Abstract
Purpose
Cell death in neurodegeneration occurs at the convergence of diverse metabolic pathways. In the retina, a common underlying mechanism involves mitochondrial dysfunction since photoreceptor homeostasis and survival are highly susceptible to altered aerobic energy metabolism. We sought to develop an assay to directly measure oxygen consumption in intact retina with the goal of identifying alterations in respiration during photoreceptor dysfunction and degeneration.
Methods
Circular punches of freshly isolated mouse retina, adjacent to the optic nerve head, were used in the microplate-based Seahorse Extracellular Flux Analyzer to measure oxygen consumption. Tissue integrity was evaluated by propidium iodide staining and live imaging. Different substrates were tested for mitochondrial respiration. Basal and maximal respiration were expressed as oxygen consumption rate (OCR) and respectively measured in Ames' medium before and after the addition of mitochondrial uncoupler, BAM15.
Results
We show that glucose is an essential substrate for retinal mitochondria. At baseline, mitochondria respiration in the intact wild-type retina was close to maximal, with limited reserve capacity. Similar OCR and limited mitochondrial reserve capacity was also observed in cone-only Nrl−/− retina. However, the retina of Pde6brd1/rd1, Cep290rd16/rd16 and Rpgrip1−/− mice, all with dysfunctional or no photoreceptors, had reduced OCR and higher mitochondrial reserve capacity.
Conclusions
We have optimized a method to directly measure oxygen consumption in acutely isolated, ex vivo mouse retina and demonstrate that photoreceptors have low mitochondrial reserve capacity. Our data provide a plausible explanation for the high vulnerability of photoreceptors to altered energy homeostasis caused by mutations or metabolic challenges.
Keywords: oxidative stress, neurodegeneration, photoreceptor homeostasis, retinal disease, mitochondrial function
Defects in mitochondrial respiration and oxidative phosphorylation are associated with pleiotropic phenotypes, which, at least in part, can be attributed to aberrant generation of reactive oxygen (O2) and nitrogen species.1–4 Mitochondrial dysfunction has been correlated with the accumulation of cellular damage in aging and in late-onset neurodegenerative diseases.5–8 Tissues with a high metabolic requirement, such as the retina, are especially vulnerable to mitochondrial damage, which is implicated as a causal factor in neuronal cell death in retinal and macular degeneration.9,10 Within the retina, rod and cone photoreceptors exhibit high energy metabolism and associated O2 consumption, primarily driven by ion transport and synaptic transmission in the dark and by cGMP turnover in the light.11–14
Quantification of O2 consumption rate (OCR) is generally adopted as a readout of overall mitochondrial function.15 Measurements of respiration have traditionally been conducted with the O2-consuming Clark-type electrode on mitochondria preparations,16 which require sample volumes in the milliliter range and continuous stirring of samples in suspension, with loss of cellular/tissue context. The Clark-type electrode has also been used to measure O2 consumption in the intact rat retina, detecting higher O2 consumption in the outer retina compared to inner retina, with downward or upward changes, respectively, during light adaptation.17 Microelectrodes that are O2-sensitive and intraretinal measurements taken at differential tissue depths18 confirmed that photoreceptors have the highest O2 consumption.18 Finally, fresh mouse retina slices in a flow culture system have been used to determine the metabolic dependence of photoreceptors on glucose as a fuel.19 A limitation of all of the above experimental conditions is that they only permit evaluation of one retina/sample at a time and are not readily amenable for simultaneous and high throughput evaluation of multiple experimental conditions.
Microplate-based assays using an extracellular flux (XF) analyzer (Seahorse XF Analyzer; Seahorse Bioscience, Billerica, MA, USA) allow parallel comparison of different samples in small microliter volumes without the need for sample suspension, continuous stirring, or large measuring volumes in the order of ∼2 mL, as is the case with typical Clark-type electrode systems. The XF analyzer (Seahorse Bioscience) has been used to determine the effects of Ca++ and oxidative stress on mitochondrial function in the photoreceptor-like cell line 661W.20 Using the same technology, mitochondrial function has been examined and compared in acutely derived sections from several brain areas of a rat model of migraine.21 Direct comparative OCR determination has not yet been accomplished in the freshly isolated mouse retina.
Here, we introduce an improved protocol for microplate-based measurement of O2 consumption in acute retina samples. Photoreceptors account for the majority of O2 consumption in the retina,22 with the entire inner retina consuming at most 17% of the amount used by photoreceptors.13 Thus, the measurement of O2 consumption in the intact retina largely reflects photoreceptor respiration. Our newly optimized method directly evaluates retinal mitochondrial function and should be valuable for assessing alterations associated with retinal dysfunction and/or disease.
Methods
Equipment and Reagents
The following equipment and supplies were used in the study: Seahorse XF24 Bioanalyzer, XF24 Islet Fluxpaks (Seahorse Bioscience); dissecting forceps and micro-scissors (Fine Science Tools, Foster City, CA, USA); fine paintbrush, biopsy puncher (Miltex, York, PA, USA).
Ames' medium (120 mM NaCl) and basic salt solution (140 mM NaCl) were buffered with 10 mM HEPES and adjusted to pH 7.35 ± 0.05 with NaOH. Osmolarity was adjusted to 285 ± 5 mOSM by adding water (Advanced Model 3320 micro-osmometer, Advanced Instruments, Norwood, MA, USA).
Animals
Mice were cared for in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals, Institute of Laboratory Animal Resources, the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The Animal Care and Use Committee of the National Eye Institute approved all mouse protocols. We maintained C57BL/6J mice, Nrlp–green fluorescent protein (GFP) mice on C57BL/6J background (after >10 backcrosses), and retinal degeneration mutant mice with Nrlp-GFP transgene in 12-hour light-dark housing conditions. Mice were used at age 1 to 3 months for optimization of experimental conditions and at postnatal day (P)28 for comparison of retinal disease phenotypes.
