Abstract
Background
Caloric restriction (CR) can increase longevity in rodents and improve memory function in humans. α-Lipoic acid (LA) has been shown to improve memory function in rats, but not longevity. While studies have looked at survival in rodents after switching from one diet to another, the underlying mechanisms of the beneficial effects of CR and LA supplementation are unknown. Here, we use RNA-seq in cerebral cortex from rats subjected to CR and LA-supplemented rats to understand how changes in diet can affect aging, neurodegeneration and longevity.
Results
Gene expression changes during aging in ad libitum-fed rats are largely prevented by CR, and neuroprotective genes are overexpressed in response to both CR and LA diets with a strong overlap of differentially expressed genes between the two diets. Moreover, a number of genes are differentially expressed specifically in rat cohorts exhibiting diet-induced life extension. Finally, we observe that LA supplementation inhibits histone deacetylase (HDAC) protein activity in vitro in rat astrocytes. We find a single microRNA, miR-98-3p, that is overexpressed during CR feeding and LA dietary supplementation; this microRNA alters HDAC and histone acetyltransferase (HAT) activity, which suggests a role for HAT/HDAC homeostasis in neuroprotection.
Conclusions
This study presents extensive data on the effects of diet and aging on the cerebral cortex transcriptome, and also emphasises the importance of epigenetics and post-translational modifications in longevity and neuroprotection.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-015-0847-2) contains supplementary material, which is available to authorized users.
Keywords: Epigenetics, lifespan, longevity, nutrigenomics, RNA-seq
Background
Ageing is arguably the major biomedical challenge of the 21st century with the incidence of age-related diseases, and neurodegenerative diseases in particular, expected to increase dramatically in the coming decades. Brain ageing frequently underlies cognitive decline and is a major risk factor for neurodegenerative conditions such as Alzheimer's disease (AD) and Parkinson's disease. Mental health is also a major concern of ageing adults. The exact molecular mechanisms underlying brain ageing, however, remain unknown [1].
A major tool available to researchers studying ageing is caloric restriction (CR), which consists of restricting food intake (without malnutrition) of organisms normally fed ad libitum (AL). CR is the only dietary intervention known to increase longevity and retard the process of ageing in several model organisms. In mice and rats, CR can increase longevity by ~30 %, delay physiological ageing and postpone or diminish the morbidity of most age-related diseases [2]. CR can also delay brain iron accumulation, preserving motor performance, and in one study increased survival in rhesus monkeys [3, 4]. However, as reported for rodent species [5, 6], there is significant variation in the mortality effects of CR feeding in different studies using rhesus monkeys [7, 8]. In elderly humans, CR has been shown to improve memory function [9].
α-Lipoic acid (LA) is an anti-oxidant which naturally occurs in many foods. It does not increase survival in rodent studies [10], but rats supplemented with LA during the AL phase of feeding when switched from CR to AL feeding at 12 months of age retained the extended longevity characteristic of CR feeding throughout life despite a return to AL feeding. This was termed a dietary memory effect [10]. These animals gained weight, indicating that although the longevity effects of CR were preserved, other effects of CR feeding were not. Conversely, animals switching from AL, with a diet supplemented with LA, to CR feeding without LA supplementation did not exhibit extended longevity, yet extended survival is a normal response of this feeding switch when LA is not included in the diet during AL feeding. This LA-induced memory effect gives us a unique opportunity to identify the mechanisms by which CR retards ageing and increases longevity without the confounding physiological effects of CR (e.g. restricted growth). Furthermore, LA can compensate for age-related, long-term memory deficits in old rats [11] and improve memory and learning in prematurely aged mice [12].
The underlying mechanisms of the beneficial effects of CR and LA supplementation are unknown. Understanding how these changes in diet and feeding regime can affect ageing, neurodegeneration and longevity is of great importance and could pinpoint key genes and pathways for further research relevant to human application [13]. The aim of this work was to study the transcriptomes, using RNA-seq, of the cerebral cortex of rats subjected to CR and LA-supplemented rats and compare them with animals maintained on an AL diet not supplemented with LA.
Using rat cerebral cortex from a previous study [10], which identified the dietary memory effect, our results show changes in expression of genes related to secretory and neuroendocrine pathways with ageing in AL-fed rats, many of which are prevented by CR. We find also that CR and LA supplementation share a largely common gene expression profile, despite the different effects on longevity. These shared genes are mainly neuroprotective and may act through prevention of excitoxicity, by down-regulation of glutamate receptors and calcium kinases, and inducing protection against oxidative stress by the up-regulation of glutathione S-transferases (GSTs) and thioredoxin. We also propose that this neuroprotective effect of CR and LA is driven by changes in histone deacetylase (HDAC) and histone acetyltransferase (HAT) homeostasis and implicate a miRNA (miR-98-3p) in this process. This was demonstrated using transfection, cell culture and HDAC/HAT activity assays. We show that genes involved in acetylation of histones are altered and may be important in the dietary memory effect of LA on longevity. Finally, we identify 137 genes that are associated with rat cohorts exhibiting diet-induced longevity. Many of these are evolutionarily conserved in mouse, worm and fly longevity networks.
Results
Using tissues from a previous study [10], we performed RNA-seq on the cerebral cortex of rats. Three basic feeding/dietary groups were established: rats fed AL throughout life; rats subjected to CR throughout life; or rats fed a LA-supplemented diet during the AL feeding phase before switching to or from CR feeding at 12 months of age. The longevity of each of these groups of animals and the number of differentially expressed (DE) genes are shown in Table 1. To establish the effect of ageing on the transcriptome and the impact of CR on age-related changes, rats were sampled at 6, 12 and 28 months of age.
Table 1.
Mean survival of rats on the various diets used in this study and the number of DE genes compared with AL feeding
| Rat cohortsa | Mean survival (days) from [10] | SE (days) | Number of DE genes compared with AL |
|---|---|---|---|
| AL | 854 | 22 | NA |
| *AL > CR | 1000 | 33 | 1028 |
| AL + LA > CR | 859 | 57 | 986 |
| *CR | 1025 | 25 | 941 |
| CR > AL | 914 | 44 | 1435 |
| *CR > AL + LA | 1009 | 34 | 1233 |
| AL + LA | 858 | 27 | 1003 |
aDiet switches are indicated by “>” and occurred at 12 months of age. Asterisks (*) indicate long-lived cohorts, all statistically significant versus AL. Benjamini-Hochberg false discovery rate <0.05; n = 6 per group (pooled two by two) for the RNA-seq; 10,620 genes expressed in total. NA not applicable, SE standard error
Changes observed in the ageing transcriptome are prevented by a calorie restricted diet in rats
We have previously published an analysis of whole transcriptome RNA-seq for an AL-fed ageing cohort (6, 12 and 28 months) [14]. In the current study we re-sequenced the AL-fed ageing cohort at a greater depth on the 5500XL SOLiD system (previously we used the SOLiD system v4), along with all the other experimental groups in this study to eliminate any platform bias. The major findings from the previous analysis were confirmed and the number of DE genes was similar, showing good replication (data not shown), but due to the updated annotation of the rat genome we found also a number of new DE genes.
Our new analysis emphasises an enrichment amongst DE genes with age for antigen presentation via MHC II (Fig. 1a; Additional file 1) and the down-regulation of heat shock proteins, as previously described [14]. However, statistically significant enrichment for synaptic vesicle cycle and vesicular transport related genes are also observed (Fig. 1a). Chromogranin B, which is important in secretion and neuroendocrine pathways, is down-regulated with age (6 versus 28 months and 12 versus 28 months), which is interesting as this gene is also down-regulated in AD [15]. Conversely, long-lived Snell’s dwarf mice show higher levels of chromogranin B [16]. Pcsk1n, an inhibitor of Pcsk1, which regulates the proteolytic cleavage of neuroendocrine peptide precursors, including chromogranin B [17], is under-expressed with age (12 versus 28 months). Moreover, down-regulation of genes related to synaptic transmission and synaptic vesicle transport (syntaxin-1b, syntaxin-binding protein 1, syntaxin-1A and Napb), post-synaptic signalling modulators (e.g. Syngap1, Shank1, Homer2 and Shank3) and a glutamate receptor (Grik4) occurs with age (6 versus 28 months) in the AL cohort. These distinct changes in secretory pathways, synaptic vesicle transport and synaptic transmission are consistent with observations of neurodegenerative diseases [18–20].
Fig. 1.
