Genomic imprinting, an inherently epigenetic phenomenon defined by parent of origin-dependent gene expression, is observed in mammals and flowering plants. In this review, Rodrigues and Zilberman review the mechanisms of plant genomic imprinting and discuss theories of imprinting evolution and biological significance in light of recent findings.
Keywords: DNA methylation, epigenetics, evolution, genomic imprinting, histone methylation, plant reproduction
Abstract
Genomic imprinting, an inherently epigenetic phenomenon defined by parent of origin-dependent gene expression, is observed in mammals and flowering plants. Genome-scale surveys of imprinted expression and the underlying differential epigenetic marks have led to the discovery of hundreds of imprinted plant genes and confirmed DNA and histone methylation as key regulators of plant imprinting. However, the biological roles of the vast majority of imprinted plant genes are unknown, and the evolutionary forces shaping plant imprinting remain rather opaque. Here, we review the mechanisms of plant genomic imprinting and discuss theories of imprinting evolution and biological significance in light of recent findings.
Genomic imprinting is defined by biased expression of a gene from one allele over the other based on the sex of the parent that contributed the allele. As such, biased expression can occur even when both alleles have identical DNA sequences. A gene that is predominantly expressed from the maternally inherited allele is referred to as a maternally expressed imprinted gene, whereas a gene that is expressed from the paternally inherited allele is referred to as a paternally expressed imprinted gene. The implication for both types of imprinted genes is that maternally and paternally inherited alleles are differentiated post-fertilization by a pre-existing mark that is epigenetic or, in other words, not directly encoded by the DNA sequence. This epigenetic mark is referred to as the “imprint” for the imprinted gene (Fig. 1).
Figure 1.
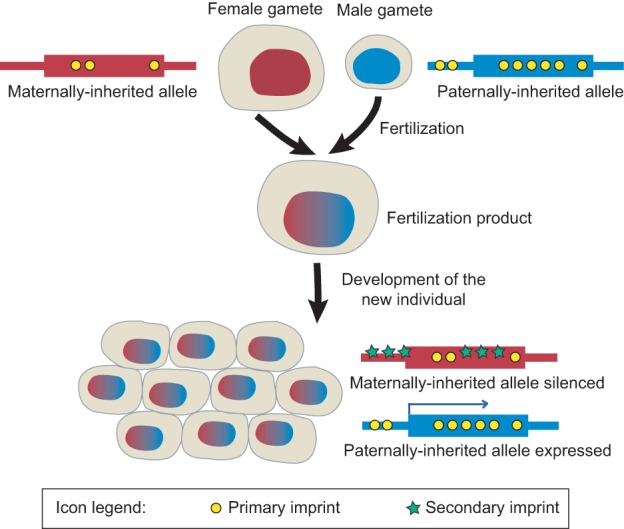
Imprinted gene expression. Maternally and paternally inherited alleles are epigenetically distinguished (or imprinted), causing differential expression in the fertilization product. Primary imprints are established before fertilization, whereas secondary imprints are guided by primary imprints after fertilization.
Although evidence of genomic imprinting was observed by mule breeders long before the development of modern genetics, the phenomenon was not formally described in plants and mammals until relatively recently (Morison and Reeve 1998). The present concept of genomic imprinting began to take shape as a result of genetic experiments in maize, which demonstrated that the R pigmentation gene is preferentially expressed when inherited from the mother (Kermicle 1970). Subsequent experiments in plants and mammals demonstrated that the paternal and maternal genomes are epigenetically distinct (Bartolomei and Ferguson-Smith 2011; Pires and Grossniklaus 2014) and led to the identification of the first mammalian imprinted genes (Barlow et al. 1991; Bartolomei et al. 1991; DeChiara et al. 1991). Further studies elucidated the mechanisms of imprinting and the functions of several imprinted genes (Huh et al. 2008; Ferguson-Smith 2011).
For a mammalian locus to be imprinted, epigenetic marks set up before the formation of the zygote (Lucifero et al. 2004; Kato et al. 2007; Kobayashi et al. 2013), referred to as primary imprints (Fig. 1; Barlow 1994), must escape chromatin reshaping mechanisms in the embryo (Mayer et al. 2000; Oswald et al. 2000; Proudhon et al. 2012). Parent of origin-dependent epigenetic differences may also appear in the zygote or later stages once maternally and paternally inherited chromosomes are in the same nucleus (Tomizawa et al. 2011). Such secondary or somatic imprints rely on the presence of a primary imprint (Fig. 1; Ferguson-Smith 2011).
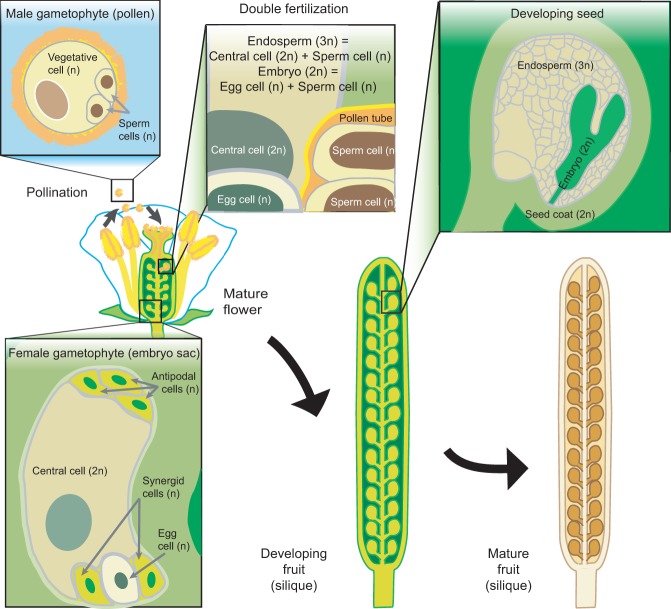
The logic of genomic imprinting in flowering plants is similar but has to be adjusted for the unique mechanism of plant reproduction. The haploid products of plant meiosis do not directly differentiate into gametes but instead divide mitotically several times to form multicellular gametophytes (Fig. 2). The male gametophyte, pollen, contains two sperm cells and a vegetative cell, which forms a pollen tube that delivers the sperm cells to the female gametophyte (Fig. 2). In flowering plants, one of these fuses with the egg to form the zygote, and the other fuses with the central cell, which is usually homodiploid, to form the triploid endosperm (Fig. 2), a transient seed tissue with nutritive and supportive functions analogous to the mammalian placenta. Although the two sperm cells are morphologically distinct in some plants, they appear to be at least somewhat functionally interchangeable (Russell 1991; Faure et al. 2003), suggesting that differential imprinting between the sperm cells is unlikely. Flowering plants therefore have three cell types (sperm, egg, and central cells) in which primary imprints can be established, with maternal imprints potentially quite different between the central and egg cells. Studies of individual plant imprinted genes (Chaudhuri and Messing 1994; Grossniklaus et al. 1998; Kinoshita et al. 1999, 2004; Luo et al. 2000; Danilevskaya et al. 2003; Gutierrez-Marcos et al. 2004, 2006; Kohler et al. 2005; Baroux et al. 2006; Gehring et al. 2006; Jullien et al. 2006a; Haun et al. 2007; Hermon et al. 2007; Makarevich et al. 2008; Tiwari et al. 2008; Fitz Gerald et al. 2009) and subsequent genome-scale investigations that identified hundreds of imprinted genes in Arabidopsis thaliana, castor bean, rice, and maize (Gehring et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Waters et al. 2011, 2013; Wolff et al. 2011; Zhang et al. 2011, 2014; Pignatta et al. 2014; Xu et al. 2014) have indeed demonstrated that the bulk of plant imprinted expression occurs in the endosperm, whereas imprinting in the embryo is rare and transient (Jahnke and Scholten 2009; Autran et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Nodine and Bartel 2012; Raissig et al. 2013; Pignatta et al. 2014). These studies also established DNA methylation as the most prevalent primary imprint (Fig. 3). Trimethylation of Lys27 of histone H3 appears to be a common secondary imprint, particularly at paternally expressed genes, but may also serve as the primary imprint at some genes (Fig. 3).
Figure 2.
Sexual reproduction in the flowering plant A. thaliana. The gametes are contained within multicellular haploid structures called gametophytes that are derived by mitosis from meiotic spores. The fusion of two haploid polar nuclei forms a diploid central cell in the female gametophyte. At the time of fertilization, the diploid female central cell and a haploid male sperm cell fuse to give rise to the endosperm, while the haploid female egg cell and haploid male sperm cell fuse to give rise to the embryo. The resulting seed is formed of the endosperm, the embryo, and a maternally derived seed coat.