Preparation of Retinal Discs for OCR Measurement
Before initiating the experiment, XF24 Islet Fluxpak mesh inserts were pretreated with 2 μg tissue adhesive (Cell-Tak; Corning Life Sciences, Tewksbury, MA, USA) using the adsorption method, as recommended by the manufacturer (Thermo Fisher Scientific, Pittsburgh, PA, USA). Briefly, Cell-Tak was neutralized with 0.1 M sodium bicarbonate supplemented with 1 N NaOH and applied to the mesh inserts. After 25 minutes of incubation, excess sodium bicarbonate was washed off with Ames' buffer containing 10 mM HEPES, pH 7.35, and the mesh insert set aside for later use.
Light adapted mice were euthanized between 12 and 2 PM by CO2 asphyxiation. Eyes were quickly enucleated and placed into ice-cold medium. For dissection, the cornea and lens were separated from the posterior segment of the eye. The scleral layer was pulled away from the neural retina leaving an intact retinal eyecup free of retinal pigment epithelium. Disks of neural retina were cut using a 1 mm diameter biopsy puncher (Miltex). Three sections equidistant from the optic nerve head were obtained from each retina and placed in ice-cold Ames' buffer containing 10 mM HEPES, pH 7.35. Using a fine paintbrush to hold the remaining vitreous humor, retinal discs were placed off-center with the ganglion cell layer side down onto pretreated XF24 Islet Fluxpak mesh inserts. Once the mesh was placed in the well, it was rotated to position the retinal disc directly under the O2 detector (Fig. 1A, XF analyzer schematic). For all experiments, retinal discs from 4 to 12 mouse retinas were collected in one dish for a specific experimental condition. Only intact retinal discs (by visual inspection) were used for measurements. Measurements of OCR from each disc were considered as independent values for statistical analysis. Buffers were kept on ice throughout the preparation process, which was completed within 1.5 hours.
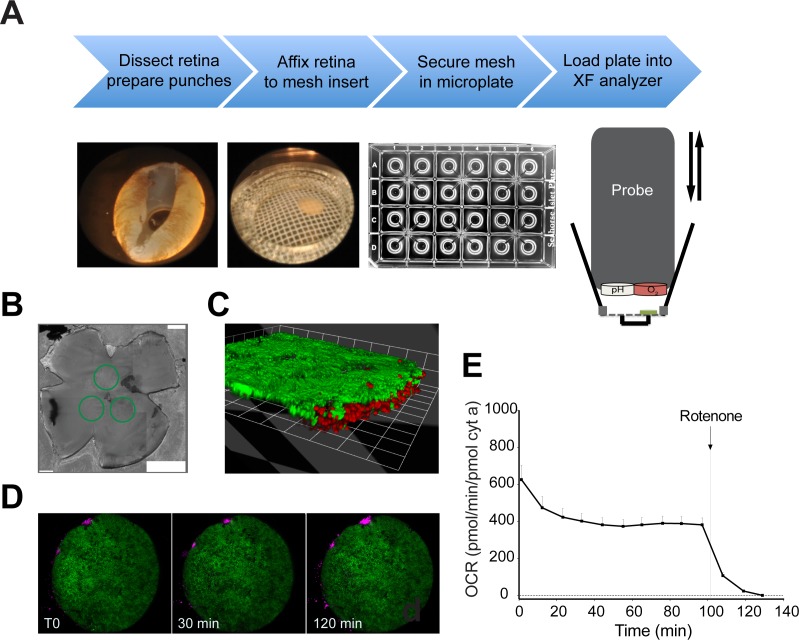
Figure 1.
(A) Workflow illustrating the preparations steps (top), dissected whole retina with one punched area, 1-mm retinal disc mounted on mesh insert, multiwell plate (Seahorse XF24 Analyzer; Seahorse Bioscience), and a schematic of the measurement chamber (bottom). A probe with O2 and CO2 (pH) fluorophore detectors is cyclically lowered and raised to each well of a 24-multiwell plate. (B) Green circles drawn on retinal flat mount highlight position from where discs were excised. (C, D) Samples of Nrlp-GFP retina stained with propidium iodide and imaged over the time interval of a typical assay. (E) Basal OCR recorded for 130 minutes to assess stability of the measurement throughout the assay. Values are normalized to cytochrome a content. The arrow indicates the time at which rotenone (1 μM final concentration) was added to the sample well. n = 15 retinal samples from 3 to 4 mice. Error bars: SEM.
Evaluation of Substrates
Several physiological substrates were tested alone or in combination to quantify respiration. A basic salt solution (140 mM NaCl, 3.1 mM KCl, 1.24 mM MgSO4, 2 mM CaCl2, 10 mM HEPES, pH 7.35) was supplemented with defined combinations of substrates (6 mM D-glucose, 0.15 mM pyruvate,1 mM lactate, 0.5 mM L-glutamine, 2 mM β-hydroxybutyrate, Sigma-Aldrich Corp., St. Louis, MO, USA) and compared to Ames' medium buffered with 10 mM HEPES. The osmolality of each solution was adjusted to 285 ± 5 mOsm (the final concentration of salts may therefore vary slightly).
