Abstract
Stromal fibroblast senescence has been linked to aging-associated cancer risk. However, density and proliferation of cancer-associated fibroblasts (CAF) are frequently increased. Loss or down-modulation of the Notch effector CSL/RBP-Jκ in dermal fibroblasts is sufficient for CAF activation and ensuing keratinocyte-derived tumors. We report that CSL silencing induces senescence of primary fibroblasts from dermis, oral mucosa, breast and lung. CSL functions in these cells as direct repressor of multiple senescence- and CAF-effector genes. It also physically interacts with p53, repressing its activity. CSL is down-modulated in stromal fibroblasts of premalignant skin actinic keratosis lesions and squamous cell carcinomas (SCC), while p53 expression and function is down-modulated only in the latter, with paracrine FGF signaling as likely culprit. Concomitant loss of CSL and p53 overcomes fibroblast senescence, enhances expression of CAF effectors and promotes stromal and cancer cell expansion. The findings support a CAF activation/stromal co-evolution model under convergent CSL/p53 control.
INTRODUCTION
Most genetic hallmarks of epithelial cancer are already present in premalignant in situ lesions that rarely progress into full-blown cancer1–3. Cellular senescence provides an intrinsic failsafe mechanism against tumor development4. However, senescence of stromal fibroblasts is associated with production of growth factors, cytokines, extracellular matrix components and degrading enzymes (Senescence Message Secretome, SMS) that promote tumor development5,6. As such, stromal cell senescence has been implicated in increased incidence of many cancer types with age5,7. However, increased density and proliferation of cancer-associated fibroblasts (CAFs), rather than senescence, are frequently observed around tumors8,9. Senescent cells can be cleared by mechanisms like macrophage activation10, and there can be selective pressure for stromal fibroblasts with CAF properties to escape senescence1,8,9,11–14.
Increased determinants of cellular senescence like p53/p21INK4Cip1 or p16Ink4a do not induce SMS production15 and mechanisms linking fibroblast senescence with CAF activation are not known. Information on transcriptional events leading from normal fibroblasts to established CAFs is also scant. CSL (RBP-Jκ, CBF-1) is a DNA binding protein with intrinsic transcription repressive function converted into transcriptional activator by activated Notch16. Mice with mesenchymal CSL gene deletion develop a skin field cancerization phenotype, with dermal fibroblast alterations preceding inflammatory infiltrates and, by 2–4 months, multifocal keratinocyte tumors17. We report here that CSL functions as direct repressor of both senescence- and CAF-determinant genes in stromal fibroblasts from various organs, with CSL-p53interactions functioning as failsafe mechanism againstcancer stromal cell evolution and expansion.
RESULTS
1) CSL as a common repressor of fibroblast senescence and CAF activation
Newborn mice with mesenchymal CSL gene deletion exhibit signs of atrophic and/or aging skin, with reduced dermal thickness and altered extra-cellular matrix17. Staining of skin sections for senescence associated-β-galactosidase activity (SA-β-Gal) showed widespread positivity of dermal cells (Fig. 1a, Supplementary Fig. 1a), which was enhanced in stroma of premalignant lesions in older mice (Fig. 1b, Supplementary Fig. 1b). Importantly for the later studies, fraction of proliferating fibroblasts was also enhanced at this stage (Fig. 1c and Supplementary Fig. 1c).
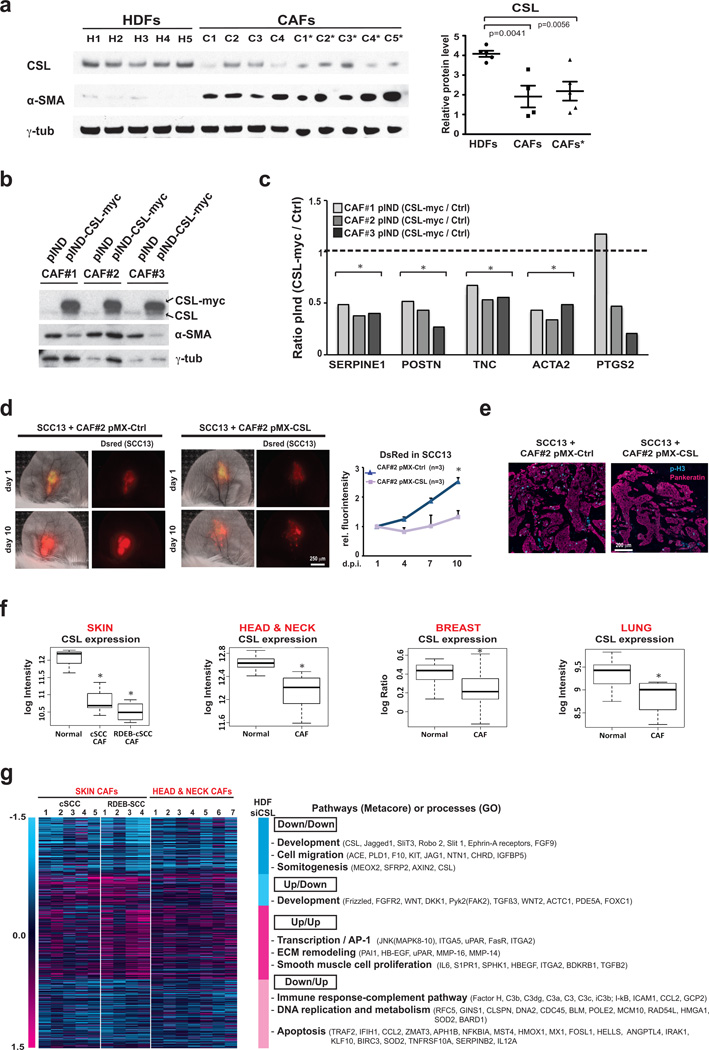
Figure 1. CSL control of stromal fibroblast senescence and CAF gene expression.
(a) Senescence Associated β-galactosidase (SA-β-Gal) staining of newborn mouse skin plus/minus mesenchymal CSL deletion (KO1/WT) with anti-vimentin immunofluorescence for fibroblast localization. (b) SA-β-Gal staining of lesions in 3-months old male mice with immunofluorescence for keratin 14 (K14), Tenascin-C (TNC), Vimentin, DAPI staining (lower panels, Supplementary Fig.1b). (c) Immunofluorescence of parallel sections for phospho-histone 3, vimentin, F4/80 and quantification of phospho-histone 3 positive fibroblasts (vimentin-positive, F4/80 negative); From left to right, n (cells) = 367, 603, 298, 453, 284, 493, assessed from 3, 4, 3, 4, 3, 4 fields, respectively; mean +/−s.d., two-tailed paired t-test. (d,e) SA-β-Gal staining (d) and clonogenicity assays (e) of second passage dermal fibroblasts from mice plus/minus CSL deletion with corresponding quantification. d: n(cells)=323(WT) and 135(KO), assessed from 4 and 7 fields respectively; e: n=3 biological replicates/condition. (f,g) SA-β–Gal (f) and clonogenicity (g) assays of HDFs plus/minus CSL silencing. f: from left to right n(cells)=118, 72, 111 assessed from 4, 6 and 6 fields, respectively; g: n=3 biological replicates/condition. Similar results were obtained with two other strains (Suppl. Fig.2b,c). (h,i) Independent HDF strains plus/minus CSL silencing by shRNA (h) and siRNA (i) analyzed by RT-qPCR; h: n(HDF strains)=8 shCSL1+2, 4 shCtrl, i: n(HDF strains)=8 siCSL1+2, 4 siCtrl. (j) HDFs plus/minus CSL silencing analyzed by immunoblotting. One blot probed for p53, p21WAF1/Cip1, CSL, γ-tubulin; another for IL6, β-actin, another for p16INK4a, γ-tubulin. Similar p21 induction was observed in two other independent experiments (Fig. 7c,d), with p21 and IL6 IF (Suppl. Fig.2h) as confirmation. For unprocessed blots see Suppl. Fig.9. (k) SA-β-Gal assays of fibroblasts from oral mucosa (HOF), breast (HBF), lung (HLF) plus/minus CSL silencing; from left to right n(cells)=207, 251, 274, 201, 206, 200, 223, 203, 200, 276, 222, 249, assessed from 4, 11, 11, 5, 8, 7, 3, 7, 10, 4, 7 and 4 fields, respectively. (l) Cells as in (k) were analyzed by RT-qPCR; n(fibroblast types)=4 shCtrl, 8 shCSL1+2. For (d-g) and (k) mean+/−s.d, two-tailed unpaired t-test; for (h-i) and (l) ratio log2(CSL/Ctrl), two-tailed one sample t-test. *p<0.05.
Dermal fibroblasts from mice plus/minus CSL deletion had initially similar attachment and proliferation rates when cultured. However, at the first and, more markedly, second passage, mutant fibroblasts exhibited senescence-associated morphology, increased SA-β-Gal activity and reduced clonogenicity (Fig. 1d,e). Similar consequences were caused by shRNA-mediated silencing of CSL in primary human dermal fibroblasts (HDFs) from individuals of different gender and age (Fig. 1f,g; Supplementary Fig. 2a–c), in parallel with overall reduction of cell proliferation (Supplementary Fig. 2d,e). Down-modulation of CSL by either shRNA or siRNA silencing induced expression of senescence-determinants like CDKN2B (p15INK4b), CDKN2A (p16INK4a), CDKN1A (p21WAF1/Cip1) and miR-34a, in parallel with SMS and CAF marker genes (Fig. 1h–j; Supplementary Fig. 2 f–h). Similar effects were elicited by CSL silencing in primary fibroblasts from oral mucosa, breast and lung (Fig. 1k, j; Supplementary Fig. 2i–j).
Notch receptor activation converts CSL from repressor into activator of transcription16. Similarly to CSL silencing, inducible expression of activated Notch1 induced cellular senescence and CAF-effector genes (Supplementary Fig. 3a–d). Endogenous Notch activation via Jagged 1 ligand exposure led to similar outcomes (Supplementary Fig. 3e).
Relative to several HDF strains, freshly derived CAFs from various skin SCCs exhibited, together with elevated alpha smooth actin marker (α-SMA) expression, low CSL levels (Fig. 2a). Exogenously increased CSL in these cells caused suppression of CAF marker genes, mirroring CSL silencing effects in HDFs (Fig. 2b,c). Findings were further validated in vivo, utilizing an ear injection/tumor imaging assay described later on. Outgrowths of fluorescently labeled skin SCC-derived cells (SCC13) expanded much less when admixed with CAFs with increased CSL versus controls, with corresponding differences in cancer cell proliferation (Fig. 2d, e).
