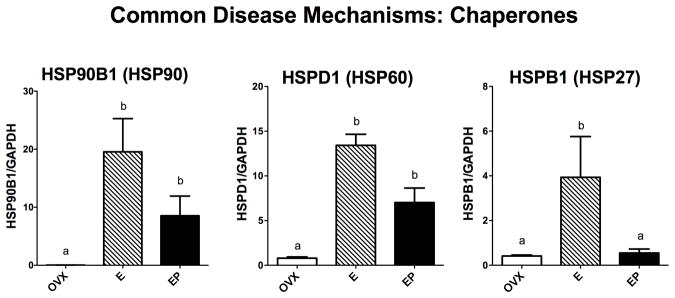
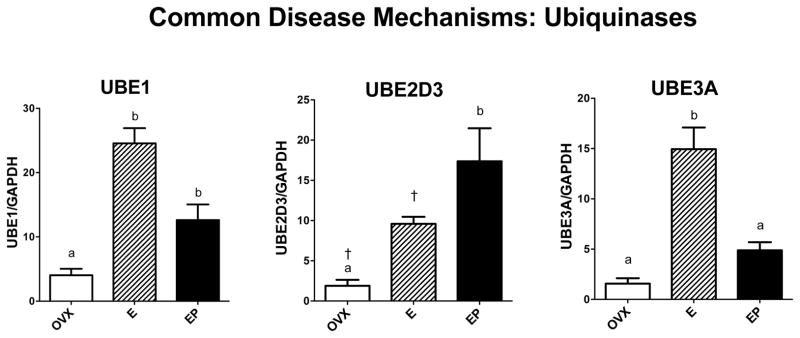
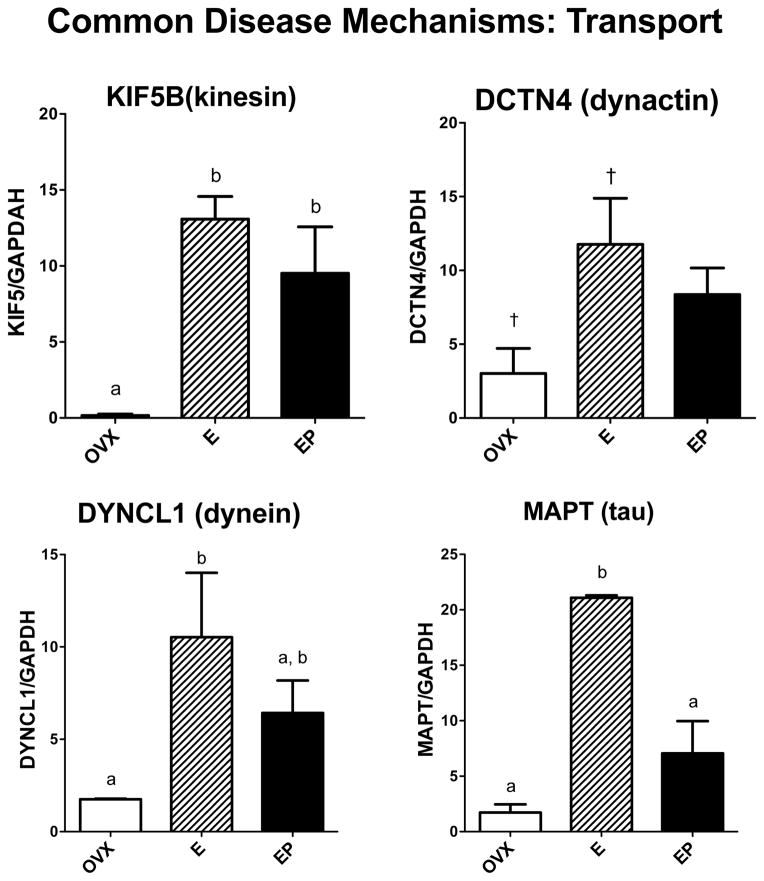
Genes related to other functions that play representative roles in protein folding, protein degradation and transport of substances from the neuronal cell body to terminal fields were examined. Sabotage of these functions is known to underlie different neurodegenerative diseases. However, none of these monkeys exhibited any symptomology of neurodegenerative disease so the gene expression data represent normal expression in the presence or absence of steroid treatment. Figure 5 illustrates mRNA expression of 3 of the 4 chaperones of the heat shock protein family that were examined. HSP90B1 mRNA levels were significantly different across groups with E treatment increased over placebo (F [2,10]=7.82; p=0.013). HSPD1 (HSP60) mRNA levels were significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=26.8; p=0.0003). HSPB1 (HSP27) mRNA levels were significantly different across groups with E treatment increased over placebo and E+P treatments (F [2,10]=5.126; p=0.037). Figure 6 illustrates mRNA expression of 3 genes examined that are representative of the 3 classes of ubiquinases, which govern protein degradation. UBE1 (activating) and UBE2D3 (conjugating) mRNA levels were significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=24.9; p=0.0004 and F [2,10]=8.94; p=0.009, respectively). UBE3A (ligase) mRNA levels were significantly different across groups with E treatment increased over placebo and E+P treatments (F [2,10]=32.8; p=0.0001). Figure 7 illustrates the 4 genes examined that relate to cytoskeletal elements underpinning transport. E increased the mRNA levels of all the examined genes, but the effect of E+P differed amongst the 4 genes. Kinesin (KIF5B) mRNA levels were significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=10.28; p=0.0062). Dynactin (DCTN4) mRNA levels exhibited a similar response with 94% confidence (F [2,10]=4.19; p=0.056). Tau (MAPT) mRNA levels were significantly different across groups with E treatment increased over placebo and E+P treatment groups (F [2,10]=24.7; p=0.0004). Dynein (DYNCL1) mRNA levels were significantly different across groups with E treatment increased over placebo (F [2,10]=4.88; p=0.041).
Figure 5.
Histograms illustrating the expression of 3 genes that code for chaperone proteins involved in protein folding. HSP90B1 and HSPD1 (HSP60) mRNAs significantly increased with E treatment over placebo and remained elevated or not different from E alone with E+P treatment. HSPB1 (HSP27) mRNAs significantly increased with E treatment over placebo and E+P treatment.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
Figure 6.