Assay Conditions and Uncoupler Concentration
Seahorse assays were set up according to the manufacturer's recommended protocol. A calibration plate was incubated with Seahorse calibration medium at 37°C overnight in a CO2-free incubator. On the day of the experiment, drugs were added at appropriate concentrations to the injection ports built into each XF24 islet plate. Thirty minutes before the completion of dissection, the calibration plate was loaded into the bioanalyzer for automatic calibration. Retina was added to 700 μL of medium in each well. After optimization, a cycle of mix (3 minutes), wait (2 minutes), and measure (5 minutes) was determined to be optimal for obtaining a robust OCR readout. Five measurements were taken to establish a basal level of respiration, followed by four measurements after the addition of a mitochondrial uncoupler, and then four final measurements following the administration of a complex I inhibitor, rotenone (1 μM final concentration, Sigma-Aldrich Corp.). The tissue was maintained at 37°C for the duration of the measurement. Drugs of interest were reconstituted in DMSO, diluted to 10X in buffered medium, and loaded into the injection ports of the XF calibration plate (final DMSO concentration, <0.01%). Mitochondrial uncouplers included carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP; Sigma-Aldrich Corp.) and (2-fluorophenyl)(6-[(2-fluorophenyl)amino](1,2,5-oxadiazolo[3,4-e]pyrazin-5-yl))amine (BAM15; Timtec, Newark, DE, USA). Oligomycin A (0.6 μM final concentration; Sigma-Aldrich Corp.) was also utilized in the assay.
Raw OCR values were computed by the Seahorse Akos algorithm.23 Residual OCR obtained after addition of rotenone (<1% of maximal OCR) was subtracted from all points. Basal OCR was taken from the last point before addition of uncoupler. The value of the highest uncoupled point was used for maximal OCR. Unless otherwise indicated, OCR was normalized to cytochrome a content.
Protein and Cytochrome A Quantification
Retinal protein was quantified in retinas isolated from each mouse model using bicinchoninic acid assay. Protein content per retinal disc was estimated after calculating the area of an individual 1 mm disc as a ratio of the measured surface area of a flat-mounted retina. Cytochrome a content in retinas was determined spectrophotometrically in a commercial spectrophotometer (Shimadzu 2700 UV-Vis; Shimadzu Corp., Kyoto, Japan). Four to eight retinas were homogenized by pipetting in 0.1 mL of ice-cold buffer containing 280 mM sucrose, 10 mM HEPES, 1mM EDTA and 1 mM EGTA, adjusted to pH 7.1 with KOH. An aliquot of the same buffer (0.9 mL) containing 1% dodecyl maltoside (Sigma-Aldrich Corp.) was added to the homogenate, followed by vortexing for 45 seconds. After centrifugation for 2 minutes at 16,000 g, the supernatant was transferred to a plastic cuvette. Potassium ferricyanide (0.1 mM) was added to ensure full oxidation of the cytochromes, and five spectra in the wavelength range of 500 to 650 nm were collected and averaged. Then, 0.5 mM of KCN was added together with 10 mM sodium ascorbate (pH 7.4) to reduce cytochrome a, and five reduced spectra were collected and averaged. The oxidized spectrum was subtracted from the reduced spectrum, and the absorbance difference at 605 nm was calculated after manually tracing a baseline from 575 and 630 nm. An extinction coefficient of 10.8 mM−1 cm−1 was used to obtain cytochrome a concentration.24 Total cytochrome a content was measured from a minimum of four retinas. The surface area of a retinal disc relative to whole retina was used to calculate the cytochrome a content/disc when normalizing OCR.
Retinal Mitochondria Isolation
Forty retinas were excised, as described above, and homogenized by pipetting. All subsequent steps were the same as elaborated previously for liver mitochondria isolation.25 Total mitochondria yield generally corresponded to ∼280 pmol of cytochrome a.
Live Retinal Imaging
Retinas were dissected from Nrlp-GFP mice, as previously described. A 1 mm diameter sample of the retina with the photoreceptor side up was mounted on the Seahorse mesh insert treated with 2 μg tissue adhesive (Cell-Tak) and transferred to a sterile chamber slide with a 1.5 mm cover slip (Lab-Tek, Electron Microscopy Science, Hatfield, PA, USA). The chamber was loaded with 3 mL Ames' medium. Prior to imaging, 10 μM of propidium iodide (Roche Diagnostics, Indianapolis, IN, USA) was added to the medium. Imaging was performed with an inverted confocal system (Zeiss 780; Carl Zeiss MicroImaging GmbH, Jena, Germany) in a 37°C heated stage to mimic the temperature conditions in the Seahorse analyzer. Consecutive z-stacks scanning through the outer nuclear layer were taken every 30 minutes for a total duration of 2 hours. Images were processed with the imaging software (LSM Zen Blue; Carl Zeiss MicroImaging GmbH).
Preparation of Retinal Sections for Histology
Mouse eyes were quickly enucleated and fixed in 4% paraformaldehyde for 10 minutes. The anterior segment of the eyecup was then removed and the tissue was fixed overnight in 4% PFA. Eyecups were embedded in 7% agarose (Sigma-Aldrich Corp.) to generate 100 μm sections using a vibratome (Leica VT1000S, Bannockburn, IL, USA). Sections were incubated in 4′, 6-diamidino-2-phenylindole (DAPI) for 10 minutes, washed and finally mounted onto slides treated with Fluoromount-G (Southern Biotech, Birmingham, AL, USA).
Statistical Analysis
All data comparing cytochrome a content and OCR from retinas of different degeneration models were analyzed by one-way ANOVA. Comparison of each experimental group to a control group was performed using post-hoc Dunnett's test. Values of P ≤ 0.05 were considered significant.