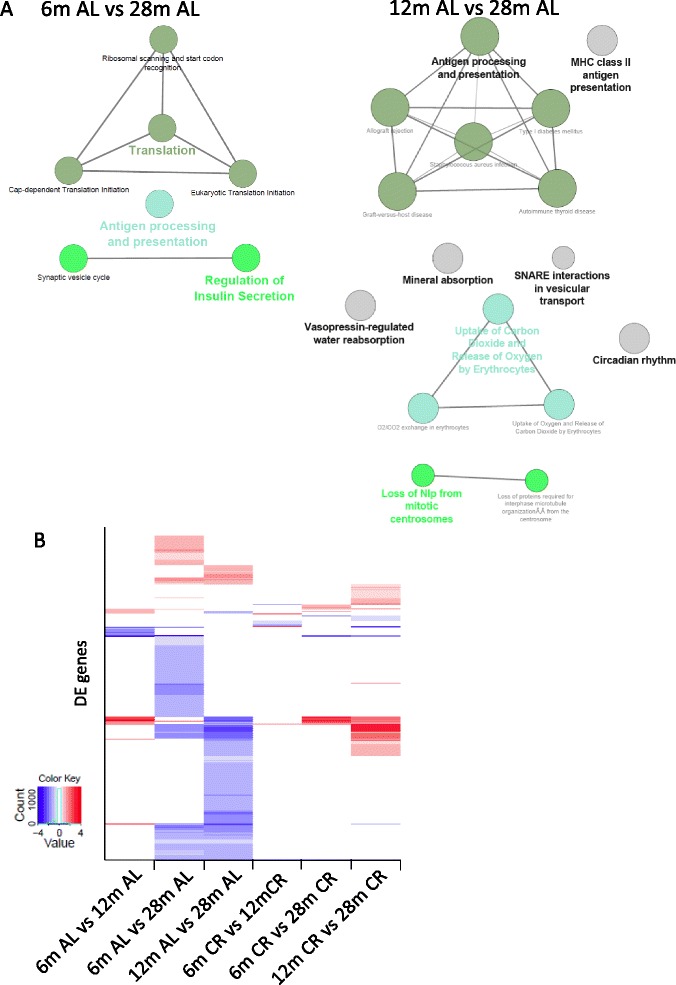
Age-related differential gene expression in rats fed AL and subjected to CR. a Two simplified networks of related, statistically significant enriched Gene Ontology (GO) terms using the Cytoscape add-on ClueGO [30, 31]. The network comparing 6 months AL (6 m AL) and 28 months AL (28 m AL) is shown on the left and 12 months AL (12 m AL) and 28 months AL (28 m AL) is shown on the right. DE genes were used to generate a GO term network. The filled coloured circles (nodes) represent each statistically significant parent GO term. The lines (edges) between the nodes show that there are overlapping genes between terms. The Cytoscape add-on ClueGO allows enrichment analysis and the collapsing of GO terms into parent categories for each comparison. Each of the terms is statistically significant (Benjamini-Hochberg correction <0.05). Colours represent shared GO terms. The different sizes of the nodes relate to how many genes fall into the terms. b Heatmap of the DE genes with age across all AL and CR datasets (Table 2); created in R using the heatmap3 package. The y-axis represents all the DE genes. Red = up-regulated, blue = down-regulated, log2 fold change reported for statistically significant DE genes. White means no statistically significant change in expression. 6 m = 6 months of age, 12 m = 12 months of age, 28 m = 28 months of age
Rev-erb-α (nuclear receptor 1D1) is a transcriptional repressor involved in circadian rhythmicity (an enriched term; Fig. 1a) and metabolism, which is down-regulated with age. Its mode of action is through the recruitment of the co-repressor NCoR, which is also down-regulated in our data, and activation of Hdac3 [21]. Nur77 (nuclear receptor 4A1), a transcription factor that is down-regulated, interacts with HAT, p300 and Hdac1, which regulates expression through acetylation [22]. These results are interesting because nuclear receptors are important in hormone homeostasis, sensing hormones and regulating the downstream expression of multiple genes through epigenetic mechanisms. All genes DE with age are presented in Additional file 2.
The majority of the gene expression changes observed with age in AL-fed rats are not present during ageing of CR rats (Fig. 1b), with the exception of Hspa1b (heat shock protein, −1.8 fold change (FC) in CR and −3.6 FC in AL groups), supporting the hypothesis that the ageing process is retarded by CR. Fewer genes are DE with age in CR than in AL (Table 2) and there are a limited number of genes which are only DE during ageing in rats subjected to CR (Fig. 1b).
Table 2.
Comparison of the number of genes differentially expressed with age in AL and CR groups
| Comparison | AL | CR |
|---|---|---|
| 6 versus 12 months | 24 (15 protein coding) | 10 (10 protein coding) |
| 6 versus 28 months | 180 (162 protein coding) | 17 (17 protein coding) |
| 12 versus 28 months | 174 (141 protein coding) | 70 (45 protein coding) |
Benjamini-Hochberg false discovery rate <0.05; n = 6 per group (pooled two by two); 10,620 genes expressed in total
Over-representation analysis (gene set enrichment analysis (GSEA), Molecular Signatures Database, Broad Institute) independently revealed that our data statistically significantly overlapped with a previous meta-analysis of ageing based on microarray studies (false discovery rate (FDR) q value = 2.12 × 10-7) [23] and with a list of genes DE after methyl binding protein knockdown (MeCP2, MB1 and MBD2) in Hela cells (1.8 × 10-4) [24]. This leads to the question: what is driving this altered gene expression profile with age?
The histone H3 methyltransferase Dot1L was downregulated with age in the AL cohort (−1.2 log FC in 6 months AL versus 28 months AL; −1.4 FC in 12 months AL versus 28 months AL). Previously, H3K79 methylation (regulated by DOT1L) was identified as a novel histone marker for ageing and cancer [25]. Tet3, an important regulator of DNA demethylation and H3K4 trimethylation [26–28], is down-regulated with age in our data (−1.2 FC in 12 months AL versus 28 months AL). Furthermore, the chromatin remodeller Chd5 (Chromodomain helicase DNA binding protein 5) was DE with age in our results and has previously been linked to ageing and AD [29].
Taken together, these results indicate that there are changes in the responsiveness of cells to hormonal signalling with age, as well as expression changes in genes related to chromatin remodelling and modification, possibly affecting transcription at various levels.
Neuroprotective genes are overexpressed in rats in response to CR and LA-supplemented diets
In order to understand the previously observed neuroprotective effects of CR and LA [3, 4, 9, 11, 12, 30], we focused on the response of rats to CR and LA-supplemented diets at 28 months of age. By directly comparing 28 months AL to 28 months CR, we found 895 DE genes enriched for glutamatergic synapse, oxidative phosphorylation and terpenoid backbone biosynthesis (Additional file 3). Furthermore, there is a 71 % overlap between genes DE in CR rats and in LA-fed rats at 28 months when compared with AL rats; this overlap is reduced to 49 % when including all the dietary variations in our study (Fig. 2b; Table 3; Additional file 4). By taking the overlapping DE genes induced by CR feeding and LA diets we generated an enrichment map based on Gene Ontology (GO) terms. Neurotrophin signalling pathway, essential in the survival of neurons through protection against excitotoxicity [31] and reported to change in the aged brain [32], is a key enriched term (Fig. 2a). Furthermore, neurotrophins protect neurons against excitotoxicity and rescue oxidative stress-mediated apoptosis by involvement of the phosphatidylinositol and mitogen-activated protein kinase (MAPK) pathways [33]. Using DAVID functional enrichment analysis we found that the majority of the down-regulated genes overlapping between CR feeding and LA diets are involved in neuroprotection (Additional file 4). Glutamate receptors and receptor activity genes were altered or enriched in our datasets, and we conducted quantitative PCR (qPCR) to confirm these findings (Fig. 2c).
Fig. 2.
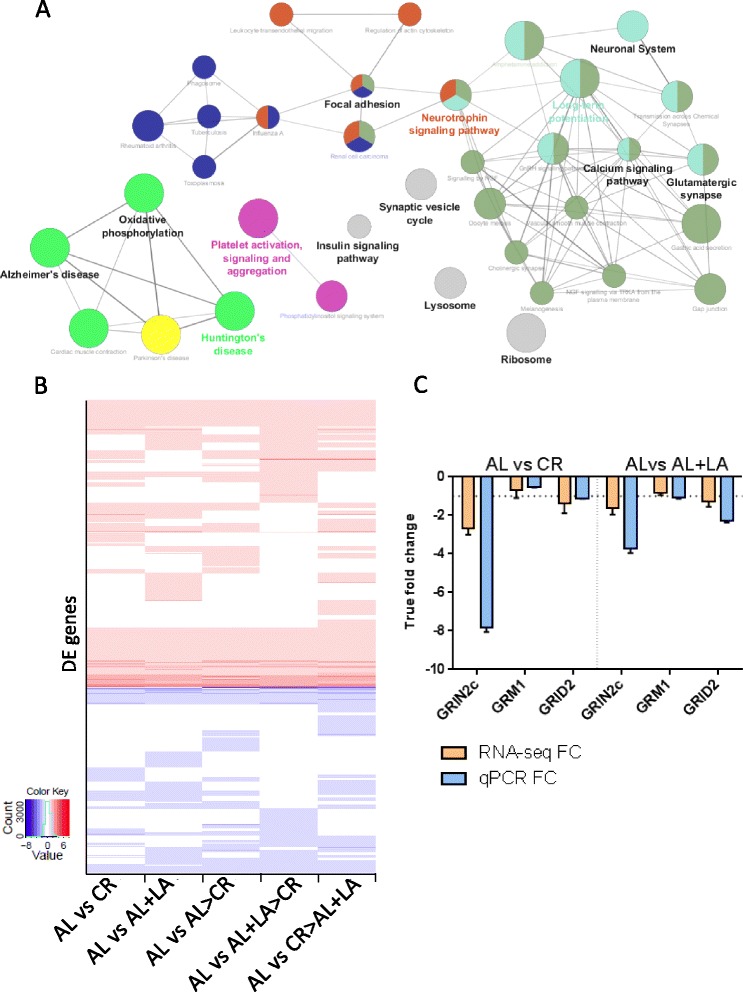
CR and LA supplementation elicit a similar neuroprotective gene expression profile. a Simplified networks of related statistically significant enriched GO terms using the Cytoscape add-on ClueGO [30, 31]. The network was made from the overlapping differentially expressed genes between all diets with CR and LA supplementation (a.k.a neuroprotective profile; see text for details). The Cytoscape add-on ClueGO allows enrichment analysis and the collapsing of GO terms into parent categories for each comparison. Each of the terms is statistically significant (Benjamini-Hochberg correction <0.05). The filled coloured circles (nodes) represent each statistically significant enriched parent GO term. The lines (edges) between the nodes show that there are overlapping genes within terms. The different sizes of the nodes relate to how many genes fall into the terms. b Heatmap of the differentially expressed genes (Table 1) across all diets with CR and LA supplementation when compared with AL; created in R using the heatmap3 package. Red = up-regulated, blue = down-regulated, log2 FC reported for statistically significant DE genes. White means no statistically significant change in expression. c Quantitative PCR confirmation of RNA-seq results, showing glutamate receptor differential expression (GRIN2c, GRM1 and GRID2) when comparing CR (left) and AL supplemented with LA (right) with AL alone. Orange bars are RNA-seq FC and blue bars are qPCR FC. The horizontal dotted line represents the one FC cutoff. The vertical dotted line separates the CR and LA supplemented comparisons. Error bars represent the standard error calculated for log2 FC as follows: (Standard error/Mean) × log2e. Hprt1, B2m and Ywhaz were used as reference genes. N = 6 per group, unpooled RNA from the RNA-seq experiment
Table 3.
