Abstract
Trophoblast cells are the first cells to differentiate from the developing mammalian embryo, and they subsequently form the blastocyst-derived component of the placenta. IFN-γ plays critical roles in activating innate and adaptive immunity, as well as apoptosis. In mice, IFN-γ is produced in the pregnant uterus, and is essential for formation of the decidual layer of the placenta and remodeling of the uterine vasculature. Responses of mouse trophoblast cells to IFN-γ appear to be selective, for IFN-γ activates MHC class I expression and enhances phagocytosis, but fails to activate either MHC class II expression or apoptosis in these cells. To investigate the molecular basis for the selective IFN-γ responsiveness of mouse trophoblast cells, IFN-γ-inducible gene expression was examined in the trophoblast cell lines SM9 and M-11, trophoblast stem cells, and trophoblast stem cell-derived giant cells. IFN-γ-inducible expression of multiple genes, including IFN regulatory factor-1 (IRF-1), was significantly reduced in trophoblast cells compared with fibroblast cells. Decreased IRF-1 mRNA expression in trophoblast cells was due to a reduced rate of IRF-1 transcription relative to fibroblast cells. However, no impairment of STAT-1 tyrosine phosphorylation or DNA-binding capacity was observed in IFN-γ-treated mouse trophoblast cells. Importantly, histone deacetylase (HDAC) inhibitors significantly enhanced IFN-γ-inducible gene expression in trophoblast cells, but not fibroblasts. Our collective studies demonstrate that IFN-γ-inducible gene expression is repressed in mouse trophoblast cells by HDACs. We propose that HDAC-mediated inhibition of IFN-γ-inducible gene expression in mouse trophoblast cells may contribute to successful pregnancy by preventing activation of IFN-γ responses that might otherwise facilitate the destruction of the placenta.
Trophoblast cells (TBCs)3 are the only blastocyst-derived cells directly exposed to maternal blood in mammalian species with hemochorial placentas, and they play multiple essential roles in successful pregnancy (1, 2). The first cells to differentiate from the growing blastocyst are primary TBCs, and they subsequently form the trophectoderm layer that encapsulates the developing embryo before implantation (2, 3). Trophectoderm attach to the uterine wall during implantation and grow invasively into the uterine tissue to establish the fetal component of the placenta. The trophoblast layer of the mouse placenta consists of several different subtypes of TBCs, each of which performs specific functions (3). The multipotent trophoblast stem (TS) cells arise from the primary and secondary trophoblast of the trophectoderm layer, and give rise to the various trophoblast subtypes (3, 4). Trophoblast giant cells, which are derived from TS cells (5–7), initiate implantation and invasion into the uterine wall (3). In addition, trophoblast giant cells invade the spiral arteries of the uterus to establish both a vascular connection between the mother and the fetus, and the outermost boundary of the fetal interface of the placenta (3). Spongiotrophoblast cells form the middle layer of the fetal component of the placenta, between the outermost giant cells and the innermost labyrinthine trophoblast layer (2, 3). Although the function of the spongiotrophoblast layer is unclear, it is thought to play a structural role, as well as produce soluble factors necessary for trophoblast function (3). Lastly, the innermost labyrinthine trophoblast layer is the site of nutrient exchange between the mother and the fetus (2, 3, 7).
In addition to regulating multiple processes necessary for normal fetal development, mouse TBCs provide a protective barrier around the semiallogeneic embryo that functions to prevent immune-mediated destruction by the maternal immune system (2, 8–10). This is in part achieved by the expression of several immunoregulatory molecules on the TBC surface. For example, mouse TBCs express complement receptor 1-related gene/protein y, which prevents deposition of the activated complement molecules C3 and C4 on the cell surface (11). Fas ligand is also expressed on the surface of mouse TBCs, and is thought to inhibit T cell-mediated inflammatory reactions (12). Furthermore, TBCs are only one of a few cell types that lack the capacity to express MHC class II Ags, either constitutively or in response to the potent MHC gene-inducing cytokine IFN-γ (13). In allogeneic mating combinations, the silencing of MHC class II gene expression in mouse TBCs is believed to be important in preventing immune-mediated rejection reactions against the semiallogeneic fetus by the maternal immune system (2, 9, 14).
The placenta contains a complex milieu of numerous hormones and cytokines that function cooperatively to ensure successful parturition. Interestingly, the proinflammatory cytokine IFN-γ is present in the pregnant uterus of several mammals, including mice, humans, and pigs (15–18). In mice, uterine IFN-γ is secreted by specialized uterine NK cells, primarily between gestation day (GD) 6 and GD16 (15, 17, 19). Elegant studies using mice deficient for IFN-γ, IFN-γRα, and NK cells demonstrated that IFN-γ is essential for pregnancy-induced remodeling of the uterine vasculature and proper formation of the decidual (maternal) layer of the placenta during murine pregnancy (19). Despite the presence of IFN-γ in the placenta, mouse TBCs appear to respond selectively to this cytokine. For instance, IFN-γ enhances phagocytosis by TBCs, and induces expression of classical MHC class I Ags on the TBC surface (8, 20, 21). In contrast, TBCs are resistant to IFN-γ-induced apoptosis and activation of MHC class II Ag expression (2, 13, 20, 22). Although mouse TBCs express IFN-γ receptors (20), the magnitude and the duration of IFN-γ responses are not well characterized in these cells.
IFN-γ signal transduction is mediated by the JAK/STAT pathway (23, 24). Binding of IFN-γ to its receptor, which consists of IFN-γR1 and IFN-γR2, leads to receptor oligomerization and activation of the receptor-associated JAK-1 and JAK-2 (23, 24). The activated JAKs phosphorylate a tyrosine residue within the intracellular domain of the IFN-γR1, which provides a docking site for monomers of STAT-1 sequestered in the cytoplasm (23, 24). STAT-1 interacts with the phosphorylated IFN-γR1 and is subsequently phosphorylated on tyrosine residue-701 by the JAKs (25). Tyrosine phosphorylation of STAT-1 leads to Src homology 2-mediated homodimerization and translocation to the nucleus (25). STAT-1 can also be phosphorylated on serine residue-727, which has been demonstrated to be necessary for optimal STAT-1 transcriptional activity through the recruitment of histone acetyltransferases (HATs) (26). Once inside the nucleus, phosphorylated dimers of STAT-1 (pSTAT-1) activate the transcription of multiple different genes that contain a IFN-γ-activating sequence (GAS) in their promoters, one of which is the gene encoding the transcription factor IFN regulatory factor-1 (IRF-1) (23, 27).
pSTAT-1 and IRF-1 are the two key transcription factors that propagate primary and secondary responses to IFN-γ, respectively, by activating transcription of genes involved in Ag processing and presentation, apoptosis, and cell cycle arrest (23). For example, IRF-1 up-regulates transcription of the MHC class I, TAP, and low molecular protein (LMP) genes involved in Ag presentation, as well as caspase genes that activate apoptosis (23). In addition, pSTAT-1 and IRF-1 cooperate with one another to activate transcription of a transcriptional cofactor termed the CIITA, as well as antiviral genes such as guanylate-binding protein (GBP) (28, 29). CIITA is the master regulator of both constitutive and IFN-γ-inducible MHC class II gene transcription (30, 31). The inability of TBCs to express MHC class II Ags, either constitutively or in response to IFN-γ, is due to the silencing of CIITA expression (32–34).
We recently demonstrated that IFN-γ-inducible gene expression is repressed in human TBCs by protein tyrosine phosphatases (PTPs) (35). Specifically, PTPs inhibit activation of the JAKs and subsequent phosphorylation of STAT-1. In this study, we demonstrate that repression of IFN-γ-inducible gene expression is conserved in mouse TBCs. However, in contrast to human trophoblast, no defects in STAT-1 tyrosine phosphorylation or DNA-binding ability were observed in mouse TBCs exposed to IFN-γ. Simultaneous treatment of mouse TBCs with IFN-γ and histone deacetylase (HDAC) inhibitors resulted in significant enhancements of IRF-1, GBP, and LMP expression, relative to treatment with IFN-γ alone. We propose that HDAC-mediated dampening of IFN-γ-inducible gene expression in mouse TBCs may be important for successful parturition.
Materials and Methods
Cell culture
M-11, SM9, and NIH-3T3 cells were cultured, as previously described (36). TS cells were a gift of T. Kunath (University of Edinburgh, Edinburgh, UK) and J. Rossant (University of Toronto, Toronto, Ontario, Canada), and were cultured as previously described by Tanaka et al. (4). TS cells were induced to differentiate into giant cells by culture in either the absence of fibroblast growth factor-4 (4), or the presence of 0.5 µM retinoic acid, as described by Yan et al. (5). Mouse IFN-γ was purchased from PBL and used at concentrations ranging from 100 to 500 U/ml. The histone deacetylase inhibitors trichostatin A (TSA; WAKO), sodium butyrate, and apicidin (Calbiochem) were reconstituted in 100% ethanol, H2O, and DMSO, respectively. M-11, SM9, and NIH-3T3 cells were exposed to various concentrations of these HDAC inhibitors for 24 h to generate a toxicity curve. Concentrations that resulted in ~80% cell viability after 24 h were subsequently used to assess the effects on IFN-γ-inducible gene expression. Thus, SM9 and M-11 cells were treated with 50 nM TSA, 1 mM sodium butyrate, and 300 nM apicidin, respectively. NIH-3T3 cells were treated with 500 nM TSA, 1 mM sodium butyrate, and 300 nM apicidin.
RNA isolation and RT-PCR
RNA isolation, reverse-transcriptase reactions, and semiquantitative RT-PCR were performed, as previously described (33, 36). Quantitative RT-PCR was performed in duplicate for each sample on 1/100 of the total cDNA using an iCycler (Bio-Rad) instrument and SYBR Green master mix (Bio-Rad), as previously described (35). Briefly, standard curves were generated using plasmids containing the respective cDNAs for each gene examined. Expression of IFN-γ-inducible genes was normalized to GAPDH gene expression, and data are represented as the ratio between values obtained for IFN-γ-inducible gene expression and GAPDH expression. Melt peak analyses were performed to ensure specificity of the reactions. Statistical analysis was performed using Student’s t test.