Results
We have modified and improved a protocol for microplate-based measurement of O2 consumption by photoreceptors in freshly dissected retina using an XF analyzer (Seahorse Biosciences). In this method, an O2/pH probe is lowered into each well of a 24-multiwell plate containing the retinal sample and filled with Ames' buffer (Fig. 1A, right panels). Changes in oxygen content (i.e., OCR), were measured at fixed intervals (see Methods section). Changes in pH in our experimental setting were negligible throughout each individual time course (90–120 minutes in duration) because of the use of HEPES-buffered solutions, maintaining an average pH > 7.2, to preserve neuronal tissue functionality. We sampled 1 mm discs of retina to adapt to the microwell configuration and preserve multilayered organization of the tissue (Fig. 1A, left panels). For each mouse retina, we collected three discs adjacent to the optic nerve head to minimize the variability in cell distribution from center to periphery (Fig. 1B). We used confocal microscopy to monitor tissue integrity of retina, isolated from Nrlp-GFP transgenic mice that express GFP under control of the promoter of the rod-specific transcription factor NRL,26 by staining with the DNA-intercalating agent propidium iodide (PI; Fig. 1C). Though a small number of dead cells were observed at the perimeter of the sample, spread of cell death was negligible over a period of 120 minutes (Fig. 1D). Furthermore, retinal basal respiration rate remained steady after the first 40 minutes, with minimal decline throughout the duration of the experiment (Fig. 1E). Rotenone (1 μM final concentration) was added at the end of the assay to identify nonmitochondrial respiration, which was small in our samples and was subtracted in the subsequent calculations (Figs. 1E, 2A, graph point 3).
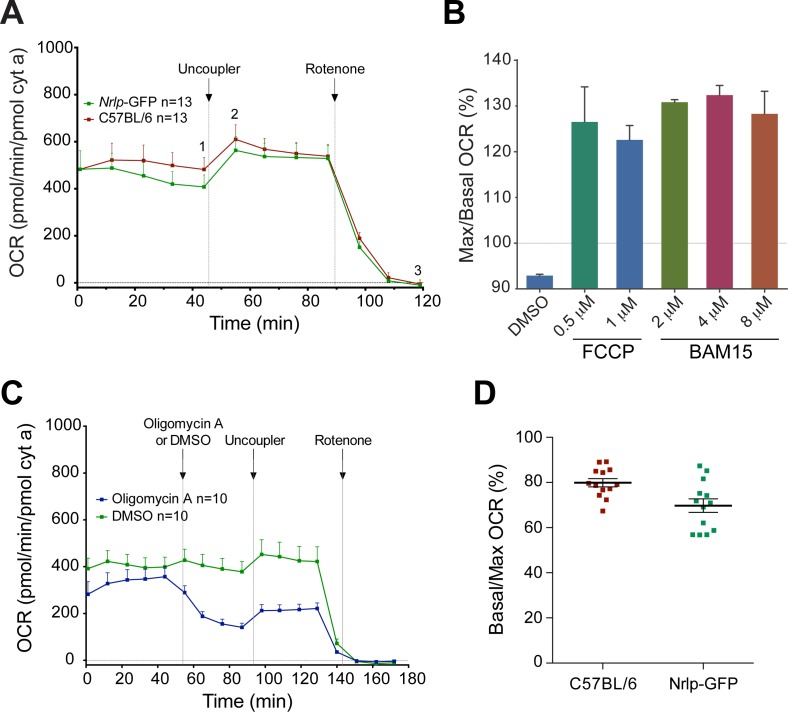
Figure 2.
(A) Comparison of OCR traces in wild-type C57BL/6 and Nrlp-GFP transgenic mice on C57BL/6 background. The OCR was normalized to mitochondrial cytochrome a content. Arrows indicate the injection of uncoupler or rotenone (1 μM final concentration) in the sample well. Number of retinal samples is shown. Error bars: SEM. (B) Ratio (%) of maximal over basal respiration measured after and before administration of titrating concentrations of mitochondrial uncouplers. (C) Comparison of OCR traces in C57BL/6 mice with or without the addition of the ATP synthase inhibitor oligomycin A. The OCR was normalized to mitochondrial cytochrome a content. Arrows indicate the injection of oligomycin A (0.6 μM final concentration), DMSO (<0.01% final concentration), uncoupler, or rotenone in the sample well. Addition of oligomycin A resulted in diminished basal OCR. Number of retinal samples is shown. Error bars: SEM. (D) Basal/maximal OCR (%) in C57BL/6 and Nrlp-GFP retina showing that mouse retina functions at 70% to 80% of its maximal respiratory capacity.
The maximal respiratory capacity of the tissue was measured after the addition of a mitochondrial uncoupler (Fig. 2A). The differential between maximal (Fig. 2A, graph point 2) and basal O2 consumption (Fig. 2A, graph point 1) is referred to as “reserve respiratory capacity.”15 We tested in parallel an established uncoupler, trifluoromethoxyphenylhydrazone (FCCP), and a recently introduced uncoupler BAM15. We found BAM15 to be more effective than FCCP in uncoupling mitochondrial proton flux with fewer off-target effects at the plasma membrane and a broader effective range with persistent cell viability up to 50 μM.27 We validated that the effect of BAM15 is less concentration-dependent (Fig. 2B) and subsequently included the same concentration of BAM15 (4 μM, final concentration) in all experiments irrespective of the degree of tissue degeneration and amount of mitochondria. Values of OCR were normalized to the amount of cytochrome a, a measure of complex IV (and mitochondrial) content and are representative of average mitochondrial respiration (Fig. 2A). Retina from Nrlp-GFP mice produced normalized OCR profiles substantially similar to those from C57BL/6 retina (Fig. 2A). We omitted the use of oligomycin A (which was originally included by the Seahorse XF manufacturer) at the start of the experiment because the inhibition of ATP synthesis resulted in >2-fold decrease in maximal respiration after uncoupling (Fig. 2C), suggesting mitochondrial damage by ATP depletion.
