Abstract
Peroxiredoxin6 (Prdx6) is one of the peroxiredoxin (Prdxs) family members that play an important role in maintaining cell homeostasis. Our previous studies demonstrated that Prdx6 was significantly associated with pig meat quality, especially meat tenderness. However, the transcriptional regulation of porcine Prdx6 remains unclear. In this study, we determined the transcription start site (TSS) of porcine Prdx6 gene by 5′ rapid-amplification of cDNA ends (5′ RACE). Several regulatory elements including CCAAT/enhancer-binding proteinβ (C/EBPβ), Myogenic Differentiation (MyoD), cAMP response element binding protein (CREB), stimulating protein1 (Sp1) and heat shock factor (HSF) binding sites were found by computational analyses together with luciferase reporter system. Overexpression and RNA interference experiments showed that C/EBPβ or CREB could up-regulate the expression of porcine Prdx6 gene at both mRNA and protein level. Electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation assays (ChIP) confirmed that C/EBPβ and CREB could interact with Prdx6 promoter. Immuoprecipitation results also showed that C/EBPβ could interact with Prdx6 in vivo. Taken together, our findings identified C/EBPβ and CREB as the important regulators of porcine Prdx6 gene expression, and offered clues for further investigation of Prdx6 gene function.
Introduction
Peroxiredoxins are a widely distributed superfamily of peroxidases with high antioxidant activity and have been received much more attention in recent years [1, 2]. Six peroxiredoxin family numbers (Prdx1-6) have been identified in mammals and classified as 1-cys, 2-cys based on the number of conserved cystines by which thiol groups are formed as catalytic center to degrade H2O2 and other peroxides [3, 4]. Prdx1-5 belongs to 2-cys peroxiredoxins and utilizes thioredoxin as a reductant. Unlike other peroxiredoxins, Prdx6 is the only 1-cysteine peroxiredoxin, and uses glutathione (GSH) rather than thioredoxin as the electron donor [5–8].
Prdx6 is widely distributed in all major organs with high expression levels in lung, kidney, liver, eye and testes [9–12]. It is a bifunctional enzyme with both GSH peroxidase and acidic Ca2+ independent phospholipase A2 activities that could participate in many physiological processes [7]. With the phospholipase A2 activity, Prdx6 can regulate phospholipid metabolism. And the GSH peroxidase activity allows Prdx6 to protect cells against oxidative stress by removing reactive oxygen species (ROSs) and limiting ROS levels. ROS are generated in cells during several normal physiologic processes and will be immediately removed by antioxidants like Prdx6. When the equilibrium status between the production and elimination of reactive oxygen species is broken, oxidative stress occur [13]. Excess ROS can result in irreversible oxidative damage to cellular lipids, DNA and proteins, and thereby caused many human diseases [13, 14]. Prdx6 has been reported to implicate in development and progression of several human diseases, like Alzheimer [15], Parkinson dementia [16], diabetes [17], cancer [18–21], cataractogenesis [22]. The formation mechanism of those diseases mostly related to oxidative stress. By analyzing the Prdx6 transcription regulation, many researchers have identified the mainly redox-active regulator of this gene in human, rat or mouse, like Nrf2 [23–25], NF-κB [26, 27], Sp1 [28], AP1 [18], HSF1 [29].
In animal production, research has shown that the Prdx6 might be a potential protein marker for meat tenderness in bovine biopsies and samples collected shortly after slaughter [30]. Meat tenderness is an important meat eating quality and has achieved much attention in modern animal production. Studies in our laboratory found that Prdx6 gene was differentially expressed in longissimus dorsi between different pig breeds (data unpublished). Besides, two SNPs (417bp C/T; 423bp A/G) were detected in the fourth exon of porcine Prdx6 gene, and were significantly associated with meat quality, especially meat tenderness [31]. The above findings indicated the new function of Prdx6 expression in pig meat quality traits. However, how the expression of Prdx6 gene is regulated is still unclear in pigs until now. In this study, the porcine Prdx6 promoter region and its transcription start site were identified and the possible transcription factors for the Prdx6 gene regulation were also investigated. We found that the transcription factor C/EBPβ and CREB could bind to their binding sites in Prdx6 promoter and regulate the expression of target genes. This study provided the insight into the basal regulation mechanisms of porcine Prdx6 gene, and the possible signaling pathway for the participation of Prdx6 gene in the adipogenesis and myogenesis.
Material and Methods
Animals, tissues and cell lines
The animal experimental procedures were approved by the Huazhong Agricultural University Institutional Animal Care and Use Committee, Wuhan, China. All pigs used in this study were from the Pig Farm of Huazhong agricultural university. The pigs were slaughtered after low voltage electrical stunning. Ten tissues including kidney, small intestine, stomach, longissimus dorsi, fat, brain, heart, liver, lung and spleen were collected from three Large White pigs at age of 4 month and then immediately frozen in liquid nitrogen and stored at -80°C. The DNA was extracted using the phenol–chloroform (Invitrogen, USA). Total RNA was extracted using the TRIzol reagent (Invitrogen, USA) and was reverse transcribed to cDNA using the PrimeScriptTM RT Reagent Kit (Takara, Japan).
Mouse myoblast cell line C2C12, mouse embryonic fibroblast-adipose like cell line 3T3-L1 and porcine kidney cell line PK15 were purchased from the Institutes for Biological Sciences Cell Resource Center, Chinese Academy of Sciences, Shanghai, China. Porcine intramuscular fat (IMF) cells were obtained according to the description in previous report [32].
Tissue expression profile analysis by RT-PCR
Using RT-PCR, ten tissues including heart, liver, spleen, lung, kidney, stomach, brain, longissimus dorsi, fat and small intestine from three 4-month-old Large White pigs were used to analyze the expression profile of Prdx6 gene. RT-PCR was performed using the CFX96 System (Bio-Rad, USA) in a total volume of 20 μl containing SusPr6DL-F/R or β-actin-F/R primers (Table 1) and SYBR-GreenⅡ mix (Takara, Japan). The gene expression results were normalized to the basal level of β-actin. The Ct (2-△△Ct) method was used to analyze the relative gene expression data.
Table 1. Primer/siRNA sequences for the experiments.