Figure 3.
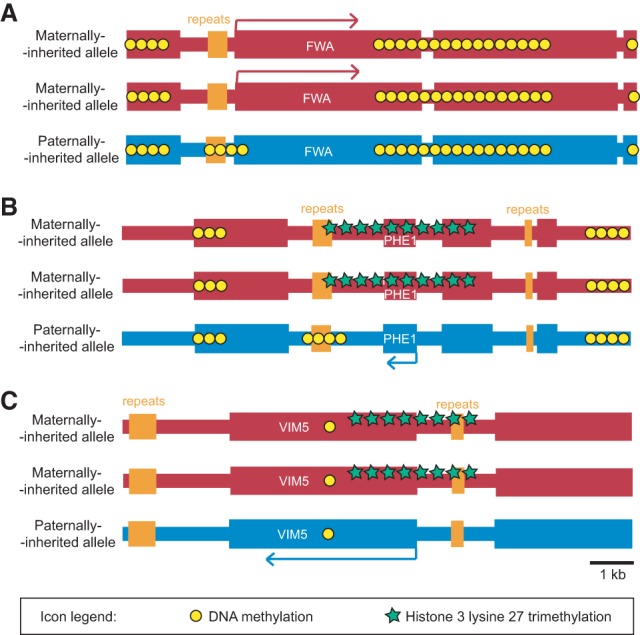
Mechanisms of imprinting in the endosperm of flowering plants. (A) At the Arabidopsis FWA locus, DEMETER (DME) activity at upstream repeats, presumed to occur in the central cell but not the sperm cells, forms a primary imprint that results in maternal-specific expression. (B) The primary imprint at the Arabidopsis PHERES1 gene is also formed by presumed central cell-specific demethylation of repeats by DME. Loss of methylation at these repeats results in histone 3 Lys27 trimethylation. This secondary imprint laid down by polycomb-repressive complex 2 (PRC2) causes silencing of the maternal loci and paternal-specific expression. (C) Arabidopsis VIM5 is an example of a gene that does not appear to rely on DME-mediated DNA demethylation for the formation of the primary imprint, as both maternal and paternal alleles are deficient in DNA methylation. Instead, histone 3 Lys27 trimethylation may form a primary imprint that results in paternal-specific expression (Hsieh et al. 2011).
Although hundreds of plant imprinted genes have been identified and much progress has been made in elucidating the epigenetic mechanisms that regulate them, the biological significance of plant imprinting remains mysterious, and the evolutionary forces that led to the emergence of imprinting have been extensively debated. In this review, we summarize the current knowledge about the mechanisms of plant imprinting and discuss the evolutionary theories and potential functional consequences of this epigenetic process.
Epigenetic mechanisms of genomic imprinting
Imprinting mechanisms have been extensively reviewed in mammals (Barlow 2011; Bartolomei and Ferguson-Smith 2011; Ferguson-Smith 2011; Adalsteinsson and Ferguson-Smith 2014; MacDonald and Mann 2014) and plants (Bauer and Fischer 2011; Raissig et al. 2011; Kohler et al. 2012; Gehring 2013; Zhang et al. 2013). We therefore keep the discussion of imprinting mechanisms fairly brief.
DNA methylation
DNA methylation is a classic mechanism of epigenetic inheritance. The most prominent type of eukaryotic DNA methylation is catalyzed by enzymes of the Dnmt1 family, called MET1 in plants (Feng et al. 2010; Law and Jacobsen 2010; Zemach et al. 2010b). These enzymes catalyze methylation of cytosines within symmetric CG dinucleotides when a cytosine on one of the DNA strands is already methylated, thereby perpetuating methylation patterns following DNA replication. DNA methylation is classically associated with the silencing of genes and transposable elements but can also promote gene activity (Makarevich et al. 2008; Shibuya et al. 2009; Jones 2012; Rigal et al. 2012; Deng and Chua 2015; Lei et al. 2015; Williams et al. 2015). Consequently, differential methylation of maternal and paternal alleles can cause imprinted expression (Fig. 3).
DNA methylation was the first imprint discovered in mammals (Bartolomei et al. 1993; Ferguson-Smith et al. 1993; Stoger et al. 1993) and in plants (Kinoshita et al. 2004) and is still the only differential mark known to be established as a primary imprint (Dickinson and Scholten 2013; MacDonald and Mann 2014). All identified parent of origin-specific DNA methylation differences in the endosperm of Arabidopsis, rice, and maize are due to site-specific hypomethylation of maternally inherited DNA (Waters et al. 2011; Zhang et al. 2011, 2014; Ibarra et al. 2012; Rodrigues et al. 2013). In Arabidopsis, this hypomethylation is catalyzed by the DEMETER (DME) DNA glycosylase, which can excise methylated cytosine from DNA (Gehring et al. 2006; Ibarra et al. 2012). DME is presumed to act in the female central cell prior to fusion with the sperm (Fig. 2; Choi et al. 2002).
DNA demethylation by DME has been associated with both maternal-specific gene activation (Kinoshita et al. 2004; Gehring et al. 2006; Hsieh et al. 2011; Ibarra et al. 2012) and maternal-specific gene repression (Makarevich et al. 2008; Hsieh et al. 2011; Ibarra et al. 2012). In maize and rice, maternal-specific DNA hypomethylation reminiscent of DME activity likewise occurs at maternally and paternally expressed imprinted genes (Gutierrez-Marcos et al. 2006; Haun et al. 2007; Hermon et al. 2007; Waters et al. 2011; Zhang et al. 2011, 2014; Rodrigues et al. 2013). Furthermore, maternally and paternally expressed genes show distinct patterns of demethylation that are similar between Arabidopsis, maize, and rice (Ibarra et al. 2012; Rodrigues et al. 2013; Zhang et al. 2014), suggesting that the locations of regulatory regions that govern activation and repression have been generally conserved across 150 million years of evolution (Chaw et al. 2004). The regulatory importance of DNA methylation is supported by evidence that perturbations of methylation patterns at individual genes abolish imprinting (Adams et al. 2000; Xiao et al. 2003; Kinoshita et al. 2004; Jullien et al. 2006b; Makarevich et al. 2008; Hsieh et al. 2011; Vu et al. 2013; Du et al. 2014). However, not all imprinted genes are clearly associated with endosperm-specific maternal DNA hypomethylation (Hsieh et al. 2011; Waters et al. 2011; Wolff et al. 2011; Zhang et al. 2011, 2014; Xu et al. 2014), suggesting either that the initial imprint is due to a yet to be discovered mechanism or that epigenetic states of parental alleles for these loci in the endosperm are poor representatives of the primary imprints present in gametes (Raissig et al. 2011).
In addition to its activity in the central cell, DME is also expressed in the pollen vegetative cell (Fig. 2; Schoft et al. 2011). DME demethylates a largely overlapping set of thousands of transposable elements in the central and vegetative cells, most of which are located far from genes and do not appear to influence gene expression (Gehring et al. 2009; Hsieh et al. 2009; Calarco et al. 2012; Ibarra et al. 2012). The vegetative cell is terminally differentiated and does not fuse with any female gametes, so the function of DME is clearly not confined to the generation of primary imprints. Instead, DME-mediated demethylation of transposons in the central and vegetative cells is proposed to generate mobile small RNA (sRNA) molecules that direct methylation of cognate transposons in the egg and sperm cells (Hsieh et al. 2009; Slotkin et al. 2009; Schoft et al. 2011; Calarco et al. 2012; Ibarra et al. 2012), thus reinforcing the silencing of transposons in the gametes. Similar transposons are also extensively hypomethylated in rice endosperm (Zemach et al. 2010a; Rodrigues et al. 2013), suggesting that transposon demethylation is a conserved feature of flowering plant sexual development. The sites of demethylation that regulate imprinted genes are commonly transposons or other repeats (Fig. 3A,B; Kinoshita et al. 2007; Gehring et al. 2009; Villar et al. 2009; Ibarra et al. 2012; Pignatta et al. 2014), indicating that imprint formation by DNA demethylation evolved from a transposon-directed silencing mechanism (Kim and Zilberman 2014). However, at least in rice endosperm, maternal-specific DNA hypomethylation of the transcribed regions of active genes is common and is strongly associated with paternally biased expression (Rodrigues et al. 2013). This suggests that demethylation that primarily regulates gene activity may have evolved in at least some plant lineages.