Shared chromatin related and epigenetic regulator genes altered by CR and LA feeding
| Ensembl ID | Associated gene name | Description | AL versus CR log2 FC | AL versus CR FDR | AL versus AL+ LA log2 FC | AL versus AL+ LA FDR |
|---|---|---|---|---|---|---|
| ENSRNOG00000001442 | Por | NADPH--cytochrome P450 reductase [Source:UniProtKB/Swiss-Prot;Acc:P00388] | 1.24 | 8.58 × 10−06 | 0.75 | 1.19 × 10−02 |
| ENSRNOG00000043098 | Mt2A | Metallothionein-2 [Source:UniProtKB/Swiss-Prot;Acc:P04355] | 1.06 | 7.02 × 10−04 | 1.30 | 4.81 × 10−05 |
| ENSRNOG00000048961 | Bhlhe41 | Bhlhe41 | 1.06 | 5.45 × 10−04 | 0.92 | 7.17 × 10−03 |
| ENSRNOG00000047374 | Gnas | GNAS isoform GNASL [Source:RefSeq peptide;Acc:NP_062005] | 0.99 | 2.69 × 10−08 | 0.90 | 1.73 × 10−05 |
| ENSRNOG00000008001 | Rab3b | Ras-related protein Rab-3B [Source:UniProtKB/Swiss-Prot;Acc:Q63941] | 0.89 | 4.16 × 10−04 | 0.57 | 3.70 × 10−02 |
| ENSRNOG00000018958 | Mt3 | Metallothionein-3 [Source:UniProtKB/Swiss-Prot;Acc:P37361] | 0.77 | 3.94 × 10−03 | 0.58 | 3.88 × 10−02 |
| ENSRNOG00000005183 | Rtf1 | RNA polymerase-associated protein RTF1 homolog [Source:RefSeq peptide;Acc:NP_001102428] | 0.58 | 4.50 × 10−03 | 0.73 | 4.32 × 10−05 |
| ENSRNOG00000006816 | Urb5 | Ubiquitin protein ligase E3 component n-recognin 5 [Source:MGI Symbol;Acc:MGI:1918040] | −0.43 | 3.64 × 10−02 | −0.44 | 2.97 × 10−02 |
| ENSRNOG00000013837 | Trp53bp1 | Transformation related protein 53 binding protein 1 [Source:MGI Symbol;Acc:MGI:1351320] | −0.50 | 1.48 × 10−02 | −0.53 | 9.34 × 10−03 |
| ENSRNOG00000005544 | Kansl1 | KAT8 regulatory NSL complex subunit 1 [Source:MGI Symbol;Acc:MGI:1923969] | −0.53 | 2.46 × 10−02 | −0.61 | 1.26 × 10−02 |
| ENSRNOG00000004289 | Kdm6a | Lysine (K)-specific demethylase 6A [Source:MGI Symbol;Acc:MGI:1095419] | −0.59 | 1.24 × 10−02 | −0.55 | 7.33 × 10−03 |
| ENSRNOG00000003220 | H3f3c | Histone H3.3 [Source:UniProtKB/Swiss-Prot;Acc:P84245] | −0.72 | 4.50 × 10−02 | −0.75 | 1.50 × 10−02 |
| ENSRNOG00000011523 | H2afy | Core histone macro-H2A.1 [Source:UniProtKB/Swiss-Prot;Acc:Q02874] | −0.73 | 2.56 × 10−03 | −0.82 | 5.60 × 10−04 |
| ENSRNOG00000015781 | Ndst3 | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 [Source:RefSeq peptide;Acc:NP_001178645] | −0.82 | 4.27 × 10−03 | −0.77 | 8.10 × 10−03 |
| ENSRNOG00000046636 | HIST1H4B | Histone H4 Osteogenic growth peptide | −0.92 | 6.85 × 10−04 | -1.27 | 8.54 × 10−08 |
| ENSRNOG00000017714 | Usp3 | Ubiquitin carboxyl-terminal hydrolase 3 [Source:RefSeq peptide;Acc:NP_001020595] | −0.99 | 1.11 × 10−02 | −0.99 | 1.37 × 10−02 |
| ENSRNOG00000006689 | Chd7 | Chromodomain helicase DNA binding protein 7 [Source:MGI Symbol;Acc:MGI:2444748] | -1.09 | 3.92 × 10−03 | −0.93 | 1.60 × 10−02 |
| ENSRNOG00000014397 | Zic2 | zinc finger protein ZIC 2 [Source:RefSeq peptide;Acc:NP_001101862] | -1.81 | 5.00 × 10−04 | -2.06 | 7.44 × 10−05 |
Benjamini-Hochberg FDR <0.05; n = 6 per group (pooled two by two)
Strong enrichment of oxidative phosphorylation (DAVID enrichment score (ES) = 5.44, FDR = 8.93 × 10-10) and AD was found in the 490 shared up-regulated genes in CR and LA diets (Fig. 2a; Additional file 4). GSTs are a family of enzymes that play an important role in detoxification and protect neurons from oxidative insult [34]. During CR feeding and LA supplemented diets, four GSTs were up-regulated (Gsta1, Gstm1, Gstp1, Gstt3). Furthermore, thioredoxins are redox proteins that respond to reactive oxygen species, therefore acting as antioxidants, and three are up-regulated in response to CR feeding and LA supplementation (Txndc17, Txnip and Txnrd1). Oxidative stress is thought to be important in ageing, in particular in energy-rich tissues such as the brain, promoting degenerative disease and accelerating ageing [35]; therefore, up-regulation of antioxidant enzymes may be neuroprotective. Overall, changes in synaptic transmission and susceptibility to glutamate excitotoxicity have been described as a potential cause of neurodegeneration [36]. Our results suggest that CR and LA feeding induce gene expression changes that prevent excitotoxicity through down-regulation of glutamate receptors and calcium kinases, and protect against oxidative stress, inducing a neuroprotective profile.
We set out to identify the genes regulating this neuroprotective profile and noticed that a large number of chromatin-related and epigenetic regulators were altered due to CR feeding and LA supplementation (Table 3). This neuroprotective profile may be driven through changes in epigenetic regulators altering the acetylation and methylation status of DNA, histones and proteins. However, there are a number of epigenetic regulators that are DE specifically in response to either CR feeding or LA supplementation, and we predicted that there would be a shared regulator of this neuroprotective profile.
In order to identify candidates for this shared regulation of the neuroprotective profile, we performed a further small RNA-seq experiment (see “Materials and methods”) to assay microRNAs (miRNAs) in our samples. We found that each diet group elicits its own specific miRNA profile (Fig. 3a), with switching from AL supplemented with LA to CR at 12 months leading to the greatest number of DE miRNAS (Table 4; for all DE miRNAs see Additional file 5). However, a single miRNA (miR-98-3p) was significantly overexpressed in all groups of CR rats and/or rats fed LA-supplemented diets (Fig. 3b). As mir-98 down-regulation in the cerebellum has been associated with AD and with neuroprotection [37, 38], we examined whether this miRNA could regulate the neuroprotective profile observed.
Fig. 3.
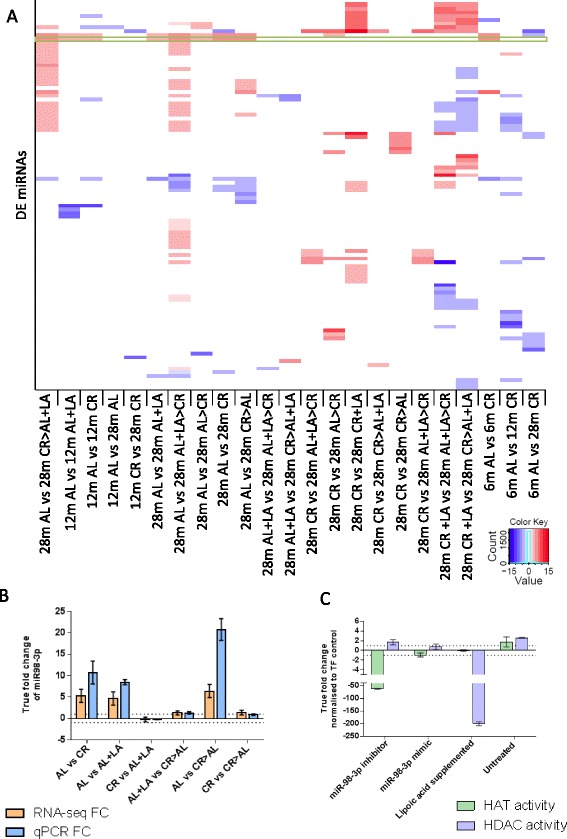
Each diet group and age elicits its own miRNA expression profile but miR-98-3p is overexpressed in all CR and LA diets and may affect HAT and HDAC activity. a Heatmap of the 175 DE miRNAs across all diets and ages; created in R using the heatmap3 package. mir-98-3p is highlighted by a green box. Red = up-regulated, and blue = down-regulated, log2 fold change reported for statistically significant DE genes. White means no statistically significant change in expression. b qPCR confirmation of RNA-seq results, showing miR98-3p differential expression in all CR and LA supplemented diets. Orange bars are RNA-seq log2 FC and blue bars are qPCR FC (corrected to provide the true FC in the sample by Ratio – 1 if the sample was up-regulated and by (–1/Ratio) + 1 for down-regulated samples). The horizontal dotted line represents the 1 FC cutoff. Errors bars represent the standard error calculated for log2 FC as follows: (Standard error/Mean) × log2e. Snord96a, Snord95 and Snord68 were used as reference genes. c The difference in HDAC and HAT activity was calculated as a ratio of the transfection (TF) control and then transformed into true FC (by Ratio – 1 if the sample was up-regulated and by (–1/Ratio) + 1 for down-regulated samples). A cutoff of 1 FC was used, represented by the dotted line. The figure shows that the miR-98-3p inhibitor significantly (p < 0.001, two-tailed t-test) decreases HAT activity when compared with HDAC activity. A miR-98-3p mimic appears not to affect HDAC/HAT balance (no statistically significant difference). LA reduces HDAC activity (p < 0.001) and the untreated cells show a slight increase in both HAT and HDAC activity, indicating that transfection might affect HDAC and HAT activity but not HDAC/HAT balance (no statistically significant difference). The error bars represent the standard deviation. Experiments were performed independently three times with n = 3 wells per treatment
Table 4.
