Abstract
MicroRNAs (miRNAs) and long-non-coding RNAs (lncRNAs) are involved in many biological processes, including viral replication. In this review, the role of miRNAs and lncRNAs in HIV replication will be discussed. The review focuses on miRNAs that target cellular proteins involved in HIV replication – proteins that mediate steps in the viral life cycle, as well as proteins of the innate immune system that inhibit HIV replication. Given the large number of miRNAs encoded in the human genome, as well as the large number of cellular proteins involved in HIV replication, the number of miRNAs identified to date that affect viral replication are certainly only the “tip of the iceberg.” The review also discusses two lncRNAs that are involved in HIV gene regulation – 7SK RNA and NEAT1 RNA. 7SK RNA is involved in HIV Tat protein stimulation of RNA polymerase II elongation of the integrated provirus, while NEAT1 RNA is involved in HIV Rev protein export of incompletely spliced viral transcripts.
MiRNAs and lncRNAs have become an active topic of research over the past decade and it is now clear that these two classes of non-coding RNAs have important functions in many biological processes, including viral replication. In the case of miRNAs, we will focus on miRNAs that regulate expression of cellular proteins involved in viral replication. Given its limited genome of ~10,000 nucleotides, HIV-1 is dependent upon cellular proteins to mediate the various steps of the viral life cycle – these cellular proteins have been termed host-dependency factors, or HDFs. A meta-analysis of genome-wide siRNA screens suggests that over 2,000 HDFs may be involved in HIV-1 replication (1). It is therefore likely that miRNA repression of key HDFs can have profound effects on HIV-1 replication. Additionally, cells express a number of proteins whose function is to counter infection – proteins of the innate immune system. This review will also discuss miRNAs that target antiviral proteins and how this class of miRNAs can enhance HIV-1 replication. Finally, two lncRNAs – 7SK RNA and NEAT1 RNA – are involved in HIV-1 gene expression, and this review will summarize our current understanding of the mechanistic role of these two lncRNAs in viral gene regulation. Before discussing miRNAs and lncRNAs, a summary of the HIV replication cycle in target cells is presented (see Figure 1).
Figure 1. Steps in HIV replication cycle regulated by cellular miRNAs.
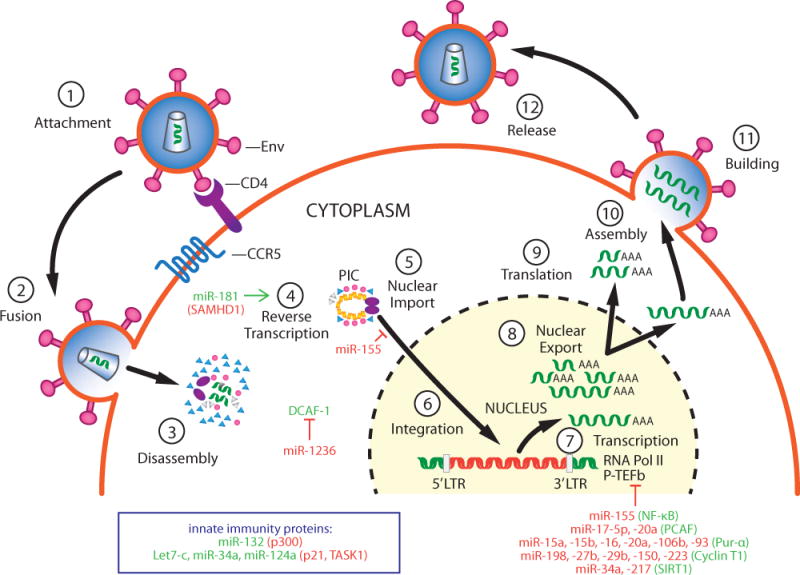
Steps in the viral replication cycle are numbered. MiRNAs that inhibit replication are shown in red and their cellular targets that promote replication are shown in green. MiRNAs that enhance viral replication are shown in green and their cellular targets (anti-viral proteins) are shown in red.
HIV-1 replication cycle
CD4+ T cells and monocytes/macrophages are the two major cell types infected by HIV-1 in vivo. Importantly, the activation or differentiation state of these cells has important effects on HIV-1 replication levels (2). Resting CD4+ T cells and monocytes do not generally support HIV-1 replication in vitro. CD4+ T cells must be activated to become highly permissive for viral replication, while monocytes must undergo the program of macrophage differentiation to become permissive for replication. The failure of resting CD4+ T cells and monocytes to support robust HIV-1 replication is due to the anti-viral action of innate immune proteins termed restriction factors, as well as to limiting levels of essential HDFs.