Nuclear run-on transcription assays
Nuclei were isolated and run-on transcription assays were performed, as previously described (37). Briefly, nuclei were purified using sucrose gradients by lysing the cells in hypotonic buffer (10 mM Tris (pH 7.4), 5 mM MgCl2, 0.4% Nonidet P-40, 0.5 mM dithiothreiotol, and 0.3 M sucrose), and pelleting the nuclei by centrifugation through a cushion of the same buffer containing 0.88 M sucrose. Run-on reactions were performed for 10 min at 26°C using 1–1.5 × 107 nuclei per sample, and 200–250 µCi of [32P]UTP per sample. Plasmids (5 µg) containing the cDNAs for IRF-1 and actin were immobilized on nitrocellulose filters and hybridized to 32P-labeled RNA for 3 days at 42°C, washed, and exposed to x-ray film (Kodak X-OMAT) with intensifying screens. The plasmid pSP65 was used as a negative control to measure background.
Western blot (WB) analysis
Whole-cell and nuclear extracts were prepared, as previously described (35). Abs to IRF-1 (sc-640), STAT-1 (sc-417), phosphotyrosine (701)-STAT-1 (sc-7988), phosphoserine (727)-STAT-1 (sc-16570-R), and upstream stimulatory factor-1 (USF-1; sc-229) were purchased from Santa Cruz Biotechnology. Abs to STAT-1 were also purchased from Millipore/Upstate Biotechnology. SDS-PAGE, protein transfer, and WB analyses were performed, as previously described (35), using the following Ab concentrations: IRF-1 (100 ng/ml); STAT-1 (333 ng/ml); USF-1 (500 ng/ml); and heat shock cognate 70 (HSC70; 1:20,000 of monoclonal 3a3 ascites (38)); phosphotyrosine (701)-STAT-1 (200 ng/ml) and phosphoserine (727)-STAT-1 (200 ng/ml). Signals were detected with the SuperSignal West Pico chemiluminescent substrate (Pierce) and subsequent exposure to Kodak Scientific Imaging film (Kodak). Relative levels of transcription factors were determined by dilution analysis of the cell extracts, followed by WB analysis. Signals were quantified using Molecular Dynamics Computing Densitometer model 300S and Image J software (39).
EMSAs
EMSAs were performed, as previously described by Briscoe et al. (40), with minor modifications. Briefly, whole-cell extracts (WCE) were prepared in lysis buffer containing 0.5% Nonidet P-40, 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 10% glycerol (v/v), 0.1 mM EDTA, 1 mM sodium orthovanadate, 1.0 mM DTT, 50 µg/ml pepstatin, 25 µg/ml aprotinin, 25 µg/ml leupeptin, and 0.5 mM PMSF. dsSTAT-1-binding oligonucleotide probes (murine IRF-1 (mIRF-1)-GAS, 5′-GATCGTGATTTCCCCGA AATGACG-3′ and human signal-inducible element-GAS, 5′-GTCGACA TTTCCCGTAAATC-3′) were end labeled with [γ-32P]ATP with the sp. act. of 3000 Ci/mmol (PerkinElmer). STAT-1-binding reactions were performed in a total volume of 20 µl, composed of binding buffer (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 0.1 mM EGTA, 5% glycerol, 1 mg/ml BSA, 0.25 mg/ml tRNA, and 2% Ficoll), 1 ng of oligonucleotide probes, and 10 µg of WCE per reaction. WCE were preincubated with binding buffer containing 150 µg/ml poly(d(I-C)) (Roche) for 15 min at room temperature before incubation with probe for an addition 15 min at room temperature. Competition assays were performed by preincubating the cell extracts with unlabeled GAS and mutant GAS competitor oligonucleotides (5′-GATCGTGATGGCTCCGAAATGACG-3′) before addition of radiolabeled oligonucleotides. STAT-1 supershift Abs (sc-417X; Santa Cruz Biotechnology) were added (4 µg/sample) following incubation with the radiolabeled oligonucleotides, and incubated for an additional 15 min at room temperature. DNA:protein complexes were resolved on 5% nondenaturing polyacrylamide gels in 0.5% TBE. The gels were dried and exposed to Kodak Scientific Imaging film (Kodak) as well as phosphor screens. The exposed phosphor screens were scanned with a Molecular Dynamics Storm 860 PhosphorImager and quantified using Image J software (39).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed essentially as previously described by Bulger et al. (41) using Abs from Upstate Biotechnology to acetylated histone 3 (H3; catalog 06-599), dimethylated H3-K4 (catalog 07-030), and trimethylated H3-K9 (catalog 07-442). Genomic DNA was purified using PCR purification kits (Invitrogen), and each input DNA sample was subjected to agarose gel electrophoresis to verify that the average size of the sonicated DNA was ~600 bp. Quantitative PCR was subsequently performed on genomic DNA, as described above for RT-PCR.
Results
IFN-γ-inducible gene expression is reduced in mouse TBCs relative to fibroblasts
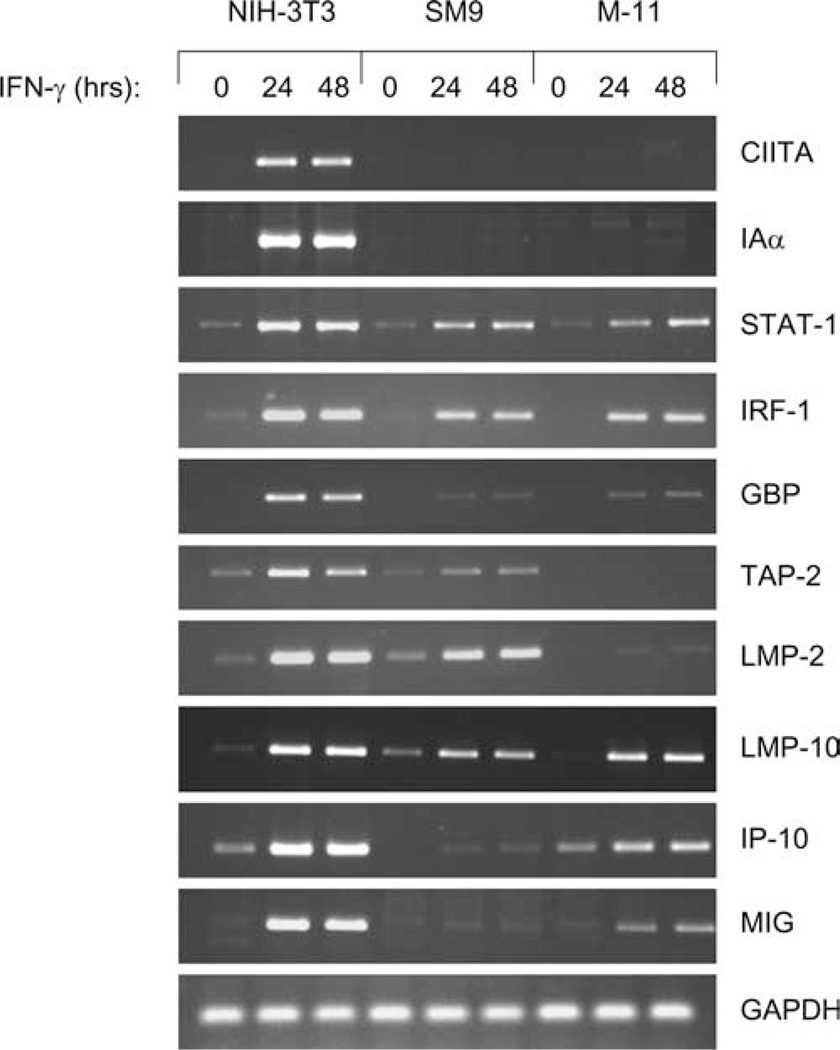
Previous studies demonstrated that mouse TBCs respond to IFN-γ by activating expression of molecules such as IRF-1 and MHC class I (21, 42, 43), but these studies did not assess whether there were quantitative differences in the levels of IFN-γ-inducible gene expression in these cells relative to fibroblasts or epithelial cells. To examine this possibility, SM9 and M-11 mouse TBCs and NIH-3T3 embryonic fibroblasts were exposed to 500 U/ml IFN-γ for 0, 24, and 48 h. RNA was isolated and subjected to semiquantitative RT-PCR for the CIITA; the MHC class II gene I-Aα; STAT-1; IRF-1; GBP; TAP-2; the immunoproteasome subunits LMP-2 and LMP-10; the chemokines IFN-γ-inducible protein-10 (IP-10) and monokine induced by IFN-γ (MIG); and the housekeeping gene GAPDH. As previously demonstrated, SM9 and M-11 cells do not express either CIITA or I-Aα mRNA following exposure to IFN-γ (Fig. 1). Higher basal levels of IRF-1 mRNA expression were detected in NIH-3T3 cells relative to SM9 and M-11 cells (Fig. 1). Several other cell types, including mouse primary embryonic fibroblasts, Colon 26 carcinoma, and 4T1 breast carcinoma cells, also exhibited higher basal expression of IRF-1 mRNA comparable to NIH-3T3 cells (data not shown). Importantly, following exposure of the cells to IFN-γ for 24 or 48 h, the expression of multiple genes (STAT-1, IRF-1, GBP, TAP-2, LMP-2, LMP-10, IP-10, and MIG) was substantially reduced in the mouse TBCs compared with NIH-3T3 fibroblasts, whereas GAPDH mRNA expression was not affected.
FIGURE 1.