Histone variants
The core histones that form the protein component of the nucleosome can be differentiated into subtypes or variants with distinct structures and functions (Talbert and Henikoff 2010). Histone variants have not yet been implicated in plant imprinting, but the specific incorporation of a particular histone H3 variant in Arabidopsis sperm cells (Ingouff et al. 2007) might distinguish the paternal allele in the endosperm or zygote.
Histone modification
Trimethylation of Lys27 of histone H3 (H3K27me3), catalyzed by polycomb-repressive complex 2 (PRC2), is the only histone modification known to correlate with imprinting in flowering plants (Kohler et al. 2005; Baroux et al. 2006; Gehring et al. 2006; Jullien et al. 2006a; Makarevich et al. 2008; Du et al. 2014; Zhang et al. 2014). Mutation of PRC2 disrupts the imprinting of many genes, most of which are paternally expressed (Hsieh et al. 2011; Wolff et al. 2011), with H3K27me3 of maternally inherited alleles correlating with DNA hypomethylation (Fig. 3B; Makarevich et al. 2008; Weinhofer et al. 2010). Perturbations of DNA methylation at some of these genes result in a loss of imprinting (Xiao et al. 2003; Makarevich et al. 2008; Hsieh et al. 2011; Du et al. 2014), suggesting that DNA methylation is the primary imprint that causes differential recognition by PRC2. Consistent with this, PRC2 activity is generally anti-correlated with DNA methylation, including in the Arabidopsis endosperm, where PRC2 targets transposons associated with DME-mediated demethylation (Weinhofer et al. 2010; Deleris et al. 2012; Makarevitch et al. 2013; Jermann et al. 2014).
However, this mechanism cannot account for PRC2-mediated regulation of the maternally expressed Arabidopsis genes MEDEA and AtFH5 (Baroux et al. 2006; Gehring et al. 2006; Jullien et al. 2006a; Fitz Gerald et al. 2009), which involves silencing of the paternal allele. Furthermore, some imprinted genes regulated by PRC2 are not marked by DNA methylation (Hsieh et al. 2011; Wolff et al. 2011; Zhang et al. 2014). For example, imprinting of a MEDEA transgene does not require DME or DNA methylation (Wohrmann et al. 2012). It is currently unclear whether H3K27me3 serves as the primary imprint at such genes, but PRC2-mediated silencing does have the requisite features. There is strong evidence that PRC2-catalyzed H3K27me3 is inherited after DNA replication and recruits PRC2, creating a self-reinforcing loop (Hansen et al. 2008; Margueron et al. 2009; Xu et al. 2010). This enables PRC2-mediated silencing of one allele while the other remains active in the same nucleus even without differentiating DNA methylation (Berry et al. 2015). Recruitment of PRC2 in the central cell or sperm—for example, by cell-specific transcription factors—could thus plausibly establish an allele-specific silent state that can be inherited by the endosperm. Unlike DME-catalyzed DNA demethylation, this mechanism could account for imprinted gene expression in the embryo (Raissig et al. 2013).
Long noncoding RNA
Imprinted long noncoding RNAs have been observed in rice and maize (Luo et al. 2011; Zhang et al. 2011). Although these molecules have not been shown to be necessary for imprinted expression of protein-coding genes, they are of potential interest because PRC2-mediated histone methylation can be targeted by long noncoding RNA in plants and animals (Heo and Sung 2011; Brockdorff 2013; Csorba et al. 2014).
sRNA
As mentioned above, sRNA molecules, particularly those that are 24 nucleotides (nt) long, target DNA methylation in plants (Matzke and Mosher 2014). DNA methylation directed by sRNA is required for imprint creation at some loci that are demethylated in the central cell (Vu et al. 2013). More generally, the role of sRNAs in imprint formation and silencing of imprinted regions is currently poorly understood. Large populations of maternally biased 24-nt sRNAs were reported in young Arabidopsis seed (Mosher et al. 2009), and some of these were implicated in regulating transription factor dosage in endosperm (Lu et al. 2012). However, many of these apparently maternally biased sRNAs might originate in the maternally derived seed coat, which lies in close proximity to the Arabidopsis endosperm (Pignatta et al. 2014), possibly explaining why their maternal bias is unaffected by mutations in DNA- and histone-modifying enzymes that are known to regulate imprinted gene expression (Mosher et al. 2011). A strong overabundance of maternally biased endosperm sRNAs has also been reported in maize (Xin et al. 2014) but not in rice, where maternally and paternally expressed sRNAs are equally common (Rodrigues et al. 2013). Some of these sRNAs are associated with the silenced alleles of imprinted genes, as is the case for paternally biased Arabidopsis sRNAs (Pignatta et al. 2014). Rice imprinted sRNAs are enriched for maternal DNA hypomethylation (Rodrigues et al. 2013), suggesting that their imprinting shares regulatory mechanisms with protein-coding genes.
The intriguing correlations between sRNAs and imprinted genes notwithstanding, it is unclear whether gene-associated imprinted sRNAs are a cause or result of imprinted gene expression, and most imprinted genes are not associated with imprinted sRNAs (Rodrigues et al. 2013; Pignatta et al. 2014; Xin et al. 2014). It is also unclear how parent of origin-specific differences in gene expression persist despite the production of trans-acting sRNAs that can, in theory, silence both alleles. A possible explanation is that low expression of DNA methyltransferases (Jullien et al. 2012) or other RNA-directed DNA methylation pathway genes (Belmonte et al. 2013) in the young endosperm of Arabidopsis renders sRNAs less effective at inducing silencing. The low levels of sRNA-mediated DNA methylation observed in rice endosperm (Zemach et al. 2010a) are consistent with this idea. Because sRNA-directed methylation is a self-reinforcing process (Kim and Zilberman 2014), it is expected to work less efficiently in trans, so that activity in the central cell and early endosperm may be predominantly in cis. It is also possible that imprinted sRNAs and gene transcripts are present in distinct compartments of the endosperm.
Mechanisms underlying the evolution of plant imprinted genes
As discussed above, the establishment of primary imprints in plants is closely linked to transposable element silencing, with transposons and other repeats serving as the sites of DNA demethylation in Arabidopsis, rice, and maize (Kinoshita et al. 2007; Gehring et al. 2009; Hsieh et al. 2009; Villar et al. 2009; Zemach et al. 2010a; Ibarra et al. 2012; Pignatta et al. 2014; Wang et al. 2015). The movement of transposable elements has therefore been widely proposed to generate new imprinted genes (McDonald et al. 2005; Wolff et al. 2011; Jiang and Kohler 2012; Gehring 2013; Vu et al. 2013). For example, DNA methylation of a transposable element that inserts near the transcriptional start site of a gene can lead to gene silencing, which can in turn be relieved by DME-catalyzed demethylation in the central cell to produce maternal-specific expression in the endosperm (Fig. 4A). This is apparently what happened to the Arabidopsis maternally expressed imprinted gene FWA (Fig. 3A) and many other imprinted genes (Kinoshita et al. 2007; Fujimoto et al. 2008; Gehring et al. 2009; Wolff et al. 2011). Although the creation of an imprinted gene from one that is biallelically expressed by a transposon insertion has yet to be explicitly demonstrated, deletions of repetitive regions have been shown to result in a loss of imprinting (Walker 1998; Villar et al. 2009). Transposon insertions can also destroy imprinting, converting an imprinted gene into a biallelically expressed one, as has been demonstrated in maize (Haun et al. 2009).
Figure 4.