The HIV-1 replication cycle begins when the virion Envelope protein binds to the CD4 receptor and CCR5 or CXCR4 co-receptor on the surface of CD4+ T cells or monocytes/macrophages (3) The virion then fuses with the cellular plasma membrane and the viral core is released into the cytoplasm, where it begins a regulated but poorly understood process of disassembly (4). As the core disassembles, the viral RNA genome is copied into double-strand cDNA by the Reverse Transcriptase enzyme packaged in the core (5). A pre-integration complex (PIC) containing the viral cDNA and Integrase enzyme (also packaged in the viral core) is then actively transported into the nucleus, where the cDNA is integrated into the human genome, with a preference for integration into genes that are being actively transcribed (6). The integrated viral genome is termed the provirus.
Following integration, cellular RNA polymerase II (Pol II) recognizes and binds to the viral promoter in the 5′ long terminal repeat (LTR) of the provirus, whereupon transcriptional initiation begins. However, the efficiency of transcription elongation is restricted by two negative factors, DSIF and NELF. The viral Tat protein functions to overcome this restriction to elongation by recruiting the general transcriptional elongation factor P-TEFb to the Pol II complex (7). P-TEFb exists in three distinct complexes that have been biochemically characterized [reviewed in (8)]. The first P-TEFb complex is the active “Core” complex composed of CDK9, CyclinT1, and BRD4. The second complex is the “Super Elongation Complex” (SEC) comprised of CDK9, Cyclin T1, ELL2, AFF4, ENL, and AF9. The third complex is the “7SK RNP” and it is composed of the 7SK lncRNA, and several proteins – CDK9, Cyclin T1, HEXIM1, LARP7, and MEPCE. Details of the 7SK lncRNA and its role in HIV gene regulation are discussed below. Cyclin T2 is encoded by a distinct gene and is also a regulatory subunit of CDK9. Cyclin T2 is also found in the 7SK RNP, but it is expressed at significantly lower levels than Cyclin T1 in most tissues and cell lines, and its abundance in the 7SK RNP is therefore considerably lower than Cyclin T1.
As illustrated in Figure 2, a current model of Tat function postulates that Tat binds to CDK9 and Cyclin T1 in either the Core or 7SK RNP complex, and Tat/CDK9/Cyclin T1 then binds to the viral TAR RNA element that forms at the 5′ end of viral transcripts. The Super Elongation Complex then assembles and CDK9 is positioned and stimulated to phosphorylate both the carboxyl terminal domain of the large subunit of Pol II and the negative factors that restrict elongation, leading to a potent stimulation of elongation.
Figure 2. Model for role of 7SK RNP in HIV Tat function.
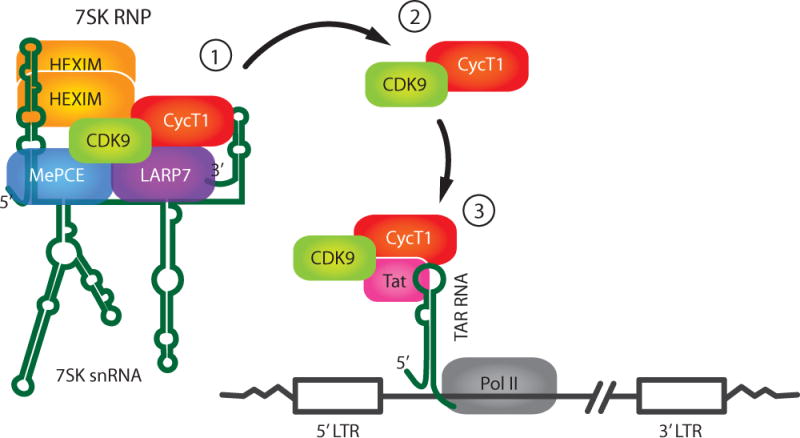
1) Cyclin T1/CDK9 are sequestered in the 7SK RNP in a catalytic inactive form. 2) Cyclin T1/CDK9 may be extracted from the 7SK RNP by Tat and this trimeric complex binds to TAR RNA formed at the 5′ end of the nascent viral transcripts (43). Alternatively, the 7SK RNP may bind to the 5′ LTR, and Cyclin T1/CDK9 are then extracted from the RNP when TAR RNA is synthesized (45). 3) Upon binding to TAR RNA, CDK9 in the Cyclin T1/CDK9/Tat complex phosphorylates several substrates in the RNA polymerase II complex, resulting in activation of transcriptional elongation.