IFN-γ-inducible gene expression is dampened in mouse TBCs. Total RNA was isolated from NIH-3T3 fibroblast cells and SM9 and M-11 TBCs exposed to 500 U/ml IFN-γ for 0, 24, and 48 h. They were subjected to semiquantitative RT-PCR with primers specific for CIITA, I-Aα, STAT-1, IRF-1, GBP, TAP-2, LMP-2, LMP-10, IP-10, MIG, and GAPDH. The data shown are representative of experiments using at least four independent preparations of RNA from each cell line.
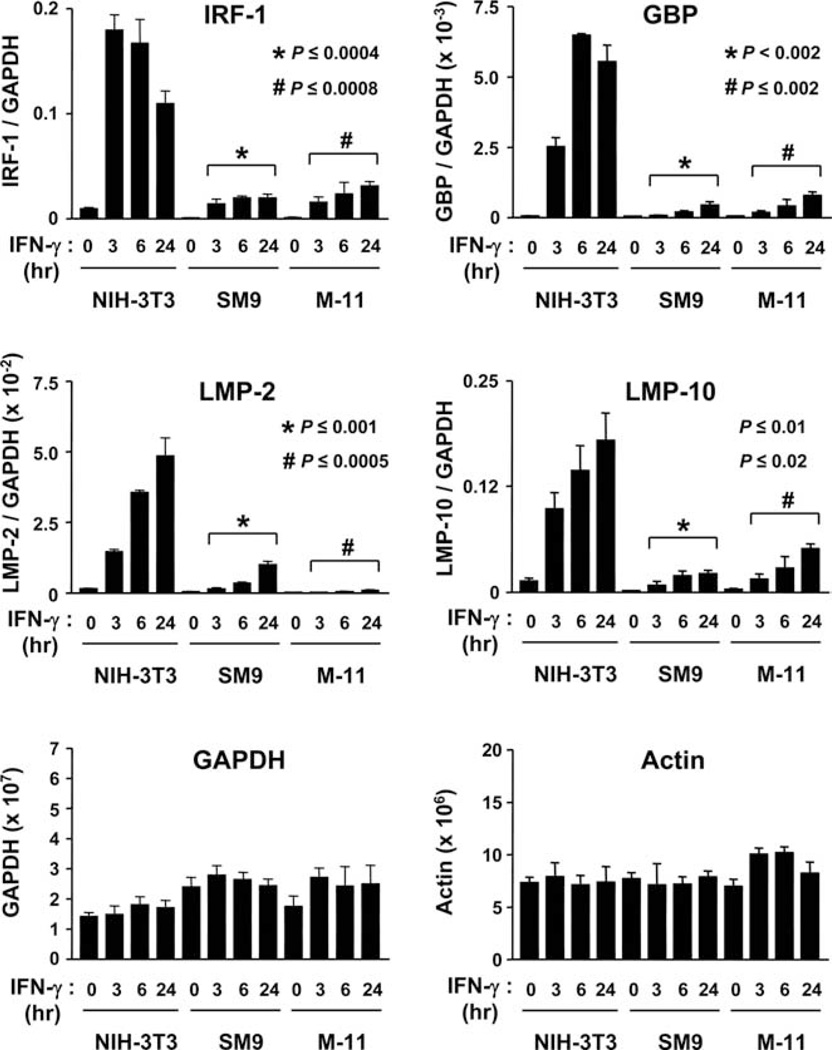
Although IFN-γ-inducible gene expression was clearly reduced at 24 and 48 h in mouse TBCs relative to fibroblasts, the kinetics of expression may differ in these cell types. To examine the kinetics and to quantify the relative differences in IFN-γ-inducible gene expression, quantitative RT-PCR was performed on RNA isolated from SM9, M-11, and NIH-3T3 cells exposed to 500 U/ml IFN-γ for 0, 3, 6, and 24 h on a subset of the genes analyzed in Fig. 1. Data are represented as the relative levels of IRF-1, GBP, LMP-2, and LMP-10 mRNA normalized to GAPDH mRNA expression. The levels of both basal and IFN-γ-inducible expression of IRF-1, GBP, LMP-2, and LMP-10 mRNA were reproducibly lower in SM9 and M-11 cells compared with NIH-3T3 cells (Fig. 2). In NIH-3T3 cells, IRF-1 mRNA expression peaked at 3–6 h post-IFN-γ treatment, followed by a ~40% reduction from peak expression by 24 h. In contrast, the highest levels of IRF-1 mRNA expression in SM9 and M-11 TBCs were detected after treatment with IFN-γ for 24 h (Fig. 2). However, the expression of IRF-1 mRNA in SM9 and M-11 cells was significantly lower at all time points examined (~13-fold reduction at 3 h, ~8-fold reduction at 6 h, and ~5-fold reduction at 24 h) compared with NIH-3T3 cells. The kinetics of GBP mRNA expression in NIH-3T3 cells were similar to IRF-1, reaching peak levels at 6 h, followed by a slight reduction at 24 h, whereas GBP levels reached a maximum at 24 h in SM9 and M-11 cells. Nevertheless, the peak expression of GBP mRNA in SM9 and M-11 cells was still ~11-fold lower relative to NIH-3T3 cells at the same time points. Likewise, significant reductions in LMP-2 and LMP-10 mRNA expression were observed in IFN-γ-treated SM9 and M-11 cells compared with NIH-3T3 cells at all time points tested (Fig. 2). Similar results were observed when NIH-3T3, SM9, and M-11 cells were exposed to 100 U/ml IFN-γ (data not shown). Taken together, our RT-PCR analyses demonstrate that the absolute levels of IFN-γ-inducible gene expression are significantly lower in SM9 and M-11 cells relative to NIH-3T3 cells, both before and after IFN-γ treatment.
FIGURE 2.
Kinetic analysis of IFN-γ-inducible gene expression in mouse TBCs. RNA was isolated from NIH-3T3, SM9, and M-11 cells exposed to 500 U/ml IFN-γ for 0, 3, 6, and 24 h and subjected to SYBR Green-based quantitative RT-PCR using primers specific for IRF-1, GBP, LMP-2, LMP-10, GAPDH, and actin. Relative copy number from 20 ng of RNA of each cell type was determined by generating a standard curve using known amounts of plasmids containing the respective cDNAs of the genes examined. The data are the average of four independent experiments and are represented as the ratio of the relative mRNA expression of each gene (i.e., IRF-1) vs GAPDH. Unpaired Student’s t test was used to compare the relative levels of IFN-γ-inducible gene expression in NIH-3T3 cells vs SM9 and M-11 cells that were exposed to IFN-γ for the same duration.
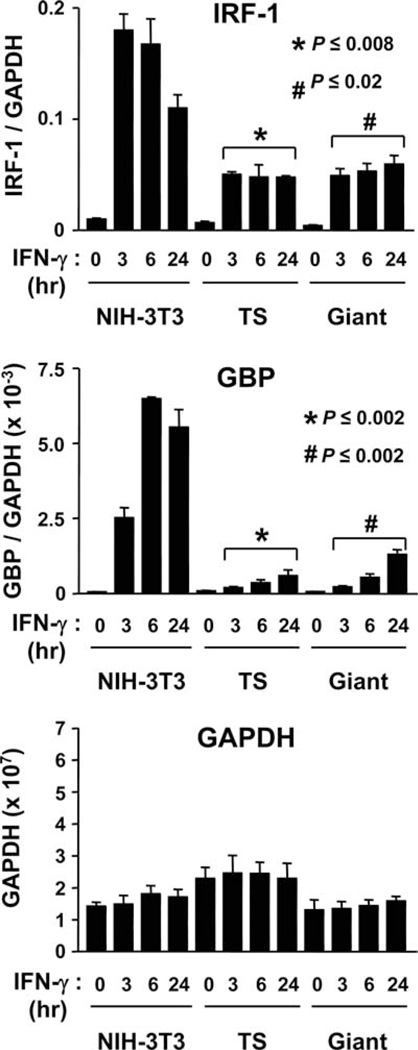
The phenotype of SM9 and M-11 cells is similar to labyrinthine trophoblast (44). To determine whether the dampening of IFN-γ-inducible gene expression observed in SM9 and M-11 cells is restricted to these cell lines, or is a feature shared by other mouse trophoblast subtypes, similar quantitative analyses were performed on TS and TS-derived trophoblast giant cells. Giant cells were derived by either incubating TS cells in the absence of fibroblast growth factor-4, or the presence of retinoic acid, as previously described (5, 6). TS cells and TS-derived giant cells were treated with IFN-γ for 0, 3, 6, and 24 h, and harvested for isolation of RNA. Quantitative RT-PCR analysis was subsequently performed for IRF-1 and GBP expression (Fig. 3). Basal expression of IRF-1 mRNA was comparable in NIH-3T3 and TS cells, but ~3.2-fold lower in trophoblast giant cells (Fig. 3). Moreover, although IRF-1 mRNA was clearly up-regulated in both TS and giant cells following exposure to IFN-γ, the absolute levels were reduced ~3-fold at all time points compared with NIH-3T3 fibroblasts. Basal expression of GBP mRNA was comparable among NIH-3T3 cells, TS, and giant cells (Fig. 3), but following exposure to IFN-γ, GBP mRNA levels were significantly lower in TS (15.7-fold reduction) and giant cells (10.6-fold reduction) compared with NIH-3T3 cells (Fig. 3). These collective results indicate that dampening of IFN-γ-inducible gene expression is conserved among multiple different trophoblast subtypes.
FIGURE 3.
Kinetic analysis of IFN-γ-inducible gene expression in mouse TS cells and giant cells. RNA was isolated from NIH-3T3, TS cells, and giant cells exposed to 500 U/ml IFN-γ for 0, 3, 6, and 24 h and subjected to SYBR Green-based quantitative RT-PCR using primers specific for IRF-1, GBP, and GAPDH, as described in Fig. 2. The data are the average of three independent experiments and are represented as the ratio of the relative mRNA expression of each gene (i.e., IRF-1) vs GAPDH. Unpaired Student’s t test was used to compare the relative levels of IFN-γ-inducible gene expression in NIH-3T3 cells vs TS and giant TBCs that were exposed to IFN-γ for the same duration.