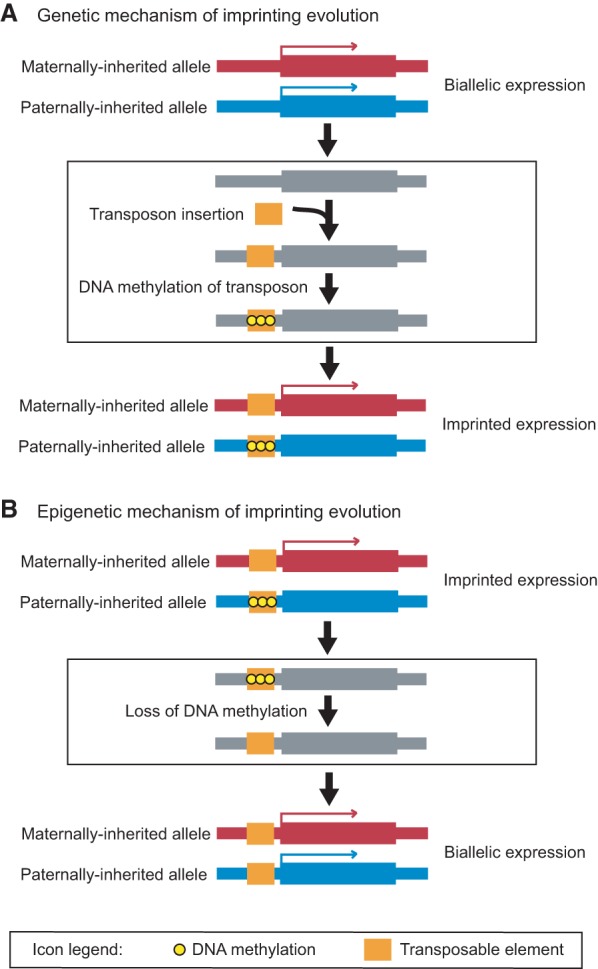
Evolution of imprinted gene expression through genetic mechanism via transposon insertion (A) and epigenetic mechanism via heritable loss of DNA methylation (B).
The relative simplicity, at least in theory, of creating imprinted genes by transposon insertions argues that ongoing transposition events provide a substantial pool of new imprinted genes for natural selection (Waters et al. 2013). This hypothesis is supported by the observation that, although many rice genes affected by LTR retrotransposons appear to evolve into pseudogenes, some genes remain functional and show evidence of selection consistent with neofunctionalization (Jiang and Ramachandran 2013). The high activity of transposons that is characteristic of plant genomes (Huang et al. 2012) may help explain why plant imprinted genes are poorly conserved (Luo et al. 2011; Waters et al. 2011, 2013).
Although transposon insertion is a straightforward mechanism for bringing a gene under the regulatory sway of DNA methylation, methylation patterns can diverge between individuals without an accompanying change of DNA sequence (Becker and Weigel 2012; Weigel and Colot 2012). This phenomenon is referred to as epigenetic mutation, and the alleles that bear differential methylation patterns are termed epigenetic alleles or epialleles. Plant DNA methylation patterns can diverge rather rapidly (Becker et al. 2011; Schmitz et al. 2011, 2013; Becker and Weigel 2012; Chodavarapu et al. 2012; Hagmann et al. 2015), and epialleles with major phenotypic consequences have been identified in a number of plant species (Weigel and Colot 2012; Zhang et al. 2012; Silveira et al. 2013). For example, stable unmethylated epialleles of FWA cause a strong delay of flowering (Soppe et al. 2000; Kankel et al. 2003). Heritable DNA methylation epialleles can therefore potentially account for some of the differences in imprinted gene expression observed within plant populations (Fig. 4B). Consistent with this possibility, differential DNA methylation has been correlated with imprinting divergence among Arabidopsis ecotypes (Pignatta et al. 2014). However, extensive intraspecies DNA methylation differences (Becker et al. 2011; Schmitz et al. 2011, 2013; Becker and Weigel 2012; Chodavarapu et al. 2012; Pignatta et al. 2014; Hagmann et al. 2015) and lack of characterization of the regulatory regions of the vast majority of plant imprinted genes complicate the assignment of any particular methylation polymorphism as the cause of an observed difference in imprinted expression. Thus, although epigenetic evolution of plant imprinted genes remains to be unambiguously demonstrated, this process quite likely contributes to imprinting divergence, at least on a relatively short time scale.
Evolutionary drivers and consequences of genomic imprinting
Monoallelic gene expression is generally disadvantageous in diploid organisms because deleterious mutations cannot be complemented by a functional homologous locus present in the same nucleus (Wilkins and Haig 2003). For example, mutations in the maternally expressed MEDEA gene result in aborted seeds even in the presence of a functional paternally inherited copy (Grossniklaus et al. 1998; Kinoshita et al. 1999; Luo et al. 2000). The prevalence of genomic imprinting therefore suggests that imprinting is associated with fitness benefits that outweigh the costs—benefits that are likely linked to the reproductive strategies of the lineages within which imprinting occurs (Jiang and Kohler 2012; Patten et al. 2014; Pires and Grossniklaus 2014; Wolf et al. 2014).
Known instances of classical genomic imprinting are confined to therian (marsupial and eutherian) mammals and flowering plants, with imprinting likely evolving at the same time as the transient extraembryonic nutritive tissues (placenta and endosperm) characteristic of each clade (Renfree et al. 2013; Pires and Grossniklaus 2014). In mammals, the origins of both imprinting and placental development are linked to regulatory novelties resulting from LTR retrotransposon activity (Renfree et al. 2013), while plant imprinting is mostly confined to the endosperm and is absent or short-lived in embryos, seedlings, and adult vegetative tissues (Jahnke and Scholten 2009; Zhang and Borevitz 2009; He et al. 2010; Autran et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Chodavarapu et al. 2012; Nodine and Bartel 2012; Raissig et al. 2013; Del Toro-De Leon et al. 2014; Pignatta et al. 2014). Given that both the placenta and endosperm serve as the interface between the maternal parent and its dependent offspring, imprinting is inextricably shaped by processes and forces governing sexual reproduction. Numerous hypotheses attempt to explain how imprinting might be associated with increased fitness. These hypotheses are not necessarily mutually exclusive; imprinting of different genes may be shaped by different selective forces, a combination of selective forces may influence the imprinting of a particular gene, and the relative impact of selective forces may vary over the evolutionary history of a lineage.
Kinship or parental conflict hypothesis
The currently predominant theory of genomic imprinting is the kinship or parental conflict hypothesis (Haig and Westoby 1989). It predicts that, in situations where the female parent contributes resources to offspring development after fertilization and can bear the offspring of multiple males, the overall fitness of males is increased if their offspring thrive at the cost of other offspring borne by the female parent, whereas the fitness of females is increased if the female is able to have as many successful offspring as possible. This drives paternal expression of genes that promote nutrient acquisition in the offspring and maternal expression of genes that restrict nutrient allocation to any one offspring. The conflict unfolds between maternally and paternally expressed genes within the offspring (Haig 2014).
In addition to the “placental habit” forming an environment in which parental conflict is theorized to occur (Crespi and Semeniuk 2004), this hypothesis receives wide support from in-depth investigations of the functions of mammalian imprinted genes (Frost and Moore 2010; Ferguson-Smith 2011) and some support from studies in plants (Huh et al. 2008; Kohler and Weinhofer-Molisch 2010; Pires and Grossniklaus 2014). Many mammalian imprinted genes affect resource acquisition from the maternal parent either by direct regulation of nutrient transfer from the placenta and embryo growth or through influence on maternal–offspring interactions (Bartolomei and Ferguson-Smith 2011). Crosses between Arabidopsis plants of different ploidy that increase paternal gene dosage result in larger seeds, whereas those that increase maternal gene dosage result in smaller seeds (Scott et al. 1998). The balance between maternal and paternal alleles in interploidy crosses also influences resource allocation in maize endosperm (Li and Dickinson 2010). Perturbations to Arabidopsis gamete DNA methylation likewise have effects on seed size that vary depending on which parental gamete was affected and are consistent with parental conflict theory (Adams et al. 2000; Xiao et al. 2006). Additionally, imprinted genes in maize, rice, and Arabidopsis are enriched for regulatory roles (Luo et al. 2011; Waters et al. 2013; Pignatta et al. 2014), with strong effects on endosperm growth resulting from loss-of-function mutations in specific genes, particularly those encoding PRC2 components (Grossniklaus et al. 1998; Kinoshita et al. 1999; Luo et al. 2000; Hermon et al. 2007). Imprinted genes also tend to show molecular signatures of the conflict between male and female fitness (Pires and Grossniklaus 2014).
However, parental conflict theory is not obviously consistent with all features of imprinting, especially in plants. First, if nutrient acquisition by the developing offspring is the main driver of imprinted expression, one might expect to observe imprinting in the growing plant embryo in addition to extraembryonic nutritive tissues, as is seen in mammals (Frost and Moore 2010; Renfree et al. 2013). Instead, imprinting in the embryo is rare and transient (Jahnke and Scholten 2009; Autran et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Nodine and Bartel 2012; Raissig et al. 2013; Del Toro-De Leon et al. 2014; Pignatta et al. 2014). It is possible that this difference between plants and mammals reflects restrictions imposed by underlying biological differences, such as two fertilization events versus one, the extent to which extraembryonic tissues regulate embryo growth, the generally much shorter developmental period that separates plant male and female gametes, and available mechanisms to differentially mark plant egg and sperm cells. Nonetheless, the preponderance of imprinting in the endosperm suggests a more intimate link with this tissue than one would predict based on parental conflict considerations.