Figure 2. CSL expression and function in CAFs.
(a) Immunoblotting of cancer associated fibroblasts (CAFs) from several skin SCCs soon after culturing (p2) and upon expansion (p4; *) in parallel with several HDF strains. Blot was probed for CSL, α-SMAand γ-tubulin with densitometric quantification; n(strains)= 5 HDF, 4 CAF, 5 CAF*, mean +/− s.e.m., two-tailed unpaired t-test. Similar results were obtained in a second independent experiment (right panel). (b) Same CAFs strains as in (a), transduced with lentiviral vector fordoxycyclin induciblemyc-tagged CSL or pIND vector control were analyzed by immunoblotting. (c) CAFs as in (b) were analyzed by RT-qPCR for indicated genes; n(CAF strains)=3 pInd-Ctrl, 3 pInd-CSL, ratio(CSL/Ctrl), two-tailed one sample t-tets. (d) DsRed2-expressing SCC13 cells admixed with CAFs (strain #2) plus/minus constitutive lentiviral CSL expression were injected in parallel into ears of 3 NOD/SCID Il2rg−/− 10-weeks-old male mice. Representative images and signal quantification relative to day 1; n=3 per condition, mean +/− s.e.m., two-tailed paired t-test at day 10. (e) Immunofluorescence for proliferation (phospho-histone 3) and epithelial (pankeratin) markers. (f) CSL levels in published gene expression profiles of CAFs from skin SCCs (from general populations (cSCC) and from patients with recessive dystrophic epidermolysis bullosa (RDEB-SCC); head/neck SCC, breast cancer and lung cancer; Skin (n=5 SCC, 4 RDEB-SCC, 3 healthy individuals), Head & Neck (n=7 SCC, 5 healthy individuals), Breast (n=23 carcinoma, 5 healthy individuals), Lung (n=4 NSCLC, 15 healthy individuals), two-class comparison with moderated t-statistic. Median, upper and lower quartiles are represented. Vertical whiskers indicate variability outside the upper and lower quartiles. (g) Heat maps of differentially expressed genes in HDFs plus/minus CSL silencing relative to data sets of CAFs from skin and head/neck SCCs. Genes modulated by CSL silencing in HDFs (> 1.4 folds) concordantly or discordantly down- or up-regulated in clinically occurring CAFs are indicated by dark and light turquoise and magenta colors, respectively. Selected pathways or processes with a statistically significant enrichment (p<0.05) are indicated along with representative genes. Complete list is provided in Supplementary Table 1. For (a) and (b) unprocessed original scans of blots are shown in Supplementary Fig. 9. *p<0.05.
In gene expression profiles of CAFs from skin18, head/neck19, breast20 and lung21 cancers, relative to normal fibroblasts from the same body sites, expression of CSL was consistently down-modulated (Fig. 2f). Published expression profiles of CAFs from skin and head/neck SCCs18,19 were compared with those of HDFs plus/minus CSL gene silencing obtained by RNA-seq analysis. 56% of differentially modulated genes in HDFs with CSL silencing were similarly regulated in the published CAF profiles, while 44% of genes were discordantly regulated (Fig. 2g, Supplementary Table 1). Functional classification showed differential distribution of affected pathways (Metacore) and processes (Gene Ontology). Development-related genes like CSL and Jagged1 were commonly down-regulated, together with genes involved in migration and somitogenesis. Commonly up-regulated were genes impinging on AP1-dependent transcription, ECM remodeling and smooth muscle cell proliferation and function, consistent with our other results and previous work17. Other development-related genes, including Wnt signaling components (Fzd, Wnt2 and DKK1) and FGFR2, were up-regulated in CAFs and down-modulated in HDFs with CSL silencing. Interestingly, genes down-modulated in CAFs and up-regulated in HDFs with silenced CSL had distinct functions, ranging from immune regulation, DNA replication, metabolism, and apoptosis (Fig. 2g). Many apoptosis-related genes, such as MX1, HMOX1, IFIH1, CCL2, SERPINB2 and IL12A, are also implicated in senescence and some are p53 targets (e.g. CCL2)22–24, consistent with our further findings on escape from p53 in established CAFs.
2) Senescence- and CAF-determinant genes as direct CSL targets
We hypothesized that CSL functions as direct repressor of senescence- and CAF-determinant genes. Bioinformatics analysis revealed putative CSL binding sites in enhancer and/or promoter regions of multiple senescence-effector genes like CDKN1A, CDKN2A, miR-34a, and IL6. Chromatin immuno-precipitation (ChIP) assays of HDFs with two different antibodies against CSL showed binding of this protein to two sites in the CDKN1A locus, at 9.7 kb and 2.3 kb from the transcription start site (TSS) (Fig. 3a and Supplementary Fig. 3f). Many predicted CSL binding sites are present in the large miR-34a locus. Among six tested sites, CSL was found to bind to one, at 14 kb from the TSS (Fig. 3a and Supplementary Fig. 3f). CSL binding was also detected at one enhancer of the CDKN2A gene and several enhancers and promoter region of the IL6 gene (Fig.3a).
Figure 3. CSL as a direct negative regulator of senescence- and CAF-effector genes.
(a) Top: maps of the predicted CSL binding sites (positions indicated in blue) relative to the Transcription Start Site (TSS) and mapped within the proximal enhancers and promoter regions of the indicated genomic loci (grey and black boxes, respectively; obtained from ENCODE information). Each map is at a different scale, as indicated by bars. Bottom: Chromatin Immunoprecipitation (ChIP) assays with two different antibodies against CSL (HM = home made; CS = Cell Signaling; grey and black bars, respectively) were performed in parallel with non-immune IgG controls for the indicated chromosomal sites (numbers) containing predicted CSL recognition sequences. Independent ChIP assays with two other HDF strains are shown in Supplementary Fig. 3f. (b) List of CAF-related and AP1 family members that were found to be targeted by CSL by ChIP-seq analysis with two different antibodies against CSL. A more detailed list of genes with specific localization of CSL binding peaks is provided in Supplementary Table 2. (c) Graphic illustration of the position of CSL binding peaks revealed by ChIP-seq analysis with two different antibodies (red and blue colors) for the indicated genes, utilizing ENCODE information for promoter and enhancer localization, as indicated by islands of Histone H3 modifications (K4m3 and K27Ac, respectively), along with the respective position of Transcription Start Site (arrow) and coding exons (black boxes). For JUNB, the entire locus, as delimited by binding peaks of the CTCF insulator, is shown. Results for 2 additional loci (MMP13 and FOSL2) are shown in Supplementary Fig. 3g.
Many genes have been implicated as CAF effectors or markers8,9,25,26. We assessed whether any are CSL targets by ChIP-seq (ChIP combined with massively parallel DNA sequencing) analysis of HDFs, utilizing the same two antibodies against CSL as for ChIP assays (Supplementary Table 2). Out of 139 CAF-related genes identified from literature searches, 77 exhibited overlapping binding peaks with the two antibodies, with statistically significant enrichment (p = 2.46495e-20) relative to total fraction of genes with CSL binding peaks at genome-wide level (5190 out of 26019, mapped with ChIPpeakAnno package of Bioconductor) (Fig. 3b and Supplementary Table 2). CSL binding to at least 40 CAF-related genes is of likely functional significance, as their expression was differentially modulated in HDFs plus/minus CSL silencing (pValue =1.26751e-15) (Supplementary Table 2). Identified CSL targets extended to all categories of CAF-related genes, including those with pro-inflammatory and/or growth modulatory functions, matrix composition/remodeling and surface markers. Genes for several AP1 family members, inducers of CAF markers under negative Notch/CSL control17, were also direct CSL targets (Fig. 3b and Supplementary Table 2). In some genes (Tenascin C and JunB), CSL binding was proximal or downstream of the TSS, while in others (PTGS2/COX-2; MMP13 and FOSL2), CSL binding occurred at upstream distant enhancers (Fig. 3c; Supplementary Fig. 3g; Supplementary Table 2).
3) CSL-p53 functional and physical interactions
The two CSL binding sites in the CDKN1A locus have closely adjoining p53 recognition sequences (sites #3 and #5), pointing to possibly close interactions between the two proteins. ChIP assays of HDFs showed binding of p53 only to the −2.3 kb site, which was consistently enhanced upon CSL silencing (Fig. 4a). p53 binding to another targets, MDM2, was affected by CSL silencing to a lesser extent (Fig. 4a), while binding to BAX, related with apoptosis 27, remained undetectable.
Figure 4. Convergent regulation and physical association of CSL and p53 proteins.
(a) ChIP assays of HDFs plus/minus CSL silencing for p53 binding to CDKN1A (sites #3,5 in Fig. 3a) and MDM2 genes. For CDKN1A, assays with two HDF strains are shown. (b) Endogenous CSL and p53 immuno-precipitations from HDFs followed by immuno-blotting. For p53 immunoprecipitation, HDFs with lentiviral p53 expression were used. Similar results with another HDF strain were obtained (Suppl. Fig. 4a, b). (c) Hela cells expressing myc-tagged CSL and p53 were immunoprecipitated with anti-myc antibodies followed by immunoblotting with anti-myc and -p53antibodies. (d) Admixed CSL and p53 proteins were immune-precipitated with anti-CSL antibodies followed by immunoblotting forCSL and p53 (green and red, respectively). Anti-CSL secondaries recognized also heavy chains (h.c.). Similar results were obtained in a second independent experiment. (e) Binding of CSL and p53 proteins measured by microscale thermophoresis (MST). Inset: thermophoretic movement of fluorescently-labeled p53. Results of second experiment and specificity controls are shown in Supplementary Fig. 4c. (f) 293T cell extracts plus/minus CSL (CSL OE) or p53 (p53 OE) overexpression were incubated with biotinylated oligonucleotide with tandem CSL and p53 binding sites of CDKN1A gene (WT) or oligonucleotide with site disruption (mt). Pull-down samples were immunoblotted with antibodies against CSL and p53 (green and red, respectively). Lower panels: fluorescence signal quantification. (g) p53 luciferase reporter activity in HDFs co-transfected with two siRNAs against CSL (pooling siRNA samples for the assay) versus scrambled controls. Shown are composite results of 2 independent experiments; n=4 biological replicates, mean +/− s.d., two tailed paired t-test. *p<0.05. (h) p53 luciferase reporter activity in 293T cells plus/minus co-transfection of p53 and CSL expressing plasmid in increasing amounts. Similar results were obtained in three other independent experiments. (i) p300 immuno-precipitation from 293T cells co-transfected with p53 and CSL expressing plasmid in increasing amounts followed by immuno-blotting for p53, p300, and CSL. Similar results were obtained in two other independent experiments as shown in Supplementary Fig. 4d. For (b-d), (f) and (i) unprocessed original scans of blots are shown in Supplementary Fig. 9.