Addition of uncoupler typically induces 2- to 3-fold increase in OCR of most cell culture and tissue preparations, including brain.27 However, the addition of 4 μM BAM15 (or 0.5 μM FCCP, data not shown) to wild type, C57BL/6 or Nrlp-GFP, retina resulted in only 20% to 25% increase above basal respiration (Figs. 2A, 2D). When we measured OCR of mitochondria isolated from the whole retina using a Clark-type electrode, maximal ADP-stimulated (state 3) rates of respiration with saturating substrates (5 mM glutamate and malate) were close to 700 pmol/min/pmol cyt a (Supplementary Figure S1). These respiration rates were similar to maximal (uncoupled) rates observed in ex vivo retinal samples (Fig. 2A) and to maximal OCR rates measured in mammalian heart mitochondria in the presence of excess substrates.28
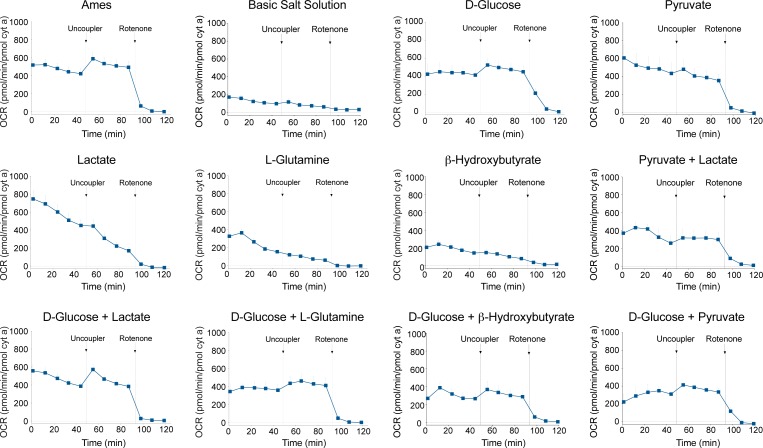
To confirm that respiration rates and reserve capacity in retinal mitochondria were not limited by substrate availability in the Ames' medium (6 mM glucose, 0.15 mM pyruvate and 0.5 mM glutamine), we evaluated single or pairs of substrates in buffered salt solution (Fig. 3). Neither lactate (1 mM), glutamine (0.5 mM), nor β-hydroxybutyrate (2 mM) was sufficient alone to maintain tissue viability. Respiration recordings in medium containing glucose alone (6 mM) or in combination with lactate (1 mM) or glutamine were comparable. Glucose with β-hydroxybutyrate or pyruvate (0.15 mM) as well as pyruvate with lactate yielded lower OCR values. Although glucose alone sustained a relatively stable basal OCR, the uncoupled maximal OCR was less than that observed with Ames' medium. Glutamine, β-hydroxybutyrate, or pyruvate did not improve maximal respiration when combined with D-glucose. However, the combination of glucose and lactate rendered uncoupled OCR values nearly identical to those obtained with Ames' medium. We therefore maintained Ames' medium for all subsequent experiments.
Figure 3.
Comparison of OCR traces in basic salt solution and with combinations of substrates (6 mM D-glucose, 0.15 mM pyruvate,1 mM lactate, 0.5 mM L-glutamine, 2 mM β-hydroxybutyrate), as indicated. We normalized OCR to mitochondrial cytochrome a content. Arrows indicate the injection of uncoupler or rotenone (1 μM final concentration) in the sample well. n = 8 retinal samples from 2 to 3 mice. Error bars: SEM.
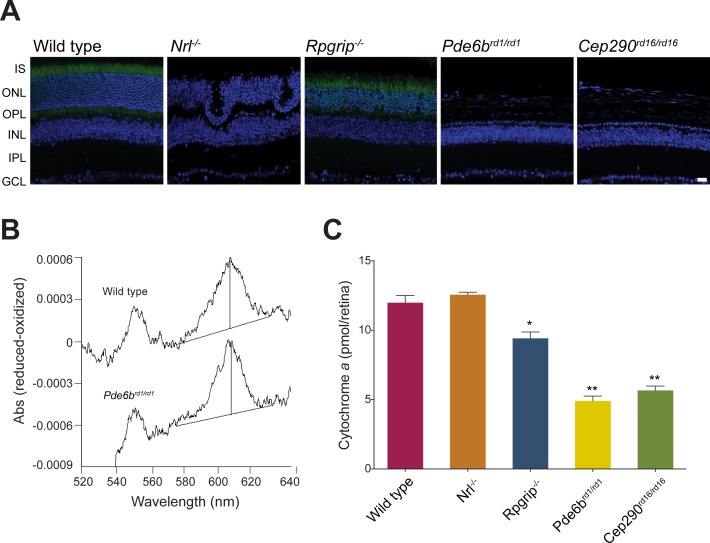
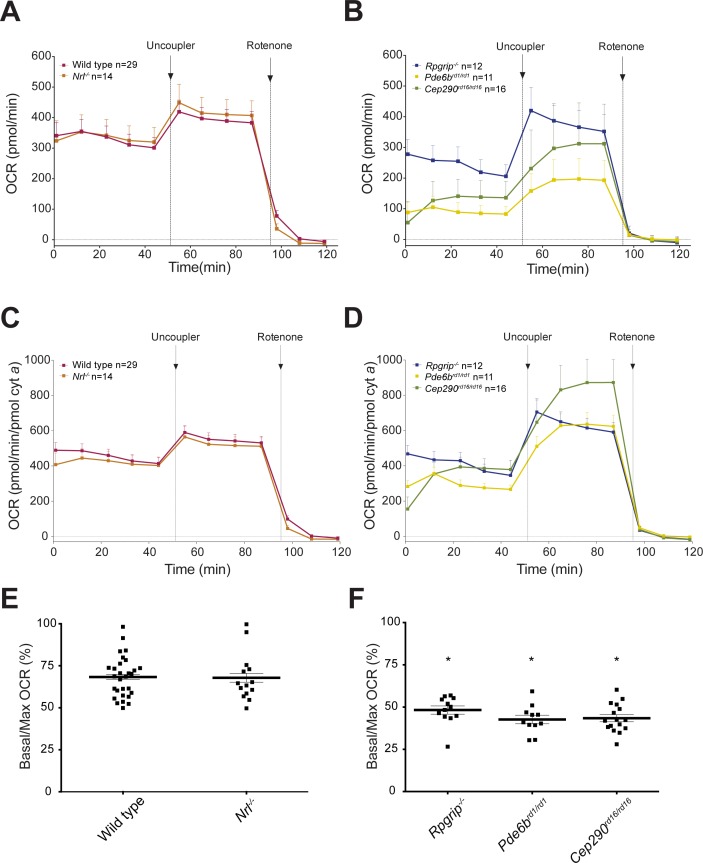
Almost 80% of the cells in the mouse neural retina are photoreceptors, and among these, rods outnumber cones by 30:1. We therefore attributed the high basal respiration rate in the retinal mitochondria to energy requirements of photoreceptors. To test this hypothesis, we applied the ex-vivo retina respiration assay to well-characterized mouse models of retinal disease (i.e., Nrl−/−, Rpgrip1−/−, Pde6brd1/rd1 and Cep290rd16/rd16) displaying distinct cell death kinetics. All lines of mice were maintained on Nrlp-GFP background to allow the visualization of rod photoreceptors (Fig. 4A). At P28, Rpgrip1−/− retina exhibits partial retention of photoreceptors,29 whereas Pde6brd1/rd1 and Cep290rd16/rd16 retina completely lack rod photoreceptors and have only few residual cones.30,31 Rod and cone ERG responses are absent in these three mouse models at P28. The Nrl−/− retina lacks the rod specific transcription factor NRL and consequently, S-cone-like photoreceptors are present instead of rods, with supranormal amplitudes of cone a- and b- waves in electroretinogram and little or no degeneration at P28.32 Inner retina neurons are viable in all 1-month-old mouse mutants used in the study. Cytochrome a content was lower at P28 in Rpgrip1−/−, Pde6b rd1/rd1, and Cep290rd16/rd16, but not in Nrl−/− retina