Another incongruence is that parental conflict is predicted to drive imprinting to the complete silencing of one allele in most cases (Wilkins and Haig 2003), but many plant imprinted genes display only moderate biases (Luo et al. 2011; Waters et al. 2013). It is possible to explain this by interpreting the apparently rapid rate of imprinting evolution in plants (Luo et al. 2011; Waters et al. 2013) as an indication that many currently imprinted genes have acquired imprinting recently and/or have not yet experienced strong selection (Waters et al. 2013). Furthermore, reactivation of paternally inherited alleles that results in incomplete maternal bias may be favored under some circumstances (Wilkins and Haig 2002). The presence of imprinted expression in self-fertilizing plants, such as Arabidopsis and domesticated rice cultivars (Bechsgaard et al. 2004; Li et al. 2006; Londo et al. 2006), is also somewhat at odds with parental conflict theory, as the resulting monogamy would not place the related maternal and paternal genomes in conflict (Jiang and Kohler 2012). However, imprinting is plausibly inherited from recent out-crossing ancestors, and the complex webs of compensating maternally and paternally biased expression would likely take a long time to unravel (Wilkins and Haig 2003).
An additional potential challenge to the parental conflict theory arises from the recent characterization of the paternally expressed Arabidopsis gene ADMETOS (Kradolfer et al. 2013). ADMETOS is overexpressed in triploid Arabidopsis seeds created through fertilization with diploid sperm. Such seeds exhibit overexpression of other imprinted genes, including maternally biased ones. Mutation of ADMETOS reduces the expression of several maternally expressed genes, suggesting that ADMETOS is a positive regulator of these genes—a function that a paternally expressed gene would not be predicted to have by parental conflict theory. However, this effect of ADMETOS appears to be restricted to seeds with unbalanced ploidy, in which key regulatory pathways such as PRC2 do not function properly (Kradolfer et al. 2013), so that ADMETOS may not promote the expression of maternally biased genes in normal diploid seeds. Furthermore, the admetos mutation may affect the expression of imprinted genes indirectly, and ADMETOS appears to promote endosperm growth by delaying cellularization (Kradolfer et al. 2013), a function consistent with parental conflict.
Perhaps the most difficult to explain in terms of parental conflict theory is the imprinted expression of the maize gene Meg1. Meg1 promotes the establishment and differentiation of the endosperm nutrient transfer cells and consequently positively regulates the uptake of maternal nutrition by the developing seed (Costa et al. 2012). Increased Meg1 dosage leads to larger embryos and endosperm. Thus, Meg1 is precisely the type of gene predicted to be imprinted but, contrary to expectation, is maternally expressed. Similarly, overexpression of the maternally biased Arabidopsis imprinted gene AtFH5 may cause overgrowth of the chalazal endosperm compartment implicated in the transfer of maternal resources (Fitz Gerald et al. 2009). Although this “wrong way” imprinting is not entirely incompatible with parental conflict (Iwasa et al. 1999; Pires and Grossniklaus 2014), maternal expression of genes that promote the transfer of maternal resources requires serious consideration of alternate hypotheses for the evolution of genomic imprinting in plants, especially since plant genomic imprinting has yet to be widely associated with nutrient regulation (Gutierrez-Marcos et al. 2012).
Coadaptation hypothesis
The coadaptation hypothesis for the evolution of genomic imprinting (Wolf and Hager 2006) was inspired by adaptive correlations between the traits of mothers and offspring in several lineages (Kolliker et al. 2000; Agrawal et al. 2001; Hager and Johnstone 2003; Lock et al. 2004) and the observation that increased investment of maternal resources does not necessarily increase fitness (Thomas et al. 2004; Dunger et al. 2007). For example, human babies with intermediate birth weight have the highest fitness (Ulizzi et al. 1981), which may favor coevolution of maternal and offspring traits that lead to birth weight optimization (Wolf and Brodie 1998). Theoretical work supporting the coadaptation hypothesis shows that selective pressure for closer integration between coadapted maternal and offspring traits can drive the evolution of maternally biased gene expression (Wolf and Hager 2006).
The coadaptation hypothesis has several limitations. It assumes that similarity between the alleles expressed in mothers and offspring will be favored and therefore can only explain the evolution of maternally expressed imprinted genes, at least in the context of plant reproduction. Because the coadaptation hypothesis relies on allele similarity rather than dosage sensitivity, substantial heterozygosity within a population is required for the selection of imprinted expression; maternally biased expression offers no benefit if the maternal and paternal alleles are identical (Haig 2014). Furthermore, no imprinted gene has yet been shown to function along the lines predicted by the coadaptation hypothesis (Haig 2014). Despite these limitations, the coadaptation hypothesis may explain the imprinting of some genes through the benefits of closer cooperation between mothers and offspring rather than conflict between maternally and paternally inherited genes within the offspring.
Coadaptation may have plausibly favored the evolution of imprinting at genes that encode components of the plant PRC2 complex. Two of the Arabidopsis genes encoding PRC2 components are imprinted and maternally expressed (Grossniklaus et al. 1998; Kinoshita et al. 1999; Luo et al. 2000; Jullien et al. 2006b), placing the complex that silences the maternal alleles of most, if not all, paternally expressed genes (Hsieh et al. 2011; Wolff et al. 2011; Makarevitch et al. 2013; Zhang et al. 2014) under maternal control. PRC2 component genes are also maternally expressed in rice and maize endosperm, but these genes either encode PRC2 components that are different from the ones imprinted in Arabidopsis or evolved imprinting independently (Danilevskaya et al. 2003; Spillane et al. 2007; Luo et al. 2009; Pires and Grossniklaus 2014). The convergent evolution of PRC2 imprinting indicates that maternal expression of PRC2 is generally favored in flowering plants. PRC2 preferentially controls transcription factors that are master regulators of development and has thousands of targets in the endosperm (Weinhofer et al. 2010; Makarevitch et al. 2013; Zhang et al. 2014; Kohler and Lafon-Placette 2015; Pu and Sung 2015). Imprinting of such a multifaceted complex is perhaps more likely explained through the integration of female and offspring traits than by restriction of nutrient allocation to the offspring, as would be predicted by parental conflict. Importantly, the imprinting of PRC2-regulated paternally expressed genes is not expected to be driven by coadaptation. Therefore, somewhat counterintuitively, imprinting of PRC2 genes and the imprinted genes controlled by PRC2 may be guided by distinct evolutionary forces.
Dosage hypothesis
As mentioned above, an implication of the parental conflict hypothesis is that the functions of imprinted genes are dose-dependent; otherwise, monoallelic expression would carry no benefit. With this in mind, the “dosage hypothesis” (Dilkes and Comai 2004) was developed as a more general explanation for the evolution of imprinted expression. This hypothesis proposes that reduced expression from one allele will be favored by natural selection if it results in a more optimal abundance of the gene product. The gene does not need to be involved in resource allocation or indeed any particular pathway, although genes near the top of regulatory hierarchies and those that encode subunits of multiprotein complexes may be affected more frequently (Birchler et al. 2001). Because optimal gene dosage is not necessarily achieved by the complete silencing of one allele, the dosage hypothesis allows for incomplete parental bias as a stable evolutionary state. These features of the hypothesis fit the observed diversity of plant imprinted genes, the prevalence of imprinted genes with regulatory functions, and the tendency for partial rather than complete parental bias (Gehring et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Waters et al. 2011, 2013; Wolff et al. 2011; Zhang et al. 2011; Pignatta et al. 2014). However, the dosage hypothesis does not obviously account for the concentration of imprinted expression in endosperm.