| Name | sequences (5′-3′) | Size (bp) | Location |
|---|---|---|---|
| D1 | AGATCTGATTGGCACGCATACAC | 1510 | (-1442∼+68) |
| D2 | AGATCTACATAGTCGGCAACGC | 1247 | (-1179∼+68) |
| D3 | AGATCTGTGGCACATAACCTTACTTT | 1041 | (-973∼+68) |
| D4 | AGATCTTGGGTCTTGCCACATT | 643 | (-575∼+68) |
| D5 | AGATCTGCCCAGGGCTGTTCTGTC | 328 | (-260∼+68) |
| D5.1 | AGATCTCGCCAAGGCCTGGCC | 215 | (-147∼+68) |
| D5.2 | AGATCTCGGCCCCGCCCATCTT | 181 | (-113∼+68) |
| D6 | AGATCTCTTGGCTGTGCTTTGGTC | 168 | (-100∼+68) |
| D7 | AGATCTCAACACCTACAAGGCTAGAAC | 84 | (-16∼+68) |
| D8 | AGATCTGTTAGCTGTCGCTGCTGG | 54 | (+15∼+68) |
| R | CCATGGCATGGCGGCAGCAGTGACGC | ||
| hsf1-F | GGGGTACCATGGATCTGCCCGTGGGC | 1557 | |
| hsf1-R | GCTCTAGACTAGGAGACAGTGGGGTC | ||
| C/EBPβ-DL-F | AAGCACAGCGACGAGTACAA | 157 | |
| C/EBPβ-DL-R | ACAGCTGCTCCACCTTCTTC | ||
| MyoD-DL-F | AGACCACTAACGCCGACCGC | 286 | |
| MyoD-DL-R | GCGTCTGAGTCACCGCTGTAGT | ||
| CREB-DL-F | CATGGAATCTGGAGCAGACAA | 109 | |
| CREB-DL-R | CTGGGCTAATGTGGCAATCT | ||
| SP1-DL-F | ACATGATGACCCAGCAGGTG | 280 | |
| SP1-DL-R | TGTGAAGCGTTTCCCACAGT | ||
| HSF1-DL-F | GGAAGCAAGAGAGCATGGAT | 123 | |
| HSF1-DL-R | TGAGCTTGTTGACGACTTTCT | ||
| SusPr6DL-F | CAGAATTTGCCAAGAGGAAT | 284 | |
| SusPr6DL-R | GTGGTAGCTGGGTAGAGGAT | ||
| β-actin-F | CCAGGTCATCACCATCGG | 158 | |
| β-actin-R | CCGTGTTGGCGTAGAGGT | ||
| M-C/EBPβ-F | CAACGCAAGGGTTATTGAATCAACTACTATCATCATC | ||
| M-C/EBPβ-R | GATGATGATAGTAGTTGATTCAATAACCCTTGCGTTG | ||
| M-MyoD-F | GTGGAAACGGATGCATATGACAATGGTAACC | ||
| M-MyoD-R | GGTTACCATTGTCATATGCATCCGTTTCCAC | ||
| siC/EBPβ-F | CCCUGAGUAAUCACUUAAATT | ||
| siC/EBPβ-R | UUUAAGUGAUUACUCAGGGTT | ||
| siCREB-F | GGACCUUUACUGCCACAAATT | ||
| siCREB-R | UUUGUGGCAGUAAAGGUCCTT | ||
| Chip-CREB-1F | CCAAGCACTTACTGACATAG | 125 | |
| Chip-CREB-1R | CTAGCCATCCTGATTTCTG | ||
| Chip-CREB-2F | CTATTGCGTTCCGCTTCCTT | 109 | |
| Chip-CREB-2R | CTCTAACCTGCCAGGATCC | ||
| Chip-C/EBPβ-F | CATAGTCGGCAACGCAAG | 159 | |
| Chip-C/EBPβ-R | GCCTCTGGGTTAATGTAG |
The transcription start site (TSS) determined by 5′ RACE was defined as +1. The bold font sequences were the restriction endonuclease digested site (AGATCT: BglⅡ; CCATGG: NcoI; GGTACC: KpnI; TCTAGA: XbaI). The mutation sites were shown by the nucleotides in bold and italic formats.
5′ RACE and Computational analysis
5′ RACE was performed using a Smarter™ RACE cDNA Amplification Kit (Clontech, USA), according to the manufacturer’s user manual. Total RNA of liver tissue from the same 4 month Large White pig was chosen as template for synthesis of Prdx6 cDNA. The gene specific primer (GSP) was synthesized by Sangon (China) as following sequence, GSP: 5′-GACCATCACGCTGTCTCCATTCTTCC-3′. The PCR products were purified and cloned into pMD-18T vector (Takara) and transformed into DH5α bacteria. Eighteen positive colonies were selected and commercially sequenced by Shanghai Sangon.
Transcription factor binding sites were predicted using the TFsearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and TESS (http://www.cbil.upenn.edu/tess). Multiple sequence alignment was performed using the ClustalW2.0 (http://www.clustal.org).
Plasmid construction
Ten Prdx6 promoter deletion fragments (D1-D8) were amplified with primers listed in Table 1. PCR products were cloned into pMD-18T vector (Takara). The recombinant plasmids were then digested with NcoI and BglⅡ (Thermo, USA), and then subcloned into luciferase reporter vector pGL3-Basic (Promega, USA) respectively.
The C/EBPβ mutated vector and MyoD mutated vector were constructed by the PCR-based site-directed mutagenesis using the KOD-Plus-Neo(Toyobo, Japan) and DpnⅠ(Thermo, USA), following the manufacturer’s protocol. The pGL3-D2 vectors containing the C/EBPβ and MyoD binding sites were chosen as template. The mutation primers M-C/EBPβ-F/R and M-MyoD-F/R were listed in Table 1. The HSF mutated D7 and Sp1 mutated D5 fragments were directly synthetized at Sangon Biotech (China) and then cloned into luciferase reporter vector pGL3-Basic as described before.
The HSF1 eukaryotic expression vector (pcDNA3.1-HSF1) was constructed. Using the cDNA (from pig liver tissue) as template, the coding sequence of HSF1 gene (NCBI No: NM_001243819.1) was amplified with hsf1-F/R primers (Table 1). The PCR products were digested with KpnI and XbaI (Thermo), and cloned into pcDNA3.1 vector (Promega) using T4 DNA Ligase (Takara). Other eukaryotic expression vectors (pcDNA3.1-C/EBPβ, CREB, MyoD and Sp1) used in over-expression experiment were from our laboratory.
Cell culture, transfection and luciferase assay
The C2C12, 3T3-L1 and PK15 cells were cultured in DMEM medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, Australia) at 37°C and 5% CO2. Cells were seeded into 24-well plate and after 24 h incubation, each well was transfected with 1.0 μg of DNA constructs and 50 ng of Renilla luciferase reporter plasmid (pRL-TK, Promega) using 2 μl lipofectamine 2000 (Invitrogen, USA). For the co-transfection groups, each well was transfected with 0.8 μg of Prdx6 promoter constructs, 0.5 μg of over-expression vector and 50 ng pRL-TK using 2 μl lipofectamine 2000. At 24–36 h after transfection, cells were lysed and assayed for the luciferase activities using dual-luciferase reporter assay system (Promega). The luciferase activity was measured using PerkinElmer 2030 Multilabel Reader (PerkinElmer) and normalized on the basis of Renilla activity.
Transfection of plasmid DNA or siRNA oligonucleotides
PK15 cells were seeded into 6-well plate and cultured at 37°C and 5% CO2. After 24 h incubation, cells of each well were transfected with 4μg of over-expression vector or pcDNA3.1 empty vector using 9 μl lipofectamine 2000. C2C12 cells were seeded into 6-well plate and cultured at 37°C and 5% CO2. When reaching 70% confluence, cells of each well were transfected with 100 pmol of siRNA using 9 μl lipofectamine 2000. The C/EBPβ and CREB siRNAs (Table 1) were purchased from genepharma (China). After transfection for 36 h, cells were collected; RNA and proteins were extracted for quantitative RT-PCR and Western Blot detection respectively. RNAs were extracted using HP Total RNA Kit (OMEGA, USA) according to the manufacturer’s instructions. The reverse transcription and RT-PCR were performed as described before. Primers used in RT-PCR were listed in Table 1.