CSL may affect p53 activity through physical interaction. Endogenous p53 and CSL association could be readily detected by co-immunoprecipitation experiments with HDFs under basal conditions and with induced p53 expression (Fig. 4b and Supplementary Fig. 4a, b). Association of p53 with CSL was confirmed by over-expression of epitope-tagged proteins (Fig. 4c). When the two purified recombinant proteins were incubated with each other, followed by co-immuno-precipitation, effective association was also detected (Fig. 4d). Microscale thermophoresis (MST)28 confirmed direct CSL-p53 binding, with affinity between 113 nM and 120 nM (Fig. 4e and Supplementary Fig. 4c).
Cell extracts pull-down assays were performed with an oligonucleotide with the CSL and p53 binding sites of the CDKN1A gene and a control with single nucleotide changes disrupting CSL and p53 recognition. The two proteins were effectively pull-down by the specific oligonucleotide, and binding of one protein was unaffected by over-expression of the other (Fig. 4f). To assess p53 transcriptional activity, cells were transfected with a p53 luciferase reporter. Basal p53 activity was increased in HDFs by silencing of CSL (Fig. 4g). Conversely, p53 induced activity was decreased by exogenous CSL expression, in dose-dependent fashion (Fig. 4h). The p300 co-activator plays a central role in p53-dependent transcription29. Co-immunoprecipitation analysis showed that association of endogenous p53 and p300 was suppressed by increasing amounts of CSL paralleling dose-dependent suppression of p53 activity (Fig. 4i and Supplementary Fig. 4d).
4) Down-modulation of p53 expression and function in CAF activation
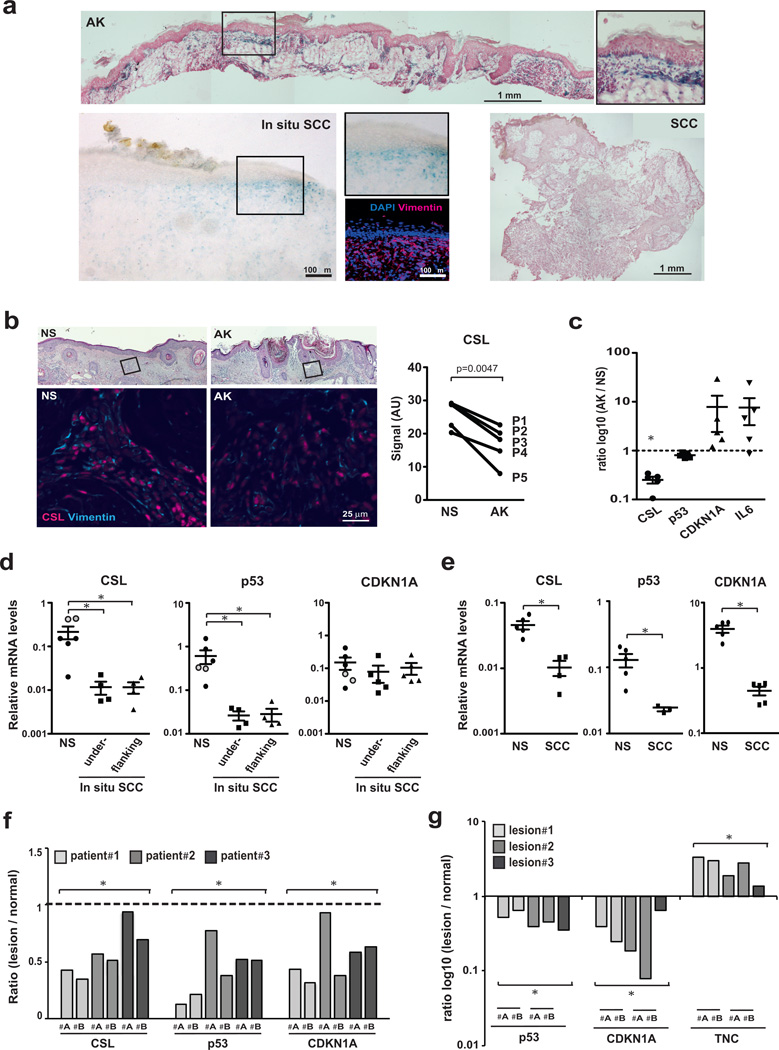
An important question is whether control of fibroblast senescence and CAF activation can be genetically dissociated. Consistent with this possibility, SA-β-Gal staining of clinically occurring premalignant keratinocytic lesions (actinic keratoses; AK) revealed senescent stromal cells in various proportions with non-senescent cells, depending on individual patients and lesion areas (Fig. 5a and Supplementary Fig. 5a). Scattered signs of stromal cell senescence were also found around in situ SCCs, while there were little or none in invasive SCCs, in which increased fibroblast density was frequently observed (Fig. 5a and Supplementary Fig. 5a–d).
Figure 5. CSL and p53 expression in premalignant (AK) versus malignant (SCC) stroma.
(a) SA-β–Gal staining of AKs, in situ and invasive skin SCCs. For in situ SCCs, immunofluorescence for vimentin and CD68, for fibroblast and macrophage localization. CD68-positive cells were only in deeper stromal regions (Supplementary Fig.5d). For additional lesions see Supplementary Fig.5a–d. (b) Double immuno-fluorescence of AKs and flanking normal skin (NS) from 5 patients for CSL and vimentin. Representative images and CSL signal quantification in vimentin-positive cells in AKs versus flanking skin. Only minority of cells double stained for vimentin and CD68 macrophage marker (Supplementary Fig.5e). n(vimentin-positive cells in P1-5)=345, 397, 310, 183, 111 (NS) and 345, 397, 310, 183, 111 (AK), mean signal intensity as individual data points, two tailed paired t-test. (c) LCM of AK-underlying stroma versus flanking normal stroma (NS) and RT-qPCR analysis; n=5 AKs and 5 flanking regions, mean of ratio log10(AK/NS) +/−s.e.m. Significance of differences in CSL expression was calculated by two-tailed one sample t-test (*p<0.0001), and of inverse relation between CSL and CDKN1A or IL6 expression by Pearson coeff. (−0.92 and −0.88, respectively; *p<0.05). (d) LCM-obtained PDGFRα positive cells (Supplementary Fig. 5j) from stroma underlying (n=5) and flanking (n=5) in situ SCC lesions and unaffected stroma (n=6) from other individuals (face- and abdomen-derived; black and grey circles) were analyzed by RT-qPCR for indicated genes; mean +/−s.e.m., two-tailed unpaired t-test, *p<0.05. (e) LCM-obtained stroma from invasive SCC (n=5) and normal skin (NS, n=5) samples from different individuals was analyzed by RT-qPCR for indicated genes; mean +/−s.e.m., two-tailed unpaired t-test, *p<0.05. (f) LCM-obtained stroma from skin SCCs (two lesion areas #A,B) and matching surgically discarded normal skin from three patients were analyzed by RT-qPCR; n=6 SCC and 3 normal regions, ratio (SCC/normal), one sample t-test, *p<0.005). (g) LCM-obtained stroma from skin lesions (two lesion areas #A, B) of 3 months old mice with mesenchymal CSL deletion (the same of Fig. 1b, c; Supplementary Fig. 1b, c) and matching stroma of unaffected skin was analyzed by RT-qPCR for indicated genes; n=5 affected and 3 unaffected regions, ratio log10(affected/unaffected), one sample t-test, *p<0.005.
Immunofluorescence and Laser Capture Microdissection (LCM) followed by RT-qPCR analysis of AK stromal tissues showed down-modulation of CSL expression and up-regulation of CDKN1A and IL6 relative to stroma of surrounding normal skin, while levels of p53 were unaffected (Fig. 5b, c and Supplementary Fig. 5e).
In situ skin SCCs are characterized by pronounced cellular atypia extending to all epidermal layers, without, as yet, cell invasion. Relative to less advanced AK lesions, more widespread stromal alterations around these lesions are likely to occur, as part of expanding cancer fields. In fact, LCM analysis of stromal fibroblasts underlying these lesions, as well as flanking skin, showed that CSL levels were consistently reduced relative to an external set of age- and site-matched skin samples from other individuals (Fig. 5d). Importantly, p53 expression was also down-regulated, with no increase of CDKN1A (Fig. 5d). Similar down-modulation of CSL and p53 was found by immunofluorescence and LCM analysis of stromal cells of invasive SCCs versus an external set of normal skin samples, with CDKN1A levels being also decreased (Fig. 5e; Supplementary Fig. 5k).
To compare gene expression levels in affected versus unaffected stroma of the same individuals, we analyzed discarded normal skin produced by closing of surgical excisions around SCCs, utilizing conditions for LCM/RT-qPCR analysis of formalin-fixed paraffin sections. Levels of CSL, p53 and CDKN1A expression were all significantly decreased in lesion-adjacent stroma versus distant unaffected skin from three different patients, confirming our other results (Fig. 5f).
For LCM, capture stromal regions were well separated from localized nests of inflammatory cells, identified by immunohistochemistry with anti-CD45 antibodies. Significant leukocyte and endothelial cell contamination was further ruled out by RT-qPCR analysis with corresponding markers (CD45 and CD31) (Supplementary Fig. 5h). In addition, for in situ SCC stromal cells and matching controls, antibodies against the fibroblast marker PDGFRα were used for labeling of cells prior to capturing (Supplementary Fig. 5j).
Findings were further validated in mouse model of mesenchymal CSL deletion, by examining skin lesions of mice at 3 months of age. Paralleling increase of proliferating fibroblasts (as shown in Fig. 1c), LCM and RT-qPCR analysis showed concomitant down-modulation of p53 and CDKN1A expression and inverse up-regulation of Tenascin C in lesion-adjacent stroma relative to distant unaffected skin (Fig. 5g). CD45 levels were slightly increased in some lesion stromal samples but not others (Supplementary Fig. 5i).