Number of differentially expressed miRNAs in all diet comparisons
| Number of miRNAS DE | |
|---|---|
| AL and CR ageing | |
| 6 months AL versus 12 months AL | 0 |
| 6 months AL versus 28 months AL | 0 |
| 12 months AL versus 28 months AL | 1 |
| 6 months CR versus 12 months CR | 16 |
| 6 months CR versus 28 months CR | 9 |
| 12 months CR versus 28 months CR | 2 |
| AL versus CR | |
| 6 months AL versus 6 months CR | 4 |
| 12 months AL versus 12 months CR | 6 |
| 28 months AL versus 28 months CR | 6 |
| LA supplementation | |
| 28 months AL versus 28 month AL + LA | 3 |
| 28 months CR versus 28 months CR + LA | 20 |
| 12 months AL versus 12 months AL + LA | 5 |
| Diet switching | |
| 28 months AL versus 28 months AL > CR | 4 |
| 28 months AL versus 28 months AL + LA > CR | 39 |
| 28 months AL versus 28 months CR > AL + LA | 26 |
| 28 months AL versus 28 months CR > AL | 14 |
| 28 months CR versus 28 months AL + LA > CR | 5 |
| 28 months CR versus 28 months AL > CR | 7 |
| 28 months CR versus 28 months CR > AL + LA | 2 |
| 28 months CR versus 28 months CR > AL | 6 |
Benjamini-Hochberg FDR <0.05; n = 6 per group; 527 miRNAs expressed in total
To consider this question and relate it to our neuroprotective profile (Fig. 2a, c) induced by CR and LA diets, we investigated the role of miR-98-3p in HAT/HDAC homeostasis because changes in HAT/HDAC homeostasis have been noted as a potential cause of neurodegeneration [39]. Using CTX TNA2 cells (rat astrocytes derived from the cerebral cortex), we transfected miR-98-3p inhibitors and mimics, and measured HDAC and HAT activity (Fig. 3c). LA-supplemented cells showed a strong inhibition of HDAC activity as predicted from the structure of LA [40]. Increasing miR-98-3p (using a mimic) did not alter HDAC or HAT activity, suggesting that LA-induced inhibition of HDAC activity is not elicited through mir-98-3p. However, inhibition of miR-98-3p did up-regulate HDAC activity and greatly inhibited HAT activity. These results suggest that miR-98-3p is a candidate regulator of the neuroprotective profile by maintaining the HAT/HDAC balance and hence acetylation equilibrium.
Defining the dietary memory effect of LA on longevity
By far the most intriguing observation of Merry et al. [10] was the fact that LA cannot increase longevity alone but rats switching from CR to AL feeding supplemented with LA at 12 months of age retained the extended longevity characteristic of CR, despite being on an AL diet. Using our RNA-seq data we set out to disentangle the dietary memory effect of LA. Table 1 shows the mean survival of these rats on various feeding and dietary combinations and the number of DE genes compared with AL feeding. Figure 4a shows how we identified the extended longevity effect of CR switching to AL with LA. Briefly, we compared the lists of DE genes from the various cohorts in order to remove the confounding effects of diet switching and LA supplementation without extended longevity (Fig. 4a). This allowed us to focus on the genes associated specifically with the dietary memory effect (all DE genes are listed in Additional file 6). Dietary memory effect genes were enriched for the GO terms neurotrophin, insulin, phosphotidylinostol, NGF and GnRH signalling (Fig. 4b).
Fig. 4.
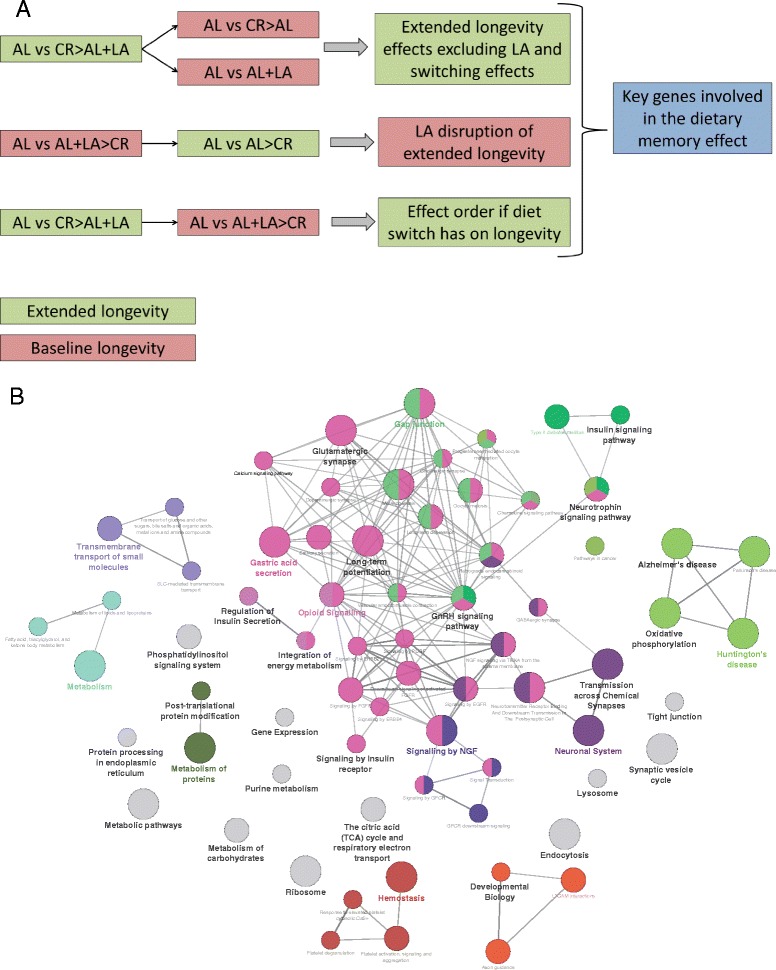
Dietary memory effect approach and networks. a The red boxes represent diet groups that have no effect on longevity (baseline longevity) and the green boxes represent diets that extend longevity. This shows how we compared the diet groups to identify the key genes involved in the dietary memory effect. First the DE gene list for AL versus CR > AL + LA (“>” = switched), which results in extended longevity, was compared with the DE gene lists for two conditions that do not result in extended longevity but do have a diet switch and LA supplementation, thereby identifying the genes for extended longevity but excluding the confounding effect of LA supplementation and diet switching. Next, DE genes from AL versus AL + LA > CR (baseline longevity) were compared with those from the extended longevity AL versus AL > CR condition, identifying genes LA induces that are important in disrupting the longevity effect of switching to CR. Finally, the effect of the order of LA supplementation (before or after diet switch) was identified. The three resulting lists were then compared to identify key genes involved in the dietary memory effect of LA. b Simplified networks of related statistically significant enriched GO terms using the Cytoscape add-on ClueGO [30, 31]. The network shows enrichment of gene function for the dietary memory effect genes (Additional file 6). The Cytoscape add-on ClueGO allows enrichment analysis and the collapsing of GO terms into parent categories for each comparison. The filled coloured circles (nodes) represent each statistically significant parent GO term. The lines (edges) between the nodes show that there are overlapping genes within each term. Each of the terms is statistically significant (Benjamini-Hochberg correction <0.05). The different sizes of the nodes relates to how many genes fall into that term
Genes involved in epigenetic regulation and specifically in the acetylation of histones are altered in the LA-supplemented long-lived groups. Examples include Nur77 (Nr4a1), which regulates gene expression through acetylation [22], and Morf4l1 (mortality like factor 1), which is involved in H2a and H4 acetylation and deacetylation (Additional file 6). These results suggest that histone acetylation is important in maintaining the CR longevity profile through LA supplementation. Further to this, other chromatin modifiers may play a role in the dietary memory effect (e.g. Bag4, Mef2c Hdac5, Bcorl1, H3f3b, Brd3, Kat6a, Hist1h1d and Chmp1a).
Genes involved in post-translational modification and metabolism of proteins are also enriched amongst genes potentially involved in the dietary memory effect (Fig. 4b). Examples include phosphatases/phosphorylases (Pp1r16a, Pygb, Ppm1f, Camk1g, Dusp8, Dusp6 and Dusp14 and a protein phosphatase inhibitor (Ppi2)) and kinases (Pacsin1, Stk32c, Cdkn1a (a.k.a. p21, a major player in cell senescence [41]), Sik1). This suggests that changes in the activity of target proteins through post-translational modification of non-histone proteins could be an important effect of LA supplementation and may play a role in its memory effect.
A previous study has noted a change in acetylation status of mitochondrial proteins with CR feeding [42]. Therefore, we also assessed non-histone protein acetylation using Western blotting (see “Materials and methods”). In our study we observed changes in global lysine acetylation status of proteins in the liver but not in the brain (Additional file 7). This suggests that post-translational modification of non-histone proteins may also be important, but possibly in a tissue- or protein-specific fashion.
Diet-induced longevity effects: 137 genes are associated with long-lived cohorts
We next wanted to define the gene expression changes specific to increased longevity. By focusing on our dietary groups with extended longevity (Table 1) we were able to pinpoint “longevity genes” that are DE specifically in such groups. We also took into account the effect of LA supplementation regardless of the longevity effect, i.e., we excluded genes showing the same trend in expression in LA-fed groups with no increase in longevity. This gave us a “longevity gene” list of 137 genes (Additional file 8) which are changed when longevity is increased by our different diets. These genes are enriched for mitochondrion, oxidative phosphorylation, AD (ES = 2.57, not FDR significant) and neuron differentiation and development (ES = 1.99, not FDR significant). We also identified Gad1 and Grik2 as being longevity associated, linking glutamate excitotoxicity to ageing and longevity. Regeneration and repair related genes were also identified (Swi5, Nrep and Smn1) in long-lived cohorts.