The nuclear export of incompletely or unspliced viral transcripts is dependent upon the viral Rev protein (9). Rev along with the CRM1 (also termed XPO1) RNA export factor binds to a highly structured RNA element, termed the Rev-Response Element (RRE), present in incompletely spliced viral transcripts. This protein/RNA complex then accesses an export pathway utilized by small nuclear RNAs and a minority of cellular mRNAs. As will be discussed below, the lncRNA NEAT1 appears to play an important role in the mechanism whereby Rev recruits incompletely spliced viral RNA for RNA export. The unspliced full-length viral RNA can either serve as mRNA to produce the Gag or GagPol polyproteins, or traffic to the plasma membrane to be incorporated into new viral particles that bud from the infected cell.
miRNAs and HIV replication
miRNAs that target HIV-1 RNA and HIV-encoded non-coding RNAs
Before discussing miRNAs that target HIV-1 HDFs, it is worth noting that considerable research activity has been involved in the study of miRNAs that directly target HIV-1 transcripts, as well as the study of non-coding RNAs that can be expressed from the viral genome. However, these two topics – miRNAs that directly target HIV-1 RNA and non-coding RNAs expressed from the viral genome – are controversial, and these issues have been thoroughly discussed in an excellent recent review (10). A number of studies identified cellular miRNAs that can directly target HIV-1 transcripts and inhibit viral replication under certain experimental conditions. However, an analysis of miRNA binding to the HIV-1 genome by the photoactivatable ribonucleotide-induced cross-linking and immunoprecipitation (PAR-CLIP) technique suggests that HIV-1 transcripts are poorly targeted by cellular miRNAs, perhaps the result of extensive RNA secondary structure (11). Given the mutation rate of HIV-1, it seems likely that HIV-1 variants can readily evolve to evade miRNAs that directly target viral RNA if such miRNAs put significant evolutionary pressure on the virus. Additionally, several studies have identified small non-coding RNAs expressed from the HIV-1 genome during infection (10). It has however been argued that these RNAs are not expressed at levels sufficient to have significant biological effects (11). The controversies regarding miRNAs that target HIV-1 transcripts and HIV-encoded non-coding RNAs may be the consequence of different experimental conditions and methods of analysis used in the various studies. Future studies are required to clarify the biological importance of these two classes of non-coding RNA.
miRNAs that target HDFs
As stated above, genome-wide siRNA screens for HIV-1 HDFs suggest that over 2,000 cellular proteins are involved in the HIV-1 replication cycle (1). There are over 1,000 distinct miRNAs encoded in the human genome and a single miRNA can potentially target ~400 distinct mRNAs (12). Therefore, there is enormous potential for miRNAs to target and repress key HDFs and thereby inhibit HIV-1 replication. Also as stated above, the activation or differentiation state of CD4+ T cells and monocyte/macrophages – the two cell types infected by HIV-1 in vivo – determine the permissivity of the cells for HIV-1 replication. As miRNAs can be differentially expressed in resting vs. activated CD4+ T cells and monocytes vs. macrophages, the physiological state of these cells influences the overall effects of miRNAs on viral replication. MiRNAs that have been shown to target critical HDFs are listed in Table 1, and these miRNAs and there targets are discussed below. However, these miRNAs are certainly only the “tip of the iceberg” of miRNAs that regulate HIV-1 replication. Steps in the HIV replication cycle that are regulated by these miRNAs are indicated in Figure 1.
Table 1.