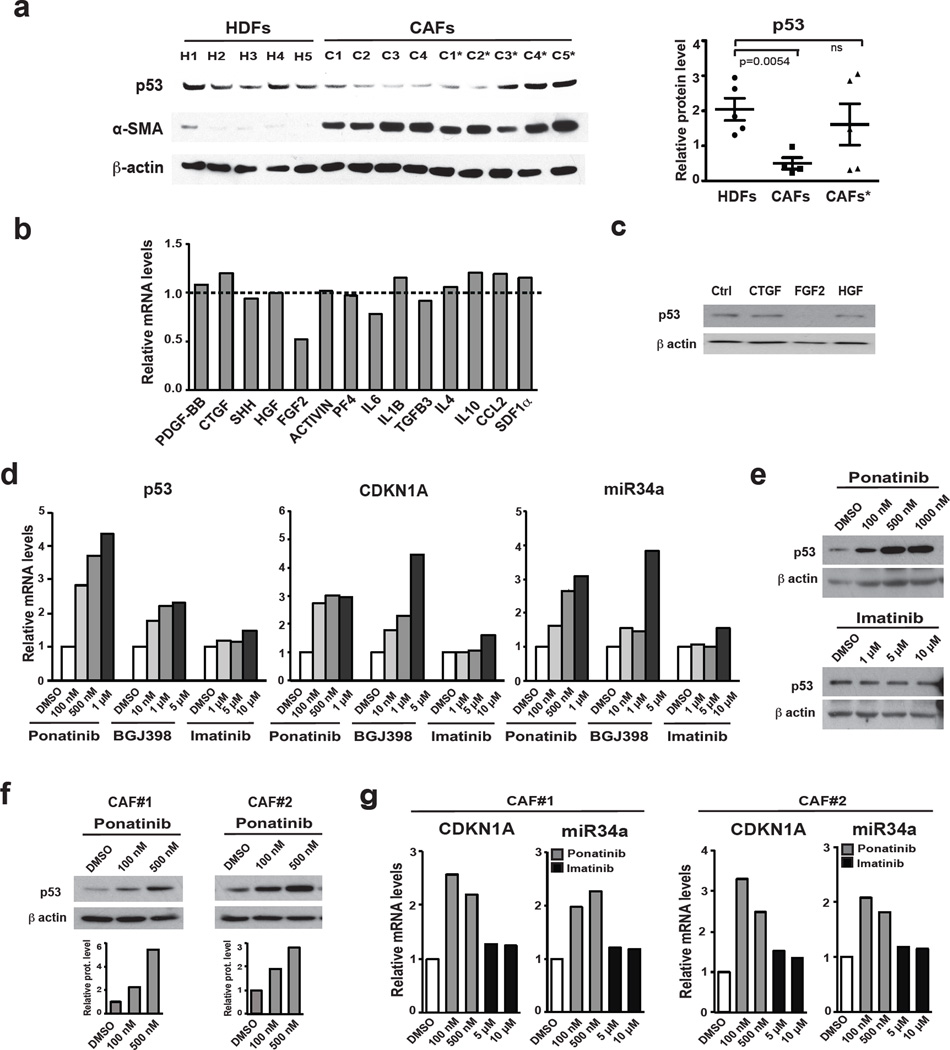
Consistent with above results, skin-derived CAFs analyzed for CSL expression (Fig. 2a) displayed also low p53 levels (Fig. 6a). Upon culturing, p53 remained poorly expressed in some CAF strains, while in others it reverted to higher levels. This suggested that the observed decrease of stromal p53 gene expression in fully established CAFs may result – at least in part – from paracrine influence of transformed keratinocytes or infiltrating inflammatory cells. A set of growth factors / cytokines known to be produced by these cells were tested for effects on stromal fibroblasts. Out of 14 factors, FGF-2 was capable of significantly down-modulating p53 expression in two different strains of HDFs, while others, like CTGF, HGF and, to a lesser extent, PF4 and IL6 exerted similar effects on only one strain (Fig. 6b, c; Supplementary Fig. 6a).
Figure 6. Modulation of p53 gene transcription and activity in dermal fibroblasts and CAFsas function of FGFR signaling.
(a) Immunoblot analysis of p53 expression in cancer associated fibroblasts (CAFs) from several skin SCCs soon after culturing (passage 2) and upon expansion (passage 4; *) in parallel with a set of HDF strains under similar growth and passage conditions. The p53 blot was re-probed with antibodies against α–SMA (left panel). Results were quantified by densitometric scanning after signal normalization for the housekeeping protein (right panel); n(strains)= 5 HDF, 4 CAF, 5 CAF*, mean +/− s.e.m., two-tailed unpaired t-test. (b) Early passage HDFs were treated with the indicated growth factors / cytokines (at the concentrations specified in methods) followed, 72 hours later, by RT-qPCR analysis of p53 expression. Results of a similar experiment with a second HDF strain of different origin are shown in Supplementary Fig. 6a. (c) Same cells as in the previous panel were treated with the indicated growth factors followed by immunoblot analysis of p53 expression. (d) Early passage HDFs were treated with a multi targeted tyrosine kinase inhibitor with potent anti-FGFR activity (Ponatinib) or a more FGFR-selective inhibitor (BGJ398) versus one devoid of such activity (Imatinib) followed, 72 hours later, by analysis of p53, CDKN1A and miR34a expression by RT-qPCR. Similar results were obtained with a second independent strain (Supplementary Fig. 6b). (e) HDFs treated as in the previous panel were analyzed for levels of p53 expression by immunoblotting, with similar results with a second HDF strain shown in Supplementary Fig. 6c. (f) Two independent CAF strains were treated with Ponatinib at the indicated concentrations for 72 hours followed by immunoblot analysis of p53 expression. (g) Similar CAF cultures treated as in the previous panel were analyzed for CDKN1A and miR-34a levels by RT-qPCR.For (a), (c) and (e-f) unprocessed original scans of blots are shown in Supplementary Fig. 9.
Mirroring these results, treatment of two different HDF strains with ponatinib, a tyrosine kinase inhibitor under clinical trial for malignancies with increased FGFR signaling, as well as BGJ398, a more selective FGFR inhibitor30, caused up-regulation of p53 expression and activity and associated cellular senescence (Fig. 6d, e; Supplementary Fig. 6b–d). Treatment with imatinib, another tyrosine kinase inhibitor with different spectrum of specificity and clinical use, exerted no such effect. Similar results were observed upon treatment of CAF strains with the inhibitors, paving the way for further studies on their possible mechanism of action and use (Fig. 6f, g; Supplementary Fig. 6e).
5) Stromal and cancer cell expansion results from compromised CSL and p53 function
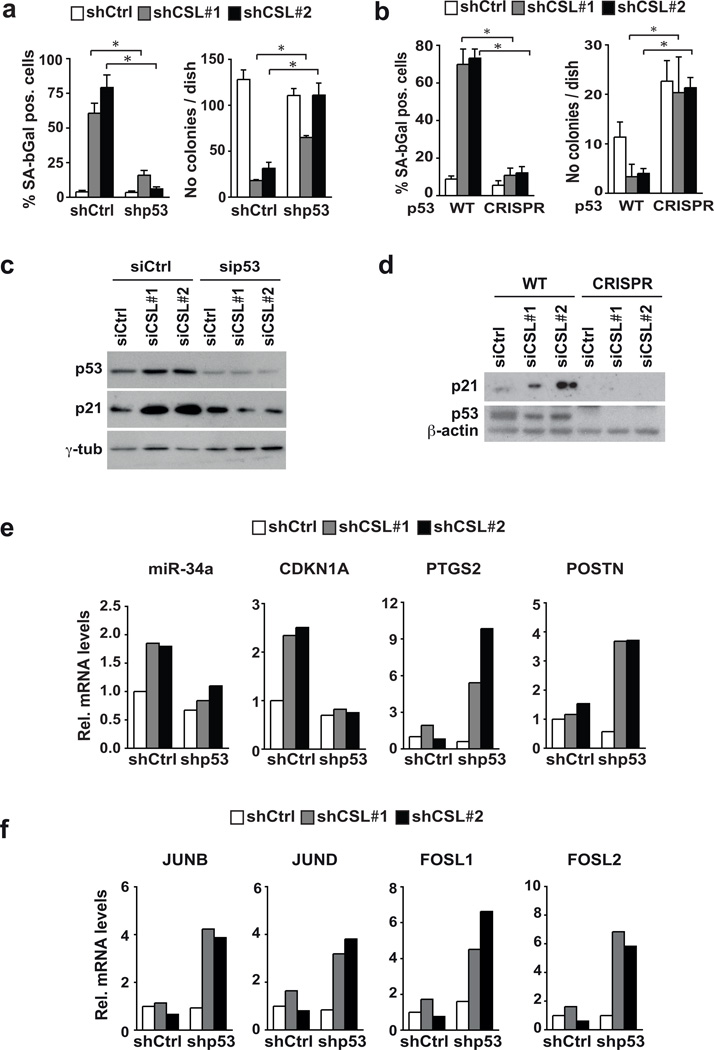
To assess whether p53 suppression is sufficient to counteract the senescence program induced by down-modulation of CSL expression, HDFs were infected with shRNA lentiviruses against these genes, individually and in combination. As an alternative approach, the p53 gene was disrupted by infection with an anti-p53 CRISPR lentivirus (Supplementary Fig. 7a). p53 gene silencing or disruption overcame reduced proliferation and senescence caused by CSL down-modulation in parallel with suppression of CDKN1A and miR-34a expression (Fig. 7a–d and Supplementary Fig. 7b). Similar counteracting effects were exerted by CDKN1A knock-down (Supplementary Fig. 7d,e).
Figure 7. Escape from senescence and enhanced CAF marker expression in stromal fibroblasts with concomitant CSL and p53 gene silencing.