Western blot
After transfection for 36 h, Cell lysates were prepared in ice-cold RIPA lysis buffer according to manufacturer’s instruction (Beyotime, China). Equal amounts of protein samples were loaded into a 10% SDS-PAGE and transferred to PVDF membrane (Millipore Billerica, MA, USA). After 1.5 h blockading within 5% nonfat dried milk, membranes were incubated overnight with primary antibodies at 4°C. After washing for 3 times, membranes were incubated with secondary antibodies for 1.5 h at room temperature. The results were visualized using the ECL Western Blotting Detection System (Bio-Rad, USA). Primary antibodies contain anti-Prdx6 (Santa Cruz, SC-134478; 1:500 dilution), anti-C/EBPβ (Santa Cruz, SC-150X; 1:500 dilution), anti-CREB (Cell Signaling Technology, #4820; 1:1000 dilution), anti-HSF1(Cell Signaling Technology, #4356P; 1:1000 dilution) and β-actin antibody (Boster, China, BM0627; 1:1000 dilution). Goat anti-rabbit IgG-HRP and goat anti-mouse IgG-HRP secondary antibodies (Boster, China; 1:3000 dilution) were used in this study.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from C2C12 cells or porcine intramuscular fat (IMF) cells with the Nuclear Extraction Kit (Active Motif, CA, USA). EMSA was performed according to the manufacturer’s instruction of Chemiluminescent EMSA Kit (Beyotime). The biotin-labeled double-stranded oligonucleotides (Sangon) containing predicted CREB or C/EBPβ binding sites were incubated with 10 μg nuclear extracts for 20 min at room temperature and were subjected to electrophoresis on 6% polyacrylamide gels. Then transferred to a nylon membrane and results were visualized using the ECL (Bio-Rad). In competition group, a 1-fold molar excess of unlabeled oligonucleotides was added to the reaction mixture prior to the addition of biotin labeled probe. For the supershift group, 2 μg C/EBPβ (Santa Cruz) or CREB (Cell Signaling Technology) antibodies was added to verify the specificity of DNA-protein interaction. The EMSAs were performed according to previous reports [33, 34].
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was carried out using the ChIP assay Kit (Beyotime) following the manufacturer’s protocol. The PK15 cells cultured in 15 cm-diameter dishes were fixed in formaldehyde for 20 min at room temperature and neutralized with glycine for 5 min. After washing with cold PBS, scraped and harvested cell lysates were sonicated to produce chromatin fragments about 200–750 bp in size. An equal amount of chromatin was immunoprecipitated overnight at 4°C with 4 μg of the following antibodies: anti-CREB, anti-C/EBPβ and Normal Rabbit IgG, and added the ProteinA+G Agarose beads (Beyotime) for further 5 h incubation. Then the beads were washed, and the bound chromatin was eluted in ChIP Elution Buffer. The DNA was treated with proteinase K for 4 h at 45°C. After purified, PCR was carried out using 3 μl DNA sample with primers specific to the Prdx6 promoter (Table 1). The first binding region of CREB was amplified by PCR as the following program: 30 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 15 s. The second binding region of CREB was amplified by PCR as the following program: 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 15 s. The binding region of C/EBPβ was amplified by PCR as the following program: 35 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 15 s. The PCR products were visualized on a 2.0% agarose gel. Total chromatin was used as input. The immunoprecipitate from Normal Rabbit IgG group was used as negative control.
Immunoprecipitation
PK15 cells cultured in 10 cm-diameter dishes were collected and lysed in nondenaturing lysis buffer (Sangon, Shanghai, China) supplemented with protease and phosphatase inhibitor cocktails. Well equal mass of lysate was incubated overnight with 2 μg either anti-Prdx6 or anti-IgG together with 25 μl Protein A+G Agarose beads. After centrifuge the beads were washed with 1ml lysates buffer for three or four times. Then the beads were added with 15 μl 2×SDS loading buffer and boiled for 10 minutes, then were loaded onto an SDS-PAGE gel for western blot analysis.
Statistical analysis
Statistical analysis was performed using Student’s t test. Data are expressed as mean ± S.D., and statistical significance was considered as p < 0.05. **P<0.01; *P<0.05; NS, not significant.
Results
Identification of transcription start site of porcine Prdx6 gene
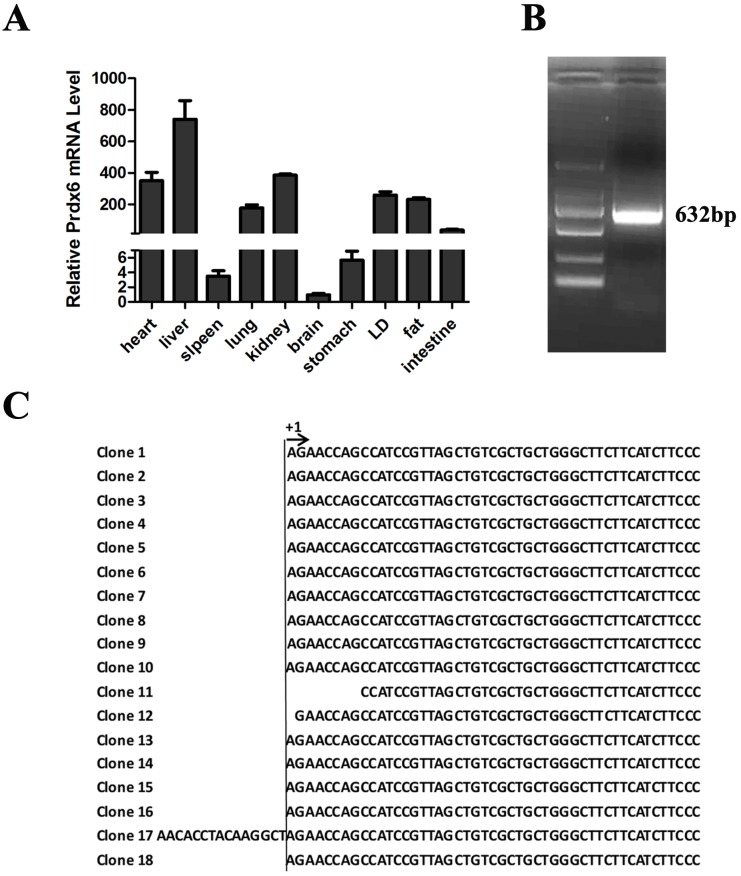
To better understand the transcription regulation of porcine Prdx6 gene, we determined the exact TSS of Prdx6 by 5′ RACE. Firstly, the tissue expression profile of Prdx6 was analyzed by real time-PCR. The Prdx6 was expressed in almost all the tissues, especially with the highest expression in liver tissue; the kidney, heart, longissimus dorsi (LD), fat and lung tissues also showed higher expression level; while a very low expression level was detected in brain tissue (Fig 1A), which was consistent with the previous report [12]. Then we selected liver tissue as the sample for 5′ RACE to determine the TSS of Prdx6. And the PCR products of 632bp were obtained (Fig 1B). After cloning and sequencing, the adenine located at 65bp before the first exon was identified as the major TSS as it appears 15 times in the total 18 clone sequencing results (Fig 1C), while some RACE clones (clone 11, 12 and 17) showed slightly different TSS which were located at the 57bp, 64bp and 80bp before the first exon, respectively. This is a common feature observed in promoter of less TATA-box [35], and Prdx6 gene belongs to the TATA-less promoter. We defined the major TSS as +1 for later illustration.
Fig 1. Identification of Prdx6 TSS using 5′ RACE.