To further investigate and validate our “longevity genes”, we mapped the orthologues of the rat genes to longevity-associated networks from mouse, Drosophila and Caenorhabditis elegans (see “Materials and methods”). Twenty-eight of our “longevity genes” were previously identified longevity-associated genes (LAGs) or partners of LAGs (Fig. 5; Additional file 9). The overlap with LAGs and their partners supports the idea that we have identified genes which have a role in longevity (overlap significantly higher than expected by chance; p = 8.37E − 03). Moreover, some of our “longevity genes” were found in multiple networks. For example, Pik3r1, which is involved in insulin resistance and AD [43], is found in the mouse, Drosophila and C. elegans networks, suggesting that it could be an important conserved modulator of longevity. NPM1, Ubr5, Nduf6, Cox4i1 and Cytochrome c oxidase subunit Va are present in at least two of the networks, suggesting that these could also be important conserved modulators of longevity.
Fig. 5.
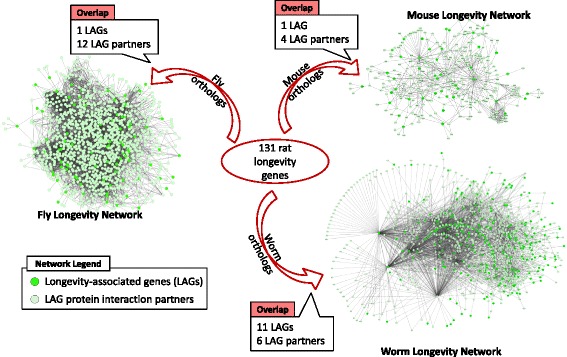
Overlap of longevity genes identified in the rat cerebral cortex with the longevity associated networks in mouse, Drosophila and C. elegans. The 28 longevity genes identified in rats in this study were found to be longevity-associated genes (LAGs) or partners of LAGs in the longevity networks for mouse, Drosophila and C. elegans (overlap significantly higher than expected by chance; p = 8.37E − 03, Fisher’s exact test). LAGs are genes which result in noticeable changes in the ageing phenotype and/or lifespan. These are identified experimentally through knockout, mutation, overexpression or RNA interference [76]. Longevity networks (previously described in detail [79]) are protein–protein interaction networks, include a core of LAGs, depicted in green in the figure, and their first order protein-interacting partners, shown in light green. The size of the network for each species is thus dependent on the protein–protein interaction data, but also on the number of LAGs available from the literature. All overlaps are higher than expected by chance (though this is not always statistically significant), with the overlap being significantly higher for worm LAGs, network partners of fly LAGs and network partners of mouse LAGs. Fisher’s exact test (one-tailed) p values for each of the overlaps are as follows: worm LAG p value = 6.11E − 05, fly LAG p value = 0.55, mouse LAG p value = 0.35, p value for LAG partners in worm longevity network = 0.09, p value for LAG partners in fly longevity network 0.027, p value for LAG partners in mouse longevity network = 0.034
Discussion
Our RNA-seq studies of the rat cerebral cortex during ageing and dietary manipulation of longevity provide insights at various levels. The cerebral cortex in ageing AL-fed rats is characterised by changes in synaptic density and vesicle transport that possibly affect synaptic transmission with increasing age. This fits with observations in humans of decreasing synaptic density after the age of 2–16 years, and a further decline between 74–90 years [44], and with models of neurodegeneration suggesting that age-related cognitive decline may be part of a continuum of neurodegenerative pathologies. We have found that key genes involved in DNA and histone methylation, transcription and chromatin re-modelling (e.g. the histone H3 methyltransferase Dot1L, Tet3 and Chd5) are differentially expressed with age in rats. In agreement with published studies [45, 46], this suggests that ageing is characterised by a global reduction in methylation, possibly driving the gene expression changes observed. Importantly, these age-related changes in gene expression are not observed with ageing when the rats are maintained on a CR feeding regime.
We show that a shared neuroprotective gene expression profile is induced by a CR feeding regime and also by LA supplementation (when fed AL). This neuroprotective signature is characterised by an altered response to oxidative stress. Glutathione (GSH) is an important antioxidant, the metabolism of which is impaired in the brain during anxiety, in memory impairment states, and age-related neurodegeneration [34]. Glrx (a member of the antioxidant system and up-regulated in this study) is increased in AD brains and may be involved in beta-amyloid toxicity [47]. Transgenic mice that overexpress thioredoxin are more resistant to inflammation [48] and show decreased mortality rates in the early part of the lifespan but maximal lifespan is not increased [49]. As LA supplementation of the diet alone does not increase longevity, it is unlikely that thioredoxins will increase longevity. They are, however, likely to contribute to a neuroprotective phenotype and it is interesting that we found them to be overexpressed. This emphasises the potential importance of anti-oxidant and diet research in improving brain health in the elderly. We also observed gene expression changes in glutamatergic activity with age and diet. Changes in glutamatergic activity have been observed previously in the rat hypothalamus and hippocampus [50] and glutamate-dependent neuroglial calcium signalling has been shown to differ between the young and the adult brain [51]. To our knowledge, ours is the first study to indicate that CR feeding and LA dietary supplementation alter the glutamatergic response in the rat cerebral cortex. It is of note that LA can potentiate glutamate receptor activity and has been suggested as a neuroprotective therapy option for AD and related dementias [52–55].
In our work, we identified a number of differentially expressed miRNAs with age and dietary intervention. miRNAs are of interest because neuronal degeneration has been linked to miRNA dysfunction in Drosophila [56] and knock-out of DICER in the forebrain of adult mice results in a neurodegenerative phenotype [57]. miRNAs are also involved in synaptic development and plasticity in mammalian neurons [58] and have been implicated in neurological conditions such as Parkinson’s disease and AD [58]. One miRNA, miR-98-3p, showed consistent overexpression in CR and LA supplementation of the diet in our study. Using transfection and HAT and HDAC assays, we demonstrated a potential role for this miRNA in maintaining HDAC and HAT balance, thereby contributing to a neuroprotective phenotype. The activity of HDACs is altered in CR rats, suggesting that global deacetylation may be important in ageing, longevity and neuroprotection [59, 60]. Also, LA exhibits HDAC inhibitor activity even if this seems to be cell type-specific [61]. Our data support previous findings indicating that epigenetic modifications have a significant role to play in ageing and dietary modulation of ageing.
Using our unique set of RNA-seq data we began to unravel the dietary memory effect and gene expression changes related to increased longevity of LA supplementation of the diet. We saw an enrichment for genes related to the insulin signalling pathway, which was first associated with ageing and longevity in C. elegans [62] and is known to modulate ageing from worms to mammals [63]. Phosphatidylinositols are part of the insulin signalling pathway [64] and are also enriched in our results. Metabolism and energy-related pathways, such as the tricarboxylic acid cycle, are also enriched. It is interesting to note that the tricarboxylic acid cycle is known to mediate the epigenetic landscape of chromatin in cancer by regulating DNA and histone methylation and it has been hypothesised that epigenetic changes with ageing could be due to an impaired Kreb’s cycle [65].
Using existing data sets on LAGs, we found that 28 of our genes specifically DE in long-lived cohorts were known LAGs. It should be noted that LAGs are identified through single gene manipulations (knockout or overexpression), followed by lifespan studies. This shows that some of the genes that can modulate lifespan, and more importantly many of their protein–protein interaction partners, are differentially expressed in response to CR feeding.
Conclusions
Additional studies are required to understand the role of epigenetics and post-translational modifications in ageing and longevity. This study presents extensive data on the effect of diet and ageing on the cerebral cortex transcriptome, serving as a starting point for future investigation into neuroprotection, longevity and dietary intervention in ageing and age-related disease. It also emphasises the importance of epigenetics and post-translational modification in ageing and longevity, and provides candidate genes for future studies.
Materials and methods
Diets
The non-supplemented CRM diet was supplied by Dietex International, Witham, UK. The LA-supplemented diet was the CRM diet enriched with a racemic mixture of DL-thioctic acid (α-lipoic acid) obtained from Fisher Scientific UK Ltd, at 1.5 g/kg by the Special Diet Systems Division of Dietex International. LA is known to have an anti-obesity effect by inhibiting food intake and retarding growth [66]; therefore, in the previous study by Merry et al. [10], it was ensured that the dose supplied to the animals did not inhibit the growth rate to induce an indirect dietary restriction effect.
Animals
A previous experiment supplied the rat brain tissue for this study [10]. All animal husbandry procedures undertaken in this study were carried out in accordance with the provisions of the United Kingdom Animals (Scientific Procedures) Act 1986. The animal study from which the tissue was generated for the current publication was conducted under the Home Office Project Licence 40/2964, held by B.J.M. The work was also reviewed by the University of Liverpool Ethical Committee for Animal Welfare. Male BN rats (SubstrainBN/SsNOlaHSD) were obtained from Harlan UK at 21–28 days of age and maintained under barrier conditions on a 12-h light:12-h dark cycle (08:00–20:00). The health status of the rats was monitored at regular intervals through the screening of sentinel animals. All animals were caged in groups of four and fed the non-supplemented CRM diet AL until 2 months of age, when they were transferred to single housing and assigned randomly to one of six dietary groups as summarised in Table 1. All rats fed AL and CR were sacrificed at 6, 12, and 28 months of age. Animals subjected to CR were fed either the CRM diet or the LA-supplemented CRM diet at 55 % the daily food intake of control rats to maintain their body weight at approximately 55 % that of age matched AL-fed animals. The diet of animals maintained on a CR regime was supplied daily as pre-weighed rations between 10:30 and 11:00 h. The daily food ration supplied to CR animals was the same for age-matched animals irrespective of whether they were fed the CRM diet or the LA-supplemented CRM diet. Six dietary feeding combinations were studied to compare the effect of AL or CR feeding with and without LA supplementation. Survival trajectories for these groups were previously determined [10]. All groups were run simultaneously and so experienced identical husbandry and housing conditions. None of the animals exhibited any signs of pathology when sacrificed. Each age group had six rats from which tissue samples were taken, flash frozen and stored at −80 °C.