(a) HDFs were infected with control versus p53 silencing retroviruses, followed by infection with either control or CSL silencing lentiviruses. Number of cells positive for SA-β-Gal activity was determined a week later (right panel). The same cells were also plated under sparse conditions, followed by determination of colony formation 10 days later (left panel). For SA-β–Gal activity n=131 (shCtrl+shCtrl), 127 (shCSL1+shCtrl), 130 (shCSL2+shCtrl), 314 (shp53+shCtrl), 151 (shp53+shCSL1), 184 (shp53+shCSL2) cells assessed from 4 fields for each condition, except for shCSL1+shCtrl and shCSL2+shCtrl from 6 and 8 fields, respectively; for clonogenicity assay n=3 biological replicates/condition. Results of a similar experiment with an independent strain of HDFs are shown in Supplementary Fig. 7b. (b) HDFs plus/minus CRISPR-mediated disruption of the p53 gene (obtained and characterized as shown in Supplementary Fig. 7a) were infected with control versus CSL silencing lentiviruses, followed by determination of SA-β–Gal activity and colony forming ability. For SA-β–Gal activity n=159 (p53-WT+shCtrl), 126 (p53-WT+shCSL1), 201 (p53-WT+shCSL2), 164 (p53-CRISPR+shCtrl), 186 (p53-CRISPR+shCSL1), 207 (p53-CRISPR+shCSL2) cells assessed from 5 fields for each condition, except for (p53-WT+shCSL1) and (p53-WT+shCSL2) assessed from 7 fields; for clonogenicity assay n=3 biological replicates/condition. (c)Same strain of HDFs as in panel (a) were transfected with siRNAs against CSL plus/minus siRNAs for p53, followed, 3 days later, by a repeat transfection. Cell extracts were analyzed 3 days later by immuno-blotting against the indicated proteins. (d) HDFs with CRISPR-mediated p53 gene disruption were transfected, in parallel with control parental cells, with siRNAs against CSL followed by immunoblot analysis of the indicated proteins. (e, f) HDFs plus/minus CSL and p53 gene silencing as in (a) were analysed by RT-qPCR for expression of the indicated genes. Results of a similar experiment with an independent strain of HDFs are shown in Supplementary Fig. 7c. For (a) and (b) mean +/− s.d., two-tailed unpaired t-test is shown; *p<0.05. For (c) and (d) unprocessed original scans of blots are shown in Supplementary Fig. 9.
Interestingly, concomitant silencing of CSL and p53 resulted in over-induction of CAF marker genes like PTSG2 (Cox-2), with a critical role in inflammation17, and POSTN (periostin), a cancer-associated stromal matrix protein of importance for recruitment of macrophages to tumors31 (Fig. 7e and Supplementary Fig. 7c). Up-regulation of these genes was also found in HDFs with CSL and CDKN1A knock-down (Supplementary Fig. 7f). Expression of AP1 family members was also enhanced in cells with concomitant CSL and p53 gene silencing (Fig. 7f).
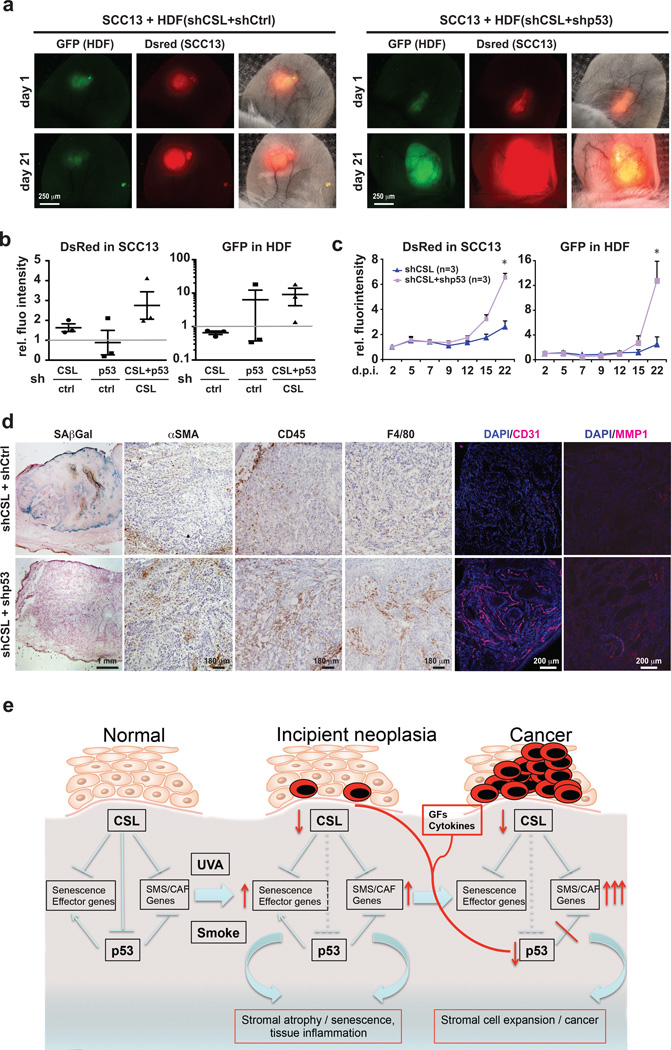
For in vivo testing, we developed an assay for cancer/stromal cell expansion, based on injections of fluorescently-labelled stromal and cancer cells into mouse ears followed by in vivo imaging. We monitored expansion of skin SCC-derived cells (SCC13) admixed with HDFs with knockdown of CSL, p53 or both. Cancer cell expansion was significantly enhanced by HDFs with dual silencing of CSL and p53 compared to CSL or p53 alone (Fig. 8a–c and Supplementary Fig. 8a). Stromal cells themselves missing both CSL and p53 expanded to a much greater extent than those with silencing of CSL alone (Fig. 8a–c; Supplementary Fig. 8a).
Figure 8. Tumor and stromal cell expansion as a result of CSL and p53 suppression.
(a) DsRed2 expressing SCC13 cells were admixed with GFP expressing HDFs with shRNA-mediated silencing of CSL or p53 individually or in combination, followed by parallel injections into mouse ears and imaging every 2–3 days under a fluorescence dissection microscope. Shown are representative images from one pair of mouse ears at the indicated times after injection. Similar images from another mouse are shown in Supplementary Fig. 8a. (b) Quantification of digital images for relative red (SCC cells) and green (stromal cells) fluorescence intensity values (intensity×surface area) for each combination of cells, at the end of the experiments (Day 21). Experimental conditions included SCC cells admixed with: (i) HDFs with individual CSL or (ii) p53 silencing versus HDFs control; (iii) HDFs with combined CSL and p53 silencing versus HDFs with CSL silencing only; for each conditions n=3 experimental and 3 control lesions (9 NOD/SCID Il2rg−/− 10-weeks-old male mice). To take into account individual animal variations, for each mouse ear pair, the signal increase in the ear injected with control cells was set to 1. Quantification was done by software (ImageJ) analysis of the digitally acquired images. (c) Quantification of changes in red and green fluorescence signal for 3 pairs of ear injections with SCC cells admixed with HDFs with combined silencing of CSL and p53 versus CSL alone; n=3 lesions per condition, mean +/− s.e.m., two-tailed paired t-test at day 22, *p>0.05. (d) SA-β-Gal assays and immuno-histochemical analysis with antibodies against the indicated markers in lesions formed by ear injections of SCC13 cells admixed with HDFs with combined silencing of CSL and p53 versus silencing of CSL alone. Similar results with lesions from an additional pair of mouse ear injections are shown in Supplementary Fig. 8b. (e) Diagrammatic illustration of the process leading from normal fibroblasts to CAFs with CSL and p53 as critical determinants, as proposed in the discussion.
Histological analysis at the end showed SA-β-Gal positive stromal cells in lesions formed by SCC13 cells admixed with HDFs with single CSL knockdown, but not in those with HDFs with combined CSL and p53 knockdown (Fig. 8d). Immunohistochemical analysis showed that lesions formed with HDFs lacking both CSL and p53 had higher levels of CAF markers (αSMA, MMP1, Tenascin C), enhanced leukocyte and macrophage infiltration (CD45 and F4/80) and angiogenesis (CD31) (Fig. 8d and Supplementary Fig. 8b).
Similar in vivo assays were repeated with HDFs with CSL plus/minus p53 silencing admixed with Head/Neck SCC Cal27 cells. Even in this case, HDFs with concomitant CSL and p53 silencing caused greater cancer cell expansion than HDFs with silencing of CSL only (Supplementary Fig. 8c). This resulted, by the end of the experiment, in neoplastic lesions with higher numbers of proliferating cancer and stromal cells (Supplementary Fig. 8d, e).
DISCUSSION
Stromal microenvironment can play a major role in tumor initiation and progression1,8,9,11–14. Together with or even preceding, chronic inflammation and angiogenesis are alterations of stromal fibroblasts leading to CAF activation32,33. Our findings indicate that at least two events are involved in transition from normal fibroblasts to CAFs: i) down-modulation of CSL expression and/or function, with cellular senescence as failsafe mechanism; ii) suppression of p53 expression and activity, with unimpeded cell expansion. Paracrine influences are likely involved in the second step (Fig. 8e). These events do not necessarily occur sequentially, as there is substantial heterogeneity of cells, with down-modulation of p53 possibly preceding or concomitant with compromised CSL function.
Fibroblast senescence is coupled with acquisition of CAF effector functions5,7. Our evidence points to CSL as underlying link, as this protein binds directly to many senescence- and CAF-determinant genes, suppressing their expression. Environmental insults, like UV irradiation and smoking, are causes of stromal cell senescence/tissue atrophy and field cancerization, a major clinical condition that can be modeled in mice by mesenchymal deletion of the CSL gene34. In ongoing work, we have found that CSL expression is inhibited in dermal fibroblasts by both UVA and smoke extract exposure through mechanisms currently under investigation. While UVA is a main cause of skin cancer34, smoke products can diffuse from surface epithelia and induce stromal cell senescence and increase cancer risk in multiple internal organs35.
Differential CSL binding to target genes is an important determinant of its cell type-specific regulatory functions36,37. Genes to which CSL binds with high affinity are induced when CSL levels decrease, with similar effects resulting from Notch activation, converting CSL from transcriptional repressor into activator36,37. Consistent with this mode of action, CSL silencing induced senescence- and CAF-effector genes in fibroblasts from various body sites but not in keratinocytes (Supplementary Fig. 1l), and similar up-regulation resulted from increased Notch activity, such as could be induced in vivo by neighboring ligand-expressing cells26.
Notch and p53 pathways interact at multiple levels, in context dependent manner38. Excitingly, we uncovered that CSL and p53 proteins physically bind, with affinity comparable to other proteins with which they pair39–41. CSL suppresses p53 association with p300, a critical determinant of its activity29. More detailed molecular events are under investigation. Here, we focused on functional significance of the findings. p53 can be involved in paracrine control of cancer development 42 and cancer-associated stromal changes11,43. Consistent with this possibility, loss of p53 not only blocked senescence caused by CSL suppression but enhanced CAF marker expression. Underlying mechanisms include up-regulation of CDKN1A, which can control transcription independently of the cell cycle44–46, and p53-induced disruption of AP1 association with basal transcription apparatus47.