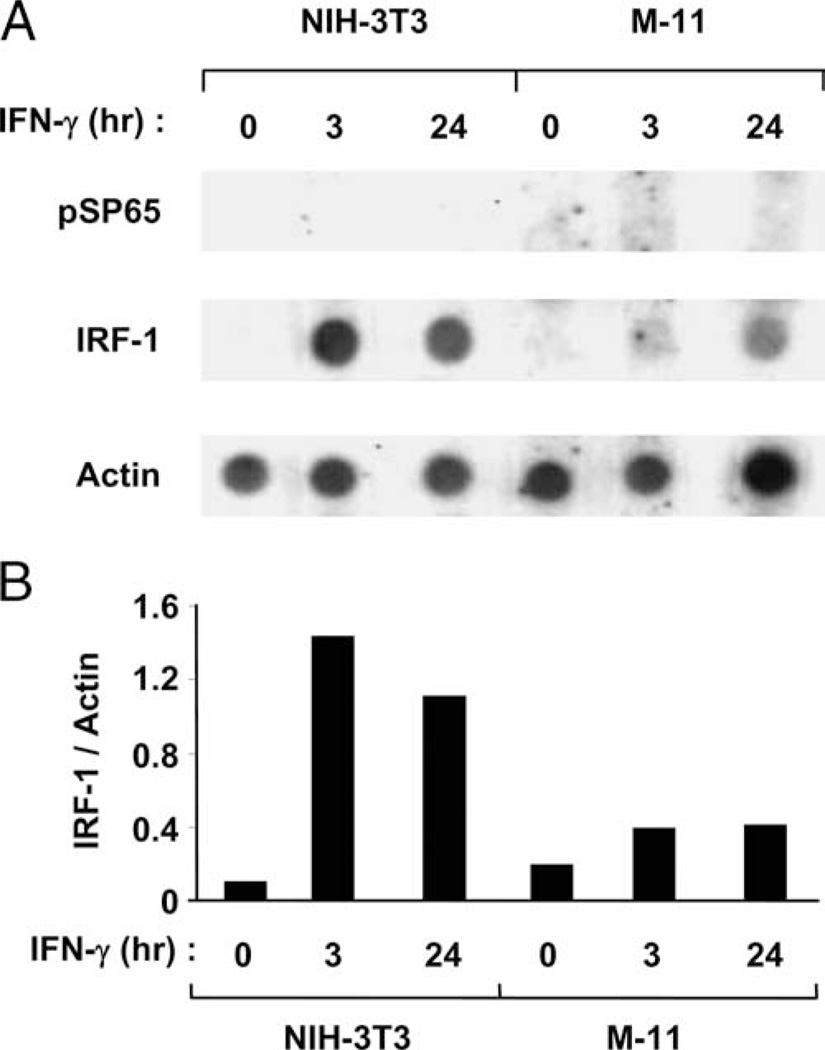
The rate of IRF-1 transcription is decreased in IFN-γ-treated M-11 cells relative to NIH-3T3 fibroblasts
The reduced expression of IFN-γ-inducible genes in mouse TBCs relative to fibroblasts can be explained by at least two possible mechanisms: decreased rates of gene transcription or reduced mRNA stability. To distinguish between these two possible mechanisms, run-on transcription assays were performed to determine the rate of IRF-1 gene transcription in M-11 and NIH-3T3 cells cultured for 0, 3, and 24 h in 500 U/ml IFN-γ (Fig. 4). The basal rate of IRF-1 transcription was not detectable above the background (compare with the signal for the control plasmid pSP65) in either cell type in these assays (Fig. 4A). Following exposure of NIH-3T3 cells to IFN-γ, the rate of IRF-1 transcription increased 14.3- and 11.0-fold at 3 and 24 h, respectively, over untreated samples (Fig. 4). However, in M-11 cells, only a 2-fold increase in the rate of IRF-1 mRNA synthesis was detected over untreated controls at both time points tested (Fig. 4). The changes in IRF-1 transcription are specific, because the rate of actin gene transcription was similar in both cell types at all three time points (Fig. 4). Thus, the IRF-1 transcription rate was reduced 3.7- and 2.7-fold at 3 and 24 h post-IFN-γ treatment in M-11 cells vs NIH-3T3 cells (Fig. 4B). These results suggest that a lower rate of transcription contributes to the dampened expression of IRF-1 mRNA in mouse TBCs exposed to IFN-γ.
FIGURE 4.
Examination of IRF-1 transcriptional rate in NIH-3T3 and M-11 cells. A, Run-on transcription assays were performed using nuclei isolated from NIH-3T3 fibroblast and M-11 TBCs exposed to 500 U/ml IFN-γ for 0, 3, and 24 h. Plasmids containing the IRF-1 and γ-actin cDNAs were used as probes for the radiolabeled RNA. The plasmid pSP65 was used as a negative control to measure background. Shown in this figure are representative data from three independent preparations of nuclei. B, The results from IRF-1 run-on transcription assay were enumerated by densitometry measurements and analysis by Image J software. The data are represented as a ratio between values obtained for IRF-1 and γ-actin.
STAT-1 phosphorylation (Tyr701/Ser727) and DNA binding are comparable among SM9, M-11, and NIH-3T3 cells
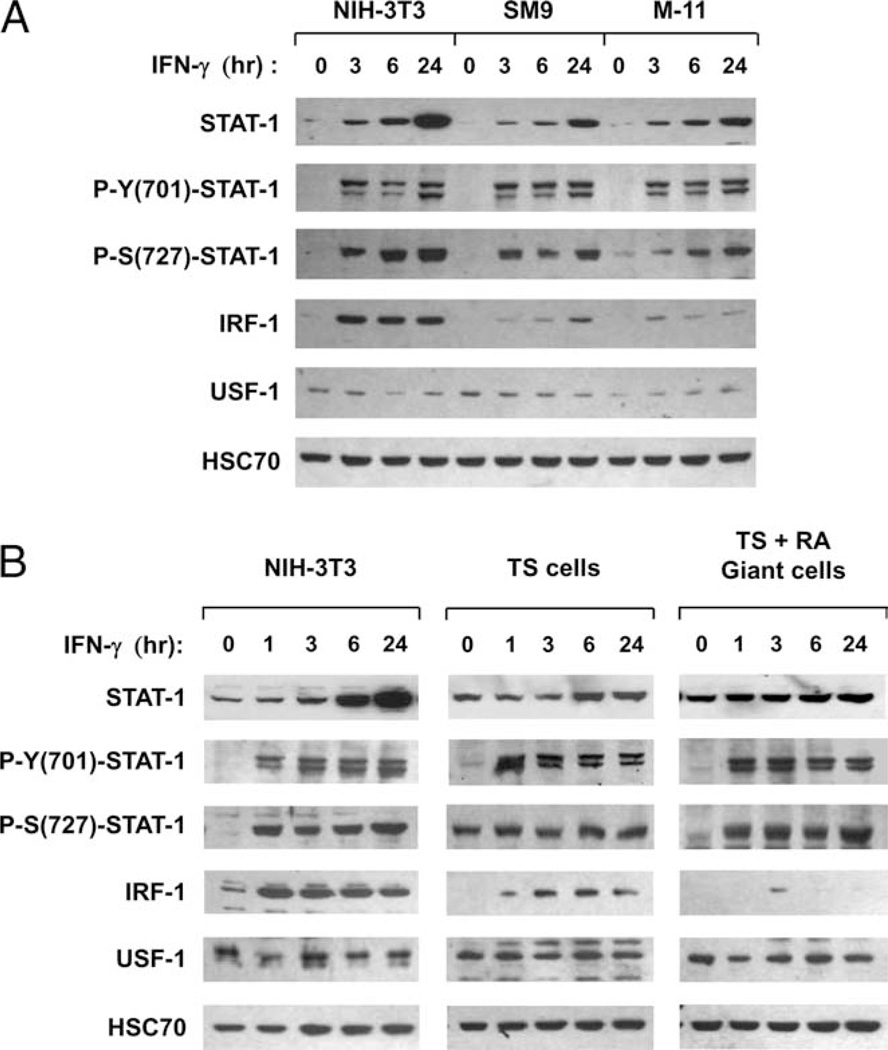
Optimal IRF-1 gene transcription in response to IFN-γ requires phosphorylation of STAT-1 at tyrosine residue-701 and serine residue-727 (23, 24). To determine whether STAT-1 is phosphorylated in mouse TBCs, WCE were isolated from NIH-3T3, SM9, and M-11 cells exposed to IFN-γ for 0, 3, 6, and 24 h, and subjected to WB analyses using Abs specific to STAT-1, phosphotyrosine (701)-STAT-1, phosphoserine (727)-STAT-1, and IRF-1 (Fig. 5A). Consistent with the results of the RT-PCR analysis, basal STAT-1 protein expression was comparable in the three cell lines, but reduced in SM9 and M-11 cells relative to NIH-3T3 cells following exposure to IFN-γ (Fig. 5A). Similar to previous studies using higher concentrations of IFN-γ (40, 45), robust STAT-1 phosphorylation at both tyrosine and serine residues was observed in NIH-3T3 cells within 3 h of IFN-γ treatment, and was sustained up to 24 h. High levels of IRF-1 protein expression were also observed in IFN-γ-treated NIH-3T3 cells at all time points examined, which is consistent with the IRF-1 mRNA expression (Fig. 5A). Interestingly, both the magnitude and duration of STAT-1 phosphorylation were comparable in IFN-γ-treated SM9 and M-11 cells to that observed in NIH-3T3 cells (Fig. 5A). However, despite robust and sustained STAT-1 phosphorylation, expression of IRF-1 protein was reduced ~5-fold in SM9 and M-11 cells relative to NIH-3T3 cells (Fig. 5A), which is consistent with the quantitative RT-PCR (qRT-PCR) analysis of IRF-1 mRNA expression. The levels of USF-1 and HSC70 were comparable at all time points in SM9, M-11, and NIH-3T3 cells, demonstrating that the differences in STAT-1 and IRF-1 protein expression were specific (Fig. 5A). Identical results were observed using nuclear extracts (data not shown).