| miRNA | Target | Effect on HIV | Reference |
|---|---|---|---|
| miR-155 | ADAM10, TNPO3 Nup153, LEDGF/p75 |
inhibits PIC nuclear import | (14) |
| miR-155 | TRIM32 | inhibits NF-κB; promotes latency | (16) |
| miR-17-5p, miR-20a | PCAF | inhibits proviral transcription | (17) |
| miR-15a, miR-15b miR-16, miR-20a miR-106b, miR-93 |
Pur-α | inhibits Tat function | (19) |
| miR-198, miR-27b miR-29b, miR-223 miR-150 |
Cyclin T1 | inhibits Tat function | (20, 21) |
| miR-34a, miR-217 | SIRT1 | inhibits Tat function | (24, 25) |
| miR-1236 | DCAF1 | inhibits Vpr function | (26) |
| miR-181 | SAMHD1 | enhances replication | (28) |
| Let7-c, miR-34a miR-124a |
p21, TASK1 | enhance replication | (29) |
| miR-132 | p300 | enhances replication | (30) |
miR-155 targets multiple HDFs involved in PIC nuclear import and also promotes HIV-1 latency
MiR-155 is known to play an important role in appropriate immune responses (13) and it is an especially significant miRNA as it affects different aspects of the HIV-1 replication cycle – nuclear import of the pre-integration complex (PIC) and transcription of the HIV provirus. In primary macrophages, miR-155 is up-regulated by agonists of Toll-like receptors (TLR) TLR3 and TLR4 and this is associated with inhibition of HIV-1 replication (14). Remarkably, miR-155 directly targets the 3′ UTR of four HDFs involved in PIC nuclear import – ADAM10, TNPO3, Nup153, and LEDGF/p75. MiR-155 therefore functions to reduce the level of multiple HDFs in TLR3/4-stimulated macrophages and therefore has strong anti-HIV activity (14).
Current anti-HIV drugs can effectively repress HIV replication in infected individuals. However, if the drugs are discontinued, a reservoir of latent HIV reactivates and rekindles infection, thus requiring lifelong administration of anti-HIV drugs. The latent reservoir largely consists of resting memory CD4+ T cells containing integrated but transcriptionally silent viruses that can reactivate following T cell activation signals (15). A recent study has implicated miR-155 in the control of HIV latency (16). The HIV 5′ LTR has binding sites for NF-κB and activation of this cellular transcription factor stimulates transcriptional initiation of the integrated provirus. The E3 ubiquitin ligase TRIM32 is involved in activation of NF-κB through degradation of the inhibitor IκBα. MiR-155 targets the 3′ UTR of TRIM32 mRNA, thereby promoting HIV-1 latency by repressing activation of NF-κB.
MiRNAs that target HDFs involved in Pol II transcription of the HIV-1 provirus
A number of miRNAs have been identified that target HDFs involved in Pol II transcription of the HIV-1 provirus. In an important study, the Benkirane laboratory demonstrated that the siRNA depletion of the miRNA processing enzymes Dicer and Drosha increased HIV-1 replication, suggesting that miRNAs in general suppress viral replication (17). In this study, the authors subsequently examined miRNAs whose expression was altered following HIV-1 infection of the Jurkat CD4+ T cell line and identified several miRNAs that were down-regulated and encoded in the polycistronic miRNA cluster miR-17/92. This cluster encodes miR-17-(5p/3p), miR-18, miR-19a, miR-20a, miR-19b-1, and miR-92-1. Subsequent analysis demonstrated that miR-17-5p and miR-20a target the 3′ UTR of the histone acetylase PCAF (KAT2B). PCAF functions to enhance Pol II transcription by acetylating core histones and thereby modifying chromatin to a configuration that is highly permissive for transcription. Inhibitors of miR-17-5p and miR-20a enhance HIV-1 replication in Jurkat T cells, while over-expression of the miRNAs decreases replication, indicating that repression of PCAF is inhibitory to HIV-1 replication.
A number of studies have identified miRNAs that negatively regulate function of the HIV Tat protein. Pur-α is a DNA- and RNA-binding protein that has multiple functions in cells, including roles in transcription and translation (18). Pur-α has been shown to enhance Tat function and remarkably six miRNAs have been shown to target the 3′ UTR of Pur-α mRNA – miR-15a, miR-15b, MiR-16, miR-20a, miR-106b, and miR-93 (19).
As discussed above, Tat binds directly to Cyclin T1 in the P-TEFb complex and this is involved in activation of transcriptional elongation of the provirus. Several miRNAs have been identified in monocytes and resting CD4+ T cells that repress expression of this key HDF and contribute to the non-permissive state of these cells. In monocytes, miR-198 directly targets the 3′ UTR of Cyclin T1 mRNA and this can inhibit HIV-1 replication in this cell type (20). Upon monocyte differentiation to macrophages, the levels of miR-198 decrease relative to Cyclin T1 mRNA and this relieves repression of Cyclin T1, contributing to HIV replication in macrophages (20). In resting CD4+ T cells, four miRNAs have been identified that repress Cyclin T1 – miR-27b, miR-29b, miR-150, and miR-223 (21). Although over-expression of any of these four miRNAs can repress Cyclin T1 protein expression, only miR-27b has been shown to directly target the Cyclin T1 mRNA 3′ UTR. The repression of Cyclin T1 by the other miRNAs is therefore likely to be indirect. Activation of resting CD4+ T cells results in a decrease in the levels of these four miRNAs, thereby relieving their repression of Cyclin T1 protein expression and contributing to robust HIV replication.