Histograms illustrating the expression of 3 genes that encode for ubiquinase proteins that designate and traffic other proteins for degradation. UBE1 mRNA significantly increased with E and E+P treatments over placebo. UBE2D3 mRNA also significantly increased with E by Student’s t-test and E+P by ANOVA over placebo. UBE3A mRNA significantly increased with E treatment over placebo and E+P treatments.
a, b – groups with the different are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
† - increased over placebo in Student’s t-test, p<0.05
Figure 7.
Histograms illustrating the expression of 3 genes that code for transport motor proteins and MAPT that codes for a cytoskeletal protein present in neurofibrillary tangles. KIF5B mRNA was significantly increased by E and E+P treatments over placebo. DCTN4 mRNA was significantly increased by E treatment over placebo in a Student’s t-test, but no differences were detected between the 3 groups by ANOVA. E alone significantly increased DYNCL1 mRNA over placebo and E+P treatment was not different from E alone. MAPT mRNA was significantly increased by E treatment compared to placebo and E+P treatments.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
† - increased over placebo in Student’s t-test, p<0.05
Disease specific
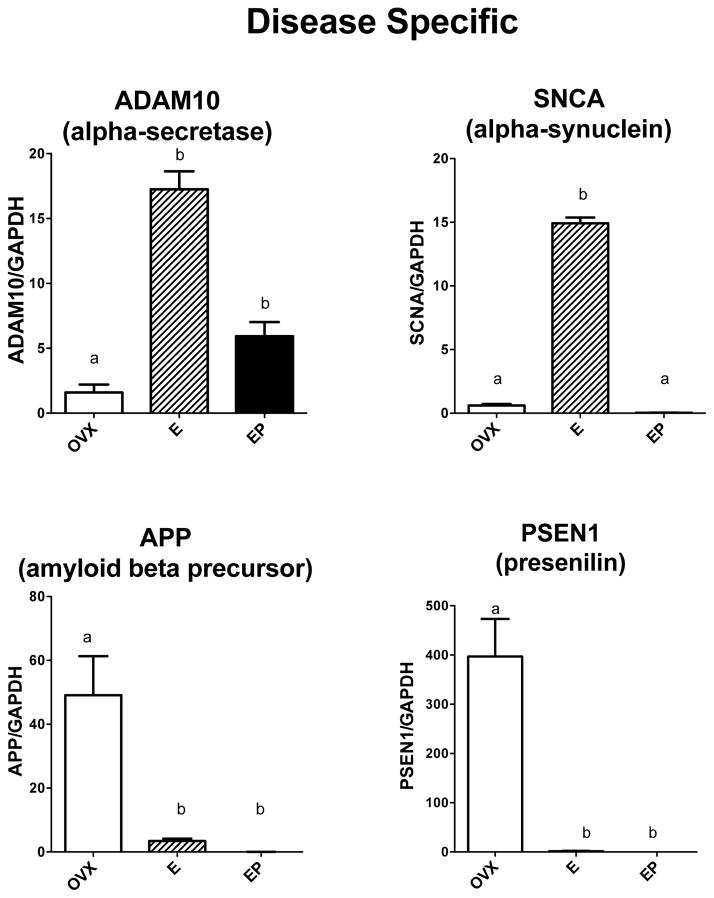
It was of further interest to examine representative expression of genes that encode proteins known to be involved in neurodegenerative pathology. Figure 8 illustrates the 4 genes that were examined. ADAM10 (α-secretase) mRNA levels were significantly different across groups with E and E+P treatments increased over placebo (F [2,10]=66.0; p<0.0001). SCNA (α-synuclein) mRNA levels were significantly different across groups with E treatment increased over placebo and E+P treatment (F [2,10]=1336; p<0.0001). In contrast, PSEN1 (presenilin, γ–secretase) and APP (amyloid beta precursor) mRNA levels were significantly suppressed by E and E+P treatments compared to placebo (F [2,10]=22.8; p=0.0005 and F [2,10]=12.92; p=0.003, respectively).
Figure 8.
Histograms illustrating the expression of 4 genes encoding proteins known to be aberrant in different NDDs. ADAM10 mRNA was significantly increased by E and E+P treatments over placebo. SNCA1 mRNA was significantly increased by E treatment over placebo and E+P. APP and PSEN1 mRNAs significantly decreased by E and E+P treatments over placebo.
a, b – groups with different letters are different at p<0.05 with Newman-Keuls posthoc pairwise comparison.
Correlation Matrix
The expression of each gene across treatment groups was correlated with all of the other genes, and the r values are summarized in Table 2. A gene correlated with its self yields a perfect correlation of r equal to 1.00. Of note, 5HT1A, NCAM, and UBE2D3 correlated poorly with the other genes. The other genes correlated with each other to a high positive degree except APP and PSEN1 exhibited a strong negative correlation with most of the other genes.
Table 2.
Correlation matrix of the genes reported in this study. Each gene correlates with its self with r = 1.00 (positive correlation), and for genes with a perfect negative correlation, r = −1.00. A correlation coefficient of zero indicates no linear relationship. Coefficients highlighted in yellow are between 0.57 > 0 > −0.54 suggesting little or no relationship between expressions of the two genes.