FIGURE 5.
Examination of IFN-γ-induced transcription factors pSTAT-1 and IRF-1 expression in mouse TBCs. A, WCE were isolated from NIH-3T3, SM9, and M-11 cells exposed to 500 U/ml IFN-γ for 0, 3, 6, and 24 h and subjected to SDS-PAGE and WB analyses, using Abs specific for STAT-1p91, phosphotyrosine (701)-STAT-1, phosphoserine (727)-STAT-1, and IRF-1. The blots were stripped and reprobed with Abs for USF-1 and HSC70 as a control for loading and the integrity of the protein extracts. WB analysis was performed on at least four independent preparations of WCE from each cell type, and the figure is representative of the data from all experiments. B, WCE from TS cells and trophoblast giant cells exposed to 500 U/ml IFN-γ for 0, 1, 3, 6, and 24 h were subjected to WB analyses, as described in A. Representative data from three independent preparations of WCE are shown in this figure.
WB analyses were also performed to examine STAT-1 phosphorylation in TS cells and TS-derived giant cells treated with IFN-γ for 0, 1, 3, 6, and 24 h. The magnitude and the duration of STAT-1 tyrosine phosphorylation in TS and giant cells exposed to IFN-γ were also comparable to NIH-3T3 cells (Fig. 5B). However, whereas STAT-1 serine phosphorylation was induced by IFN-γ in giant cells, it was reproducibly detected in both untreated and IFN-γ-treated TS cells (Fig. 5B). In addition, minor differences in IRF-1 expression were also observed between TS and giant cells. Despite the presence of pSTAT-1, IRF-1 protein expression was almost undetectable in IFN-γ-treated giant cells, with low amounts being detected only at 3 h. Higher amounts of IRF-1 protein were detected in TS cells compared with giant cells at all time points following IFN-γ treatment. The minor differences in IRF-1 protein expression between TS and giant cells were not reflected at the mRNA level (Fig. 3), suggesting that posttranscriptional mechanism(s) may mediate these differences. Nonetheless, the levels of IRF-1 protein expression were still substantially lower in IFN-γ-treated TS cells compared with NIH-3T3 cells (Fig. 5B). Importantly, robust, sustained STAT-1 phosphorylation and IRF-1 protein expression were also observed in primary mouse embryonic fibroblasts exposed to IFN-γ, in a pattern identical with NIH-3T3 cells, indicating that these results portray an accurate reflection of the fibroblast phenotype (data not shown).
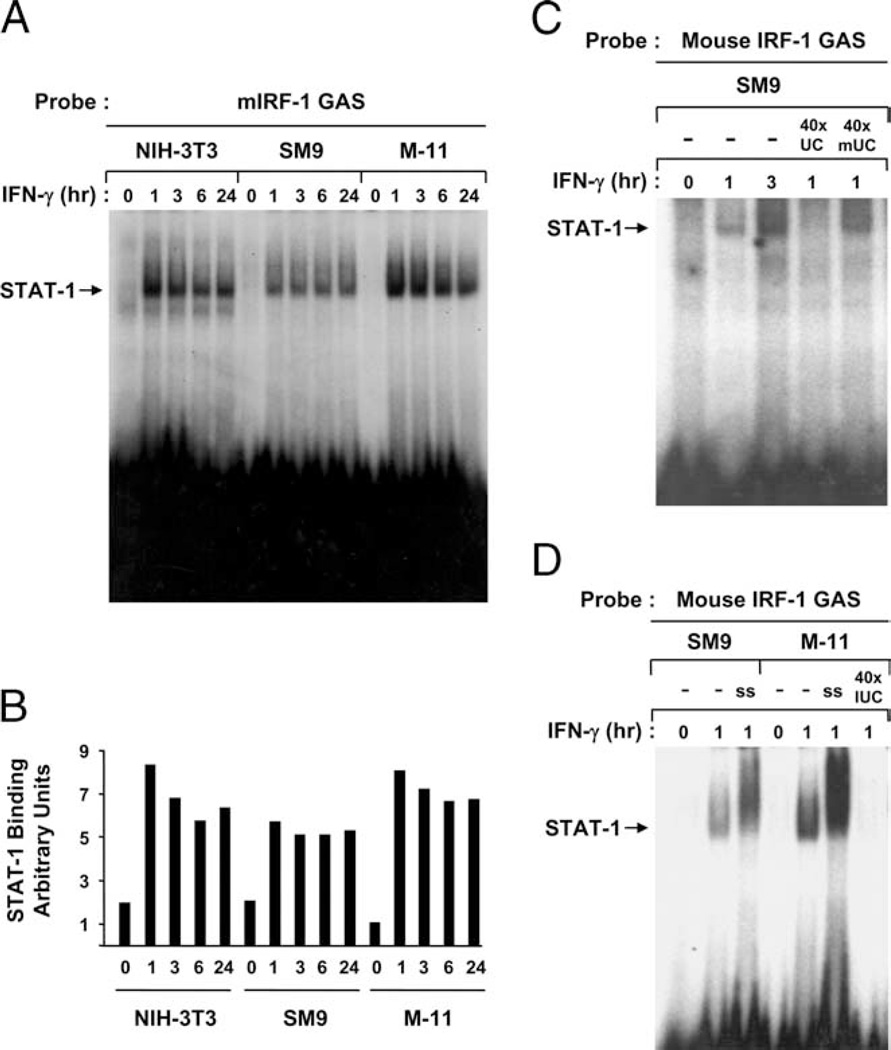
To determine whether phosphorylated STAT-1 from IFN-γ-treated mouse TBCs can bind DNA, the WCE used in WB assays were subjected to EMSA using 32P-labeled DNA probes corresponding to the GAS within the mIRF-1 promoter. STAT-1-binding activity was not detected in any of the cells under basal conditions (Fig. 6). However, consistent with the kinetics of STAT-1 phosphorylation, STAT-1 binding to GAS was detectable within 1 h of IFN-γ treatment in NIH-3T3, SM9, and M-11 cells, and the levels of binding activity were sustained through 24 h (Fig. 6, A and B). Excess unlabeled mIRF-1 GAS, but not mutated mIRF-1 GAS, competed for pSTAT-1 binding (Fig. 6C), demonstrating the specificity of factor binding. Preincubation of the WCE with Abs to STAT-1 supershifted the DNA: protein complex, confirming that the GAS-binding factor from IFN-γ-treated SM9 and M-11 cells is STAT-1 (Fig. 6D). Identical results were obtained using oligonucleotides corresponding to the STAT-1-binding sequence from the human signal-inducible element (data not shown). Taken together, these results indicate that the reduced levels of IFN-γ-inducible gene expression in TBCs relative to fibroblasts are not due to impaired STAT-1 phosphorylation or DNA-binding ability.
FIGURE 6.
Examination of STAT-1 DNA-binding activity in mouse TBCs exposed to IFN-γ. A, WCE were prepared from NIH-3T3, SM9, and M-11 cells incubated with IFN-γ for 0, 1, 3, 6, and 24 h, and subjected to EMSA using radiolabeled oligonucleotides corresponding to GAS from the mouse IRF-1 promoter. The gels were exposed to phosphor screens and scanned with a Storm scanner to generate the figure shown, which is representative of three independent experiments. B, The phosphor screen was scanned, and the intensity of the bands was quantified by Image J software. C, Competition experiments were performed using 40× excess of unlabeled GAS double-stranded oligonucleotides (40× UC), or unlabeled mutated GAS oligonucleotides (40× mUC) that are incapable of being bound by STAT-1. They were incubated with SM9 cell extracts before radiolabeled GAS oligonucleotides. D, A total of 4 µg of STAT-1 supershift Abs (ss) was added immediately after WCE isolated from SM9 and M-11 cells were incubated with the radiolabeled GAS oligonucleotides.