Imprinting under relaxed selection
So far, we have discussed several hypotheses under which genomic imprinting is favored by natural selection. However, it is quite possible that the imprinted expression of many plant genes arose under weak, if any, selection. The imprinting status of plant genes appears to evolve quite rapidly; there is substantial intraspecific imprinting variation (Waters et al. 2013; Pignatta et al. 2014) and almost no overlap between the imprinted genes of Arabidopsis, rice, and maize (Luo et al. 2011; Waters et al. 2011, 2013). Most of the examined Arabidopsis imprinted genes were subjected to recent gene duplication events, have increased rates of amino acid changes, and appear to have acquired expression specificity to flowers or seeds compared with their more broadly expressed nonimprinted homologs (Wolff et al. 2011; Qiu et al. 2014). Inactivating mutations of many imprinted genes have no apparent influence on seed development (Berger et al. 2012; Wolff et al. 2015). This is consistent with imprinted genes undergoing neofunctionalization following gene duplication (Spillane et al. 2007; Bai and Settles 2014), whereas the low intraspecies and interspecies rates of conservation imply frequent loss of imprinting during evolution. Imprinting may thus be a transient state that facilitates neofunctionalization by reducing selection against mutations of silent alleles while accelerating selection of expressed alleles, in essence rendering imprinting as a form of diversifying selection (Beaudet and Jiang 2002; Bai and Settles 2014).
One can envision how this might work by considering the FWA gene. FWA is DNA methylated and silenced in most tissues and is specifically activated in the central cell by DME-mediated demethylation, resulting in maternal-specific expression in the endosperm (Fig. 3A; Kinoshita et al. 2004). Loss of FWA methylation leads to ectopic expression and strongly delayed transition to reproductive growth and flowering (Soppe et al. 2000; Kinoshita et al. 2004, 2007), whereas inactivating fwa mutations do not have an obvious phenoype (Soppe et al. 2000). If FWA were expressed throughout the plant and if delayed flowering were deleterious under a given set of environmental conditions, then mutations that inactivate FWA would be favored. DNA methylation of FWA accomplishes the same end while preserving gene expression in the seed and providing an opportunity for evolution of a new function. In this view, the imprinting of most genes in the seed has no fitness benefit except perhaps via the silencing of these genes in other tissues (Berger et al. 2012), with imprinted genes generally either degenerating into pseudogenes or acquiring new functions and biallelic expression. The lack of imprinted gene conservation implies that long-term persistance of neofunctionalized plant imprinted genes is rare.
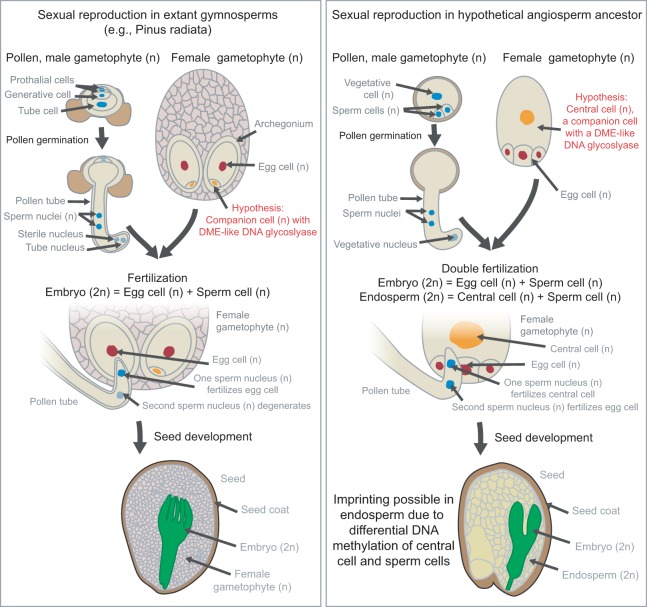
Because the two main mechanisms known to be responsible for plant imprinting—DME-catalyzed DNA demethylation and PRC2-catalzyed histone methylation—have functions unrelated to imprinting, selection on imprinted genes is unnecessary to explain the maintenance of imprinting mechanisms. In fact, the activity of DME in the male vegetative cell (Fig. 2), where it demethylates some of the same genes that are imprinted in the endosperm (Schoft et al. 2011; Ibarra et al. 2012), may be a hint about the evolutionary origins of imprinting in flowering plants. Gymnosperms such as conifers have no endosperm, the function of which is performed by the haploid female gametophyte (Fig. 5; Baroux et al. 2002). How the endosperm evolved in flowering plants remains a mystery, but a leading hypothesis postulates that the female gametophyte became sexualized (Nowack et al. 2007). If DME-type activity in the female gametophyte—for example, in the ventral canal cell that undergoes fertilization is some gymnosperms (Friedman and Floyd 2001)—predates the evolution of the endosperm, imprinting and the endosperm may have arisen simultaneously upon the evolution of double fertilization in the ancestor of flowering plants (Fig. 5). This would explain the predominance of imprinted expression in the endosperm without recourse to the argument that imprinting in the endosperm is especially favored by natural selection.
Figure 5.
Model for the simultaneous evolution of imprinting and endosperm in the ancestor of angiosperms (flowering plants). Angiosperms emerged from a gymnosperm (nonflowering seed plant) lineage, evolving flowers, fruits, and endosperm. Endosperm potentially evolved through the sexualization of a female gamete companion cell, such as the ventral canal cell (Friedman and Floyd 2001; Rudall 2006), via fusion with one of the two sperm cells of pollen. Activity of a DME-like enzyme in the sexualized female gamete companion cell would give rise to an endosperm with DNA methylation-based imprints in the ancestor of modern angiosperms.
Imprinting as a post-zygotic barrier to hybridization
As discussed above, imprinting may have important consequences for plant evolution by promoting genetic neofunctionalization. Another proposed evolutionary consequence of imprinting is the facilitation of speciation following polyploidization (Schatlowski and Kohler 2012). Crosses between polyploid plants and their diploid relatives alter the usual balance between maternal and paternal chromosomes. If dosage-sensitive genes are expressed differently from maternal and paternal alleles, either maternal or paternal excess can reduce offspring fitness. Therefore, genomic imprinting may pose a reproductive barrier between plants of different ploidy, leading to reproductive isolation and speciation (Haig and Westoby 1991). This hypothesis is supported by the long-standing observation that interploidy crosses tend to fail due to aberrant endosperm development (Kohler and Kradolfer 2011; Schatlowski and Kohler 2012; Birchler 2014). More direct evidence was provided by the demonstration that mutation of the paternally expressed Arabidopsis imprinted gene ADMETOS rescues seed abortion caused by excess paternal ploidy (Kradolfer et al. 2013). The finding that the dosage of a single imprinted gene is crucial for the success of triploid seeds may indicate that the majority of imprinted genes is not involved in interploidy interactions. However, because mutation of ADMETOS normalizes the expression of many imprinted genes in triploid endosperm, it is quite possible that ADMETOS has such a powerful effect through the regulation of imprinted genes in general (Kradolfer et al. 2013). This conclusion is supported by the recent finding that mutations in three other paternally expressed Arabidopsis genes rescue triploid seeds, with one of these, suvh7, also normalzing the expression of other imprinted genes (Wolff et al. 2015).
Concluding remarks
Five years ago, about a dozen imprinted genes had been identified in plants, with handfuls detected in any one species (Berger and Chaudhury 2009). Maternal-specific DNA demethylation and histone methylation by PRC2 were known to control the differential activation and silencing of maternal and paternal alleles of some of these genes, but whether these regulatory paradigms were reflective of the bulk of plant imprinting was unclear (Huh et al. 2008). Since then, hundreds of new imprinted genes have been discovered, and the generality of the mechanisms of imprinted gene regulation has been firmly established (Gehring et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Waters et al. 2011, 2013; Wolff et al. 2011; Zhang et al. 2011, 2014; Ibarra et al. 2012; Rodrigues et al. 2013; Du et al. 2014; Pignatta et al. 2014; Xu et al. 2014). Many mechanistic mysteries remain, including the existence of primary imprints other than DNA methylation, the plasticity of epigenetic marks during early seed development, the biogenesis and functions of imprinted sRNAs in endosperm, and the targeting specificity of DME-like enzymes. Nevertheless, the current understanding of the core mechanisms responsible for plant imprinting appears to be fairly robust. In contrast, our understanding of the evolution and biological significance of plant imprinting is in its infancy. Very few plant imprinted genes have known functions, which is perhaps to be expected given the recent order of magnitude expansion in the number of identified imprinted genes. Furthermore, none of the proposed theories for the natural selection of imprinted gene expression are fully consistent with all of the evidence. Imprinting of some genes, such as those encoding PRC2 components, is clearly very important, yet we must consider the sobering possibility that the imprinting of most plant genes is of little or no fitness benefit. Given this state of affairs, elucidation of the biological functions of plant imprinted genes should be a priority.