(A) The Prdx6 mRNA expression profile was analyzed by RT-PCR. (B) 5′ RACE PCR product of 632bp was visualized in 1.5% agarose gel. (C) Eighteen sequencing results of Prdx6 5′ RACE clones are listed and the major TSS adenine was defined as +1.
Identification of crucial transcriptional factors controlling Prdx6 gene expression
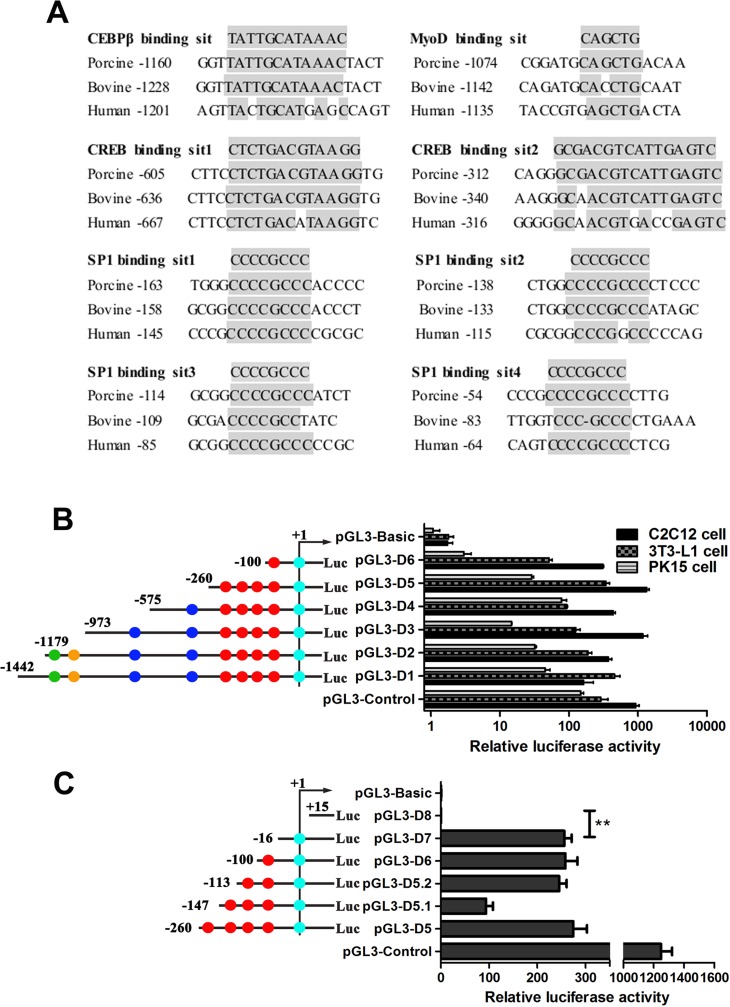
Using the TFSEARCH and TESS website analysis, a series of transcription factor binding sites related to muscle or fat development were found in the porcine Prdx6 promoter region, such as C/EBPβ (-1156/-1146), MyoD (-1068/-1063), CREB (-601/-588; -308/-293). Besides, four GC-boxes (Sp1-response element) were found at the proximal region (-161/-153; -136/-128; -112/-104; -52/-44). By alignment of multiple sequences, higher similarities of those factor binding sites were found among pig, bovine and human (Fig 2A). This suggested that Prdx6 gene might be regulated by multiply transcription factors.
Fig 2. The porcine Prdx6 gene promoter was analyzed by computational analyses together with luciferase reporter system.
(A) Conserved sequences of transcription factor binding sites of Prdx6 promoter. The aligned region is conserved among pig, human and bovine. (B) Six promoter constructs were transfected into C2C12, 3T3-L1 and PK15 cell lines, and assayed for luciferase activity. The pGL3-Control and pGL3-Basic were the positive and negative control respectively. Green, orange, blue, red and bright blue dots on the left panel represented the C/EBPβ, MyoD, CREB, Sp1 and HSF binding sites predicted on TFsearch or TESS websites, respectively. Data are expressed as means ± SD of three replicates. (C) Four further deletions (D5.1, D5.2, D7, and D8) were transfected into C2C12 cells and assayed for luciferase activity.
To determine the crucial promoter motifs, six promoter deletions (D1-D6) were linked to the pGL3-basic vector, and their luciferase activity was analyzed in PK15, C2C12 and 3T3-L1 cell lines (Fig 2B). The promoter activity was with the highest expression in C2C12 cells and the lowest expression in PK15 cells, which may reflect the difference of transfection efficiency in different cell lines and the transcriptional regulation in different species. Compared with the basal level of pGL3-basic, every deletion showed significant promoter activity in each cell line. Generally, D5 deletion had the highest promoter activity; the shortest D6 deletion had the lowest one; while the promoter activities of other deletions were between them. This experiment further indicated that the regulation of Prdx6 gene is complex, and it might be regulated by multiple transcription elements. For example, the fragment from -575bp to -973bp of D3 deletion may contain crucial cis elements which are important for Prdx6 gene expression in C2C12 cells since the luciferase activity of D3 deletion is much higher than that of D4 deletion in C2C12 cells. The D5 and D6 deletions may also contain crucial cis elements which are vital for Prdx6 transcriptional initiation since the shortest two deletions have significant luciferase activity compared with negative control. We then constructed four deletion fragments (D5.1, D5.2, D7 and D8) to further analyze the transcriptional activity of Prdx6 promoter. The results showed that the minimal transcription activity region was located at -16bp~+15bp containing one HSF binding site (Fig 2C).
C/EBPβ and CREB increase Prdx6 gene expression
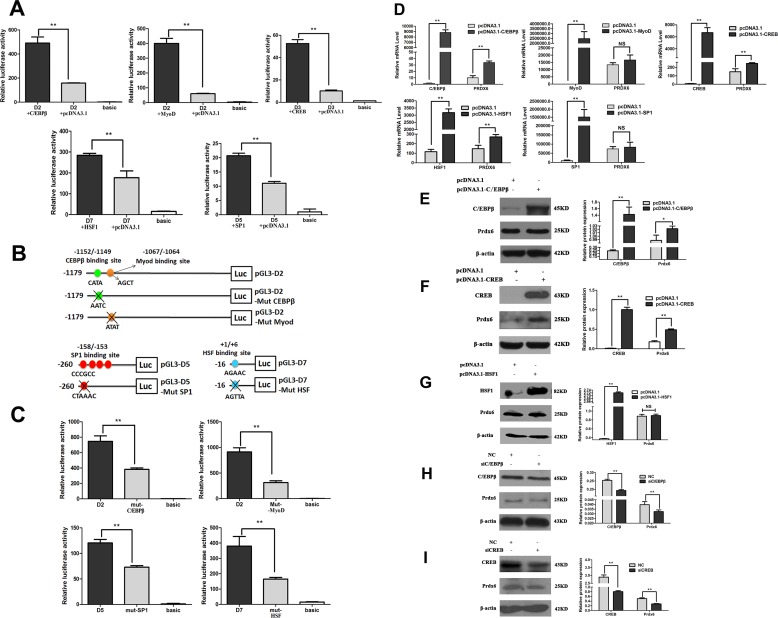
Co-transfection experiments were performed to identify the transcription factor regulating the Prdx6 gene transcription. As shown in Fig 3A, co-transfection of Prdx6 deletion fragments with over-expression vectors of each transcription factor could remarkably increase the luciferase activities compared with the co-transfection with pcDNA3.1 empty vector. Then we mutated the C/EBPβ, MyoD, Sp1 and HSF binding sites in the Prdx6 deletion plasmids (Fig 3B), and found sharply decreased luciferase activity of the mutated group compared with the wild-type group (Fig 3C). To determine which factor can indeed affect the Prdx6 transcription at mRNA level, five transcription factors were transfected into PK15 cells. The RT-PCR results revealed that C/EBPβ, CREB and HSF could improve Prdx6 transcription level, while MyoD and Sp1 had no effects (Fig 3D). Prdx6 protein level was also increased after transient transfection with pcDNA3.1-C/EBPβ or pcDNA3.1-CREB (Fig 3E and 3F), but nearly no change after transfected with the pcDNA3.1-HSF1 vector (Fig 3G). Inhibition of C/EBPβ or CREB by siRNA could decrease Prdx6 protein level (Fig 3H and 3I). Those data indicated that C/EBPβ and CREB increase Prdx6 expression both at mRNA and protein level.