| FEV | TPH2 | SERT | 5HT1A | CRF-R1 | CRF-R2 | UCN1 | NLGN3 | NTRK2 | SNAP25 | NCAM | GRIN2A | GRIA2 | GRM1 | NSB1 | NTHL1 | LIG4 | GTF2H5 | RAD23 | APEX1 | DISS1 | HSP90 | HSP60 | HSP27 | UBEA5 | UBE2D3 | UBE3A | KIF5 | DYNCL1 | DCTN4 | MAPT | ADAM10 | SCNA | APP | PSEN1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEV | 1.00 | ||||||||||||||||||||||||||||||||||
| TPH2 | 0.71 | 1.00 | |||||||||||||||||||||||||||||||||
| SERT | 0.88 | 0.96 | 1.00 | ||||||||||||||||||||||||||||||||
| 5HT1A | -0.17 | 0.57 | 0.33 | 1.00 | |||||||||||||||||||||||||||||||
| CRF-R1 | −0.74 | −1.00 | −0.97 | −0.54 | 1.00 | ||||||||||||||||||||||||||||||
| CRF-R2 | 0.94 | 0.44 | 0.67 | −0.48 | −0.48 | 1.00 | |||||||||||||||||||||||||||||
| UCN1 | 0.88 | 0.96 | 1.00 | 0.31 | −0.97 | 0.68 | 1.00 | ||||||||||||||||||||||||||||
| NLGN3 | 0.99 | 0.61 | 0.81 | −0.29 | −0.65 | 0.98 | 0.82 | 1.00 | |||||||||||||||||||||||||||
| NTRK2 | 1.00 | 0.75 | 0.90 | −0.11 | −0.78 | 0.92 | 0.91 | 0.98 | 1.00 | ||||||||||||||||||||||||||
| SNAP25 | 1.00 | 0.65 | 0.84 | −0.25 | −0.69 | 0.97 | 0.84 | 1.00 | 0.99 | 1.00 | |||||||||||||||||||||||||
| NCAM | 0.13 | 0.79 | 0.58 | 0.96 | −0.76 | −0.21 | 0.57 | −0.01 | 0.18 | 0.04 | 1.00 | ||||||||||||||||||||||||
| GRIN2A | 0.56 | 0.98 | 0.89 | 0.73 | −0.97 | 0.25 | 0.88 | 0.45 | 0.60 | 0.49 | 0.89 | 1.00 | |||||||||||||||||||||||
| GRIA2 | 0.94 | 0.91 | 0.99 | 0.19 | −0.93 | 0.77 | 0.99 | 0.88 | 0.95 | 0.90 | 0.47 | 0.82 | 1.00 | ||||||||||||||||||||||
| GRM1 | 1.00 | 0.69 | 0.86 | −0.20 | −0.72 | 0.95 | 0.87 | 0.99 | 1.00 | 1.00 | 0.09 | 0.53 | 0.92 | 1.00 | |||||||||||||||||||||
| NSB1 | 1.00 | 0.65 | 0.83 | −0.25 | −0.68 | 0.97 | 0.84 | 1.00 | 0.99 | 1.00 | 0.04 | 0.49 | 0.90 | 1.00 | 1.00 | ||||||||||||||||||||
| NTHL1 | 0.97 | 0.86 | 0.97 | 0.08 | −0.88 | 0.83 | 0.97 | 0.93 | 0.98 | 0.95 | 0.36 | 0.74 | 0.99 | 0.96 | 0.95 | 1.00 | |||||||||||||||||||
| LIG4 | 0.97 | 0.86 | 0.97 | 0.08 | −0.88 | 0.83 | 0.97 | 0.93 | 0.98 | 0.95 | 0.36 | 0.74 | 0.99 | 0.96 | 0.95 | 1.00 | 1.00 | ||||||||||||||||||
| GTF2H5 | 0.87 | 0.97 | 1.00 | 0.34 | −0.98 | 0.66 | 1.00 | 0.80 | 0.90 | 0.83 | 0.60 | 0.89 | 0.99 | 0.85 | 0.83 | 0.96 | 0.96 | 1.00 | |||||||||||||||||
| RAD23 | 1.00 | 0.72 | 0.88 | −0.16 | −0.75 | 0.94 | 0.89 | 0.99 | 1.00 | 1.00 | 0.13 | 0.57 | 0.94 | 1.00 | 1.00 | 0.97 | 0.97 | 0.87 | 1.00 | ||||||||||||||||
| APEX1 | 0.96 | 0.88 | 0.98 | 0.11 | −0.90 | 0.82 | 0.98 | 0.92 | 0.97 | 0.94 | 0.39 | 0.76 | 1.00 | 0.95 | 0.93 | 1.00 | 1.00 | 0.97 | 0.96 | 1.00 | |||||||||||||||
| DISS1 | 0.96 | 0.49 | 0.71 | −0.44 | −0.52 | 1.00 | 0.72 | 0.99 | 0.94 | 0.98 | −0.16 | 0.30 | 0.80 | 0.97 | 0.98 | 0.86 | 0.86 | 0.70 | 0.96 | 0.85 | 1.00 | ||||||||||||||
| HSP90 | 0.94 | 0.90 | 0.99 | 0.17 | −0.92 | 0.78 | 0.99 | 0.89 | 0.96 | 0.91 | 0.45 | 0.80 | 1.00 | 0.93 | 0.91 | 1.00 | 1.00 | 0.98 | 0.95 | 1.00 | 0.81 | 1.00 | |||||||||||||
| HSP60 | 0.92 | 0.93 | 1.00 | 0.24 | −0.95 | 0.73 | 1.00 | 0.86 | 0.94 | 0.88 | 0.51 | 0.84 | 1.00 | 0.91 | 0.88 | 0.99 | 0.99 | 0.99 | 0.92 | 0.99 | 0.77 | 1.00 | 1.00 | ||||||||||||
| HSP27 | 1.00 | 0.66 | 0.84 | −0.24 | −0.69 | 0.97 | 0.85 | 1.00 | 0.99 | 1.00 | 0.05 | 0.49 | 0.91 | 1.00 | 1.00 | 0.95 | 0.95 | 0.83 | 1.00 | 0.94 | 0.98 | 0.91 | 0.89 | 1.00 | |||||||||||
| UBEA5 | 0.95 | 0.90 | 0.98 | 0.15 | −0.91 | 0.79 | 0.99 | 0.90 | 0.97 | 0.92 | 0.43 | 0.79 | 1.00 | 0.94 | 0.92 | 1.00 | 1.00 | 0.98 | 0.95 | 1.00 | 0.82 | 1.00 | 1.00 | 0.92 | 1.00 | ||||||||||
| UBE2D3 | 0.11 | 0.77 | 0.57 | 0.96 | −0.75 | −0.23 | 0.56 | −0.02 | 0.16 | 0.03 | 1.00 | 0.88 | 0.45 | 0.08 | 0.02 | 0.35 | 0.35 | 0.58 | 0.12 | 0.38 | −0.18 | 0.43 | 0.49 | 0.03 | 0.41 | 1.00 | |||||||||