Acknowledgments
We thank Robert Fischer for comments on the manuscript, and Anna Koltunow and Brian Dilkes for helpful discussions. This work was supported by National Institutes of Health grant GM069415 and National Science Foundation grant IOS-1025890 to D.Z.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.269902.115.
References
- Adalsteinsson BT, Ferguson-Smith AC. 2014. Epigenetic control of the genome-lessons from genomic imprinting. Genes 5: 635–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ. 2000. Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502. [DOI] [PubMed] [Google Scholar]
- Agrawal AF, Brodie ED III, Brown J. 2001. Parent–offspring coadaptation and the dual genetic control of maternal care. Science 292: 1710–1712. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. 2011. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. [DOI] [PubMed] [Google Scholar]
- Bai F, Settles AM. 2014. Imprinting in plants as a mechanism to generate seed phenotypic diversity. Front Plant Sci 5: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP. 1994. Imprinting: a gamete's point of view. Trends Genet 10: 194–199. [DOI] [PubMed] [Google Scholar]
- Barlow DP. 2011. Genomic imprinting: a mammalian epigenetic discovery model. Ann Rev Genet 45: 379–403. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87. [DOI] [PubMed] [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U. 2002. Evolutionary origins of the endosperm in flowering plants. Genome Biol 3: reviews1026–reviews1026.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U. 2006. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 7: 1663–1673. [DOI] [PubMed] [Google Scholar]
- Bauer MJ, Fischer RL. 2011. Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol 14: 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet AL, Jiang YH. 2002. A rheostat model for a rapid and reversible form of imprinting-dependent evolution. Am J Hum Genet 70: 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard J, Bataillon T, Schierup MH. 2004. Uneven segregation of sporophytic self-incompatibility alleles in Arabidopsis lyrata. J Evol Biol 17: 554–561. [DOI] [PubMed] [Google Scholar]
- Becker C, Weigel D. 2012. Epigenetic variation: origin and transgenerational inheritance. Curr Opin Plant Biol 15: 562–567. [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Muller J, Koenig D, Stegle O, Borgwardt K, Weigel D. 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249. [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. 2013. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci 110: E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Chaudhury A. 2009. Parental memories shape seeds. Trends Plant Sci 14: 550–556. [DOI] [PubMed] [Google Scholar]
- Berger F, Vu TM, Li J, Chen B. 2012. Hypothesis: selection of imprinted genes is driven by silencing deleterious gene activity in somatic tissues. Cold Spring Harb Symp Quant Biol 77: 23–29. [DOI] [PubMed] [Google Scholar]
- Berry S, Hartley M, Olsson TS, Dean C, Howard M. 2015. Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. Elife 4: e07205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. 2014. Interploidy hybridization barrier of endosperm as a dosage interaction. Front Plant Sci 5: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Brockdorff N. 2013. Noncoding RNA and Polycomb recruitment. Rna 19: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo JA, Becker JD, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Messing J. 1994. Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc Natl Acad Sci 91: 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. 2004. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M. 2012. Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci 109: 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. 2002. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42. [DOI] [PubMed] [Google Scholar]
- Costa LM, Yuan J, Rouster J, Paul W, Dickinson H, Gutierrez-Marcos JF. 2012. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr Biol 22: 160–165. [DOI] [PubMed] [Google Scholar]
- Crespi B, Semeniuk C. 2004. Parent–offspring conflict in the evolution of vertebrate reproductive mode. Am Nat 163: 635–653. [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C. 2014. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111: 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Hermon P, Hantke S, Muszynski MG, Kollipara K, Ananiev EV. 2003. Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859. [DOI] [PubMed] [Google Scholar]
- Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE. 2012. Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet 8: e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro-De Leon G, Garcia-Aguilar M, Gillmor CS. 2014. Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627. [DOI] [PubMed] [Google Scholar]
- Deng S, Chua NH. 2015. Inverted-repeat RNAs targeting FT intronic regions promote FT expression in Arabidopsis. Plant Cell Physiol 56: 1667–1678. [DOI] [PubMed] [Google Scholar]
- Dickinson H, Scholten S. 2013. And baby makes three: genomic imprinting in plant embryos. PLoS Genet 9: e1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Comai L. 2004. A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Luo M, Zhang R, Finnegan EJ, Koltunow AM. 2014. Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites. Plant J 79: 232–242. [DOI] [PubMed] [Google Scholar]
- Dunger DB, Petry CJ, Ong KK. 2007. Genetics of size at birth. Diabetes Care 30: S150–S155. [DOI] [PubMed] [Google Scholar]
- Faure JE, Rusche ML, Thomas A, Keim P, Dumas C, Mogensen HL, Rougier M, Chaboud A. 2003. Double fertilization in maize: the two male gametes from a pollen grain have the ability to fuse with egg cells. Plant J 33: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci 107: 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith AC. 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12: 565–575. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. 1993. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362: 751–755. [DOI] [PubMed] [Google Scholar]
- Fitz Gerald JN, Hui PS, Berger F. 2009. Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136: 3399–3404. [DOI] [PubMed] [Google Scholar]
- Friedman WE, Floyd SK. 2001. Perspective: the origin of flowering plants and their reproductive biology—a tale of two phylogenies. Evolution 55: 217–231. [DOI] [PubMed] [Google Scholar]
- Frost JM, Moore GE. 2010. The importance of imprinting in the human placenta. PLoS Genet 6: e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Kinoshita Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Nasrallah ME, Shimizu KK, Kudoh H, Kakutani T. 2008. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet 4: e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M. 2013. Genomic imprinting: insights from plants. Annu Rev Genet 47: 187–208. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. 2006. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S. 2011. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana Seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. 1998. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O'Sullivan DM, Wormald M, Perez P, Dickinson HG. 2004. Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Dal Pra M, Scholten S, Kranz E, Perez P, Dickinson HG. 2006. Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38: 876–878. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Constancia M, Burton GJ. 2012. Maternal to offspring resource allocation in plants and mammals. Placenta 33: e3–e10. [DOI] [PubMed] [Google Scholar]
- Hager R, Johnstone RA. 2003. The genetic basis of family conflict resolution in mice. Nature 421: 533–535. [DOI] [PubMed] [Google Scholar]
- Hagmann J, Becker C, Muller J, Stegle O, Meyer RC, Wang G, Schneeberger K, Fitz J, Altmann T, Bergelson J, et al. 2015. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet 11: e1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. 2014. Coadaptation and conflict, misconception and muddle, in the evolution of genomic imprinting. Heredity 113: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Westoby M. 1989. Parent-specific gene expression and the triploid endosperm. Am Nat 134: 147–155. [Google Scholar]
- Haig D, Westoby M. 1991. Genomic imprinting in endosperm—its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos Trans R Soc Lond B Biol Sci 333: 1–13. [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- Haun WJ, Laoueille-Duprat S, O'Connell MJ, Spillane C, Grossniklaus U, Phillips AR, Kaeppler SM, Springer NM. 2007. Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J 49: 325–337. [DOI] [PubMed] [Google Scholar]
- Haun WJ, Danilevskaya ON, Meeley RB, Springer NM. 2009. Disruption of imprinting by Mutator transposon insertions in the 5′ proximal regions of the Zea mays Mez1 locus. Genetics 181: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. 2010. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Hermon P, Srilunchang KO, Zou J, Dresselhaus T, Danilevskaya ON. 2007. Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol 64: 387–395. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. 2011. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD. 2012. Active transposition in genomes. Annu Rev Genet 46: 651–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer RL. 2008. Cellular programming of plant gene imprinting. Cell 132: 735–744. [DOI] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. 2007. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Mochizuki A, Takeda Y. 1999. The evolution of genomic imprinting: abortion and overshoot explain aberrations. Evol Ecol Res 1: 129–150. [Google Scholar]
- Jahnke S, Scholten S. 2009. Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol 19: 1677–1681. [DOI] [PubMed] [Google Scholar]
- Jermann P, Hoerner L, Burger L, Schubeler D. 2014. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci 111: E3415–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kohler C. 2012. Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot 63: 4713–4722. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Ramachandran S. 2013. Genome-wide survey and comparative analysis of LTR retrotransposons and their captured genes in rice and sorghum. PLoS One 8: e71118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F. 2006a. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F. 2006b. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. 2012. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. 2003. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280. [DOI] [PubMed] [Google Scholar]
- Kermicle JL. 1970. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66: 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zilberman D. 2014. DNA methylation as a system of plant genomic immunity. Trends Plant Sci 19: 320–326. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. 1999. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJ, Koornneef M, Kakutani T. 2007. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J 49: 38–45. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. 2013. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 23: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Kradolfer D. 2011. Epigenetic mechanisms in the endosperm and their consequences for the evolution of flowering plants. Biochim Biophys Acta 1809: 438–443. [DOI] [PubMed] [Google Scholar]