Reports of genetic mutations of p53 in cancer-associated fibroblasts were questioned by later studies12. An important form of p53 regulation is at the level of gene transcription48–53. In contrast to less advanced actinic keratosis lesions, p53 gene expression is down-regulated in stromal fibroblasts of invasive and in situ skin SCCs, with paracrine influences of other cells as likely culprit. FGF-2 caused consistent down-modulation of p53 expression in HDFs, with more limited effects exerted by CTGF and HGF. The findings are consistent with previous reports of negative control of p53 gene transcription by ERK/AP1 activation50,54,55, and point to a possible, more selective mode of regulation connected with FGFR signaling. Besides activating mutations of FGFR 1, 2 or 3 genes, up-regulation of FGF ligands occurs frequently in several cancer types56. These factors can also be up-regulated in cancer associated fibroblast themselves 17, with possible autocrine mechanism reinforcing p53 down-modulation in these cells. FGFR inhibitory compounds are currently in clinical trials30 and, besides cancer cells, our findings suggest that they may also impact surrounding fibroblasts. By this or other approaches, restoration of CSL and p53 activity in these cells may serve to counteract the cancer-stroma co-evolution process.
ONLINE METHODS
Mice and human samples
Characterization of mice with mesenchymal CSL/RBP-Jκ deletion was previously reported 17,57. All mouse work was performed according to the Swiss guidelines and regulations for the care and use of laboratory animals with approved protocol from the Canton de Vaud veterinary office. Discarded human tissue samples for primary cell preparation were obtained from the Department of Surgery, or Department of Pediatrics, Lausanne University Hospital (Switzerland) and Department of Dermatology, Massachusetts General Hospital (Boston, MA) with institutional approvals and informed consent as part of institutional requirements. Parts not needed for diagnosis of excised skin actinic keratosis and SCC samples were obtained from the Departments of Dermatology, University of Tübingen (Germany) and Massachusetts General Hospital (Boston, MA), with institutional approvals and informed consent as part of institutional requirements. These included also tumor-free excess skin from routine Mohs surgery of facial skin cancer (“dog ears” resulting from bunching of skin at the end of an incision after wound closure)
Cell manipulations
Conditions for culturing of cells, viral infection, siRNA-mediated gene silencing, RT-qPCR and Chromatin Immunoprecipitation (ChIP) were previously as reported 17,58,59. For derivation of CAFs, surgically excised discarded skin SCCs samples were cut in 1–2mm pieces after removal of fat excess, followed by incubation in 2 ml of PBS containing 0.25 mg/ml of Liberase TL (Roche) for 40 min at 37°C with gentle shaking. After equal volume addition of FBS to stop the enzymatic digestion, the dissociated tissue was passed through a 10 ml syringe attached to a 70 µm sieve. Cells from the flow through were centrifuged, washed three times with DMEM 10% FBS and seeded in a 10 cm tissue culture dish. Adherent cells were expanded for characterization as with HDFs. Skin SCC13 60 and oral Cal27 SCC cells 61 were originally reported and subsequently checked for presence of oncogenic mutations 62 (https://cansar.icr.ac.uk/cansar/cell-lines/CAL-27/mutations/). They have been routinely tested for mycoplasma and have not been authenticated for STR profiling. They were infected with a DsRed2 expressing lentivirus and puromycin selected. Human THP 1 monocytic leukemic cells were differentiated into adherent macrophages (MΦ) by treatment with 10 µM 12-O-tetra-decanoylphorbol-13-acetate (TPA) plus/minus activation by overnight treatment with 1 µg/ml lipopolysaccharide (LPS). Human umbilical vein endothelial cells (HUVEC) and mouse primary lung endothelial cells were kindly provided by Dr. Tatiana Petrova. Human WI-38 diploid lung fibroblasts cell line was purchased from ATCC. None of the used human cell lines is listed in the NCBI Biosample Database of misidentified cell lines.
HDFs were infected with lenti- and retro-viruses at high titer, sufficient to infect the vast majority of HDFs so that similar results were obtained in experiments with and without antibiotic resistance selection: results in Fig. 7a, e, f and Suppl. Fig. 7b, c, d were obtained without selection, elsewhere else with selection. The list of shRNA vectors, siRNAs, primers and antibodies is provided in Supplementary Table 3. Transient transfection and promoter activity assays were also as described 57 using the pG13-p53 Luc plasmid as reporter 63.
CAFs strains stably infected with a doxycyclin inducible lentiviral vector for myc-tagged CSL in parallel with empty vector control were treated for 5 days with Doxycyclin (500 ng/ml) prior to RT-qPCR and immunoblot analysis. For in vivo assays, CAFs (strain#2) were infected with a lentiviral vector for constitutive CSL expression in parallel with empty vector control, followed, 7 days after infection, by injection into mice with admixed DsRed-expressing SCC13 cells.
For disruption of the p53 gene, the 20 nt CRISPR RNA sequence (ACTTCCTGAAAACAACGTTC) targeting the end of exon 2 of human TP53 gene, was selected as guide RNA (sgRNA) from the published genome-scale sgRNA library on the basis of being close to the beginning of the coding region and predicted minimal off-target probability 64. The corresponding oligonucleotide was cloned into lentiCRISPR vector followed by lentivirus production in 293T as previously described 64. HDFs were infected with the resulting virus, following, 24 hours after infection, by selection for the virally-transduced puromycin resistance gene. 7 days after selection, stably transduced cells (approximately 1000 cells, i.e. 5% of the originally infected cells) were pooled and expanded prior to further analysis in comparison with parental HDFs. The same lentiCRISPR vector was used to target p53 in two other HDF strains with similar efficacy.
Treatment of HDFs with Jagged-1 ligand was as previously reported 57, using Rabbit Anti-Human IgG (Sigma-l2011) plus/minus rhJagged1 Fc chimera (R&D systems; 1277-JG) for coating of dishes. HDFs were treated with the following growth factors / cytokines at the indicated concentrations every 24 hours for 3 days: SHH at 2 µg/ml, CTGF at 200 ng/ml, HGF, FGF-2, ACTIVIN-A, PF4/CXCL4, IL-6, MCP1/CCL2, IL-10 and SDF1/CXCL12 at 100 ng/ml, PDGF-BB at 50 ng/ml, IL-4 at 25 ng/ml, IL-1 β and TGF- β3 at 10 ng/ml final concentration.
Plasmid constructs
Plasmid encoding for inducible activated form of Notch1 (pIND-NICD) was previously described 59. To make doxycycline inducible Myc-tagged RBPJk plasmid, PCR product using gateway primers from pcDNA-RBPJk 65 was cloned into pENTR-D-TOPO entry vector (Life Tecnologies) and transferred through Gateway LR Clonase system (Life Tecnologies) into pINDUCER20 destination vector. RBPJk-Myc was further subcloned into pMX-neo using BamHI and SalI cut for PCR product and BamHI and XhoI cut for the plasmid.
Cell assays
Clonogenicity, Alamar blue and senescence β-galactosidase (SA-β-Gal) activity assays, by use of a commercially available chromogenic assay kit (Cell Signalling), were as reported 62.
For senescence, second passage mouse dermal fibroblasts were processed for SA-β-Gal staining 3 days after plating (5x103 cells/60 mm dish), while human fibroblasts were assayed 9 days after shRNA-mediated gene silencing. β-Gal activity assays at optimal pH (pH 4) were performed as control for β-galactosidase activity. Cells were counter-stained with Nuclear Fast Red and a minimum of 100 cells from at least 4 different fields per condition were counted, using digitally acquired images with Leica DM2000 microscope.
For clonogenicity assays, second passage mouse dermal fibroblasts plus/minus CSL gene deletion and human fibroblasts plus/minus shRNA-mediated silencing of CSL, individually or together with p53, were plated at clonal density in triplicate dishes (103 cells/60 mm dish) and cultivated for 2 weeks, followed by colony quantification.
For proliferation, human fibroblasts plus/minus CSL silencing were tested, 9 days after infection, by a 4 hours incorporation assay with 5-bromo-2`-deoxyuridine (BrdU; Amersham), followed by immunofluorescence analysis with anti-BrdU antibody (BD Biosciences) according to the manufacturer’s instructions. Percentage of BrdU-positive cells was evaluated in triplicate wells by digitally captured images and Image J software analysis, using DAPI staining for total cells identification. For Alamar blue fluorescence intensity assays, HDFs were plated, 2 days after shRNA-mediated CSL silencing, onto 96-well plates (1000 cells/well; triplicate wells per condition) followed by fluorescence intensity determination with a fluorimeter (Molecular Devices).
For gene expression, human fibroblasts were collected for RT-qPCR analysis 9 days after shRNA CSL gene silencing and 3 days after siRNA transfection. RNA samples were analyzed in triplicate with gene-specific primers and 36β4 for normalization.
For p53 activity assays, HDFs were co-transfected in triplicate dishes with a p53 luciferase reporter plasmid (PG13-luc; 3 µg/well) and two siRNAs against CSL versus scrambled controls. 293T cells were transfected in triplicate wells with the same p53 reporter (PG13-luc; 3 µg/well) plus/minus a p53 expressing vector (2 µg/well) and increasing amount of a CSL expressing plasmids (0.25, 0.5, 1, 2 µg). In all cases, luciferase activity was determined 48 hours later, using total protein amount for normalization.
Tissue analysis
Senescence β-galactosidase (SA-β-Gal) activity assays of tissue sections was performed by use of a commercially available chromogenic assay kit (Cell Signalling) as for cultured cells.
Laser Capture Microdissection (LCM), using an Arcturus XT microdissection system (Applied Biosystems), and ensuing RNA preparation and analysis were performed as before 17,66; for RNA purification from paraffin sections RNeasy FFPE Kit (Qiagen) was used. For immunofluorescence-guided LCM, frozen blocks of normal skin and in situ SCCs were freshly cut and immediately fixed with 75% ethanol for 30 seconds. After a brief blocking procedure (in 10% donkey serum in nuclease-free PBS for 2 min), sections were incubated with a mixture of FITC-conjugated antibodies against PDGFRα and propidium iodide for 2 min, followed by quick rinsing with PBS. The air-dried sections were then used to fluorescence-guided LCM using an Arcturus XT microdissection system as before. RNA samples were analyzed in triplicate with gene-specific primers and β-actin for normalization, except for paraffin samples in which 36β4 was used for normalization.