Cotreatment of mouse TBCs with IFN-γ and HDAC inhibitors enhances the expression of IFN-γ-inducible genes
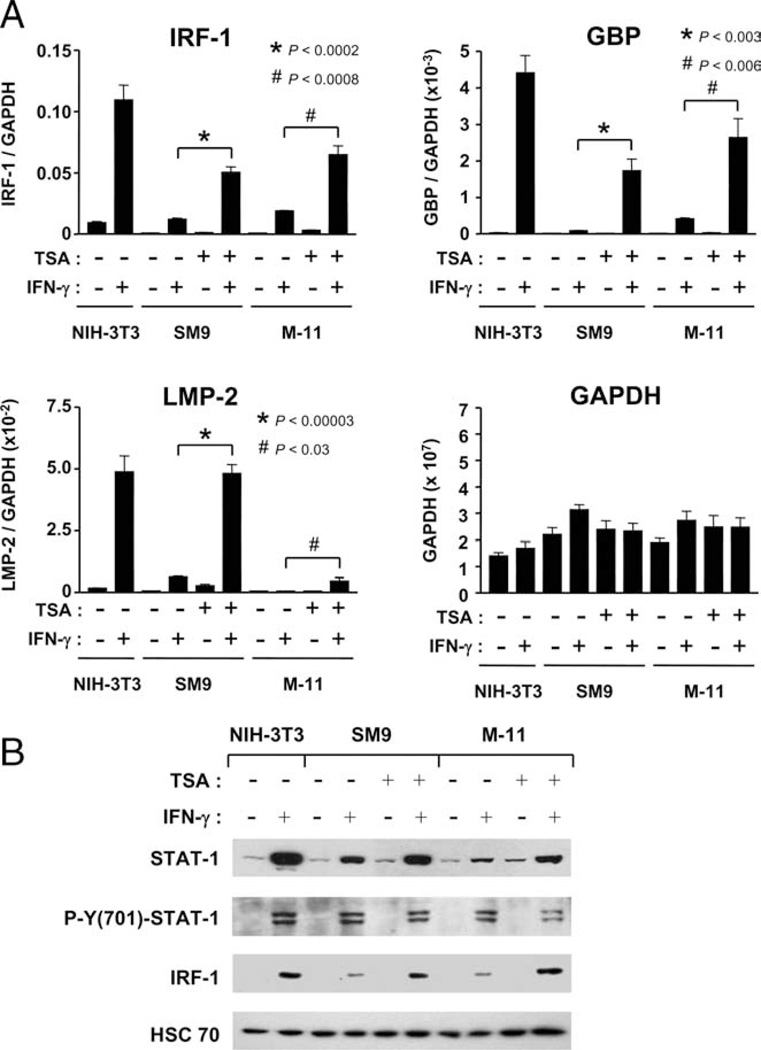
We previously reported that simultaneous treatment of SM9 and M-11 cells with IFN-γ and the HDAC inhibitor TSA alleviated silencing of CIITA transcription (36), but that study did not assess whether the effects of TSA are specific for CIITA, or whether it has a broader impact on IFN-γ-inducible gene expression. Therefore, qRT-PCR was performed to examine IRF-1, LMP-2, and GBP mRNA expression in SM9 and M-11 cells treated with IFN-γ alone, TSA alone, or in combination for 24 h (Fig. 7A). With IFN-γ treatment alone, 97- and 191-fold inductions of IRF-1 mRNA levels were observed in SM9 and M-11 cells, respectively, over untreated samples (Fig. 7A). Enhancements in IRF-1 mRNA levels were also detected in SM9 (5-fold) and M-11 cells 24-fold) treated with TSA alone relative to the untreated samples. However, simultaneous treatment of SM9 and M-11 cells with both agents enhanced IRF-1 mRNA expression 422- and 668-fold, respectively, over the untreated samples, indicating that the effects of these agents were synergistic rather than additive (Fig. 7A). Moreover, the IFN-γ/TSA combination treatment increased the expression of IRF-1 mRNA in TBCs 4.3- and 3.5-fold, respectively, over IFN-γ treatment alone. Similar synergistic enhancements in the levels of GBP and LMP-2 mRNA expression were also observed in SM9 and M-11 cells treated with IFN-γ and TSA compared with the untreated controls (Fig. 7A). Furthermore, significant enhancements of GBP (24.6-fold in SM9; 6.6-fold in M-11) and LMP-2 (8.2-fold in SM9; 65.8-fold in M-11) mRNA expression were also observed in mouse TBCs exposed to IFN-γ and TSA vs IFN-γ alone.
FIGURE 7.
Effects of IFN-γ and TSA on pSTAT-1/IRF-1 protein expression and IFN-γ-inducible gene transcription in mouse TBCs. A, RNA was isolated from SM9 and M-11 cells exposed to 500 U/ml IFN-γ, 50 nM TSA, or the combination, for 24 h and subjected to SYBR Green-based quantitative RT-PCR using primers specific for IRF-1, GBP, LMP-2, and GAPDH, as described for Fig. 2. The data are the average of four independent experiments and are represented as the ratio of the relative mRNA expression of each gene (i.e., IRF-1) vs GAPDH. Unpaired Student’s t test was used to compare the levels of mRNA expression in SM9 and M-11 cells treated with the combination of IFN-γ/TSA vs IFN-γ alone. B, WCE were prepared from NIH-3T3, SM9, and M-11 cells that were treated as described in A, and subjected to WB analyses, using Abs specific for STAT-1, phosphotyrosine (701)-STAT-1, and IRF-1. The blots were stripped and reprobed with Abs for HSC70 as a control for loading and the integrity of the protein extracts. Representative data from three independent preparations of WCE are shown.
To determine whether the enhanced expression of IRF-1 mRNA in SM9 and M-11 cells cotreated with IFN-γ and TSA led to a corresponding increase in protein expression, WB analysis was performed on WCE from SM9 and M-11 cells treated with IFN-γ, TSA, or the combination for 24 h (Fig. 7B). As previously observed, STAT-1 phosphorylation was clearly detectable in SM9 and M-11 cells treated with IFN-γ for 24 h, whereas IRF-1 protein expression was reduced relative to NIH-3T3 cells (Fig. 7B). TSA treatment alone or in combination with IFN-γ had no detectable effect on STAT-1 phosphorylation in SM9 and M-11 cells (Fig. 7B). Although an increase in IRF-1 mRNA expression was detected in SM9 and M-11 cells treated with TSA by qRT-PCR, a corresponding increase was not detected at the protein level. However, simultaneous treatment of SM9 and M-11 cells with IFN-γ and TSA resulted in a 3.8-fold enhancement of IRF-1 protein expression compared with IFN-γ treatment alone (Fig. 7A). Similar results were obtained in experiments using two other chemically distinct HDAC inhibitors, sodium butyrate and apicidin (data not shown). In contrast, simultaneous treatment of NIH-3T3 cells with IFN-γ and TSA, using concentrations ranging from 50 to 250 nM, resulted in little change in IFN-γ-inducible gene expression, whereas co-treatment with 500 nM-1 mM TSA led to minor decreases (data not shown). Taken together, our results strongly suggest that HDACs dampen the expression of IFN-γ-inducible genes at the level of transcription in mouse TBCs.
promoters in IFN-γ-treated TBCs
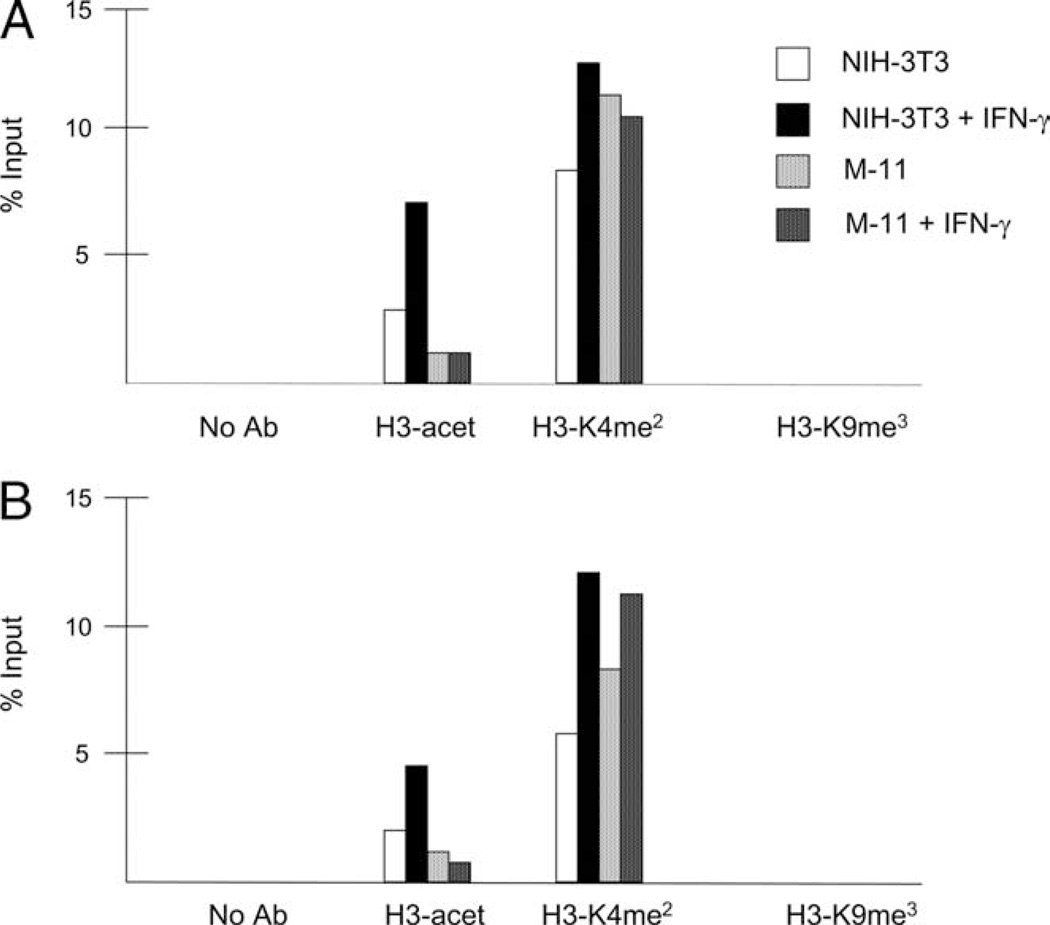
The demonstration that treatment of TBCs with HDAC inhibitors significantly enhances IFN-γ-inducible IRF-1 and CIITA expression suggests that histone acetylation at these promoters is deficient in these cells. To test this possibility, M-11 and NIH-3T3 cells were cultured in 500 U/ml IFN-γ for 0 and 3 h and subjected to ChIP assays using Abs to acetylated H3. The following histone modifications were also examined: dimethylated H3-K4, which correlates with permissive chromatin (46, 47), and methylated H3-K9, which is associated with repressive chromatin (46, 47). Data are represented as the percentage of input chromatin. Basal levels of H3 acetylation were detected at both the IRF-1 promoter and CIITA promoter IV (pIV) in NIH-3T3 and M-11 cells (Fig. 8). Stimulation with IFN-γ resulted in significantly increased levels of H3 acetylation at the IRF-1 and CIITA promoters in NIH-3T3 cells (Fig. 8). However, the levels of H3 acetylation did not appreciably change at these promoters in IFN-γ-treated M-11 cells (Fig. 8). Interestingly, dimethylated H3-K4 (H3-K4me2) was observed at the IRF-1 and CIITA promoters at similar levels in untreated NIH-3T3 and M-11 cells (Fig. 8). IFN-γ did not appreciably alter the levels of dimethylated H3-K4 at the IRF-1 promoter, but this histone mark increased at CIITA pIV in NIH-3T3 and M-11 cells exposed to IFN-γ for 3 h. Trimethylated H3-K9 (H3-K9me3) was not detected above background levels at the IRF-1 or CIITA promoters in either NIH-3T3 or M-11 cells, irrespective of whether they were cultured with or without IFN-γ (Fig. 8). Taken together, these results suggest that the ability to acetylate histones at IFN-γ-inducible promoters is compromised in mouse TBCs. Moreover, they suggest that silencing of transcription from CIITA pIV in mouse TBCs does not correlate with trimethylated H3-K9 at this locus.