Fig 3. C/EBPβ and CREB could up-regulate the Prdx6 expression.
(A) The Prdx6 deletions were co-transfected with relative overexpressed vector or pcDNA3.1 empty vector. The promoter activity was measured and the results were expressed as means± SD of three replicates. (B) Schematic structure of site-direct mutants of Prdx6 promoter linked to pGL3-Basic vector. (C) Wild-types and mutants of the Prdx6 deletions were transfected into C2C12 cells, and luciferase activity was detected and the results were expressed as means± SD of three replicates. (D) The Prdx6 mRNA expression level was detected by RT-PCR after overexpression of each transcription factor into PK15 cells, data were showed as means± SD of three replicates. (E-G) The transfection efficiency of C/EBPβ, CREB or HSF1 overexpression vector and their effect on target Prdx6 protein expression level were determined by western blot. (H-I) The interference efficiency of Knockdown of C/EBPβ or CREB and their effect on target Prdx6 protein expression level were determined by western blot. Quantification results of western blot represented by ratio of C/EBPβ, CREB, HSF1 or Prdx6 to β-actin protein expression level (Image J software).
CREB could bind to the first CRE site of Prdx6 promoter in vivo and in vitro
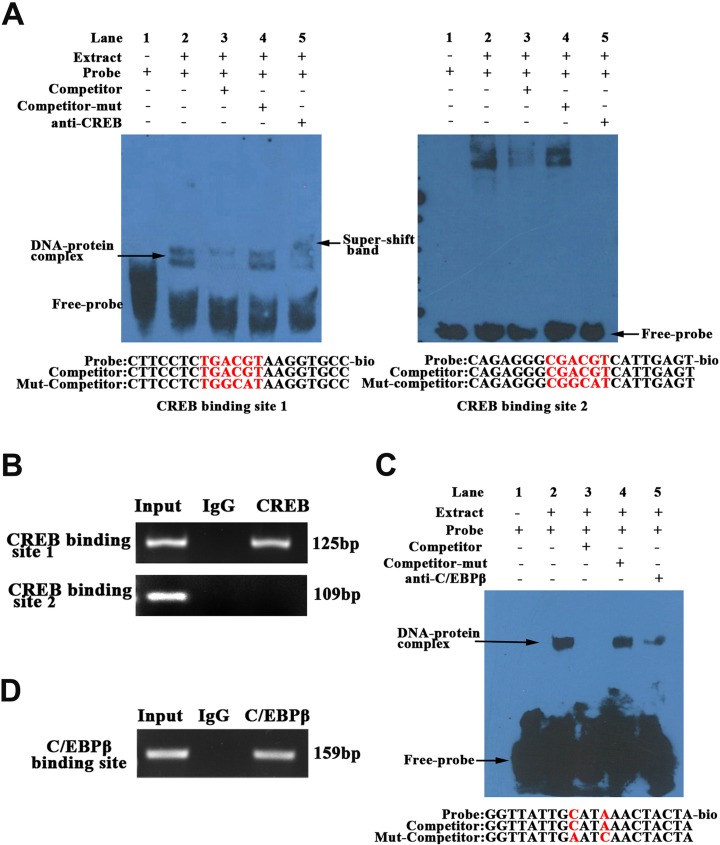
The EMSAs were performed with nuclear extracts from C2C12 cells to determine whether CREB bind to Prdx6 promoter (Fig 4A). For the first CREB binding site (-601/-588), the DNA-protein complex were obviously present in the biotin-labeled probe (Lane2) and mutation-competitor (Lane4) groups, whereas no complexes were observed in the absence of nuclear extracts (Lane1). The cold-competitor group (Lane3) displayed decreased intensity of the complex. The specific anti-CREB antibody was added to the mixture of Lane5 and caused the weakness of DNA-protein complex and the appearance of super-shift band. For the second CREB binding site (-308/-293), no specific DNA-protein complex was detected (Fig 4A right). These results indicated that CREB could bind to the first CREB binding site of Prdx6 promoter.
Fig 4. C/EBPβ and CREB bind to Prdx6 promoter region in vitro and in vivo.
(A) Binding of CREB with Prdx6 promoter was detected by EMSA with C2C12 cell nuclear extracts. Probes of CREB binding sites were labeled with biotin and the mutated nucleotides were depicted in red color. Lane1 was a blank control that did not add nuclear extracts; Lane2 was experimental group. For Lane3 and 4, a 1-fold excess of unlabeled or mutant probe was added to the reaction complex. The Lane5 was added 2μg anti-CREB antibody in the complex. The DNA-protein complex and the super-shift bands were indicated by arrows (B) ChIP assay of CREB binding to the Prdx6 promoter in PK15 cells. (C) The interaction of C/EBPβ with Prdx6 promoter was detected by EMSA with the porcine IMF cell nuclear extracts. The Lane1-5 experimental groups are similar to figure 4A. (D) Binding of C/EBPβ on the Prdx6 promoter was determined by ChIP assay.
ChIP assay was preformed to confirm the binding of CREB to the Prdx6 promoter in vivo. Chromatin fragments from lysates of PK15 cells were precipitated with a CREB antibody or control IgG (Fig 4B). The immunoprecipitates were analyzed by PCR with primers specific to the Prdx6 promoter harboring the CREB binding site. The results also revealed that only the first CRE site of Prdx6 promoter could bind to CREB in vivo. These results suggested that the CREB promotes the expression of Prdx6 gene through binding to the first rather than the second CRE site. This also explained why the luciferase activity of D3 deletion (with two CRE site) was significantly higher than that of D4 deletion (with only the second CRE site) detected in C2C12 cells (P = 0.003) (Fig 2B). No significant difference of Luciferase activity between D3 and D4 detected in 3T3-L1 cells (P = 0.052) may due to the relatively lower expression of CREB in this cell line. In PK15 cells, a completely opposite result was obtained, which may result from the different transcriptional regulation mechanism existed in this cell line.