SIRT1, a class III histone deacetylase that requires NAD+ as a cofactor, negatively regulates NF-κB by deacetylation of RelA/p65 (22). Additionally, SIRT1 can negatively regulate Tat by its deacetylase activity, as Tat is acetylated at lysine 50 and this has been shown to be important for Tat activity (23). Several miRNAs have been identified that target SIRT1 and this can stimulate HIV-1 proviral transcription through increasing NF-κB and Tat activity. MiR-217 and miR-34a can repress SIRT1 (24, 25).
miR-1236 targets DCAF1
DCAF1 (also termed VPRBP1) associates with the HIV-1 Vpr protein to form E3 ubiquitin ligase complexes, and this is thought to target antiviral proteins for proteasome-mediated proteolysis and thereby enhance viral replication. In monocytes, miR-1236 has recently been shown to target the 3′ UTR of DCAF1 mRNA and thereby inhibit HIV-1 replication in this cell type (26).
MiRNAs that target anti-viral proteins
In contrast to miRNAs that target HDFs and inhibit HIV replication, several miRNAs have been identified that target anti-viral proteins and stimulate HIV replication. SAMHD1 is an anti-HIV protein that inhibits reverse transcription through both phosphohydrolase and nuclease activities (27). A recent study has shown that miR-181 targets the 3′ UTR in SAMHD1 mRNA and therefore conferring to this miRNA a potential positive role in HIV replication (28). Additionally, a recent study found that let-7c, miR-34a, and miR-124a target the 3′ UTRs of p21 and TASK1 mRNAs – proteins that have been implicated in inhibition of HIV replication (29). Finally, miR-132 has been shown to repress MeCP2, a transcriptional regulatory protein and this is thought to stimulate HIV replication through down-regulation of the transcriptional co-activator p300 and effects on the host innate immune response to infection (30). Interestingly, miR-132 is up-regulated during infections of Kaposi’s sarcoma herpesvirus, herpes simplex virus-1, and human cytomegalovirus, suggesting that several viruses have acquired the ability to up-regulate this miRNA and thereby repress innate immunity (31).
lncRNAs and HIV gene regulation
7SK RNA
7SK RNA is an abundant 331 nucleotide snRNA transcribed by RNA polymerase III. Almost most 15 years ago, Cyclin T1 and CDK9 were found to be associated with 7SK RNA (32, 33). Subsequent biochemical studies identified three additional proteins associated with 7SK and Cyclin T1/CDK9 – HEXIM1, MEPCE, and LARP7 (34–38). HEXIM1 binds to structural elements within 7SK, while MEPCE is a methylphosphate capping enzyme that binds to the 5′ end of 7SK RNA. LARP7 binds to the 3′ end of 7SK and stabilizes the RNA. Although 7SK RNA/Cyclin T1/CDK9/HEXIM1/MEPCE/LARP7 exists as a distinct biochemical entity (8), additional proteins have been found associated with the 7SK RNP – SRSF2(also termed SC35) and DDX21 (39, 40). These proteins are involved in RNA polymerase II transcriptional elongation, but their roles in HIV Tat function remain to be elucidated.
The kinase activity of CDK9 is repressed in the 7SK RNP, and activation of transcriptional elongation by P-TEFb is thought to require the extraction of Cyclin T1/CDK9 from the 7SK RNP. Tat has been shown to be capable of extracting Cyclin T1 and CDK9 from the 7SK RNP to activate HIV proviral transcription (41). Additionally, activation of the PI3K/Akt pathway, or the ERK pathway through T-cell receptor signaling, has been shown to release Cyclin T1/CDK9 from the 7SK RNP and activate Tat-dependent elongation of the HIV provirus (42, 43). Some chromatin-immunoprecipitation (ChIP) experiments suggest that Cyclin T1/CDK9 free from the 7SK RNP are recruited to the HIV provirus to activate elongation (43), while other ChIP experiments suggest that the entire 7SK RNP is recruited directly to the HIV provirus where Tat extracts Cyclin T1/CDK9 as TAR RNA is synthesized (44, 45). Additionally work will be required to clarify the apparent discrepancies between these ChIP experiments.