| UBE3A | 0.99 | 0.80 | 0.93 | −0.04 | −0.82 | 0.89 | 0.94 | 0.97 | 1.00 | 0.98 | 0.25 | 0.66 | 0.97 | 0.99 | 0.98 | 0.99 | 0.99 | 0.93 | 0.99 | 0.99 | 0.91 | 0.98 | 0.96 | 0.98 | 0.98 | 0.23 | 1.00 | ||||||||
| KIF5 | 0.79 | 0.99 | 0.99 | 0.48 | −1.00 | 0.54 | 0.98 | 0.70 | 0.82 | 0.73 | 0.71 | 0.95 | 0.95 | 0.77 | 0.73 | 0.91 | 0.91 | 0.99 | 0.79 | 0.93 | 0.58 | 0.95 | 0.97 | 0.74 | 0.94 | 0.70 | 0.86 | 1.00 | |||||||
| DYNCL1 | 0.90 | 0.95 | 1.00 | 0.28 | −0.96 | 0.70 | 1.00 | 0.84 | 0.92 | 0.86 | 0.55 | 0.86 | 1.00 | 0.89 | 0.86 | 0.98 | 0.98 | 1.00 | 0.90 | 0.99 | 0.74 | 0.99 | 1.00 | 0.86 | 0.99 | 0.53 | 0.95 | 0.98 | 1.00 | ||||||
| DCTN4 | 0.86 | 0.97 | 1.00 | 0.37 | −0.98 | 0.64 | 1.00 | 0.78 | 0.88 | 0.81 | 0.62 | 0.91 | 0.98 | 0.84 | 0.81 | 0.96 | 0.96 | 1.00 | 0.86 | 0.97 | 0.68 | 0.98 | 0.99 | 0.81 | 0.97 | 0.60 | 0.92 | 0.99 | 1.00 | 1.00 | |||||
| MAPT | 0.99 | 0.81 | 0.94 | −0.01 | −0.84 | 0.88 | 0.95 | 0.96 | 0.99 | 0.97 | 0.28 | 0.68 | 0.98 | 0.98 | 0.97 | 1.00 | 1.00 | 0.94 | 0.99 | 0.99 | 0.90 | 0.98 | 0.97 | 0.97 | 0.99 | 0.26 | 1.00 | 0.87 | 0.96 | 0.93 | 1.00 | ||||
| ADAM10 | 0.99 | 0.82 | 0.94 | −0.01 | −0.84 | 0.88 | 0.95 | 0.96 | 0.99 | 0.97 | 0.28 | 0.68 | 0.98 | 0.98 | 0.97 | 1.00 | 1.00 | 0.94 | 0.99 | 0.99 | 0.90 | 0.98 | 0.97 | 0.97 | 0.99 | 0.27 | 1.00 | 0.88 | 0.96 | 0.93 | 1.00 | 1.00 | |||
| SCNA | 0.99 | 0.62 | 0.81 | −0.29 | −0.65 | 0.98 | 0.82 | 1.00 | 0.98 | 1.00 | 0.00 | 0.45 | 0.88 | 1.00 | 1.00 | 0.93 | 0.93 | 0.80 | 0.99 | 0.92 | 0.99 | 0.89 | 0.86 | 1.00 | 0.90 | −0.02 | 0.97 | 0.70 | 0.84 | 0.79 | 0.96 | 0.96 | 1.00 | ||
| APP | −0.54 | −0.98 | −0.88 | −0.74 | 0.97 | −0.23 | −0.87 | −0.43 | −0.59 | −0.47 | −0.90 | −1.00 | −0.80 | −0.52 | −0.47 | −0.73 | −0.73 | −0.89 | −0.55 | −0.75 | −0.28 | −0.79 | −0.83 | −0.48 | −0.78 | −0.89 | −0.65 | −0.95 | −0.85 | −0.90 | −0.67 | −0.67 | −0.43 | 1.00 | |
| PSEN1 | −0.59 | −0.99 | −0.90 | −0.70 | 0.98 | −0.29 | −0.90 | −0.48 | −0.63 | −0.52 | −0.87 | −1.00 | −0.84 | −0.57 | −0.52 | −0.77 | −0.77 | −0.91 | −0.60 | −0.79 | −0.34 | −0.82 | −0.86 | −0.53 | −0.81 | −0.87 | −0.69 | −0.96 | −0.88 | −0.92 | −0.71 | −0.71 | −0.49 | 1.00 | 1.00 |
Further comparison
A Benjamimi-Hochberg test for multiple comparisons was conducted on the qRT-PCR data using a freeware FDR calculator. Thirty-six p-values were entered (36 graphed genes). The False Discovery Rate Procedure indicated that the critical ‘p’ was equal to 0.0388 using a false discovery rate of 0.05. This substantiated the majority of our individual results. Eight genes were considered false positives. Genes that we suggest exhibited trends, were considered false positives including SERT, NLGN3, NTRK2, and PCNA. Only 4 genes that we found significantly different individually, LIG4, UCN1, DCTN4 and DYNCL1 were considered false positives with ‘p’ from 0.0414 to 0.0488. On the other hand, the small ‘n’ raises the risk that rejecting these genes as ‘not regulated’ by E or E+P could be a type 2 statistical error. Therefore, they are included in the discussion.
Effect Size
Cohen’s d analysis was used to determine the effect size of the treatments in the absence of statistical differences. The effect size of the E treatment and the E+P treatment in comparison to placebo treatment for the genes that exhibited trends is shown in Table 3. E treatment had a large effect on SERT, NLGN3, and NTRK expression. E+P treatment had a large effect on SERT, NTRK and PCNA expression.
Table 3.