C/EBPβ could bind to the Prdx6 promoter in vivo and in vitro
By EMSA and ChIP assays, we detected the interaction of C/EBPβ with Prdx6 promoter in vitro and in vivo (Fig 4C and 4D). The specific DNA-protein complex was found in biotin-labeled probe group (Lane2), and was attenuated after addition of cold-competitor probe (Lane3), while the mutated competitor probe failed to abolish complex formation (Lane4). The specific complex did not appear without nuclear extracts in Lane1. Although there is no super-shift band appearance in lane5, the complex band was also diminished after addition of C/EBPβ antibody (Fig 4C). Therefore, we infer that the super-shift band may exist but were not exhibited due to the less sensitivity. ChIP results showed that the specific DNA fragments containing the C/EBPβ binding site of Prdx6 promoter could be amplified in the anti-C/EBPβ group immunoprecipitates rather than the IgG group (Fig 4D). The results indicated that C/EBPβ could bind to the Prdx6 promoter and increase its expression. In 3T3-L1 and PK15 cells, the luciferase activities of D1 and D2 deletions (containing C/EBPβ binding site) were significantly higher than those of D3 deletion (no C/EBPβ binding site) (Fig 2B), which was consistent with our EMSA and ChIP results. In C2C12 cells, the opposite results were found, because the C/EBPβ was higher expressed in fat cells like 3T3-L1 rather than C2C12 cells.
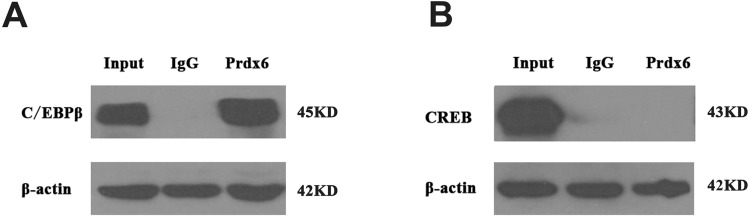
Prdx6 could interact with C/EBPβ in vivo
Although the transcription regulation mechanism of porcine Prdx6 gene by CREB and C/EBPβ have been illustrated, the interaction of Prdx6 protein with CREB and C/EBPβ proteins were still not clear in vivo. To address this, the immunoprecipitation was performed in PK15 cells. Proteins from the PK15 cells were collected and immunoprecipitated by Prdx6 antibody or IgG antibody. The results showed that compared with IgG group, C/EBPβ protein was detected in the immunoprecipitates of anti-Prdx6 group (Fig 5A), whereas CREB could not be detected (Fig 5B), demonstrating that Prdx6 could interact with C/EBPβ and might participate in its biological function.
Fig 5. Prdx6 could interact with C/EBPβ in vivo.
Immunoprecipitation was performed in PK15 cells using anti-Prdx6 and anti-IgG antibodies. Total Protein was used as input. The protein from Normal rabbit IgG group was used as negative control. (A) Detection of C/EBPβ protein in immunoprecipitation. (B) Detection of CREB protein in immunoprecipitation.
Discussion
In this study we determined the core promoter motifs and key regulators controlling the transcription of porcine Prdx6 gene. By 5′ RACE, we analyzed the Prdx6 transcription start site for the first time and found that it is different from the computational prediction. The TSS of Prdx6 is an adenine located at 65bp before the first exon. Bioinformatic analysis and experiments revealed some important cis element binding sites including C/EBPβ, MyoD, CREB, Sp1, and HSF. Our data indicated that only C/EBPβ and CREB could up-regulated the Prdx6 expression.
The CCAAT/enhancer-binding proteinβ (C/EBPβ) is a basic leucine zipper DNA-binding factor and could recognize a consensus DNA sequence (TTNNGNAAT) in target gene [36]. In present study, one C/EBPβ binding sites (TTGCATAAACT) was identified in the porcine Prdx6 promoter, which was located at -1156/-1146 bp. Co-transfection of Prdx6 promoter deletion with a construct over-expressing C/EBPβ significantly increased Prdx6 promoter activity (about 3-fold, P<0.01, Fig 3A). After mutated the C/EBPβ binding site of Prdx6 promoter, the luciferase activity was significantly lower than that of wild type vector (Fig 3C). Over-expression experiment results showed the expression of the Prdx6 gene could be up-regulated by C/EBPβ (Fig 3D and 3E). Many studies proved that C/EBPβ is an important early regulator of adipogenesis that could modulate the key adipogenic transcription factors C/EBPα, PPARγ and SREBP1 [37, 38]. By EMSA and ChIP assays, we also found the C/EBPβ could bind to Prdx6 promoter and promote its expression. The important function of Prdx6 is scavenging and limiting reactive oxygen species (ROS) level. Previous reports indicated that oxidative stress and the generation of ROS could regulate the adipogenesis. For example, the hypoxia-inducible factor HIF1 which can inhibit PPARγ was regulated by ROS [39]. ROS have been investigated to facilitate adipogenesis by accelerating mitotic clonal expansion during 3T3-L1 cell differentiation [40]. So it is possible that the C/EBPβ affects adipogenesis through promoting the expression of Prdx6 and thereby leads to the down-regulation of ROS levels. Besides, C/EBPβ could interact with Prdx6 protein (Fig 5A), suggesting the possibility that Prdx6 might participate in the biological function of C/EBPβ in adipogenesis. However, this possibility still needs more research.
The cAMP response element binding protein (CREB) is a cellular transcription factor that recognizes and binds to the cAMP response element (CRE, TGACGTCA) of target gene promoter [41]. By regulating the expression of its target genes CREB affect several physiological process like cell proliferation, apoptosis, brain circuits [42], and is also involved in responses to oxidative stress [43]. The thioredoxin and redox factor 1 were regulated by CREB under certain stimuli [44]. Here we found that the porcine Prdx6 is also regulated by CREB. Prdx6 promoter region contains two CREB binding sites (-601/-588; -308/-293), and only the first one (-601/-588) could bind with CREB and affect the Prdx6 promoter activity by EMSA, ChIP and over-expression experiments (Fig 3F; Fig 4A and 4B). Previous studies have proved that CREB plays a vital role in muscle differentiation and regeneration. For example, Zuloaga et al proved that the activated CREB by insulin-like growth factor-1(IGF-1) regulates myostatin expression and further inhibits muscle differentiation [45]. Stewart et al found that the CREB is activated by muscle injury and promotes muscle regeneration [46]. CREB-induced up-regulation of C/EBPβ is required in infiltrating macrophages for induction of M2-specific genes expression and muscle regeneration [47]. The present study demonstrated the Prdx6 could interact with C/EBPβ, which might provide a new insight of Prdx6 function in muscle differentiation and regeneration through CREB/C/EBPβ cascade. Moreover, ROS is an important signal molecule that plays an essential role in muscle differentiation [48, 49]. Since its important role in regulating ROS level, we hypothesized that Prdx6 participate in muscle development by eliminating ROS.
The present study also analyzed several other potential regulators of porcine Prdx6 gene, such as Sp1, MyoD and HSF. Prdx6 promoter contains four conserved GC-boxes but no TATA-box. According to the previous study, the TATA-less promoter is likely to be regulated by GC-box and its response element Sp1 [50]. A conserved E-box (-1068/-1063, CAGCTG), the bHLH (basic helix loop helix) transcription factor MyoD binding site, was also found in the Prdx6 promoter. Although the HSF binding site (+1/+5, AGAAC) of Prdx6 promoter was not conserved among species, it showed remarkable effect in initiating promoter activity (Fig 2C). Co-transfection and site direct mutation assays suggesting the possibility that expression of the Prdx6 gene might be regulated by Sp1, MyoD and HSF1. However, overexpression of Sp1 and MyoD in the PK15 cells did not change the mRNA expression level of Prdx6 (Fig 3D). HSF1 could up-regulate the mRNA level of Prdx6 but have no effect at the protein level of Prdx6 (Fig 3D and 3G). The possible reason maybe two following points: (1) we just analyzed the activating effect of transcription factors to certain promoter fragment of Prdx6 gene, but ignored the complex transcriptional regulation system that may exist in Prdx6 promoter. For Prdx6 promoter, there is a complex of transcription sites for of the different transcription factors in vivo and the transcriptional regulation is dependent on this complex and not from any single individual transcriptional factor. (2) The porcine PK15 cells used in this paper were different from previous study, in which the human LEC cells were used and the Sp1 had been proved to be an activator of Prdx6 [27]. Therefore, we infer that the regulation of Prdx6 gene regulated by Sp1 is different among species.