FIGURE 8.
Histone modifications at the IRF-1 and CIITA promoters in mouse fibroblasts and TBCs. NIH-3T3 and M-11 cells were cultured for 0 and 3 h with 500 U/ml IFN-γ, and subjected to ChIP assays using Abs specific for acetylated H3, acetylated H3-lysine 9 (K9), dimethylated H3-K4 (H3-K4me2), and trimethylated H3-K9 (H3-K9me3). Quantitative PCR was performed in duplicate for each sample using primers specific for the IRF-1 promoter (A) and CIITA pIV (B). Results for each histone modification are represented as the percentage of input, and are the average of at least two independent experiments.
Discussion
We previously demonstrated that IFN-γ signal transduction is dampened in human TBCs by PTPs (35). Our current studies demonstrate that inhibition of IFN-γ-inducible gene expression is conserved between human and mouse TBCs, but the mechanisms are distinct. Specifically, PTPs inhibit JAK and STAT-1 activation in human TBCs (35), whereas HDACs repress IFN-γ-inducible transcription in mouse TBCs. The necessity of using different mechanisms for repression between human and mouse TBCs is currently unclear. However, there are also key differences between human and mouse pregnancies, such as the duration of the gestation period and the degree of trophoblast invasion into the uterine wall (2). Perhaps these differences play a role in the differential mechanisms used by human and mouse TBCs in repressing responses to IFN-γ.
IRF-1 plays a central role in IFN-γ-induced up-regulation of GBP, LMP-2, and LMP-10 transcription (48–50). Therefore, the reduced expression of these genes in IFN-γ-treated mouse TBCs relative to fibroblast cells can be explained, at least in part, by the reduced levels of IRF-1 protein. Because transcription of LMP-2 in the absence of IFN-γ is mediated by monomers of STAT-1 complexed with IRF-1 (51), the lower basal level of IRF-1 in mouse TBCs compared with NIH-3T3 cells may also directly contribute to the reduced basal expression of LMP-2. Furthermore, basal transcription of LMP-10 is mediated by another member of the IRF protein family, IRF-2 (49), the transcription of which is also regulated by IRF-1 (52, 53). Therefore, the lower basal level of IRF-1 in mouse TBCs may indirectly affect the baseline expression of LMP-10 by regulating IRF-2 levels.
We previously reported that IFN-γ-inducible CIITA transcription is silenced in mouse TBCs (33, 36), and our current studies extend these observations to demonstrate that transcription of multiple IFN-γ-responsive genes is inhibited in these cells. The current study demonstrates that this phenomenon is not due to defects in STAT-1 activation or DNA-binding ability. Moreover, the histone mark trimethylated H3-K9, which is associated with a repressive chromatin structure, was not detected at the IRF-1 or CIITA promoters in M-11 TBCs. However, H3 acetylation was deficient at these promoters in mouse TBCs exposed to IFN-γ. Furthermore, HDAC inhibitors significantly increased IFN-γ-inducible gene expression in mouse TBCs. STAT-1 has been shown to recruit HATs to target promoters, but recent studies indicate that STAT-1 also physically interacts with class I HDACs in vitro (54). Based on these collective results, inefficient recruitment of STAT-1 to target promoters may be responsible for the reduced rate of IFN-γ-inducible gene transcription in TBCs. Alternatively, STAT-1 may be targeted successfully to the IRF-1 and CIITA promoters, but the trans activation capacity of phosphorylated STAT-1 may be compromised by interactions with class I HDACs that block recruitment of HATs by STAT-1, and therefore prevent histone acetylation and remodeling of the promoter chromatin structure. This hypothesis is consistent with the effects of HDAC inhibitor (HDACi), the reduced levels of histone acetylation at the IRF-1 and CIITA promoters in TBCs, and our observation that the levels of the HAT CREB binding protein are reduced in SM9 and M-11 cells compared with NIH-3T3 cells (J. Choi and S. Murphy, unpublished data). HDACs may also inhibit acetylation of transacting factors necessary for STAT-1-mediated transcriptional activation.
Our studies demonstrating that several chemically distinct HDACi enhance IFN-γ-inducible gene expression in mouse TBCs directly contrast with previous reports showing that class I HDACs are required for transcriptional activation of both type I and type II IFN-responsive genes in several different human tumor cell lines (54–57). Studies by Nusinzon and Horvath (54) and Chang et al. (55) demonstrated that class I HDACs contribute to activation of IFN-γ-inducible gene transcription in human tumor cells, and suggest that the HDACs mediate their functions at a step downstream of STAT-1 activation and in vivo binding to target promoters. In contrast, Klampfer et al. (57) reported that HDACi repress IFN-γ signaling by blocking activation of the JAKs, and therefore STAT-1 phosphorylation and activation. Although these studies conflict regarding the underlying molecular mechanisms responsible, they collectively suggest that HDACs are required for transcriptional activation of IFN-γ-responsive genes in human tumor cells. However, one key difference between our current studies and the previous reports is that the latter analyzed transformed human cells of nontrophoblastic origin. Thus, there may be fundamental differences between human tumor cells and mouse TBCs in the role that HDACs play in regulating IFN-γ-inducible gene transcription. We found that although cotreatment of NIH-3T3 embryonic fibroblasts with IFN-γ and 50–250 nM TSA had little to no effect on IRF-1 gene expression relative to IFN-γ treatment alone, higher TSA doses (500 nM-1 mM) led to modest reductions in expression. Taken together, these results suggest that the enhancement of IFN-γ-inducible gene expression by HDACi may be relatively specific to mouse TBCs. One potential explanation for the differential effects of HDACi on IFN-γ responses in mouse TBCs may be the expression of novel tissue-specific HDAC(s), or accessory factors that mediate HDAC function. Precedence for this proposal comes from studies showing that respiratory syncytial virus infection induces expression of Bcl-3, which subsequently interacts simultaneously with HDAC-1 and STAT-1, thereby inhibiting STAT-1 activity (58).
Modulation of IFN-γ and IFN-αβ signaling through disruption of the JAK/STAT-1 pathway is used by a multitude of pathogens to evade host immune responses, by a wide variety of strategies that include the following: 1) degrading the IFN-γRs or the JAKs, 2) blocking JAK activation, 3) inhibiting STAT-1 function by several distinct mechanisms, and 4) repressing the function of IRF-1 (59–62). Mutations or deletions of the viral genes responsible for modulating the functions of STAT-1 or IRF-1 result in attenuation of viral infection, which directly demonstrates the importance of these transcription factors in mediating antiviral immune responses (59). Importantly, IFN-γ also plays an essential role in the immunosurveillance of tumors; therefore, disruption of the JAK/STAT pathway in tumors provides an important mechanism of tumor immunoevasion (63). Thus, inhibiting the JAK-STAT pathway to prevent optimal IFN-γ-responsive gene expression is a common feature of pathogens, certain tumor cells, and TBCs.
IFN-γ is expressed at detectable levels in the pregnant mouse uterus from GD6 to GD18 (15, 19), but mouse TBCs are resistant to IFN-γ-mediated activation of apoptosis (20), and MHC class II Ags, the latter due to silencing of CIITA expression (33, 36). Our collective studies demonstrate that dampening of IFN-γ-inducible gene expression is conserved in human and mouse TBCs, although the molecular mechanisms mediating these effects are distinct. Based on these collective results, we propose that this phenomenon plays a critical role for successful pregnancy by preventing deleterious IFN-γ responses that would otherwise interfere with placental development and function. Inhibition of IRF-1 expression in TBCs exposed to IFN-γ may have an especially important role in pregnancy, for IRF-1 activates transcription of genes involved in cell cycle arrest, apoptosis, and Ag presentation (23, 24, 27). Aberrant activation of the JAK/STAT pathway in TBCs could therefore result in disruption of the trophoblast layer, and placental pathology.
Acknowledgments
We thank Drs. Janet Rossant and Tilo Kunath for providing us with TS cells, and Paul Soloway for primary mouse fibroblasts. We also thank Dr. Michael Bulger for help in performing ChIP assays. We are grateful to Drs. Edith Lord, Richard K. Miller and Chris Stodgell, and Kelly Cycon and Elizabeth Sorenson for critical reading of the manuscript.
Footnotes
This work was supported by grants from the National Institutes of Health (R01 HD37464) and the Roswell Park Cancer Institute Alliance, and Roswell Park Cancer Center Support Grant P30 CA 16056. J.C.C. was supported by National Cancer Institute Predoctoral Training Grant 55640201.
Abbreviations used in this paper: TBC, trophoblast cell; ChIP, chromatin immunoprecipitation; GAS, IFN-γ-activating sequence; GBP, guanylate-binding protein; GD, gestation day; H3, histone 3; HAT, histone acetyltransferase; HDAC, histone deacetylase; HSC70, heat shock cognate 70; IP-10, IFN-γ-inducible protein-10; IRF-1, IFN regulatory factor-1; LMP, low molecular protein; MIG, monokine induced by IFN-γ; mIRF-1, murine IRF-1; pSTAT-1, phosphorylated dimers of STAT-1; PTP, protein tyrosine phosphatase; qRT-PCR, quantitative RT-PCR; TS, trophoblast stem; TSA, trichostatin A; USF-1, upstream stimulatory factor-1; WB, Western blot; WCE, whole-cell extract; pIV, promoter IV; HDACi, HDAC inhibitor
Disclosures
The authors have no financial conflict of interest.