compared to wild type, reflecting a reduced number of mitochondria in degenerating retina (Figs. 4B, 4C). Overall (un-normalized) OCR was reduced in all samples in which photoreceptors had been lost (Rpgrip1−/−, Pde6b rd1/rd1, and Cep290rd16/rd16), but not in cone only Nrl−/− retina (Figs. 5A, 5B). Upon normalization, basal OCR was comparable in wild type and all degenerating models except Pde6brd1/rd1, in which it was decreased (Figs. 5C, 5D). Notably, the addition of uncoupler induced a delayed response in Pde6brd1/rd1 and Cep290rd16/rd16 retina (no photoreceptors), but not in Rpgrip1−/− retina (with reduced and dysfunctional photoreceptors; Figs. 5B, 5D). Furthermore, there were significantly lower ratios of basal/maximal respiration in Rpgrip1−/−, Pde6b rd1/rd1, and Cep290rd16/rd16 retina compared to wild type (Figs. 5E, 5F). Basal/maximal OCR in cone only Nrl−/− retina was comparable to wild type at P28 (Fig. 5E). However, consistent with the other degenerating models, overall basal OCR and basal/maximal ratio were reduced in Nrl−/− retina by age 3 and 6 months (Supplementary Figure S2), when fewer photoreceptors are present in the ONL.32
Figure 4.
(A) Representative microphotographs of retinal sections from adult P28 wild type and mutant mice transgenic for Nrlp-GFP. Green fluorescent protein is expressed in rod photoreceptors under control of the Nrl promoter. Nuclei are labeled with DAPI. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer. Scale bar: 20 μm. (B) Spectra showing ascorbate-reduced minus ferricyanide-oxidized difference obtained by pooling wild type (n = 4) or Pde6b rd1/rd1 (n = 8) retinas. (C) Cytochrome a content in the mouse retina of control and retinal disease mutants (pools of 4–8 retinas). *P < 0.0001. **P < 0.01 compared to wild type, with 1-way ANOVA and Dunnett's post hoc test.
Figure 5.
(A, B) The OCR traces from control and degenerating retinas of mutant mice, as indicated. The OCR is not normalized against cytochrome a content. Arrows indicate the injection of uncoupler or rotenone (1 μM final concentration) in the sample well. Number of retinal samples is shown. Error bars: SEM. (C, D) OCR recordings of retina from retinal disease mutants normalized to pmol cytochrome a/sample. Number of retinal samples is shown. Error bars: SEM. (E, F) Larger residual respiratory capacity in retinal disease mutants undergoing degeneration was evidenced by a lower basal/maximal (%) OCR. Error bars: SEM; *P < 0.0001 compared to wild type, with 1-way ANOVA and Dunnett's post hoc test.
Discussion
Energy demand in the central nervous system and the retina is especially high, making mitochondria homeostasis essential for neuronal health and survival. We hypothesized that altered mitochondria oxidative phosphorylation is an early common step in the degeneration cascade initiated by a variety of mutations in blinding retinal diseases. Testing this hypothesis requires measuring mitochondria function in intact retina of different mouse models at one time. Thus, we established a simple and reproducible method to determine and compare mitochondria respiration ex vivo using the XF analyzer (Seahorse Bioscience) microplate-based assay. Measurements of OCR on acutely isolated retinal punches provided a sensitive and reproducible method of assessing mitochondria function in retinas from several degeneration mouse models in parallel, with minimal use of animals. In addition to animal models of retinal disease, our optimized protocol may be applied to postmortem human retina for functional comparison of regions with distinct photoreceptor distribution. We also envisage its use for screening of small molecules targeted to mitochondria in the context of neurodegenerative diseases.