Analysis of genes that exhibited non-significant trends with Cohen’s d -test. Cohen’s d > 0.8 indicates a large effect of the treatment. Cohen’s’ d < 0.2 indicates less than a small effect of treatment.
| Gene | Treatment | d | r |
|---|---|---|---|
| SERT | E | 1.22 | 0.52 |
| E+P | 1.94 | 0.69 | |
| NLGN3 | E | 1.19 | 0.57 |
| E+P | 0.18 | 0.09 | |
| NTRK | E | 1.41 | 0.57 |
| E+P | 1.82 | 0.67 | |
| PCNA | E | 0.05 | 0.02 |
| E+P | 1.69 | 0.64 |
Steroid hormone concentrations achieved by implants
Serum E concentrations in the placebo group equaled <30 pg/ml and serum P concentrations equaled 0.15 ± 0.05 ng/ml. Both hormones often fell below the limit of assay sensitivity. In the E-treated animals, the average serum E concentration during the final two years of the study was 118.8 ± 15.9 pg/ml, and at the time of tissue collection, E equaled 94.3 ± 20.5 pg/ml. P concentrations in the E-only treated animals equaled 0.18 ± 0.06 ng/ml. In the E+P treated animals, E concentrations were similar to the E-only treated animals and P concentrations equaled 11.33 ± 0.79 ng/ml. Although the serum E concentrations were somewhat higher than those observed in the very early follicular phase or achieved by vaginal rings in women (60–70 pg/ml), the treatment was identical to that used in earlier studies with young adult females. Moreover, the achieved concentration produced consistent experiment-to-experiment increases in TPH2 mRNA, our standard for efficacy in the dorsal raphe. Nonetheless, the serum E concentration was far below the concentrations observed during the ovulatory surge (400–1000 pg/ml) in macaques.
Other Comparisons
Limited gene expression data from the hypothalamus has also been published from this same set of animals, and the old animals showed marked elevation of progestin receptor (PR) and other mRNAs with E treatment over placebo, indicating that hypothalamic neurons also remained responsive to E (Eghlidi and Urbanski, 2015). There was a correlation between circadian activity parameters and performance in the spatial maze (Haley, et al., 2009) and correlation between performance in the spatial maze and immune responses (Haley, et al., 2011). Preliminary analysis indicates better performance in the Delayed Response test by the E-treated animals compared to the placebo-treated animals (Renner, et al., 2010). In addition, the aged placebo-treated females exhibited a reduced response to the vaccination compared to the aged E-treated animals (Engelmann, et al., 2010).
Discussion
The serotonin neural system projects to all forebrain areas and impacts the functions that are affected by ovarian steroid loss. Therefore, understanding the actions of ovarian steroids in serotonin neurons in macaques has been a major focus of this laboratory. We found robust effects of E or E+P on gene expression in laser-captured serotonin neurons of adult, short-term Ovx macaques with only 1 month of treatment. This study addresses age and treatment length in a clinically relevant model, using older macaques (≥18 years old at the start of treatment) that were Ovx to induce surgical menopause and then treated with E or E+P for ~4 years. Also, a small block of tissue containing the dorsal raphe was used instead of laser captured serotonin neurons.
Altogether, there were 4 variables that differed between this study and previous studies: (1) age, (2) length of treatment (3) unintended higher serum P concentrations and (4) preparation extracted for qRT-PCR. All animals were maintained on standard low fat, low sugar monkey chow. In these old, chronically treated animals, the mRNA expression (increase or decrease) pattern in the E-treated group was very similar to that observed in laser captured serotonin neurons from young adult, acutely treated animals for the majority of genes.. Nonetheless, the block preparation contained numerous other cell types that can utilize the same common genes, but the genes may be regulated differently than in serotonin neurons. Therefore, there was more variance within groups. Also, the expression of each mRNA in relation to GAPDH was 10–100x lower than in the laser captured serotonin neuron preparation. Based upon our study of blocks versus laser-captured preparations (Bethea and Reddy, 2008), we attribute much of the variance observed in this study to the block preparation. Unfortunately, it was not possible to perform laser capture on serotonin neurons of these old animals.
The cyclic supplemention of P in the E+P-treated group yielded different responses across the genes examined in this study. Different mRNAs in the E+P-treated group were greater than, equal to, or less than the group treated with E alone. We have observed these three different responses across different genes in immunohistochemical assays, in situ hybridization assays, microarray assays and in qRT-PCR outcomes of young adult, acutely treated animals as well. The different responses may derive from the presence or absence of E-inducible PRs, the presence or absence of a PR-response element on the gene in question, or different protein-protein interactions and cofactors available for different genes. Overall, we have observed more variance in the E+P treated groups than in the other groups. In addition, the optimal ratio of E to P for normal biological responses in macaques was established as approximately 1/50. With other ratios, progestins are often antagonistic to estrogens. (Neumann, 1978). The serum P concentrations achieved by the oral micronized P in this study are higher than the range obtained in studies of acutely treated, young adults or normally attained in menstrual cycles. Many of the mRNAs exhibited suppression with supplemental P compared to E alone. When this differed from results in young adults, we attribute this to the high P, which antagonizes the effect of E.
Although this study focused on the serotonin system and expression in the dorsal raphe, gene expression in other areas of the brain is of significant concern. Currently, funding is not available, but interested investigators are very welcome to other parts of the brain and should contact the corresponding author.
Serotonin Regulation
E probably elevated serotonin neurotransmission in old surgically menopausal macaques; or more likely, E prevented a decline in serotonin neurotransmission after Ovx. E increased FEV (Pet1) and TPH2 mRNAs, which would cause an increase in the serotonin phenotype and increased production of serotonin. E also increased CRHR2 and UCN1 mRNAs. CRHR2 is considered anxiolytic; it binds UCN1 and causes a release of serotonin. CRHR1 is the anxiolytic receptor that binds CRH and decreases serotonin release. E and E+P decreased CRHR1 expression, which would further increase serotonin. Therefore, E supported the serotonin neuronal phenotype (Hendricks, et al., 2003), increased serotonin synthesis (Kloiber, et al., 2010) and increased serotonin release via regulation of CRH receptors (Pernar, et al., 2004).
5HT1A mRNA regulation was the opposite of that found in young adults (Lu and Bethea, 2002, Pecins-Thompson and Bethea, 1998) using ISH and receptor binding assays. This may be due to the presence of differential regulated autoreceptors and postsynaptic receptors in the block of tissue.