Cortex dissection and RNA and miRNA extraction
To minimise thawing and therefore degradation of RNA, the cerebral cortex was removed from the whole brain on a solid CO2 base under a dissecting microscope. The cerebral cortex was cut into small pieces to aid RNA extraction.
RNA was extracted from the cerebral cortex using Qiagen's TissueLyser II and RNeasy lipid tissue kit. The quality of the extracted RNA was assessed using the Agilent 2100 Bioanalyser; all RNA integrity numbers (RINs) were above 8, indicating that good quality RNA had been extracted. Each sample was split in two for either whole transcriptome or small RNA analysis. For the whole transcriptome analysis the samples were pooled two by two (leaving three samples per age/diet group). Ribosomal RNA was removed from the pooled samples using a Eukaryote ribominus kit (Invitrogen) and confirmed with the Agilent 2100 Bioanalyser. Ribosomal removal, rather than poly(A) selection, allows certain non-coding RNAs without poly(A) tails to be included in the sequencing.
The miRNA fraction was enriched using the miRVana kit. The enriched miRNA samples were then pooled as with the whole transcriptome samples.
cDNA library preparation and SOLiD sequencing
The whole transciptome library preparation protocol was carried out according to the manufacturer’s instructions. The RNA was fragmented and cleaned up using spin columns (Invitrogen) and SOLiD RNA adapters were then hybridised and ligated to the samples. Reverse transcription was performed to generate cDNA. The cDNA was purified, size selected, amplified and then purified again (as detailed in the SOLiD protocol). The size distribution of the cDNA library was assessed using the Bioanalyser. The samples were then subjected to emulsion PCR and sequenced in the Centre for Genomic Research at the University of Liverpool (https://www.liverpool.ac.uk/genomic-research/) using the SOLiD system 5500xl to generate 75-bp forward reads.
The miRNA library preparation protocol was carried out according to the manufacturer’s instructions. The samples were then subjected to emulsion PCR and sequenced in the Centre for Genomic Research at the University of Liverpool using the SOLiD system v4 to generate 50-bp forward reads.
Whole transcriptome data mapping and differential expression analysis
The RNA-seq results from the SOLiD system were output as colour space fasta and quality files, and these files were mapped to the Ensembl release 71 rat reference genome (Rnor_5.0, March 2012) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml); the reads were filtered by quality using the –e phred quality setting (-e 400), multiple mapping reads were not allowed and the best option was used with mismatches limited to 2. For each sample approximately 36 million reads were generated. On average, 23 million reads per sample were mapped to the reference genome (approximately 63 % of reads generated were mapped; this figure is relatively low because of the conservative options specified in Bowtie but ensures that the highest quality and most robust alignments are reported). All data have been submitted to Gene Expression Omnibus (GEO) under the accession GSE57110. The differential expression analysis was carried out as in our previous study [14]. In brief, raw counts per gene were estimated by the Python script HTSeq count (http://www-huber.embl.de/users/anders/HTSeq/) and used by EdgeR [67] to estimate DE in pair-wise comparisons. Counts per million (cpm) were calculated and only genes with 1 cpm in at least three samples were included in the analysis. Trimmed mean of M-values (TMM) normalisation of the sequenced libraries was performed to remove effects due to differences in library size. EdgeR generates a FC for each gene; p values and the Benjamin-Hochberg FDR were calculated to statistically test the measured DE. As in previous studies, no effect of size cutoff was used, as ageing-related changes tend to be subtle [23].
Enrichment analysis and heatmap generation
To create heatmaps of DE genes, R and the R package heatmap3 were used along with the log2 FC output from EdgeR. To assess the biological significance of gene expression changes we used the Cytoscape plug-in ClueGO to perform an enrichment analysis on each individual comparison and then, using a built-in algorithm, the GO terms were collapsed based on related terms and statistical significance in order to give a simplified network [68, 69]. Further enrichment analysis was performed by GSEA [70, 71] and DAVID [72].
Whole transcriptome qPCR validation
To generate cDNA for qPCR, 3.5 μg of total RNA was reverse transcribed using Superscript III First-strand synthesis system for RT-PCR (Invitrogen, Paisley, UK). The Roche Universal Probe Library was used to design primers with sequences obtained from Ensembl. All primers were designed to cross an exon–exon boundary. The specificity of the primers was checked using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A reference gene experiment was conducted to identify the most stably expressed genes in the cerebral cortex with age and diet (data not shown); Hprt1 (Additional file 10), B2m (Rn00560865_m1, Applied Biosystems) and Ywhaz (Rn00755072_m1 Applied Biosystems) were the most stably expressed in all conditions. The reference genes were used to normalise the qPCR results. Additional file 10 shows the primers, amplicons, and probes used. The qPCR assays were all performed in triplicate using a TaqMan™ ABI PRISM 7500 fast (Applied Biosystems, Foster City, CA, USA) in 96-well plate format. A 20-ml reaction volume was used per well, consisting of 10 μl Taqman 2× PCR master mix (Universal PCR Mastermix, Applied Biosystems), 0.2 μl each of 20 mM forward and reverse primers, 0.2 μl of 10 mM probe (Exiqon, Roche Diagnostics Ltd), 0.2 μl distilled water and 9.2 μl of cDNA or water for the negative controls. The amplification was performed as follows: 2 min at 50 °C, 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The efficiency of the assays was between 93 % and 107 % and the R2 values were >0.98. The ΔΔcT method was used to measure expression. The data were further corrected by the efficiency of the standard curve for each gene. Log2 FC was calculated and compared with the RNA-seq results in order to confirm the expression results. The standard error was calculated for log2 FC as follows: (Standard error/Mean) × log2e. For qPCR the relative quantification value was used to calculate standard error. For RNA-seq, raw reads converted into relative values were used to calculate standard errors.
miRNA mapping
The RNA-seq results from the SOLiD system were output as colour space fasta and quality files. Using a custom script the base space adapter sequences was converted into colour space. A freely available Python script (cutadapt; https://cutadapt.readthedocs.org/en/stable/) was used to remove the adapter sequence and output a cfastq file. The freely available stand-alone Java program miRanalyzer [73] was used for mapping and analysis (http://bioinfo2.ugr.es/miRanalyzer/standalone.html). Using the Perl scripts provided by miRanalyzer, the reads were grouped and then mapped using Bowtie. miRanalyzer maps to a Bowtie index file for the rat; it then removes reads mapping to RefSeq (known mRNA) and Rfam (other known non-coding RNAs), leaving the user with reads that are likely to be miRNAs. New novel miRNAs are identified by miRanalyzer, detailing the secondary structure and number of times predicted by the five algorithms it uses. The results are also split into files for uniquely mapping and ambiguously mapping reads. This results in very clean results which are uniquely mapped.
miRNA differential expression analysis
The differential expression analysis was carried out using a range of custom Python scripts, conditional quantile normalisation [74] and EdgeR. Conditional quantile normalisation and Lowess normalisation have been shown to be most appropriate for miRNA analysis [75]. The known miRNAs were analysed separately from the novel predictions. Only reads with one count per million in at least three samples were included in the analysis. After normalisation and exclusion of low read counts, the differential expression analysis was carried out using EdgeR’s generalised linear model and tag-wise dispersion. A FDR cutoff of 0.05 was used.
miRNA qPCR
Validation of the miRNA differential expression results was carried out using the miScript PCR system (Qiagen). Reverse transcription using the miScript II RT kit was carried out according to the manufacturer’s instructions: 4 μl 5× miScript HiSpec buffer, 2 μl 10× miScript nucleics mix, 2 μl miScript reverse transcriptase mix into a total volume of 20 μl with 10 ng to 2 μg of template miRNA. This was incubated for 60 min at 37 °C and then 5 min at 95 °C. The resulting cDNA was diluted by adding 200 μl of distilled water. The miScript SYBR Green PCR Kit was used for the qPCR reaction. All samples were performed in triplicate in a 96-well plate format, using 2.5 μl cDNA, 12.5 μl 2× quantiTECT SYBR, 2.5 μl 10× miScript universal primers, 2.5 μl miScript primer assay, 5 μl RNase free water per well. Six small nucleolar RNAs (snoRNAs) provided and validated by Qiagen were used as the reference genes. Subsequently, a reference gene stability experiment ascertained that only three reference genes were required (Snord96a, Snord95 and Snord68).
LAG analysis
The comparison with known mouse, fly and worm LAGs was done using data from the Human Ageing Genomics Resources GenAge database, using build 16 [76]. Protein–protein interaction data for the construction of longevity networks and for the analysis of LAG partners were retrieved from the BioGRID database [77, 78], release 3.1.83. The construction of longevity networks has been described in detail previously [79]. Briefly, the networks include LAGs as a core set, and their first order interaction partners. Only the largest connected component is kept in the network.
In order to compare the rat “longevity genes” with genes from other species, orthologs were obtained using the InParanoid7 database [80]. Exclusion of inparalogs was done for the default threshold score of 0.05.
Cell culture
CTX TNA2 cells were purchased from HPA cultures (UK). The cells were maintained in DMEM (Life Tech) with high glucose, L-glutamine, phenol red, sodium pyruvate. The DMEM was supplemented with 10 % foetal bovine serum and penicillin-streptomycin-glutamine. The cells were sub-cultured three times a week. Passages 12–14 were used for the experiments.