In this study, we have demonstrated that: (1) photoreceptor mitochondria perform at their maximal respiratory capacity, with limited reserve of 20% to 25%; (2) respiration is comparable in rods and cone-like photoreceptors from the Nrl−/− mouse; (3) like in other central nervous system neurons, reserve respiratory capacity is higher in neurons of the inner retina than in photoreceptors; and (4) mitochondria respiration is altered in Rpgrip1−/− retina, consistent with functional (ERG) photoreceptor failure despite the presence of most photoreceptor cells.
The limited reserve capacity of retinal mitochondria indicates a state of high metabolic stress, defined as the ratio of basal to maximal (uncoupled) respiration, similar to what has been reported for mouse heart and liver.33 The high basal rates imply that small reductions in mitochondrial ATP production could have a strong negative effect on retinal energy homeostasis. Our direct measurement of photoreceptor residual respiratory capacity can explain the quick fall of b-wave recording in rabbit retina immediately after removal of glucose that had been attributed to low retinal energy reserves.11 Respiration recordings in medium containing glucose alone or in combination with glutamine or lactate were consistent with the notion that glucose is both necessary and sufficient to maintain retinal tissue viability during short-term culture.19,34 Previous work indicates that outer retinal O2 consumption is higher in the dark because of the need to maintain the ion current.11,17 However, we were not able to examine the impact of dark-adaptation in the retina since our assay uses LED illuminated sensors that have excitation wavelength between 470 and 530 nm.
In mice and primates, cones contain two and ten times as many mitochondria than rods, respectively,35,36 and mouse cones appear to have qualitatively more complex IV, as judged by activity-dependent histochemistry of fixed retinas.36 This evidence has prompted the hypotheses that cones have higher energy demand than rods35 or that cone mitochondria are less efficient than rod mitochondria.36 We used Nrl−/− mouse as a model of cone-only retina in which all rods are converted to cone-like photoreceptors in the absence of the rod-specific transcription factor NRL.37,38 Under our experimental conditions, rod and cone-like photoreceptors have similar cytochrome a content, basal and maximal respiration, and comparably limited residual respiration capacity. Whether cone-like photoreceptors in Nrl−/− mouse can be matched to wild-type cones with regard to energetic metabolism remains to be established.
Adult (P28) Pde6brd1/rd1 and Cep290rd16/rd16 retina are devoid of photoreceptors, thereby suggesting that oxygen consumption reflects mitochondrial function in the inner retina (including ganglion cells). Our results remarkably reproduce previous work,17 which demonstrated that inner segments of photoreceptors contain 55% to 65% of retinal mitochondria and that the outer and inner retina consume 54% and 46% of retinal O2 respectively, in the light adapted rat. In our study, Pde6brd1/rd1 and Cep290rd16/rd16 retina contained approximately only 40% of cytochrome a compared to wild type. In addition, the photoreceptor mitochondria in intact wild type retina exhibited basal OCR of 400 pmol/min/pmol cyt a, whereas average basal OCR of inner retina mitochondria in the three degenerating retinas was 340 pmol/min/pmol cyt a. Thus, the sum basal OCR of outer retina (wt) and inner retina (degenerating) is 740 pmol/min/pmol cyt a, with a calculated contribution of 54% by the outer retina and 46% by the inner retina. Furthermore, since basal mitochondrial respiration in Pde6brd1/rd1 and Cep290rd16/rd16 retina was close to 50% of uncoupled respiration, we conclude that inner retina neurons have a larger reserve capacity and are subjected to a lower mitochondrial stress relative to photoreceptors.
Based on studies in 661W cell line, it has been proposed that changes in mitochondrial reserve capacity are predictors of incipient photoreceptor death.20 Higher respiratory reserve capacity in Rpgrip1−/− retina suggests that the OCR recorded is mostly accounted for by neurons in the inner retina and that mitochondria in the residual photoreceptor population are dysfunctional. The presence of dysfunctional mitochondria in photoreceptors correlates with the lack of ERG response in Rpgrip1−/− retina.39 Taken together, our data strongly support the use of mitochondrial OCR as an early biomarker of retinal dysfunction, before the onset of overt degeneration.
Direct quantification of mitochondrial OCR in intact retina samples, as reported here, reveals that photoreceptors have very little reserve capacity to generate ATP, leaving them especially vulnerable to small changes in energy homeostasis. Our studies thus provide a plausible explanation for the early and highly penetrant photoreceptor degeneration phenotype in diseases that are associated with mutations in distinct widely expressed genes.40,41
Supplementary Material
Acknowledgments
The authors thank Jessica Chang for initiating this study, and Shan Chen, Luyi Adesanya, and Megan Kopera for technical assistance.
Supported by the Intramural Research Program of the National Eye Institute, National Institutes of Health (EY000473, EY000474). The authors alone are responsible for the content and writing of the paper.