Synapses and Glutamate Receptors
Much of adult plasticity relies on proliferating dendritic spines and regulated insertion of glutamate receptors. The development of new synapses on dendritic spines and the expression of glutamate receptors have been linked (McKinney, et al., 1999; Srivastava, et al., 2008). E alone increased the expression of 4 genes that code for selected synapse assembly proteins as determined with ANOVA or Cohen’s d. The variable effect of supplemental P was also observed in acutely treated, young adults (Bethea and Reddy, 2015).
The increase in NMDA2A and AMPA2 mRNAs by E and E+P also suggests that synapses were increased by E replacement and supplemental P was neutral. The expression of these two types of receptors is particularly important in the ontogeny of mature dendritic spines (Fortin, et al., 2011; Yoshihara, et al., 2009). E stimulated GRM1 mRNA, but the suppression of GRM1 mRNA by supplemental P differs from results in young adults. Altogether, the data support an ongoing supportive effect of E on dendritic spine proliferation and excitatory synapse assembly.
DNA repair
In young adult Ovx macaques, the absence of ovarian steroids led to serotonin neuron degeneration and death in the absence of any overt trauma to the brain as demonstrated by DNA fragmentation detected with a TUNEL assay (Lima and Bethea, 2009). DNA repair minimally requires lesion recognition, single strand excision, lesion removal, gap-filling synthesis, and finally ligation (Dantuma, et al., 2009). In this study, E alone increased mRNA expression in 8 of 16 genes coding for DNA repair enzymes. The addition of P to the E regimen suppressed the effect of E to varying degrees, but 7 mRNAs, were still expressed above the level exhibited by the placebo group. Thus, chronic hormone therapy with E or E+P maintained expression of DNA repair factors in the dorsal raphe of old macaques, which is essential for maintaining neural function. The monkey substantia nigra also exhibited a decline in tyrosine hydroxylase-positive neurons after Ovx (Leranth et al., 2000).
Underlying mechanisms of Neurodegenerative Diseases (NDDs)
The common NDDs observed in aging populations have not been observed in female macaques that do not live much past 30 years of age, and they produce ovarian hormones for almost their entire life. However, macaques accumulate lipofuscin, a product of free radical-induced lipid peroxidation that correlates with cellular aging (Terman and Brunk, 1998). A number of NDDs involve translation of normal genes whose proteins subserve important cellular functions under normal conditions, but the proteins are mis-folded (chaperones), mis-processed (ubiquinases), or subverted from normal function in various ways (transport) (Bucciantini, et al., 2002). Since depression, a hallmark of serotonin dysfunction often precedes overt NDD symptoms; it was of interest to determine expression of representative genes in these categories in serotonin neurons. E or E+P administration caused a significant increase in mRNA expression related to protein folding, degradation, and transport in the old macaques in a manner similar to young macaques (Bethea and Reddy, 2015). Continuing these functions in serotonin and other neurons is vital for healthy ageing.
Disease Specific
A number of proteins have been specifically implicated in neuropathies, but they play pivotal roles in normal functioning. E induction of ADAM10 mRNA, which would improve correct processing of APP, with suppression of APP mRNA, the amyloid precursor protein, and PSEN1 mRNA, the component of 7gamma;-secretase that produces Aβ, was found in young adult and old monkeys. This data is consistent with epidemiological observations that ovarian steroids may delay the onset of Alzheimer’s (Henderson, 2006).
It is essential to keep in mind that diet plays a large role in the effects of E in serotonin neurons. We found that a high fat diet blocked the positive effect of E on genes coding for proteins that promote serotonin neurotransmission and decrease norepinephrine transmission (Bethea et al., 2015). This data suggests that clinical populations that are obese and/or eat a high fat and sugar diet may not respond to E therapy, and we are currently testing this hypothesis in old macaques.
Correlations
There was strong correlation between several genes from the same or different categories in this study. It is attractive to speculate that the steroid receptor mechanisms operate on the correlated genes in a like manner. It is also possible that the correlated genes are impacting one another.
Conclusions
Old rhesus macaques that are Ovx and administered E or E+P for ~4 years have improved expression of genes that govern serotonin neurotransmission, synapse assembly, glutamate receptors, DNA repair, chaperones, ubiquinases, transport and select neuropathologies compared to placebo-treated controls. Except for the confound of excess P replacement, old animals responded to hormone therapy like young animals; and chronic therapy in old animals produced similar outcomes as acute therapy in younger adult animals. Altogether, the data reveal mechanisms that could improve serotonin function and decrease neuropathologies in women who initiate bio-identical hormone therapy during the peri-menopause and continue it for at least 4 years.
Highlights.
Old ovariectomized rhesus macaques treated with continuous estradiol plus or minus cyclic progesterone
qRT-PCR for gene expression in the serotonergic dorsal raphe
Protective effect of estradiol on genes related to serotonin transmission, plasticity and neurodegeneration
Variable effect of cyclic progesterone
Acknowledgments
We are deeply grateful to the dedicated staff of the Division of Animal Resources including the staff of the Departments of Surgery and Pathology for their expertise and helpfulness in all aspects of monkey management and care. This work was supported by NIH grants MH62677 to CLB; AG19100, AG26472, AG29612 to HFU, and P51 OD011092 for the operation of ONPRC.
Footnotes
Disclosures
The authors have no financial disclosures to make.
The work was supported by NIH grants as stated in the acknowledgements.