References
- 1.Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 2.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 3.Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice: a review. Placenta. 2005;26(Suppl. A):S3–S9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 5.Yan J, Tanaka S, Oda M, Makino T, Ohgane J, Shiota K. Retinoic acid promotes differentiation of trophoblast stem cells to a giant cell fate. Dev. Biol. 2001;235:422–432. doi: 10.1006/dbio.2001.0300. [DOI] [PubMed] [Google Scholar]
- 6.Hemberger M, Hughes M, Cross JC. Trophoblast stem cells differentiate in vitro into invasive trophoblast giant cells. Dev. Biol. 2004;271:362–371. doi: 10.1016/j.ydbio.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 7.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 2007;304:567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Petroff MG. Immune interactions at the maternal-fetal interface. J Reprod. Immunol. 2005;68:1–13. doi: 10.1016/j.jri.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Trowsdale J, Betz AG. Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nat. Immunol. 2006;7:241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 10.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Mao D, Holers VM, Palanca B, Cheng AM, Molina H. A critical role for murine complement regulator crry in fetomaternal tolerance. Science. 2000;287:498–501. doi: 10.1126/science.287.5452.498. [DOI] [PubMed] [Google Scholar]
- 12.Hunt JS, Vassmer D, Ferguson TA, Miller L. Fas ligand is positioned in mouse uterus and placenta to prevent trafficking of activated leukocytes between the mother and the conceptus. J Immunol. 1997;158:4122–4128. [PubMed] [Google Scholar]
- 13.Murphy SP, Choi JC, Holtz R. Regulation of major histocompatibility complex class II gene expression in trophoblast cells. Reprod. Biol. Endocrinol. 2004;2:52. doi: 10.1186/1477-7827-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt JS, Orr HT. HLA and maternal-fetal recognition. FASEB J. 1992;6:2344–2348. doi: 10.1096/fasebj.6.6.1544544. [DOI] [PubMed] [Google Scholar]
- 15.Ashkar AA, Croy BA. Interferon-γ contributes to the normalcy of murine pregnancy. Biol. Reprod. 1999;61:493–502. doi: 10.1095/biolreprod61.2.493. [DOI] [PubMed] [Google Scholar]
- 16.Paulesu L, Romagnoli R, Cintorino M, Ricci MG, Garotta G. First trimester human trophoblast expresses both interferon-γ and interferon-γ-receptor. J Reprod. Immunol. 1994;27:37–48. doi: 10.1016/0165-0378(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 17.Platt JS, Hunt JS. Interferon-γ gene expression in cycling and pregnant mouse uterus: temporal aspects and cellular localization. J Leukocyte Biol. 1998;64:393–400. doi: 10.1002/jlb.64.3.393. [DOI] [PubMed] [Google Scholar]
- 18.Cencic A, La Bonnardiere C. Trophoblastic interferon-γ: current knowledge and possible role(s) in early pig pregnancy. Vet. Res. 2002;33:139–157. doi: 10.1051/vetres:2002003. [DOI] [PubMed] [Google Scholar]
- 19.Ashkar AA, Di Santo JP, Croy BA. Interferon γ contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy. J Exp. Med. 2000;192:259–270. doi: 10.1084/jem.192.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albieri A, Hoshida MS, Gagioti SM, Leanza EC, Abrahamsohn I, Croy A, Ashkar AA, Bevilacqua E. Interferon-γ alters the phagocytic activity of the mouse trophoblast. Reprod. Biol. Endocrinol. 2005;3:34. doi: 10.1186/1477-7827-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshida MS, Gorjao R, Lima C, Daher S, Curi R, Bevilacqua E. Regulation of gene expression in mouse trophoblast cells by interferon-γ. Placenta. 2007;28:1059–1072. doi: 10.1016/j.placenta.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Mattsson R. The non-expression of MHC class II in trophoblast cells. Am. J. Reprod. Immunol. 1998;40:383–384. doi: 10.1111/j.1600-0897.1998.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 23.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 24.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 25.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 26.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 27.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 28.Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-γ requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 29.Piskurich JF, Linhoff MW, Wang Y, Ting JP. Two distinct γ interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor β. Mol. Cell. Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell. Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu. Rev. Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 32.Morris AC, Riley JL, Fleming WH, Boss JM. MHC class II gene silencing in trophoblast cells is caused by inhibition of CIITA expression. Am. J. Reprod. Immunol. 1998;40:385–394. doi: 10.1111/j.1600-0897.1998.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 33.Murphy SP, Tomasi TB. Absence of MHC class II antigen expression in trophoblast cells results from a lack of class II transactivator (CIITA) gene expression. Mol. Reprod. Dev. 1998;51:1–12. doi: 10.1002/(SICI)1098-2795(199809)51:1<1::AID-MRD1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Van den Elsen PJ, van der Stoep N, Vietor HE, Wilson L, van Zutphen M, Gobin SJ. Lack of CIITA expression is central to the absence of antigen presentation functions of trophoblast cells and is caused by methylation of the IFN-γ inducible promoter (PIV) of CIITA. Hum. Immunol. 2000;61:850–862. doi: 10.1016/s0198-8859(00)00159-2. [DOI] [PubMed] [Google Scholar]
- 35.Choi JC, Holtz R, Petroff MG, Alfaidy N, Murphy SP. Dampening of IFN-γ-inducible gene expression in human choriocarcinoma cells is due to phosphatase-mediated inhibition of the JAK/STAT-1 pathway. J Immunol. 2007;178:1598–1607. doi: 10.4049/jimmunol.178.3.1598. [DOI] [PubMed] [Google Scholar]
- 36.Holtz R, Choi JC, Petroff MG, Piskurich JF, Murphy SP. Class II transactivator (CIITA) promoter methylation does not correlate with silencing of CIITA transcription in trophoblasts. Biol. Reprod. 2003;69:915–924. doi: 10.1095/biolreprod.103.017103. [DOI] [PubMed] [Google Scholar]
- 37.Murphy SP, Gorzowski JJ, Sarge KD, Phillips B. Characterization of constitutive HSF2 DNA-binding activity in mouse embryonal carcinoma cells. Mol. Cell. Biol. 1994;14:5309–5317. doi: 10.1128/mcb.14.8.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carr VM, Murphy SP, Morimoto RI, Farbman AI. Small subclass of rat olfactory neurons with specific bulbar projections is reactive with monoclonal antibodies to the HSP70 heat shock protein. J Comp. Neurol. 1994;348:150–160. doi: 10.1002/cne.903480109. [DOI] [PubMed] [Google Scholar]
- 39.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 40.Briscoe J, Rogers NC, Witthuhn BA, Watling D, Harpur AG, Wilks AF, Stark GR, Ihle JN, Kerr IM. Kinase-negative mutants of JAK1 can sustain interferon-γ-inducible gene expression but not an antiviral state. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 41.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattsson R, Holmdahl R, Scheynius A, Bernadotte F, Mattsson A, Van der Meide PH. Placental MHC class I antigen expression is induced in mice following in vivo treatment with recombinant interferon-γ. J Reprod. Immunol. 1991;19:115–129. doi: 10.1016/0165-0378(91)90012-f. [DOI] [PubMed] [Google Scholar]
- 43.Zuckermann FA, Head JR. Expression of MHC antigens on murine trophoblast and their modulation by interferon. J Immunol. 1986;137:846–853. [PubMed] [Google Scholar]
- 44.Rasmussen CA, Pace JL, Banerjee S, Phillips TA, Hunt JS. Trophoblastic cell lines generated from tumor necrosis factor receptor-deficient mice reveal specific functions for the two tumor necrosis factor receptors. Placenta. 1999;20:213–222. doi: 10.1053/plac.1998.0356. [DOI] [PubMed] [Google Scholar]
- 45.Qing Y, Costa-Pereira AP, Watling D, Stark GR. Role of tyrosine 441 of interferon-γ receptor subunit 1 in SOCS-1-mediated attenuation of STAT1 activation. J Biol. Chem. 2005;280:1849–1853. doi: 10.1074/jbc.M409863200. [DOI] [PubMed] [Google Scholar]
- 46.Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 47.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 48.Briken V, Ruffner H, Schultz U, Schwarz A, Reis LF, Strehlow I, Decker T, Staeheli P. Interferon regulatory factor 1 is required for mouse Gbp gene activation by γ interferon. Mol. Cell. Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foss GS, Prydz H. Interferon regulatory factor 1 mediates the interferon-γ induction of the human immunoproteasome subunit multicatalytic endopeptidase complex-like 1. J Biol. Chem. 1999;274:35196–35202. doi: 10.1074/jbc.274.49.35196. [DOI] [PubMed] [Google Scholar]
- 50.White LC, Wright KL, Felix NJ, Ruffner H, Reis LF, Pine R, Ting JP. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harada H, Takahashi E, Itoh S, Harada K, Hori TA, Taniguchi T. Structure and regulation of the human interferon regulatory factor 1 (IRF-1) and IRF-2 genes: implications for a gene network in the interferon system. Mol. Cell. Biol. 1994;14:1500–1509. doi: 10.1128/mcb.14.2.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 54.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I, Levy DE. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J Biol. Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 57.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol. Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 58.Jamaluddin M, Choudhary S, Wang S, Casola A, Huda R, Garofalo RP, Ray S, Brasier AR. Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-κB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J Virol. 2005;79:15302–15313. doi: 10.1128/JVI.79.24.15302-15313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 60.Takaoka A, Yanai H. Interferon signalling network in innate defense. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 61.Zhu H, Nelson DR, Crawford JM, Liu C. Defective Jak-Stat activation in hepatoma cells is associated with hepatitis C viral IFN-α resistance. J Interferon Cytokine Res. 2005;25:528–539. doi: 10.1089/jir.2005.25.528. [DOI] [PubMed] [Google Scholar]
- 62.Zimring JC, Goodbourn S, Offermann MK. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]