Disclosure: K. Kooragayala, None; N. Gotoh, None; T. Cogliati, None; J. Nellissery, None; T.R. Kaden, None; S. French, None; R. Balaban, None; W. Li, None; R. Covian, None; A. Swaroop, None
References
- 1. Wright AF,, Jacobson SG,, Cideciyan AV,, et al. Lifespan and mitochondrial control of neurodegeneration. Nat Genet. 2004; 36: 1153–1158. [DOI] [PubMed] [Google Scholar]
- 2. Bossy-Wetzel E,, Schwarzenbacher R,, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004; (suppl 10): S2–S9. [DOI] [PubMed]
- 3. Orrenius S,, Gogvadze V,, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol. 2007; 47: 143–183. [DOI] [PubMed] [Google Scholar]
- 4. Scheibye-Knudsen M,, Fang EF,, Croteau DL,, Wilson DM,, III,, Bohr VA. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015; 25: 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duarte JM,, Schuck PF,, Wenk GL,, Ferreira GC. Metabolic disturbances in diseases with neurological involvement. Aging Dis. 2014; 5: 238–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lopez-Otin C,, Blasco MA,, Partridge L,, Serrano M,, Kroemer G. The hallmarks of aging. Cell. 2013; 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balaban RS,, Nemoto S,, Finkel T. Mitochondria oxidants, and aging. Cell. 2005; 120: 483–495. [DOI] [PubMed] [Google Scholar]
- 8. Lin MT,, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006; 443: 787–795. [DOI] [PubMed] [Google Scholar]
- 9. Vlachantoni D,, Bramall AN,, Murphy MP,, et al. Evidence of severe mitochondrial oxidative stress and a protective effect of low oxygen in mouse models of inherited photoreceptor degeneration. Hum Mol Genet. 2011; 20: 322–335. [DOI] [PubMed] [Google Scholar]
- 10. Jarrett SG,, Boulton ME. Consequences of oxidative stress in age-related macular degeneration. Mol Aspects Med. 2012; 33: 399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ames A III,, Li YY,, Heher EC,, Kimble CR. Energy metabolism of rabbit retina as related to function: high cost of Na+ transport. J Neurosci. 1992; 12: 840–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okawa H,, Sampath AP,, Laughlin SB,, Fain GL. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr Biol. 2008; 18: 1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu DY,, Cringle SJ. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog Retin Eye Res. 2001; 20: 175–208. [DOI] [PubMed] [Google Scholar]
- 14. Linton JD,, Holzhausen LC,, Babai N,, et al. Flow of energy in the outer retina in darkness and in light. Proc Natl Acad Sci U S A. 2010; 107: 8599–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brand MD,, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011; 435: 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chance B,, Williams GR. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J Biol Chem. 1955; 217: 383–393. [PubMed] [Google Scholar]
- 17. Medrano CJ,, Fox DA. Oxygen consumption in the rat outer and inner retina: light- and pharmacologically-induced inhibition. Exp Eye Res. 1995; 61: 273–284. [DOI] [PubMed] [Google Scholar]
- 18. Yu DY,, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005; 80: 745–751. [DOI] [PubMed] [Google Scholar]
- 19. Chertov AO,, Holzhausen L,, Kuok IT,, et al. Roles of glucose in photoreceptor survival. J Biol Chem. 2011; 286: 34700–34711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perron NR,, Beeson C,, Rohrer B. Early alterations in mitochondrial reserve capacity; a means to predict subsequent photoreceptor cell death. J Bioenerg Biomembr. 2013; 45: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fried NT,, Moffat C,, Seifert EL,, Oshinsky ML. Functional mitochondrial analysis in acute brain sections from adult rats reveals mitochondrial dysfunction in a rat model of migraine. Am J Physiol Cell Physiol. 2014; 307: C1017–C1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buono RJ,, Sheffield JB. Changes in distribution of mitochondria in the developing chick retina. Exp Eye Res. 1991; 53: 187–198. [DOI] [PubMed] [Google Scholar]
- 23. Gerencser AA,, Neilson A,, Choi SW,, et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009; 81: 6868–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balaban RS,, Mootha VK,, Arai A. Spectroscopic determination of cytochrome c oxidase content in tissues containing myoglobin or hemoglobin. Anal Biochem. 1996; 237: 274–278. [DOI] [PubMed] [Google Scholar]
- 25. Aponte AM,, Phillips D,, Hopper RK,, et al. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J Proteome Res. 2009; 8: 2679–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akimoto M,, Cheng H,, Zhu D,, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006; 103: 3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kenwood BM,, Weaver JL,, Bajwa A,, et al. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol Metab. 2014; 3: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mootha VK,, Arai AE,, Balaban RS. Maximum oxidative phosphorylation capacity of the mammalian heart. Am J Physiol. 1997; 272: H769–775. [DOI] [PubMed] [Google Scholar]
- 29. Zhao Y,, Hong DH,, Pawlyk B,, et al. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci U S A. 2003; 100: 3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bowes C,, Li T,, Danciger M,, Baxter LC,, Applebury ML,, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990; 347: 677–680. [DOI] [PubMed] [Google Scholar]
- 31. Chang B,, Khanna H,, Hawes N,, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006; 15: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roger JE,, Ranganath K,, Zhao L,, et al. Preservation of cone photoreceptors after a rapid yet transient degeneration and remodeling in cone-only Nrl-/- mouse retina. J Neurosci. 2012; 32: 528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phillips D,, Covian R,, Aponte AM,, et al. Regulation of oxidative phosphorylation complex activity: effects of tissue-specific metabolic stress within an allometric series and acute changes in workload. Am J Physiol Regul Integr Comp Physiol. 2012; 302: R1034–R1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winkler BS. Buffer dependence of retinal glycolysis and ERG potentials. Exp Eye Res. 1986; 42: 585–593. [DOI] [PubMed] [Google Scholar]
- 35. Perkins GA,, Ellisman MH,, Fox DA. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol Vis. 2003; 9: 60–73. [PubMed] [Google Scholar]
- 36. Hoang QV,, Linsenmeier RA,, Chung CK,, Curcio CA. Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis Neurosci. 2002; 19: 395–407. [DOI] [PubMed] [Google Scholar]
- 37. Mears AJ,, Kondo M,, Swain PK,, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001; 29: 447–452. [DOI] [PubMed] [Google Scholar]
- 38. Daniele LL,, Lillo C,, Lyubarsky AL,, et al. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005; 46: 2156–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Won J,, Gifford E,, Smith RS,, et al. RPGRIP1 is essential for normal rod photoreceptor outer segment elaboration and morphogenesis. Hum Mol Genet. 2009; 18: 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wright AF,, Chakarova CF,, Abd El-Aziz MM,, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010; 11: 273–284. [DOI] [PubMed] [Google Scholar]
- 41. Veleri S,, Lazar CH,, Chang B,, Sieving PA,, Banin E,, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015; 8: 109–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.