The data contained in the manuscript being submitted have not been previously published, have not been submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
All authors have reviewed the contents of the manuscript being submitted, approve of its contents and validate the accuracy of the data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Albert PR, Vahid-Ansari F, Luckhart C. Serotonin-prefrontal cortical circuitry in anxiety and depression phenotypes: pivotal role of pre- and post-synaptic 5-HT1A receptor expression. Frontiers Behav Neurosci. 2014;8:199. doi: 10.3389/fnbeh.2014.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin Z, Gueorguieva R, Cappiello A, Czarkowski KA, Stiklus S, Anderson GM, Naftolin F, Epperson CN. Estradiol and tryptophan depletion interact to modulate cognition in menopausal women. Neuropsychopharmacol. 2006;31:2489–97. doi: 10.1038/sj.npp.1301114. [DOI] [PubMed] [Google Scholar]
- Araragi N, Lesch KP. Serotonin (5-HT) in the regulation of depression-related emotionality: insight from 5-HT transporter and tryptophan hydroxylase-2 knockout mouse models. Current Drug Targets. 2013;14:549–70. doi: 10.2174/1389450111314050005. [DOI] [PubMed] [Google Scholar]
- Bethea CL. Regulation of progestin receptors in raphe neurons of steroid-treated monkeys. Neuroendocrinology. 1994;60:50–61. doi: 10.1159/000126719. [DOI] [PubMed] [Google Scholar]
- Bethea CL, Lu NZ, Gundlah C, Streicher JM. Diverse actions of ovarian steroids in the serotonin neural system. Frontiers Neuroendocrinol. 2002;23:41–100. doi: 10.1006/frne.2001.0225. [DOI] [PubMed] [Google Scholar]
- Bethea CL, Pau FK, Fox S, Hess DL, Berga SL, Cameron JL. Sensitivity to stress-induced reproductive dysfunction linked to activity of the serotonin system. Fertil Steril. 2005;83:148–55. doi: 10.1016/j.fertnstert.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Bethea CL, Reddy AP. Effect of ovarian hormones on survival genes in laser captured serotonin neurons from macaques. J Neurochem. 2008;105:1129–43. doi: 10.1111/j.1471-4159.2008.05213.x. [DOI] [PubMed] [Google Scholar]
- Bethea CL, Reddy AP. Effect of ovarian hormones on genes promoting dendritic spines in laser-captured serotonin neurons from macaques. Mol Psychiatry. 2010;15:1034–44. doi: 10.1038/mp.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea CL, Reddy AP. Effect of ovarian steroids on gene expression related to synapse assembly in serotonin neurons of macaques. J Neurosci Res. 2012a;90:1324–34. doi: 10.1002/jnr.23004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea CL, Reddy AP. Ovarian steroids increase glutamatergic related gene expression in serotonin neurons of macaques. Mol Cell Neurosci. 2012b;49:251–62. doi: 10.1016/j.mcn.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea CL, Reddy AP. Ovarian steroids regulate gene expression related to DNA repair and neurodegenerative diseases in serotonin neurons of macaques. Mol Psychiatry. 2015 doi: 10.1038/mp.2014.178. E pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner RM, Slayden OD. Cyclic changes in the primate oviduct and endometrium. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2. Raven Press, Ltd; New York: 1994. pp. 541–69. [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Burger HG, Dudley EC, Robertson DM, Dennerstein L. Hormonal changes in the menopause transition. Recent Prog Horm Res. 2002;57:257–75. doi: 10.1210/rp.57.1.257. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair. 2009;8:449–60. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Diaconescu AO, Kramer E, Hermann C, Ma Y, Dhawan V, Chaly T, Eidelberg D, McIntosh AR, Smith GS. Distinct functional networks associated with improvement of affective symptoms and cognitive function during citalopram treatment in geriatric depression. Human Brain Mapping. 2011;32:1677–91. doi: 10.1002/hbm.21135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JL, Urbanski HF. Neuroendocrine changes in the aging reproductive axis of female rhesus macaques (Macaca mulatta) Biol Reprod. 2006;75:539–46. doi: 10.1095/biolreprod.106.051839. [DOI] [PubMed] [Google Scholar]
- Engelmann F, Barron A, Urbanski H, Neuringer M, Kohama SG, Park B, Messaoudi I. Accelerated immune senescence and reduced response to vaccination in ovariectomized female rhesus macaques. Age. 2011;33:275–89. doi: 10.1007/s11357-010-9178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson CN, Amin Z, Ruparel K, Gur R, Loughead J. Interactive effects of estrogen and serotonin on brain activation during working memory and affective processing in menopausal women. Psychoneuroendocrinol. 2012;37:372–82. doi: 10.1016/j.psyneuen.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin DA, Srivastava T, Soderling TR. Structural modulation of dendritic spines during synaptic plasticity. Neuroscientist. 2012;18:326–41. doi: 10.1177/1073858411407206. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Girdler SS, Meltzer-Brody SE, Stika CS, Thurston RC, Clark CT, Prairie BA, Moses-Kolko E, Joffe H, Wisner KL. Ovarian hormone fluctuation, neurosteroids, and HPA axis dysregulation in perimenopausal depression: A novel heuristic model. Am J Psychiatry. 2015;172:227–36. doi: 10.1176/appi.ajp.2014.14070918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlah C, Kohama SG, Mirkes SJ, Garyfallou VT, Urbanski HF, Bethea CL. Distribution of estrogen receptor beta (ERbeta) mRNA in hypothalamus, midbrain and temporal lobe of spayed macaque: continued expression with hormone replacement. Mol Brain Res. 2000;76:191–204. doi: 10.1016/s0006-8993(99)02475-0. [DOI] [PubMed] [Google Scholar]
- Gundlah C, Lu NZ, Mirkes SJ, Bethea CL. Estrogen receptor beta (ERbeta) mRNA and protein in serotonin neurons of macaques. Mol Brain Res. 2001;91:14–22. doi: 10.1016/s0169-328x(01)00108-5. [DOI] [PubMed] [Google Scholar]