- Kohler C, Lafon-Placette C. 2015. Evolution and function of epigenetic processes in the endosperm. Front Plant Sci 6: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Weinhofer-Molisch I. 2010. Mechanisms and evolution of genomic imprinting in plants. Heredity 105: 57–63. [DOI] [PubMed] [Google Scholar]
- Kohler C, Page DR, Gagliardini V, Grossniklaus U. 2005. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30. [DOI] [PubMed] [Google Scholar]
- Kohler C, Wolff P, Spillane C. 2012. Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63: 331–352. [DOI] [PubMed] [Google Scholar]
- Kolliker M, Brinkhof MW, Heeb P, Fitze PS, Richner H. 2000. The quantitative genetic basis of offspring solicitation and parental response in a passerine bird with biparental care. Proc Biol Sci 267: 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D, Wolff P, Jiang H, Siretskiy A, Kohler C. 2013. An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26: 525–535. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu JK. 2015. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci 112: 3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Dickinson HG. 2010. Balance between maternal and paternal alleles sets the timing of resource accumulation in the maize endosperm. Proc Biol Sci 277: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. 2006. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170: 185–193. [DOI] [PubMed] [Google Scholar]
- Lock JE, Smiseth PT, Moore AJ. 2004. Selection, inheritance, and the evolution of parent–offspring interactions. Am Nat 164: 13–24. [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. 2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci 103: 9578–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang C, Baulcombe DC, Chen ZJ. 2012. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci 109: 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. 2004. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 13: 839–849. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. 2000. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci 97: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES. 2009. Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2: 711–723. [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A. 2011. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald WA, Mann MR. 2014. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev 81: 126–140. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Kohler C. 2008. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912. [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM. 2013. Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502. [DOI] [PubMed] [Google Scholar]
- McDonald JF, Matzke MA, Matzke AJ. 2005. Host defenses to transposable elements and the evolution of genomic imprinting. Cytogenet Genome Res 110: 242–249. [DOI] [PubMed] [Google Scholar]
- Morison IM, Reeve AE. 1998. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum Mol Genet 7: 1599–1609. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. 2009. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Tan EH, Shin J, Fischer RL, Pikaard CS, Baulcombe DC. 2011. An atypical epigenetic mechanism affects uniparental expression of Pol IV-dependent siRNAs. PLoS One 6: e25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2012. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, Schnittger A. 2007. Bypassing genomic imprinting allows seed development. Nature 447: 312–315. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478. [DOI] [PubMed] [Google Scholar]
- Patten MM, Ross L, Curley JP, Queller DC, Bonduriansky R, Wolf JB. 2014. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity (Edinb) 113: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M. 2014. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. ELife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Grossniklaus U. 2014. Different yet similar: evolution of imprinting in flowering plants and mammals. F1000prime Rep 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Duffie R, Ajjan S, Cowley M, Iranzo J, Carbajosa G, Saadeh H, Holland ML, Oakey RJ, Rakyan VK, et al. 2012. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol Cell 47: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Sung ZR. 2015. PcG and trxG in plants—friends or foes. Trends Genet 31: 252–262. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Liu SL, Adams KL. 2014. Frequent changes in expression profile and accelerated sequence evolution of duplicated imprinted genes in Arabidopsis. Genome Biol Evol 6: 1830–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U. 2011. Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Bemer M, Baroux C, Grossniklaus U. 2013. Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet 9: e1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Suzuki S, Kaneko-Ishino T. 2013. The origin and evolution of genomic imprinting and viviparity in mammals. Philos Trans R Soc Lond B Biol Sci 368: 20120151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M, Kevei Z, Pelissier T, Mathieu O. 2012. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J 31: 2981–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D. 2013. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci 110: 7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ. 2006. How many nuclei make an embryo sac in flowering plants? Bioessays 28: 1067–1071. [DOI] [PubMed] [Google Scholar]
- Russell SD. 1991. Isolation and characterization of sperm cells in flowering plants. Annu Rev Plant Physiol Plant Mol Biol 42: 189–204. [Google Scholar]
- Schatlowski N, Kohler C. 2012. Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J Exp Bot 63: 6059–6067. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. 2013. Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, et al. 2011. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci 108: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG. 1998. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Shibuya K, Fukushima S, Takatsuji H. 2009. RNA-directed DNA methylation induces transcriptional activation in plants. Proc Natl Acad Sci 106: 1660–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira AB, Trontin C, Cortijo S, Barau J, Del Bem LE, Loudet O, Colot V, Vincentz M. 2013. Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet 9: e1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. 2000. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802. [DOI] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueille-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. 2007. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448: 349–352. [DOI] [PubMed] [Google Scholar]
- Stoger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73: 61–71. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2010. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275. [DOI] [PubMed] [Google Scholar]
- Thomas F, Teriokhin AT, Budilova EV, Brown SP, Renaud F, Guegan JF. 2004. Human birthweight evolution across contrasting environments. J Evol Biol 17: 542–553. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Schulz R, Ikeda Y, Dytham L, Bravo J, Mathers L, Spielman M, Guzman P, Oakey RJ, Kinoshita T, et al. 2008. Maternally expressed PAB C-terminal, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. 2011. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulizzi L, Gravina MF, Terrenato L. 1981. Natural selection associated with birth weight. II. Stabilizing and directional components. Ann Hum Genet 45: 207–212. [DOI] [PubMed] [Google Scholar]
- Villar CB, Erilova A, Makarevich G, Trosch R, Kohler C. 2009. Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol Plant 2: 654–660. [DOI] [PubMed] [Google Scholar]
- Vu TM, Nakamura M, Calarco JP, Susaki D, Lim PQ, Kinoshita T, Higashiyama T, Martienssen RA, Berger F. 2013. RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development 140: 2953–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL. 1998. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xia H, Zhang Y, Zhao S, Zhao C, Hou L, Li C, Li A, Ma C, Wang X. 2015. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may). BMC genomics 16: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM. 2011. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM. 2013. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci 110: 19639–19644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Colot V. 2012. Epialleles in plant evolution. Genome Biol 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Kohler C. 2010. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. 2002. Parental modifiers, antisense transcripts and loss of imprinting. Proc Biol Sci 269: 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. 2003. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet 4: 359–368. [DOI] [PubMed] [Google Scholar]
- Williams BP, Pignatta D, Henikoff S, Gehring M. 2015. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrmann HJ, Gagliardini V, Raissig MT, Wehrle W, Arand J, Schmidt A, Tierling S, Page DR, Schob H, Walter J, et al. 2012. Identification of a DNA methylation-independent imprinting control region at the Arabidopsis MEDEA locus. Genes Dev 26: 1837–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED III. 1998. The coadaptation of parental and offspring characters. Evolution 52: 299–308. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R. 2006. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol 4: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Oakey RJ, Feil R. 2014. Imprinted gene expression in hybrids: perturbed mechanisms and evolutionary implications. Heredity 113: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Kohler C. 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Jiang H, Wang G, Santos-Gonzalez J, Kohler C. 2015. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. ELife doi: 10.7554/eLife.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. 2003. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901. [DOI] [PubMed] [Google Scholar]
- Xiao W, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. 2006. Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol 142: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yang R, Yao Y, Ma C, Peng H, Sun Q, Wang X, Ni Z. 2014. Dynamic parent-of-origin effects on small interfering RNA expression in the developing maize endosperm. BMC Plant Biol 14: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. 2010. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci 107: 19266–19271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dai M, Li F, Liu A. 2014. Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res 42: 6987–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. 2010a. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci 107: 18729–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010b. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. 2009. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Guan H, Hu X, Jiao Y, Song W, et al. 2011. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci 108: 20042–20047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cheng Z, Qin R, Qiu Y, Wang JL, Cui X, Gu L, Zhang X, Guo X, Wang D, et al. 2012. Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chaudhury A, Wu X. 2013. Imprinting in plants and its underlying mechanisms. J Genet Genomics 40: 239–247. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xie S, Dong X, Zhao X, Zeng B, Chen J, Li H, Yang W, Zhao H, Wang G, et al. 2014. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res 24: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]