Because the kinase activity of CDK9 is repressed by 7SK, it was originally postulated that the 7SK RNA is involved in establishing latent HIV infection in resting CD4 T cells through negative regulation of CDK9. However, phosphorylation of the CDK9 T-loop (Thr186) is largely absent in resting CD4+, and T-loop phosphorylation is required for both CDK9 catalytic activity and the assembly of the 7SK RNP (46, 47). Additionally, the levels of Cyclin T1 are generally quite low in resting CD4+ T cells (48, 49). 7SK RNP levels in resting cells that support latent HIV infection are consequently extremely low and unlikely to make a significant contribution to HIV latency (48–50). Additionally, analysis of a primary CD4+ T cell model of latency demonstrated that the amount of Cyclin T1/CDK9 in the 7SK RNP is greatly reduced as HIV latency is established, and when latent virus is reactivated, the amount of the 7SK RNP is greatly increased (51). Thus, the expression levels of the 7SK RNP in resting and activated CD4+ T cells appear incompatible with a regulatory role in establishing and maintaining HIV latency.
NEAT1 RNA
NEAT1 RNA is transcribed by RNA polymerase II and exists as two isoforms, NEAT1_1 (3.7 kb) and NEAT_2 (23 kb), also termed MENɛ and MENβ, respectively. NEAT1_1 and NEAT_2 are synthesized from a single promoter but differ in their 3′ ends; NEAT1 contains a polyadenylated 3′ end, while NEAT1_2 contains a non-polyadenylated 3′ end that is generated by the tRNA biogenesis machinery (52). Importantly, NEAT1 RNA is essential for the formation of nuclear structures termed paraspeckles (53). Paraspeckles contain more than 30 nuclear proteins, including p54nrb, PSF, and Matrin3, and these three proteins have been shown to be involved in HIV Rev nuclear export of incompletely spliced viral RNAs (54, 55). Furthermore, paraspeckles have been implicated in retaining incompletely spliced viral transcripts in the nucleus, where they are either degraded or traffic to the nucleolus and access a Rev-dependent nuclear export pathway (54, 56). A recent study demonstrated that siRNA depletion of NEAT1 enhances nuclear export of incompletely spliced HIV RNAs (57).
The Rev export pathway utilizes the export factor CRM1 (XPO1) and its co-factor RAN-GTP. A paraspeckle protein termed RBM14 has recently been shown to co-immunoprecipitate with CRM1 and Rev and be involved in export of viral transcripts (58). These recent findings of paraspeckle proteins and Rev suggest the rather speculative model shown in Figure 3 for nuclear export of incompletely spliced HIV transcripts. In this model, unspliced viral transcripts are retained in paraspeckles through the RNA binding proteins p54nrb, PSF, and RBM14. CRM1 associates with RBM14, either directly or indirectly, and this may lead to the trafficking of viral RNA to the nucleolus, where the Rev/XPO1/RAN-GTP complex binds to the RRE in viral transcripts. This protein-RNA complex is then exported to the cytoplasm, where the hydrolysis of RAN-GTP to RAN-GDP releases viral transcripts for either translation, or in the case of full-length viral RNA, for packaging in viral particles that assemble at the plasma membrane.
Figure 3. Model for role of NEAT1 RNA and paraspeckles in HIV Rev function.
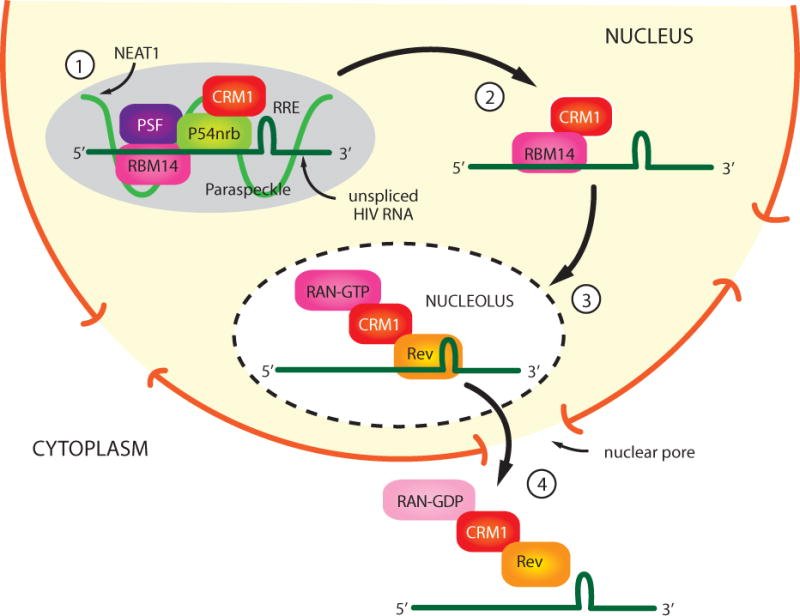
1) NEAT1 RNA is required for the formation of paraspeckle structures in the nucleus. PSF, p54nrb, and RBM14 are protein components of paraspeckles, and unspliced HIV transcripts are sequestered in paraspeckles through binding to PSF, p54nrb, and RBM14. 2) The association of RBM14 with CRM1 allows Rev/CRM1 to recruit unspliced viral transcripts to the nucleolus. 3) A nuclear export complex containing Rev/CRM1/RAN-GTP are bound to unspliced viral transcripts, and this complex traffics through nuclear pores to the cytoplasm. 4) The hydrolysis of RAN-GTP to RAN-GDP results in the releases of viral transcripts in the cytoplasm.