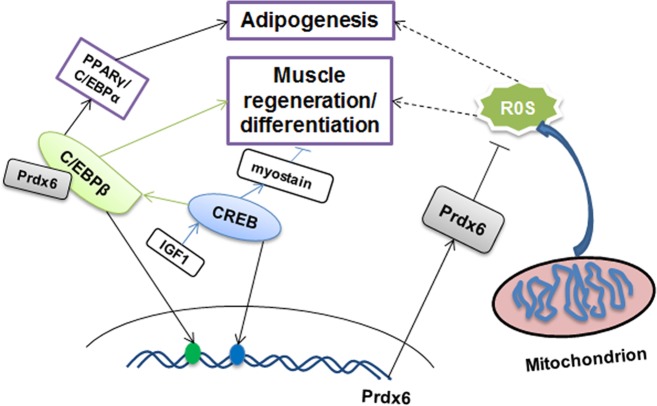
In conclusion, we have identified the transcription start site and important cis elements of porcine Prdx6 and studied the transcription regulators affecting Prdx6 transcription. The CREB and C/EBPβ were found for the first time to transactivate Prdx6 gene expression by binding to the relevant promoter region. We also found the C/EBPβ could interact with Prdx6. A hypothetical model was proposed that Prdx6 might participate in adipogenesis or myogenesis by interacting with C/EBPβ or scavenging ROS level (Fig 6). The present study will help to better understand the basal transcriptional regulation mechanism and provide clues for further investigation of Prdx6 gene function.
Fig 6. Model for mechanisms of Prdx6 participates in adipogenesis, muscle differentiation or regeneration.
C/EBPβ or CREB-mediated up-regulation of Prdx6 may participate in adipogenesis, muscle differentiation or regeneration through two pathways: (1), Prdx6 inhibits adipogenesis and muscle differentiation by scavenging ROS; (2), Prdx6 interacts with C/EBPβ to participate in adipogenesis and muscle regeneration.
Acknowledgments
We are very grateful to Dr. Yunxia Zhang for technical support on this study.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was financially supported by the Key National High Technology Development Project of China (project no. 2011AA100301), the National Key Technology Support Program of China (grant no. 2014BAD20B01), the Agricultural Innovation Fund of Hubei Province (grant no. 2007-620), the Fundamental Research Funds for the Central Universities (grant no. 2014PY038) and the Key Project of Science and Technology of Zhejiang Province (grant no. 2014C02010).
References
- 1. Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J. 2009; 276(9): p. 2469–77. 10.1111/j.1742-4658.2009.06985.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinkmann C, Brixius K. Peroxiredoxins and sports: new insights on the antioxidative defense. J Physiol Sci. 2013; 63(1): p. 1–5. 10.1007/s12576-012-0237-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chae H, Kim I, Kim K, Rhee S. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. Journal of Biological Chemistry 1993, 268(22):16815–16821. [PubMed] [Google Scholar]
- 4. Kang SW, Chae HZ, Seo MS, Kim K, Baines IC, Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-α. Journal of Biological Chemistry. 1998; 273(11): p. 6297–6302. [DOI] [PubMed] [Google Scholar]
- 5. Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid Hydroperoxides Are Substrates for Non-selenium Glutathione Peroxidase. Journal of Biological Chemistry. 1999; 274(30): p. 21326–21334. [DOI] [PubMed] [Google Scholar]
- 6. Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc Natl Acad Sci U S A. 2004; 101(11): p. 3780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000; 275(37): p. 28421–7. [DOI] [PubMed] [Google Scholar]
- 8. Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007; 104(12): p. 4886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005; 38(11): p. 1422–32. [DOI] [PubMed] [Google Scholar]
- 10. Kim T-S, Dodia C, Chen X, Hennigan BB, Jain M, Feinstein SI, et al. Cloning and expression of rat lung acidic Ca2+-independent PLA2 and its organ distribution. American Journal of Physiology-Lung Cellular and Molecular Physiology. 1998; 274(5): p. L750–L761. [DOI] [PubMed] [Google Scholar]
- 11. Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB life. 2001; 52(1): p. 35–41. [DOI] [PubMed] [Google Scholar]
- 12. Singh AK, Shichi H. A Novel Glutathione Peroxidase in Bovine Eye: SEQUENCE ANALYSIS, mRNA LEVEL, AND TRANSLATION. Journal of Biological Chemistry. 1998; 273(40): p. 26171–26178. [DOI] [PubMed] [Google Scholar]
- 13. Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007; 39(1): p. 44–84. [DOI] [PubMed] [Google Scholar]
- 14. Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009; 16(7): p. 1040–52. 10.1038/cdd.2009.49 [DOI] [PubMed] [Google Scholar]
- 15. Power JH, Asad S, Chataway TK, Chegini F, Manavis J, Temlett JA, et al. Peroxiredoxin 6 in human brain: molecular forms, cellular distribution and association with Alzheimer's disease pathology. Acta Neuropathol. 2008; 115(6): p. 611–22. 10.1007/s00401-008-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yun HM, Choi DY, Oh KW, Hong JT. PRDX6 Exacerbates Dopaminergic Neurodegeneration in a MPTP Mouse Model of Parkinson's Disease. Mol Neurobiol. 2014; 1–10. [DOI] [PubMed] [Google Scholar]
- 17. El Eter E, Al Masri A, Habib S, Al Zamil H, Al Hersi A, Al Hussein F, et al. Novel links among peroxiredoxins, endothelial dysfunction, and severity of atherosclerosis in type 2 diabetic patients with peripheral atherosclerotic disease. Cell Stress Chaperones. 2014; 19(2): p. 173–81. 10.1007/s12192-013-0442-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jo M, Yun HM, Park KR, Hee Park M, Myoung Kim T, Ho Pak J, et al. , Lung tumor growth-promoting function of peroxiredoxin 6. Free Radic Biol Med. 2013; 61: p. 453–63. 10.1016/j.freeradbiomed.2013.04.032 [DOI] [PubMed] [Google Scholar]
- 19. Rolfs F, Huber M, Gruber F, Bohm F, Pfister HJ, Bochkov VN, et al. Dual role of the antioxidant enzyme peroxiredoxin 6 in skin carcinogenesis. Cancer Res. 2013; 73(11): p. 3460–9. 10.1158/0008-5472.CAN-12-4369 [DOI] [PubMed] [Google Scholar]
- 20. Liu FJ, Wang XB, Cao AG. Screening and functional analysis of a differential protein profile of human breast cancer. Oncol Lett. 2014; 7(6): p. 1851–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seibold P, Hall P, Schoof N, Nevanlinna H, Heikkinen T, Benner A, et al. , Polymorphisms in oxidative stress-related genes and mortality in breast cancer patients—potential differential effects by radiotherapy? Breast. 2013; 22(5): p. 817–23. 10.1016/j.breast.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 22. Kubo E, Fatma N, Akagi Y, Beier DR, Singh SP, Singh DP. TAT-mediated PRDX6 protein transduction protects against eye lens epithelial cell death and delays lens opacity. American Journal of Physiology-Cell Physiology. 2008; 294(3): p. C842–C855. 10.1152/ajpcell.00540.2007 [DOI] [PubMed] [Google Scholar]