Intradermal Ear Injection and in vivo imaging
Mouse ear injections of cells were carried out in 10-weeks-old male NOD/SCID mice with interleukin-2 receptor gamma chain null mutation (Il2rg−/−), with only slight modifications of a recently described procedure 67. SCC13 cells (1×105) expressing the DsRed2 fluorescence marker were admixed with equal numbers of GFP expressing HDFs with shRNA-mediated silencing of CSL or p53, individually or in combination. Alternatively, for the in vivo experiment of supplementary Fig. 8c–e, DsRed2-expressing oral Cal27 SCC cells were admixed with HDFs labeled with the PHK67 green fluorescent cell membrane dye (Sigma-Aldrich), according to the manufacturer’s instructions (4x10−6 M of dye for 1x106 cells). In all cases, cells were concentrated by centrifugation and re-suspended in 3 µl of sterile HBSS solution per ear injection using a 33-gauge micro-syringe (Hamilton). Live images were taken every 2–3 days with a fluorescence stereomicroscope (Leica M205F) starting 24 hours post-injection. Quantification of red (DsRed2-SCC) and green (GFP-HDF) fluorescence intensity values (intensity×surface area) was achieved by ImageJ software analysis of digitally acquired images using the Lasso Toll to focus on the regions of interest. Mice were sacrificed 3 weeks after injection for direct tissue analysis.
Immunodetection techniques
Conditions for immuno-blotting, immuno-fluorescence and immuno-histochemistry were as described previously 17,66. Antibodies are listed in Supplementary Table 4 (including additional information for the validation profile from 1DegreeBio website).
Affinity purified anti-CSL polyclonal antibody was made in rabbit (Eurogentech, Seraign, Belgium) by immunization against two epitopes, one from the BTD region of CSL (aa 209–224 : LDDDESEGEEFVRDG) and the other from the CTD region (aa 426–440: FTYTPEPGPRPHCSA), both sequences being conserved in all seven CSL isoforms. After serum heat inactivation, antibodies were affinity purified on sepharose columns (Activated CH Sepharose 4B, GE Heelthcare) coupled with the immunizing peptides. Fractions were eluted by pH 2.5 Glycine into Tris buffer solution for pH neutralization. Protein eluted fractions (as determined by Bradford) were pooled and concentrated by ultrafiltration to 2 mg/ml in PBS (Amicon Ultra-15 30KD, Millipore). Validation of antibody specificity was assessed by IP/WB analysis of cells +/− over-expression of myc-tagged CSL, and immuno-staining using neutralizing peptide for signal neutralization.
Immunofluorescence images were acquired with a Zeiss LSM700 meta laser-scanning microscope. For quantitation of immunofluorescence nuclear signal, digitally acquired images for each color channel were separately imported into Image J software and converted into binary images. For single cell quantification of signal, the numbers of pixels (representing the intensity of the fluorescence signal) were measured using the function “Measurement” for multiple region of interest” (ROIs) on previously selected nuclei, on the basis of DAPI staining. For single cell CSL signaling measurements on tissue sections, only vimentin positive and CD68 negative cells were taken into account. For cytoplasmic signal quantification, fluorescence intensity was measured in the acquired fields and normalized to number of cells. All measurements were exported as Excel Microsoft files.
Co-immunoprecipitation experiments
Cells were lysed in NP-40 buffer and 500 µg total protein extracts were incubated overnight at 4°C with 10 µg primary antibodies against the CSL (home made; affinity purified) and p53 (DO1) proteins or the Myc-epitope tag (Zymed) in parallel with corresponding non immune IgGs. This was followed by addition of packed ProteinA/G magnetic beads (25 µl per sample; Novex) and further incubation for 4 hours at 4°C. Beads were washed 5 times with NP-40 buffer, eluted in 60 µl 2x SDS sample buffer at 95°C for 5 minutes and analyzed by gel electrophoresis and immuno-blotting.
For purified proteins, 100 ng of recombinant CSL/RBP-Jκ and p53 proteins, purchased from Origene, Rockville, MD (RBPJ transcript variant 1; p53 transcript variant 1) were incubated with each other in equal amounts for 20 minutes at room temperature in TBS buffer and 0.05% Tween, followed by standard co-immunoprecipitation assays as above.
For p300 association, 293T cells were co-transfected with a p53 (3 µg) and a CSL expressing plasmid in increasing amounts (0, 1.5, 3 µg; in 10 cm dishes), followed, 24 hours later, by immuno-precipitation of p300 and immuno-blotting as before.
Microscale Thermophoresis
Purified recombinant p53, labeled with RED-NHS Protein Labeling Kit (Nanotemper, Munich, Germany), was incubated at a constant concentration (900 nM) with two-fold serial dilutions of CSL (from 9 µM to 0.75 nM) in standard MST buffer. Equal volumes of proteins were mixed by pipetting and incubated for 20 minutes at room temperature. Mixtures were included into standard-treated glass capillaries and loaded into the instrument (Monolith NT.115, NanoTemper, Munich, Germany). Measurement protocol times were as follows: Fluorescence before 5s, MST on 30s, fluorescence after 5s, delay 25s. Both LED power and laser power were kept at 50%. Kd values were determined with the NanoTemper analysis tool.
DNA-oligo pull-down assay
A double stranded oligo-nucleotide corresponding to the CDKN1A promoter region with wild type CSL and p53 recognition sequences, and one with two nucleotide substitutions disrupting such sequences (as shown in Supplementary Table 3) were commercially obtained (Miltenyi) in biotinylated form. 300 µg of total protein extracts from 293T cells (obtained by a freeze-thawing method) were incubated with 1 µg oligos for 20 minutes at room temperature in 500 µl binding buffer containing 12 mM HEPES (pH 7.9), 4mM Tris (pH 7.9), 150 mM KCl, 1mM EDTA, 12% glycerol, 1mM DTT and 10 µg poly(dI-dC) competitor. This was followed by addition of 30 µl streptavidin-magnetic beads (Pierce), which had been pre-treated in 500 µl binding buffer with 1mg/ml BSA, 50 µg poly(dI-dC), 50 µg of sheared salmon sperm DNA for 30 minutes at room temperature. After 2 hours of incubation at 4°C, beads were washed 3 times with binding buffer and the protein-DNA complexes were eluted with 30 µl 2x SDS sample buffer at 95°C for 5 minutes, followed by gel electrophoresis and immuno-blotting analysis of the recovered material.
Transcriptome analysis
HDFs were infected with two different siRNA against CSL in parallel with a control virus. Total RNA was extracted 3 days post-transfection using the directZol RNA miniprep kit (Zymo Research) with on-column DNase treatment. RNA quality was verified on the bioanalyzer (Agilent) with RIN > 8. Four µg of total RNA were used for library preparation using the Truseq kit (Illumina). Single read was done on the Illumina HiSeq 2000 sequencer at the Genomic Technologies Facility (Lausanne University). Reads have been trimmed by using Trimmomatic (v0.22) and then mapped to the human hg19 reference genome by using TopHat (v2.0.8b). Gene expression levels have been evaluated by using HTSeq package (release 0.5.4p1). Data can be retrieved from GEO # GSE65473.
Published gene expression profiles of CAFs from skin SCCs (derived from 5 patients of the general populations (cSCC) and from 4 patients with recessive dystrophic epidermolysis bullosa (RDEB-SCC) in comparison with dermal fibroblasts from 3 healthy individuals; GSE37738), head & neck SCCs (derived from 7 patients and compared with oral mucosa fibroblasts from 5 healthy individuals, GSE38517), breast cancer (derived from 23 patients and compared with breast fibroblasts from 5 healthy individuals, GSE29270) and lung cancer (derived from 4 patients with non-small cell lung cancer in comparison with lung fibroblasts from 15 unaffected individuals GSE22862), were analyzed for normalized expression levels of CSL. Results were expressed as log intensity or log ratio values depending on the microarray platform.
For comparative transcriptomic analysis, gene expression profiles in skin and head/neck SCC CAFs versus normal fibroblasts, one single color Agilent dataset (GEO GSE37738, for skin CAFs) and one Affymetrix dataset (GEO GSE38517, for oral CAFs) were used. Agilent data were background subtracted (normexp method) and normalized with the quantile function, while Affymetrix data were processed with the RMA algorithm. Log2 ratios of the individual tumors versus mean values of control tissues were calculated separately for each data set. Boxplots were created with R v2.13, using normalized expression values of CSL in skin (GSE37738), head/neck (GSE38517), lung (GSE22862) and breast (GSE29270) CAFs versus correspondent normal fibroblasts. Pathways or processes with a statistically significant distribution (p<0.05) among the concordantly and discordantly regulated groups of genes as identified by Metacore and Gene Ontology gene family classification are provided in Supplementary Table 1.
ChIP and ChIP-Seq
ChIP assays with antibodies against CSL or p53 versus non-immune IgG control were performed as previously described 58 followed by determination of binding enrichment for the indicated sites; primer sequences and antibodies are given in Supplementary Table 3 and 4, respectively.
For ChIP-Seq analysis, immunoprecipitated DNA from HDFs was processed as for ChIP assays 58, using 5 µg of homemade and 10 µl of commercially available (Cell Signaling) antibodies against CSL per 106 cells. The ChIPed DNA was quantified by fluorometry on the Qubit system (Invitrogen). A total of 10 ng DNA were used for library preparation using NEBNext® ChIP-Seq Library Prep Reagent Set for Illumina, as recommended by the manufacturer. Burrows-Wheeler Aligner [http://bio-bwa.sourceforge.net/] was used for fastq files alignments and, for peak detection, MACS software [http://liulab.dfci.harvard.edu/MACS/] with default parameters. The Integrative Genomics Viewer (IGV) [http://www.broadinstitute.org/igv] was used for graphic illustration of ChIP-Seq peaks, and ENCODE data (http://genome.ucsc.edu/ENCODE/) for information on chromatin organization. ChIP-Seq data are deposited in the public repository (GSE59942).