Conclusion
Given the large number of miRNAs encoded in the human genome (>1,000) and the ability of a single miRNA to target multiple mRNAs, there surely are many more miRNAs to be identified that target cellular proteins involved in HIV replication. As more miRNAs of relevance to HIV are identified, a challenge for the future will be to identify those miRNAs that repress key cellular proteins that regulate genetic pathways that are especially significant to HIV infection. Pathways such as an enhanced immune response or maintenance of latent HIV may be regulated by distinct miRNAs in CD4+ T cells and monocytes/macrophages. Specific inhibition of such miRNAs may provide a clinical benefit, and these putative miRNAs have potential as therapeutic targets.
Although the role of the 7SK RNP in HIV Tat function has been extensively studied, important questions remain. It is unclear whether the entire 7SK RNP is recruited to the integrated proviral genome (43), or whether Cyclin T1/CDK9 are released from the RNP prior to recruitment to the provirus (45). Determination of the structure of the 7SK RNA to ≤ 25 Angstroms is feasible by cryoEM technology (59) and such a structural determination will provide insight into how Tat and cellular proteins extract Cyclin T1/CDK9 from the RNP. Mechanisms involved in down-regulation of the 7SK RNP as activated CD4+ T cells return to a resting state are poorly understood (51), and elucidation of these mechanisms may provide insight into how HIV latency is established. Finally, mechanisms whereby NEAT1 RNA and paraspeckles are involved in HIV Rev function remain to be elucidated, as the model shown in Figure 2 is quite speculative. Limiting Rev function is involved in HIV latent infection (60) and mechanistic insight into NEAT1 RNA and paraspeckles may suggest therapeutic approaches to reactivate latent virus.
Acknowledgments
Work in the Author’s laboratory related to this review was supported by the National Institutes of Health (Baylor-UTHouston CFAR P30 AI036211, R21 AI14335, P50 GM103297). I thank Jason Kimata for critical comments on the manuscript.
References
- 1.Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang K, Rice AP. MicroRNA-mediated restriction of HIV-1 in resting CD4+ T cells and monocytes. Viruses. 2012;4:1390–1409. doi: 10.3390/v4091390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss RA. Thirty years on: HIV receptor gymnastics and the prevention of infection. BMC Biol. 2013;11:57. doi: 10.1186/1741-7007-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambrose Z, Aiken C. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology. 2014:454–455. 371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014:454–455. 328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeang KT. Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev. Biology (Basel) 2012;1:165–174. doi: 10.3390/biology1020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaminathan G, Navas-Martin S, Martin-Garcia J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol. 2014;426:1178–1197. doi: 10.1016/j.jmb.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. MBio. 2013;4:e000193. doi: 10.1128/mBio.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zhang Z. Computational Biology in microRNA. Wiley Interdiscip Rev RNA. 2015 doi: 10.1002/wrna.1286. [doi] [DOI] [PubMed] [Google Scholar]
- 13.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 14.Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog. 2012;8:e1002937. doi: 10.1371/journal.ppat.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siliciano JD, Siliciano RF. Recent developments in the search for a cure for HIV-1 infection: targeting the latent reservoir for HIV-1. J Allergy Clin Immunol. 2014;134:12–19. doi: 10.1016/j.jaci.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Ruelas DS, Chan JK, Oh E, Heidersbach AJ, Hebbeler AM, Chavez L, Verdin E, Rape M, Greene WC. MicroRNA-155 Reinforces HIV Latency. J Biol Chem. 2015 doi: 10.1074/jbc.M115.641837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 18.Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen CJ, Jia YH, Tian RR, Ding M, Zhang C, Wang JH. Translation of Pur-alpha is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J. 2012;26:4755–4764. doi: 10.1096/fj.12-209023. [DOI] [PubMed] [Google Scholar]