- Haley GE, Landauer N, Renner L, Weiss A, Hooper K, Urbanski HF, Kohama SG, Neuringer M, Raber J. Circadian activity associated with spatial learning and memory in aging rhesus monkeys. Exp Neurol. 2009;217:55–62. doi: 10.1016/j.expneurol.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley GE, Urbanski HF, Kohama SG, Messaoudi I, Raber J. Spatial memory performance associated with measures of immune function in elderly female rhesus macaques. Eur Geriatric Med. 2011;2:117–21. doi: 10.1016/j.eurger.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson VW. Estrogen-containing hormone therapy and Alzheimer’s disease risk: understanding discrepant inferences from observational and experimental research. Neuroscience. 2006;138:1031–9. doi: 10.1016/j.neuroscience.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–47. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Kloiber S, Kohli MA, Brueckl T, Ripke S, Ising M, Uhr M, Menke A, Unschuld PG, Horstmann S, Salyakina D, Muller-Myhsok B, Binder EB, Holsboer F, Lucae S. Variations in tryptophan hydroxylase 2 linked to decreased serotonergic activity are associated with elevated risk for metabolic syndrome in depression. Mol Psychiatry. 2010;15:736–47. doi: 10.1038/mp.2008.142. [DOI] [PubMed] [Google Scholar]
- Leranth C, Roth RH, Elsworth JD, Naftolin F, Horvath TL, Redmond DE., Jr Estrogen is essential for maintaining nigrostriatal dopamine neurons in primates: implications for Parkinson’s disease and memory. J Neurosci. 2000;20:8604–9. doi: 10.1523/JNEUROSCI.20-23-08604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima FB, Bethea CL. Ovarian steroids decrease DNA fragmentation in the serotonin neurons of non-injured rhesus macaques. Mol Psychiatry. 2009;15:657–68. doi: 10.1038/mp.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu NZ, Bethea CL. Ovarian steroid regulation of 5HT1A receptor binding and G protein activation in female monkeys. Neuropsychopharmacol. 2002;27:12–24. doi: 10.1016/S0893-133X(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Maki PM, Freeman EW, Greendale GA, Henderson VW, Newhouse PA, Schmidt PJ, Scott NF, Shively CA, Soares CN. Summary of the National Institute on Aging-sponsored conference on depressive symptoms and cognitive complaints in the menopausal transition. Menopause. 2010;17:815–22. doi: 10.1097/gme.0b013e3181d763d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay SM, Brambilla DJ, Posner JG. The normal menopause transition. Maturitas. 2008;61:4–16. doi: 10.1016/j.maturitas.2008.09.005. [DOI] [PubMed] [Google Scholar]
- McKinney RA, Capogna M, Durr R, Gahwiler BH, Thompson SM. Miniature synaptic events maintain dendritic spines via AMPA receptor activation. Nat Neurosci. 1999;2:44–9. doi: 10.1038/4548. [DOI] [PubMed] [Google Scholar]
- Mittelman-Smith MA, Williams H, Krajewski-Hall SJ, McMullen NT, Rance NE. Role for kisspeptin/neurokinin B/dynorphin (KNDy) neurons in cutaneous vasodilatation and the estrogen modulation of body temperature. Proc Natl Acad Sci U S A. 2012;109:19846–51. doi: 10.1073/pnas.1211517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F. The physiological action of progesterone and the pharmacological effects of progestogens--a short review. Postgrad Med J. 1978;54(Suppl 2):11–24. [PubMed] [Google Scholar]
- Pecins-Thompson M, Bethea CL. Ovarian steroid regulation of 5HT1A autoreceptor messenger ribonucleic acid expression in the dorsal raphe of rhesus macaques. Neuroscience. 1998;89:267–77. doi: 10.1016/s0306-4522(98)00326-1. [DOI] [PubMed] [Google Scholar]
- Pernar L, Curtis AL, Vale WW, Rivier JE, Valentino RJ. Selective activation of corticotropin-releasing factor-2 receptors on neurochemically identified neurons in the rat dorsal raphe nucleus reveals dual actions. J Neurosci. 2004;24:1305–11. doi: 10.1523/JNEUROSCI.2885-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp PR, Morrison JH, Roberts JA. Cyclic estrogen replacement improves cognitive function in aged ovariectomized rhesus monkeys. J Neurosci. 2003;231:5708–5714. doi: 10.1523/JNEUROSCI.23-13-05708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner L, Weiss A, Landauer N, Kohama SG, Urbanski H, Voytko ML, Neuringer M. Long-term effects of estrogen replacement on spatial working memory in rhesus monkeys. 40th Annual Meeting of the Society for Neuroscience; Nov 13–17; San Diego, CA. 2010. [Google Scholar]
- Rivera HM, Bethea CL. Ovarian steroids increase spinogenetic proteins in the macaque dorsal raphe. Neuroscience. 2012;208:27–40. doi: 10.1016/j.neuroscience.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarti CD, Chiantera A, Graziottin A, Ognisanti F, Sidoli C, Mincigrucci M, Parazzini F, Gruppo di Studio Iper A. Hormone therapy and sleep quality in women around menopause. Menopause. 2005;12:545–51. doi: 10.1097/01.gme.0000172270.70690.5e. [DOI] [PubMed] [Google Scholar]
- Srivastava DP, Woolfrey KM, Jones KA, Shum CY, Lash LL, Swanson GT, Penzes P. Rapid enhancement of two-step wiring plasticity by estrogen and NMDA receptor activity. Proc Natl Acad Sci U S A. 2008;105:14650–5. doi: 10.1073/pnas.0801581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollstorff M, Munakata Y, Jensen AP, Guild RM, Smolker HR, Devaney JM, Banich MT. Individual differences in emotion-cognition interactions: emotional valence interacts with serotonin transporter genotype to influence brain systems involved in emotional reactivity and cognitive control. Frontiers Human Neurosci. 2013;7:327. doi: 10.3389/fnhum.2013.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, Brunk UT. Lipofuscin: mechanisms of formation and increase with age. APMIS. 1998;106:265–76. doi: 10.1111/j.1699-0463.1998.tb01346.x. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, De Roo M, Muller D. Dendritic spine formation and stabilization. Curr Opin Neurobiol. 2009;19:146–53. doi: 10.1016/j.conb.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Voytko ML. The effects of long-term ovariectomy and estrogen replacement therapy on learning and memory in monkeys. Behav Neurosci. 2000;114:1078–1087. [PubMed] [Google Scholar]
- Voytko ML. Estrogen and the cholinergic system modulate visuospatial attention in monkeys. Beh Neurosci. 2002;116:187–197. doi: 10.1037//0735-7044.116.2.187. [DOI] [PubMed] [Google Scholar]