Statistical analysis
Data are presented as mean +/− s.d., mean +/− s.e.m., or ratio among experimental groups and controls as indicated in the figure legends. For gene expression and functional testing assays, statistical significance of differences between experimental groups and controls was assessed by one-/two-tails un-paired or paired t-test and one-/two-sided one sample t-test as indicated in the figure legends, except for Fig. 5c where a Pearson correlation test was performed. *p value < 0.05 was considered as statistically significant, unless otherwise indicated.
For comparative transcriptomic analysis, two-class comparison analysis was performed using the moderated t-statistic available within the limma package (Smyth, G. K. (2004). Linear models and empirical Bayes methods were used for assessing differential expression in microarray experiments. Gene Ontology enrichment analysis was performed with the Fisher exact test, using the Database for Annotation, Visualization and Integrated Discovery (DAVID ) v6.7. Data meet the assumptions of the used tests; variation within each group of data has not been estimated.
For ear injection/tumorigenicity assays, sample size of adequate statistical power was ensured by minimizing the individual animal variability issue through parallel ear injections of the same animals with control versus experimental combinations of cells. No statistical method was used to predetermine sample size.
No exclusion criteria were adopted for animal studies and samples collection, except for RT-qPCR analysis in case of undetectable amplification signal.
No randomization was used and the researchers involved in the study were not blinded during sample obtainment or data analysis.
Reproducibility of experiments
For RT-qPCR and ChIP-PCR analysis as well as SA-β-Gal and clonogenicity assay, individual experiments on primary cell strains are shown, with repeated experiments on other independent primary strains shown in either the same figure or supplementary figures as indicated in the corresponding figure legends. RT-qPCR results in Figs. 1h, i and Fig. 2c are shown, respectively, with four HDF and three CAF independent strains. For Figs. 1f, 6b, d, e, 7a, e and Suppl. Fig. 3e similar results were obtained with a second independent strain (as shown in Suppl. Fig. 2b, 6a, b, c, 7b and c, respectively); for Figs. 1g and 3a results with two other independent strains are shown in Suppl. Fig. 2b,c and 3f. Findings in Figs. 1g, j, 7a,c and Suppl. Fig. 3c were also validated by alternative approaches in Suppl. Figs. 2d–e, 2h, 7b,d and Suppl. Fig. 3e, respectively. For IP-WB in Fig. 4b similar results were obtained with another HDF strain (Suppl. Fig.4a, b); for Fig. 4d and i similar results were obtained, respectively, in one and two other independent experiments (Suppl. Fig. 4d).
Luciferase reporter activity in Fig. 4g is a composite result of 2 independent experiments; for Fig. 4h similar results were obtained in two other independent experiments (statistics data source). Additional SA-β-Gal tissue staining, immunofluorescence, immunohistochemistry and mouse ears pictures for Fig. 1a–c, 5a, 8a and d are shown in Suppl. 1a–c, 5a–c, 8a, b, respectively. In Figs. 1c, d, f, 2d, 5b, 8a and Suppl. Figs. 1h, 5e, 5k and 8c representative images are shown together with signal quantification from multiple samples as specified in figure legends.
Accession numbers
RNA-seq and ChIP-Seq data sets generated for this study are available from the NCBI GEO public repository GSE65473 and GSE59942, respectively. Published gene expression profiles of CAFs from skin SCCs GSE37738 18, head & neck SCCs GSE38517 19, breast cancer GSE29270 20 and lung cancer GSE22862 21 were analyzed for CSL expression levels.
Supplementary Material
ACKOWLEDGEMENTS
We thank R. Bernards, R. Agami,W. Raffoul, J.P. Rival, J.M. Joseph, U. Just, F. Martinon and T. Petrova for vectors or cells, F. De Sousa e Melo and B. Hu for early contributions, K. Harshman for RNA-seq and ChIP-seq analysis, C. Pasche and T. Proust for technical help. Work was supported by grants from Swiss National Science Foundation (310030_156191/1), National Institute of Health (R01AR039190; R01AR064786; the content not necessarily representing the official views of NIH), European Research Council (26075083),OncoSuisse (OCS-2922-02-2012 and KFS-3301-08-2013). P. Ostano was supported by a grant from Lauretana S.P.A.
Footnotes
AUTHORS CONTRIBUTIONS
M.G.P., C.L., D.A.L., D.E.K., P.B., S.H.J., S.G., E.M., K.L. performed work and contributed to analysis of results. P.O., U.A. and P.P. conducted bioinformatics. W.H. and V.N. provided clinical samples. W.K. and M.A.S. contributed to ear injection experiments. C.B. contributed experimental reagents and insights. G.P.D. designed the study. M.G.P., K.L. and G.P.D. wrote the manuscript.
REFERENCES
- 1.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatenby R. Perspective: Finding cancer's first principles. Nature. 2012;491:S55. doi: 10.1038/491s55a. [DOI] [PubMed] [Google Scholar]
- 3.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campisi J, Andersen JK, Kapahi P, Melov S. Cellular senescence: a link between cancer and age-related degenerative disease? Semin Cancer Biol. 2011;21:354–359. doi: 10.1016/j.semcancer.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 7.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 9.Polanska UM, Orimo A. Carcinoma-associated fibroblasts: non-neoplastic tumour-promoting mesenchymal cells. J Cell Physiol. 2013;228:1651–1657. doi: 10.1002/jcp.24347. [DOI] [PubMed] [Google Scholar]
- 10.Hoenicke L, Zender L. Immune surveillance of senescent cells--biological significance in cancer- and non-cancer pathologies. Carcinogenesis. 2012;33:1123–1126. doi: 10.1093/carcin/bgs124. [DOI] [PubMed] [Google Scholar]
- 11.Bar J, Moskovits N, Oren M. Involvement of stromal p53 in tumor-stroma interactions. Semin Cell Dev Biol. 2010;21:47–54. doi: 10.1016/j.semcdb.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg RA. Coevolution in the tumor microenvironment. Nat Genet. 2008;40:494–495. doi: 10.1038/ng0508-494. [DOI] [PubMed] [Google Scholar]
- 15.Coppe JP, et al. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu B, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 2012;149:1207–1220. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng YZ, et al. Fibroblast-derived dermal matrix drives development of aggressive cutaneous squamous cell carcinoma in patients with recessive dystrophic epidermolysis bullosa. Cancer Res. 2012;72:3522–3534. doi: 10.1158/0008-5472.CAN-11-2996. [DOI] [PubMed] [Google Scholar]
- 19.Costea DE, et al. Identification of two distinct carcinoma-associated fibroblast subtypes with differential tumor-promoting abilities in oral squamous cell carcinoma. Cancer Res. 2013;73:3888–3901. doi: 10.1158/0008-5472.CAN-12-4150. [DOI] [PubMed] [Google Scholar]
- 20.Hosein AN, et al. Breast carcinoma-associated fibroblasts rarely contain p53 mutations or chromosomal aberrations. Cancer Res. 2010;70:5770–5777. doi: 10.1158/0008-5472.CAN-10-0673. [DOI] [PubMed] [Google Scholar]
- 21.Navab R, et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2011;108:7160–7165. doi: 10.1073/pnas.1014506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med. 2013;210:2057–2069. doi: 10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novakova Z, et al. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene. 2010;29:273–284. doi: 10.1038/onc.2009.318. [DOI] [PubMed] [Google Scholar]
- 24.Yoon IK, et al. Exploration of replicative senescence-associated genes in human dermal fibroblasts by cDNA microarray technology. Experimental gerontology. 2004;39:1369–1378. doi: 10.1016/j.exger.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinz B, et al. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 28.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nature communications. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 29.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 30.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 31.Zhou W, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015 doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 33.Quante M, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dotto GP. Multifocal epithelial tumors and field cancerization: stroma as a primary determinant. J Clin Invest. 2014;124:1446–1453. doi: 10.1172/JCI72589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salem AF, et al. Cigarette smoke metabolically promotes cancer, via autophagy and premature aging in the host stromal microenvironment. Cell Cycle. 2013;12:818–825. doi: 10.4161/cc.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castel D, et al. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JE, Macdonald RJ. Notch-independent functions of CSL. Curr Top Dev Biol. 2011;97:55–74. doi: 10.1016/B978-0-12-385975-4.00009-7. [DOI] [PubMed] [Google Scholar]
- 38.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9:587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chene P. Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol Cancer Res. 2004;2:20–28. [PubMed] [Google Scholar]
- 40.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooks T, et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer cell. 2013;23:634–646. doi: 10.1016/j.ccr.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19:1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dotto GP. p21WAF1/Cip1 : more than a break to the cell cycle? Biochim. Biophys. Acta. 2000;87483:1–14. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 46.Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol. 2013;25:52–58. doi: 10.1097/CCO.0b013e32835b639e. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, Zeng XR, Wenger L, Firestein GS, Cheung HS. P53 down-regulates matrix metalloproteinase-1 by targeting the communications between AP-1 and the basal transcription complex. J Cell Biochem. 2004;92:258–269. doi: 10.1002/jcb.20044. [DOI] [PubMed] [Google Scholar]
- 48.Boggs K, Reisman D. C/EBPbeta participates in regulating transcription of the p53 gene in response to mitogen stimulation. J Biol Chem. 2007;282:7982–7990. doi: 10.1074/jbc.M611675200. [DOI] [PubMed] [Google Scholar]
- 49.Bruno T, et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006;10:473–486. doi: 10.1016/j.ccr.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Kolev V, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 52.Reisman D, Loging WT. Transcriptional regulation of the p53 tumor suppressor gene. Semin Cancer Biol. 1998;8:317–324. doi: 10.1006/scbi.1998.0094. [DOI] [PubMed] [Google Scholar]
- 53.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 54.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 55.Stepniak E, et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malchers F, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014;4:246–257. doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 57.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, et al. Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev. 2010;24:1519–1532. doi: 10.1101/gad.1886910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Restivo G, et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011;30:4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 61.Gioanni J, et al. Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: establishment, characterization and response to cytotoxic treatment. European journal of cancer & clinical oncology. 1988;24:1445–1455. doi: 10.1016/0277-5379(88)90335-5. [DOI] [PubMed] [Google Scholar]
- 62.Wu X, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature. 2010;465:368–372. doi: 10.1038/nature08996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 64.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu L, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 66.Lefort K, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. Embo J. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kilarski WW, et al. Intravital immunofluorescence for visualizing the microcirculatory and immune microenvironments in the mouse ear dermis. PLoS ONE. 2013;8:e57135. doi: 10.1371/journal.pone.0057135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.