- 20.Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang K, Sung TL, Rice AP. Regulation of cyclin T1 and HIV-1 Replication by microRNAs in resting CD4+ T lymphocytes. J Virol. 2012;86:3244–3252. doi: 10.1128/JVI.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell. 2003;12:167–176. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HS, Chen XY, Wu TC, Sang WW, Ruan Z. MiR-34a is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation through the SIRT1/NFkappaB pathway. FEBS Lett. 2012;586:4203–4207. doi: 10.1016/j.febslet.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HS, Wu TC, Sang WW, Ruan Z. MiR-217 is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation by down-regulation of SIRT1. Biochim Biophys Acta. 2012;1823:1017–1023. doi: 10.1016/j.bbamcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Ma L, Shen CJ, Cohen EA, Xiong SD, Wang JH. miRNA-1236 inhibits HIV-1 infection of monocytes by repressing translation of cellular factor VprBP. PLoS One. 2014;9:e99535. doi: 10.1371/journal.pone.0099535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sze A, Olagnier D, Lin R, van GJ, Hiscott J. SAMHD1 host restriction factor: a link with innate immune sensing of retrovirus infection. J Mol Biol. 2013;425:4981–4994. doi: 10.1016/j.jmb.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Jin C, Peng X, Liu F, Cheng L, Lu X, Yao H, Wu H, Wu N. MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro. Biochem Biophys Res Commun. 2014;452:760–767. doi: 10.1016/j.bbrc.2014.08.151. [DOI] [PubMed] [Google Scholar]
- 29.Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. J Cell Sci. 2015;128:1607–1616. doi: 10.1242/jcs.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang K, Liu H, Rice AP. miR-132 enhances HIV-1 replication. Virology. 2013;438:1–4. doi: 10.1016/j.virol.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 34.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 36.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–253. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YK, Mbonye U, Hokello J, Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD. Transition Step during Assembly of HIV Tat:P-TEFb Transcription Complexes and Transfer to TAR RNA. Mol Cell Biol. 2012;32:4780–4793. doi: 10.1128/MCB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNamara RP, McCann JL, Gudipaty SA, D’Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013;5:1256–1268. doi: 10.1016/j.celrep.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 47.Ramakrishnan R, Dow EC, Rice AP. Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4(+) T lymphocytes. J Leukoc Biol. 2009;86:1345–1350. doi: 10.1189/jlb.0509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung TL, Rice AP. Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. HDAC inhibitors that release Positive Transcription Elongation Factor b (P-TEFb) from its Inhibitory Complex also activate HIV Transcription. J Biol Chem. 2013;288:14400–14407. doi: 10.1074/jbc.M113.464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haaland RE, Herrmann CH, Rice AP. Increased association of 7SK snRNA with Tat cofactor P-TEFb following activation of peripheral blood lymphocytes. AIDS. 2003;17:2429–2436. doi: 10.1097/00002030-200311210-00004. [DOI] [PubMed] [Google Scholar]
- 51.Budhiraja S, Famiglietti M, Bosque A, Planelles V, Rice AP. Cyclin T1 and CDK9 T-Loop Phosphorylation Are Downregulated during Establishment of HIV-1 Latency in Primary Resting Memory CD4+ T Cells. J Virol. 2013;87:1211–1220. doi: 10.1128/JVI.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yedavalli VS, Jeang KT. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology. 2011;8:61. doi: 10.1186/1742-4690-8-61. doi:1742-4690-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daelemans D, Costes SV, Lockett S, Pavlakis GN. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol. 2005;25:728–739. doi: 10.1128/MCB.25.2.728-739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596–12. doi: 10.1128/mBio.00596-12. doi:mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budhiraja S, Liu H, Couturier J, Malovannaya A, Qin J, Lewis DE, Rice AP. Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 Rev function. J Virol. 2015;89:3557–3567. doi: 10.1128/JVI.03232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi P, Wang Z, Feng Q, Pintilie GD, Foulds CE, Lanz RB, Ludtke SJ, Schmid MF, Chiu W, O’Malley BW. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol Cell. 2015;57:1047–1058. doi: 10.1016/j.molcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]