Transfection
The confluent cells were sub-cultured and diluted 1 in 10 in antibiotic-free media and then counted using a haemocytometer. The cells were diluted to 1 × 104 and 100 μl of cell solution was added to each well of a 96-well plate. The cells were incubated overnight at 37 °C, 5 % CO2 to allow them to attach. The transfection was performed using the Dharmacon miRNA system. Mimics and inhibitors for miR-98-3p, positive controls, including a miRNA mimic that inhibits GAPDH and a miRNA mimic that increases miR-16, and two negative controls (mimic and inhibitor) were used to assess the transfection efficiency and viability by comparing with a transfection reagent-only control and an untreated control. The viability was assessed using the Alamar Blue assay and by 96 hours the mimic was at 115 % viability compared with the transfection control, and the inhibitor was at 86 % viability compared with the transfection control (see Additional file 11 for results). The mRNA was extracted using the cells-to-CT kit (Life Tech) according to the manufacturer’s protocol. The miRNA was extracted using the miRvana kit as above. cDNA synthesis and qPCR were performed as above using Actb as the reference gene for the mRNA experiment. The transfection efficiency was ascertained by qPCR by measuring GAPDH and miR-16 knock down. GAPDH was reduced 4.2-fold and miR-16 was reduced 5.75-fold (see Additional file 11 for qPCR results).
Nuclear extraction from cells and HAT and HDAC activity assay
The EpiQuik Nuclear Extraction Kit was used according to the manufacturer’s instructions to extract the nuclear proteins from transfected cells.
The Epigenase HDAC activity direct assay kit and the EpiQuik HAT activity kit (Cambridge Biosciences) were used to assess HAT and HDAC activity in transfected cells. The kit is an antibody-based assay with a colourmetric readout, assessed by the Multiskan microplate reader at 450 nm.
Western blotting
Frozen liver tissue was defrosted and homogenised in an approximately equal volume of protein extraction buffer (50 mM Tris-HCl, pH 6.8, 86 nM 2-mercaptoethanol, 2 % sodium dodecyl sulfate (SDS) and general protease inhibitors (cocktail, Sigma, category no. P8340), centrifuged at 13,000 rpm for 15 min and the supernatant containing the soluble protein collected. The protein concentration of the supernatant was determined using a bicinchoninic acid assay according to the supplier's protocol (Abcam, Sigma) adapted to a 96-well plate format. Absorbance at 562 nm was measured and protein concentrations were back-calculated from the standard curve using the SpectroMax software (Molecular Dynamics). Samples were diluted to 5 mg protein ml−1.
SDS PAGE was carried out on a large BIO-RAD kit using a 7.5 % or 12 % SDS-polyacrylamide gel and containing 1.5 M Tris-HCl (pH 8.8). Two SDS-PAGE runs were carried out simultaneously in a standard 0.1 % SDS, Tris-Glycine running buffer. The mini gels were electrophoresed overnight at 45 V. Proteins were then transferred at 350 mA (4 hours) in transfer buffer (20 % methanol, Tris-Glycine buffer) onto a nitrocellulose membrane (Amersham Hybond ECL). Membranes were blocked with 5 % dried skimmed milk (Marvel) dissolved in Tris-buffered saline (TBS; pH 8.8). Membranes were then washed twice for 10 min in changes of TBS before being incubated for 1 hour with a 1/5000 dilution of mouse anti-acetylated lysine antibody (Ac-K-103, Cell Signalling Technologies). Membranes were incubated with the secondary horseradish peroxidise-conjugated anti-mouse antibody (Amersham ECL) at a 1/10,000 dilution in blocking solution (50 ml per membrane) for 1–2 hours. Visualisation of the protein bands was by enhanced chemiluminescence (ECL). The gels were stripped and re-probed with beta actin as a loading gel control.
The developed films were analysed using ChemiImager v4.4 software to give an arbitrary quantitative value to allow comparison of band intensity. This was adjusted to account for background, area and absolute protein concentration (by normalising to the loading gel control intensity).
Data availability
All data have been submitted to GEO under the accession GSE57110.
Ethics approval
The animal study from which the tissue was generated for the current publication was conducted under the Home Office Project Licence 40/2964, held by B.J.M. The work was also reviewed by the University of Liverpool Ethical Committee for Animal Welfare.
Acknowledgements
This project was funded by a BBSRC grant (BB/H008497/1) to JPM. The tissue samples were generated as part of a BBSRC award (ERA164170) to BJM. RT was supported by a Marie Curie FP7-PEOPLE-2011-IEF Fellowship within the 7th European Community Framework Programme. Thanks to the other group members, especially Michael Keane for useful discussions and assistance and Chris Lofthouse for assistance with the Western blots. Thanks to staff at the Liverpool Centre for Genomic Research for their technical support, especially John Kenny. We also thank Ben Saer and Toryn Poolman at the University of Manchester for their technical assistance and support. Final thanks go to an anonymous reviewer whose comments were incredibly helpful in the editing of this manuscript as well as to four other anonymous reviewers.
Abbreviations
- AD
Alzheimer's disease
- AL
Ad libitum
- CR
Caloric restriction
- DE
Differentially expressed
- FDR
False discovery rate
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GSEA
Gene set enrichment analysis
- GST
Glutathione S-transferase
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- LA
α-Lipoic acid
- LAG
Longevity-associated gene
- miRNA
MicroRNA
- PCR
Polymerase chain reaction
- qPCR
Quantitative PCR
- SDS
sodium dodecyl sulfate
Additional files
All the DAVID enrichments for each comparison. (XLSX 2288 kb)
All genes DE with age. (XLSX 44 kb)
Enriched GO terms as a network generated from genes differentially expressed between AL and CR. (PDF 64 kb)
The overlap between genes DE when feeding CR and LA dietary supplementation at 28 months of age when compared with AL feeding the non-supplemented diet. It also shows the DAVID enrichment of the overlaps. (XLSX 375 kb)
All DE miRNAs for all diets and ages. (XLSX 17 kb)
All genes altered in three comparisons. First, the extended longevity effect of CR switching to AL with lipoic acid. Next, the inhibitory effect lipoic acid had on extended longevity when switching from AL to CR feeding. Finally, the genes specific to CR feeding when switched to AL feeding with lipoic acid supplementation in comparison with the opposite switch which does not induce increased longevity. The DAVID enrichments are also included. (XLSX 157 kb)
Western blots for lysine acetylation in liver and in brain in multiple diet groups. (PDF 146 kb)
The 137 longevity genes and the DAVID enrichments. (XLSX 48 kb)
The 28 longevity genes that have been identified as longevity-associated genes (LAGs) or partners of LAGs. (XLSX 10 kb)
The primers, amplicons, and probes used in the qPCRs. (XLSX 9 kb)
The transfection efficiencies and viability after transfection with mimics and inhibitors. (XLSX 62 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SHW conceived the design, conducted the RNA-seq, qPCR, HAT/HDAC assays, miRNA mimic and inhibitor cell culture experiments, analysed the data and wrote the manuscript. SVD, TC, and RT contributed to the analysis of data and preparation of the manuscript. AO conducted the Western blotting, performed the analysis and revised the manuscript. BJM conceived the design, interpreted the data, assisted in the experiments and critically revised the manuscript. JPM conceived the design, interpreted the data and critically revised the manuscript. All authors read and approved the final manuscript.
References
- 1.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–22. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Kastman EK, Willette AA, Coe CL, Bendlin BB, Kosmatka KJ, McLaren DG, et al. A calorie-restricted diet decreases brain iron accumulation and preserves motor performance in old rhesus monkeys. J Neurosci. 2012;32:11897–904. doi: 10.1523/JNEUROSCI.2553-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendlin BB, Canu E, Willette A, Kastman EK, McLaren DG, Kosmatka KJ, et al. Effects of aging and calorie restriction on white matter in rhesus macaques. Neurobiol Aging. 2011;32:2319. doi: 10.1016/j.neurobiolaging.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rikke BA, Liao C-Y, McQueen MB, Nelson JF, Johnson TE. Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp Gerontol. 2010;45:691–701. doi: 10.1016/j.exger.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper JM, Leathers CW, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–9. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman R, Anderson R, Johnson S, Kastman E. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–21. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witte AV, Fobker M, Gellner R, Knecht S, Flöel A. Caloric restriction improves memory in elderly humans. Proc Natl Acad Sci U S A. 2009;106:1255–60. doi: 10.1073/pnas.0808587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merry BJ, Kirk AJ, Goyns MH. Dietary lipoic acid supplementation can mimic or block the effect of dietary restriction on life span. Mech Ageing Dev. 2008;129:341–8. doi: 10.1016/j.mad.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Stoll S, Hartmann H, Cohen SA, Müller WE. The potent free radical scavenger alpha-lipoic acid improves memory in aged mice: putative relationship to NMDA receptor deficits. Pharmacol Biochem Behav. 1993;46:799–805. doi: 10.1016/0091-3057(93)90204-7. [DOI] [PubMed] [Google Scholar]
- 12.Poon HF, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, Morley JE, et al. Proteomic analysis of specific brain proteins in aged SAMP8 mice treated with alpha-lipoic acid: implications for aging and age-related neurodegenerative disorders. Neurochem Int. 2005;46:159–68. doi: 10.1016/j.neuint.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 13.de Magalhães JP, Wuttke D, Wood SH, Plank M, Vora C. Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol Rev. 2012;64:88–101. doi: 10.1124/pr.110.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood SH, Craig T, Li Y, Merry B, de Magalhães JP. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age (Dordr) 2013;35:763–76. doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechner T, Adlassnig C, Humpel C, Kaufmann WA, Maier H, Reinstadler-Kramer K, et al. Chromogranin peptides in Alzheimer’s disease. Exp Gerontol. 2004;39:101–13. doi: 10.1016/j.exger.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Alderman JM, Flurkey K, Brooks NL, Naik SB, Gutierrez JM, Srinivas U, et al. Neuroendocrine inhibition of glucose production and resistance to cancer in dwarf mice. Exp Gerontol. 2009;44:26–33. doi: 10.1016/j.exger.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laslop A, Weiss C, Savaria D, Eiter C, Tooze SA, Seidah NG, et al. Proteolytic processing of chromogranin B and secretogranin II by prohormone convertases. J Neurochem. 1998;70:374–83. doi: 10.1046/j.1471-4159.1998.70010374.x. [DOI] [PubMed] [Google Scholar]