- 23. Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009; 46(2): p. 146–53. 10.1016/j.freeradbiomed.2008.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chowdhury I, Fisher AB, Christofidou-Solomidou M, Gao L, Tao JQ, Sorokina EM, et al. Keratinocyte growth factor and glucocorticoid induction of human peroxiredoxin 6 gene expression occur by independent mechanisms that are synergistic. Antioxid Redox Signal. 2014; 20(3): p. 391–402. 10.1089/ars.2012.4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang C-F, Zhang L, Ma S-R, Zhao Z-L, Wang W-M, He K-F, et al. Clinical Significance of Keap1 and Nrf2 in Oral Squamous Cell Carcinoma. PLoS ONE. 2013; 8(12): p. e83479 10.1371/journal.pone.0083479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fatma N, Kubo E, Takamura Y, Ishihara K, Garcia C, Beebe DC, et al. Loss of NF-kappaB control and repression of Prdx6 gene transcription by reactive oxygen species-driven SMAD3-mediated transforming growth factor beta signaling. J Biol Chem. 2009; 284(34): p. 22758–72. 10.1074/jbc.M109.016071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chhunchha B, Fatma N, Kubo E, Rai P, Singh SP, Singh DP. Curcumin abates hypoxia-induced oxidative stress based-ER stress-mediated cell death in mouse hippocampal cells (HT22) by controlling Prdx6 and NF-kappaB regulation. Am J Physiol Cell Physiol. 2013; 304(7): p. C636–55. 10.1152/ajpcell.00345.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chhunchha B, Fatma N, Bhargavan B, Kubo E, Kumar A, Singh DP. Specificity protein, Sp1-mediated increased expression of Prdx6 as a curcumin-induced antioxidant defense in lens epithelial cells against oxidative stress. Cell Death Dis. 2011; 2: p. e234 10.1038/cddis.2011.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxidants & redox signaling. 2011; 15(3): p. 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia X, Veiseth-Kent E, Grove H, Kuziora P, Aass L, Hildrum KI, et al. Peroxiredoxin-6—a potential protein marker for meat tenderness in bovine longissimus thoracis muscle. J Anim Sci. 2009; 87(7): p. 2391–9. 10.2527/jas.2009-1792 [DOI] [PubMed] [Google Scholar]
- 31. Liu Y, Wu W-J, Zuo B, Ren Z-Q, Xiong Y-Z. Polymorphism in coding region of pig PRDX6 gene and its genetic effects analysis. Hereditas (Beijing). 2011; 33(7): p. 743–748. [DOI] [PubMed] [Google Scholar]
- 32. Zhou G X, Wang S B, Wang Z G, Zhu X T, Shu G, Liao W Y, et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Science. 2010; 86(2):440–450. 10.1016/j.meatsci.2010.05.031 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y-X, Li WT, Zhu M-F, Li Y, Xu Z-Y, Zuo B. FHL3 differentially regulates the expression of MyHC isoforms through interactions with MyoD and pCREB. Cellular Signalling. 2016; 28: 60–73 [DOI] [PubMed] [Google Scholar]
- 34. Xia X-L, Yan C, Wu W-J, Zhou Y, Hou L-M, Zuo B, et al. Characterization of the porcine peptidylarginine deiminase type VI gene (PADI6) promoter: Sp1 regulates basal transcription of the porcine PADI6. Gene. 10.1016/j.gene.2015.09.042 [DOI] [PubMed] [Google Scholar]
- 35. Fuchs NV, Kraft M, Tondera C, Hanschmann KM, Lower J, Lower R. Expression of the Human Endogenous Retrovirus (HERV) Group HML-2/HERV-K Does Not Depend on Canonical Promoter Elements but Is Regulated by Transcription Factors Sp1 and Sp3. Journal of Virology. 2011; 85(7): p. 3436–3448. 10.1128/JVI.02539-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. J Johnson P, Williams S. CCAAT/enhancer binding (C/EBP) proteins. Liver gene expression. 1994; 231. [Google Scholar]
- 37. Payne VA, Au WS, Lowe CE, Rahman SM, Friedman JE, O'Rahilly S, et al. , C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem J. 2010; 425(1): p. 215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010; 318(1–2): p. 10–4. 10.1016/j.mce.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gummersbach C, Hemmrich K, Kroncke KD, Suschek CV, Fehsel K, Pallua N. New aspects of adipogenesis: radicals and oxidative stress. Differentiation. 2009; 77(2): p. 115–20. 10.1016/j.diff.2008.09.009 [DOI] [PubMed] [Google Scholar]
- 40. Lee H, Lee YJ, Choi H, Ko EH, Kim JW. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J Biol Chem. 2009; 284(16): p. 10601–9. 10.1074/jbc.M808742200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sands WA, Palmer TM. Palmer, Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008; 20(3): p. 460–6. [DOI] [PubMed] [Google Scholar]
- 42. Carlezon WA Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005; 28(8): p. 436–45. [DOI] [PubMed] [Google Scholar]
- 43. Bedogni B, Pani G, Colavitti R, Riccio A, Borrello S, Murphy M, et al. , Redox regulation of cAMP-responsive element-binding protein and induction of manganous superoxide dismutase in nerve growth factor-dependent cell survival. J Biol Chem. 2003; 278(19): p. 16510–9. [DOI] [PubMed] [Google Scholar]
- 44. Bai J, Nakamura H, Kwon Y-W, Hattori I, Yamaguchi Y, Kim Y-C, et al. , Critical roles of thioredoxin in nerve growth factor-mediated signal transduction and neurite outgrowth in PC12 cells. The Journal of neuroscience. 2003; 23(2): p. 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuloaga R, Fuentes EN, Molina A, Valdes JA. The cAMP response element binding protein (CREB) is activated by insulin-like growth factor-1 (IGF-1) and regulates myostatin gene expression in skeletal myoblast. Biochem Biophys Res Commun. 2013; 440(2): p. 258–64. 10.1016/j.bbrc.2013.09.067 [DOI] [PubMed] [Google Scholar]
- 46. Stewart R, Flechner L, Montminy M, Berdeaux R. CREB is activated by muscle injury and promotes muscle regeneration. PloS one. 2011; 6(9): p. e24714 10.1371/journal.pone.0024714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, et al. , A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009; 106(41): p. 17475–80. 10.1073/pnas.0908641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007; 26(1): p. 1–14. [DOI] [PubMed] [Google Scholar]
- 49. Won H, Lim S, Jang M, Kim Y, Rashid MA, Jyothi KR, et al. , Peroxiredoxin-2 Upregulated by NF-κB Attenuates Oxidative Stress During the Differentiation of Muscle-Derived C2C12 Cells. Antioxidants & Redox Signaling. 2012; 16(3): p. 245–261. [DOI] [PubMed] [Google Scholar]
- 50. Blake M, Jambou R, Swick A, Kahn J, Azizkhan J. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Molecular and Cellular Biology. 1990; 10(12): p. 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.