- 18.Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–82. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Chung CY, Koprich JB, Siddiqi H, Isacson O. Dynamic changes in presynaptic and axonal transport proteins combined with striatal neuroinflammation precede dopaminergic neuronal loss in a rat model of AAV alpha-synucleinopathy. J Neurosci. 2009;29:3365–73. doi: 10.1523/JNEUROSCI.5427-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millecamps S, Julien J-P. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–76. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- 21.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–9. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 22.Kang S-A, Na H, Kang H-J, Kim S-H, Lee M-H, Lee M-O. Regulation of Nur77 protein turnover through acetylation and deacetylation induced by p300 and HDAC1. Biochem Pharmacol. 2010;80:867–73. doi: 10.1016/j.bcp.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 23.de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–81. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Serra L, Ballestar E, Ropero S, Setien F, Billard L-M, Fraga MF, et al. Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene. 2008;27:3556–66. doi: 10.1038/sj.onc.1211022. [DOI] [PubMed] [Google Scholar]
- 25.Kim W, Kim R, Park G, Park J-W, Kim J-E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J Biol Chem. 2012;287:5588–99. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang J, Kalantry S, Rao A. PGC7, H3K9me2 and Tet3: regulators of DNA methylation in zygotes. Cell Res. 2013;23:6–9. doi: 10.1038/cr.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–56. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langemeijer SMC, Aslanyan MG, Jansen JH. TET proteins in malignant hematopoiesis. Cell Cycle. 2009;8:4044–8. doi: 10.4161/cc.8.24.10239. [DOI] [PubMed] [Google Scholar]
- 29.Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH, et al. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lettieri Barbato D, Baldelli S, Pagliei B, Aquilano K, Ciriolo MR. Caloric restriction and the nutrient-sensing PGC-1α in mitochondrial homeostasis: new perspectives in neurodegeneration. Int J Cell Biol. 2012;2012:759583. doi: 10.1155/2012/759583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen TL, Kim CK, Cho J-H, Lee K-H, Ahn J-Y. Neuroprotection signaling pathway of nerve growth factor and brain-derived neurotrophic factor against staurosporine induced apoptosis in hippocampal H19-7 cells. Exp Mol Med. 2010;42:583. doi: 10.3858/emm.2010.42.8.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prenderville JA, Kennedy PJ, Dinan TG, Cryan JF. Adding fuel to the fire: the impact of stress on the ageing brain. Trends Neurosci. 2014;38:13–25. doi: 10.1016/j.tins.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Skaper SD, Floreani M, Negro A, Facci L, Giusti P. Neurotrophins rescue cerebellar granule neurons from oxidative stress-mediated apoptotic death: selective involvement of phosphatidylinositol 3-kinase and the mitogen-activated protein kinase pathway. J Neurochem. 2002;70:1859–68. doi: 10.1046/j.1471-4159.1998.70051859.x. [DOI] [PubMed] [Google Scholar]
- 34.Björk K, Saarikoski ST, Arlinde C, Kovanen L, Osei-Hyiaman D, Ubaldi M, et al. Glutathione-S-transferase expression in the brain: possible role in ethanol preference and longevity. FASEB J. 2006;20:1826–35. doi: 10.1096/fj.06-5896com. [DOI] [PubMed] [Google Scholar]
- 35.Tchaikovskaya T, Fraifeld V, Urphanishvili T, Andorfer JH, Davies P, Listowsky I. Glutathione S-transferase hGSTM3 and ageing-associated neurodegeneration: relationship to Alzheimer’s disease. Mech Ageing Dev. 2005;126:309–15. doi: 10.1016/j.mad.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 36.Salińska E, Danysz W, Łazarewicz JW. The role of excitotoxicity in neurodegeneration. Folia Neuropathol. 2005;43:322–39. [PubMed] [Google Scholar]
- 37.Babenko O, Kovalchuk I, Metz GA. Epigenetic programming of neurodegenerative diseases by an adverse environment. Brain Res. 2012;1444:96–111. doi: 10.1016/j.brainres.2012.01.038. [DOI] [PubMed] [Google Scholar]
- 38.Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–9. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–50. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–9. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Magalhães JP, Chainiaux F, de Longueville F, Mainfroid V, Migeot V, Marcq L, et al. Gene expression and regulation in H2O2-induced premature senescence of human foreskin fibroblasts expressing or not telomerase. Exp Gerontol. 2004;39:1379–89. doi: 10.1016/j.exger.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liolitsa D, Powell J, Lovestone S. Genetic variability in the insulin signalling pathway may contribute to the risk of late onset Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2002;73:261–6. doi: 10.1136/jnnp.73.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peter RH. Synaptic density in human frontal cortex — Developmental changes and effects of aging. Brain Res. 1979;163:195–205. doi: 10.1016/0006-8993(79)90349-4. [DOI] [PubMed] [Google Scholar]
- 45.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–7. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 46.Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akterin S, Cowburn RF, Miranda-Vizuete A, Jiménez A, Bogdanovic N, Winblad B, et al. Involvement of glutaredoxin-1 and thioredoxin-1 in beta-amyloid toxicity and Alzheimer’s disease. Cell Death Differ. 2006;13:1454–65. doi: 10.1038/sj.cdd.4401818. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida T, Nakamura H, Masutani H, Yodoi J. The involvement of thioredoxin and thioredoxin binding protein-2 on cellular proliferation and aging process. Ann N Y Acad Sci. 2005;1055:1–12. doi: 10.1196/annals.1323.002. [DOI] [PubMed] [Google Scholar]
- 49.Pérez VI, Cortez LA, Lew CM, Rodriguez M, Webb CR, Van Remmen H, et al. Thioredoxin 1 overexpression extends mainly the earlier part of life span in mice. J Gerontol A Biol Sci Med Sci. 2011;66:1286–99. doi: 10.1093/gerona/glr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal S, Poddar MK. Long-term exposure of variable dietary protein-to-carbohydrate ratio: effect on brain regional glutamatergic activity with age. Neurochem Res. 2008;33:952–61. doi: 10.1007/s11064-007-9460-4. [DOI] [PubMed] [Google Scholar]
- 51.Sun W, McConnell E, Pare J-F, Xu Q, Chen M, Peng W, et al. Glutamate-dependent neuroglial calcium signaling differs between young and adult brain. Science. 2013;339:197–200. doi: 10.1126/science.1226740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holmquist L, Stuchbury G, Berbaum K, Muscat S, Young S, Hager K, et al. Lipoic acid as a novel treatment for Alzheimer’s disease and related dementias. Pharmacol Ther. 2007;113:154–64. doi: 10.1016/j.pharmthera.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Hager K, Kenklies M, McAfoose J, Engel J, Münch G. Alpha-lipoic acid as a new treatment option for Alzheimer’s disease--a 48 months follow-up analysis. J Neural Transm Suppl. 2007;189–93. [DOI] [PubMed]
- 54.Maczurek A, Hager K, Kenklies M, Sharman M, Martins R, Engel J, et al. Lipoic acid as an anti-inflammatory and neuroprotective treatment for Alzheimer’s disease. Adv Drug Deliv Rev. 2008;60:1463–70. doi: 10.1016/j.addr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N, Landreh M, Cao K, Abe M, Hendriks G-J, Kennerdell JR, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–23. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hébert SS, Papadopoulou AS, Smith P, Galas M-C, Planel E, Silahtaroglu AN, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–69. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 58.Barbato C, Giorgi C, Catalanotto C, Cogoni C. Thinking about RNA? MicroRNAs in the brain. Mamm Genome. 2008;19:541–51. doi: 10.1007/s00335-008-9129-6. [DOI] [PubMed] [Google Scholar]
- 59.Chung KW, Kim DH, Park MH, Choi YJ, Kim ND, Lee J, et al. Recent advances in calorie restriction research on aging. Exp Gerontol. 2013;48(10):1049–53. doi: 10.1016/j.exger.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 60.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2012;339:211–4. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Mark K, Chen JS, Steliou K, Perrine SP, Faller DV. Alpha-lipoic acid induces p27Kip-dependent cell cycle arrest in non-transformed cell lines and apoptosis in tumor cell lines. J Cell Physiol. 2003;194:325–40. doi: 10.1002/jcp.10205. [DOI] [PubMed] [Google Scholar]
- 62.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 63.Wuttke D, Connor R, Vora C, Craig T, Li Y, Wood S, et al. Dissecting the gene network of dietary restriction to identify evolutionarily conserved pathways and new functional genes. PLoS Genet. 2012;8:e1002834. doi: 10.1371/journal.pgen.1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matuoka K, Chen KY, Takenawa T. A positive role of phosphatidylinositol 3-kinase in aging phenotype expression in cultured human diploid fibroblasts. Arch Gerontol Geriatr. 2003;36:203–19. doi: 10.1016/S0167-4943(02)00151-6. [DOI] [PubMed] [Google Scholar]
- 65.Salminen A, Kauppinen A, Hiltunen M, Kaarniranta K. Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process. Ageing Res Rev. 2014;16:45–65. doi: 10.1016/j.arr.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Kim M-S, Park J-Y, Namkoong C, Jang P-G, Ryu J-W, Song H-S, et al. Anti-obesity effects of alpha-lipoic acid mediated by suppression of hypothalamic AMP-activated protein kinase. Nat Med. 2004;10:727–33. doi: 10.1038/nm1061. [DOI] [PubMed] [Google Scholar]
- 67.Robinson M, Mccarthy D, Chen Y, Smyth GK. edgeR: differential expression analysis of digital gene expression data user’s guide. 2011. https://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf. [DOI] [PMC free article] [PubMed]
- 68.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 72.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 73.Hackenberg M, Sturm M, Langenberger D, Falcón-Pérez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37(Web Server issue):W68–76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–16. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;18(6):1279–88. doi: 10.1261/rna.030916.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, et al. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2012;41:D1027–33. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tacutu R, Budovsky A, Fraifeld VE. The NetAge database: a compendium of networks for longevity, age-related diseases and associated processes. Biogerontology. 2010;11:513–22. doi: 10.1007/s10522-010-9265-8. [DOI] [PubMed] [Google Scholar]
- 80.Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38(Database issue):D196–203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data have been submitted to GEO under the accession GSE57110.


