Models that incorporate both quantitative image features and conventional prognostic factors were found to stratify patients according to overall survival more effectively than models that incorporate conventional prognostic factors alone.
Abstract
Purpose
To determine whether quantitative imaging features from pretreatment positron emission tomography (PET) can enhance patient overall survival risk stratification beyond what can be achieved with conventional prognostic factors in patients with stage III non–small cell lung cancer (NSCLC).
Materials and Methods
The institutional review board approved this retrospective chart review study and waived the requirement to obtain informed consent. The authors retrospectively identified 195 patients with stage III NSCLC treated definitively with radiation therapy between January 2008 and January 2013. All patients underwent pretreatment PET/computed tomography before treatment. Conventional PET metrics, along with histogram, shape and volume, and co-occurrence matrix features, were extracted. Linear predictors of overall survival were developed from leave-one-out cross-validation. Predictive Kaplan-Meier curves were used to compare the linear predictors with both quantitative imaging features and conventional prognostic factors to those generated with conventional prognostic factors alone. The Harrell concordance index was used to quantify the discriminatory power of the linear predictors for survival differences of at least 0, 6, 12, 18, and 24 months. Models were generated with features present in more than 50% of the cross-validation folds.
Results
Linear predictors of overall survival generated with both quantitative imaging features and conventional prognostic factors demonstrated improved risk stratification compared with those generated with conventional prognostic factors alone in terms of log-rank statistic (P = .18 vs P = .0001, respectively) and concordance index (0.62 vs 0.58, respectively). The use of quantitative imaging features selected during cross-validation improved the model using conventional prognostic factors alone (P = .007). Disease solidity and primary tumor energy from the co-occurrence matrix were found to be selected in all folds of cross-validation.
Conclusion
Pretreatment PET features were associated with overall survival when adjusting for conventional prognostic factors in patients with stage III NSCLC.
© RSNA, 2015
Introduction
Physicians have long observed that patients with the same type and stage of cancer often respond differently to the same treatments and ultimately have a wide range of outcomes (1). The concept of “personalized medicine” proposes that patients would benefit from having medical decisions tailored according to characteristics inherent to each individual patient and their underlying disease and not based on population-based risk assessments such as staging. The invention of positron emission tomography (PET) with fluorodeoxyglucose has enabled the quantification of tumor metabolism by means of standardized uptake value (SUV) measurements and is now widely used in clinical practice for a variety of solid tumors (2). A number of publications have demonstrated the prognostic value of tumor PET SUV measurements in non–small cell lung cancer (NSCLC) (3–5). Furthermore, pretreatment PET is now considered the standard of care for patients with NSCLC; therefore, any prognostic information from these scans can be extracted without additional cost, time, or radiation dose to the patient (6). Most researchers have investigated the prognostic ability of PET metrics such as mean SUV, maximum SUV, and metabolic tumor volume but did not quantify metrics indicative of fluorodeoxyglucose uptake distribution and heterogeneity. Many of these investigations have demonstrated the utility of SUV measurements obtained either after treatment or by measuring changes from pre- to posttreatment scans (4,7). However, prognostic models that solely use pretreatment scans may ultimately be more useful because clinicians could then use this information to make decisions with regard to upfront treatment. Recently, studies have suggested that the examination of spatial heterogeneity with computed tomography (CT) and the use of SUVs within solid tumors (particularly NSCLC) may provide prognostic information (8–12). Although these studies have generated compelling early evidence that heterogeneous tumors may lead to inferior outcomes, they have lacked adjustment for known prognostic factors and the use of proper validation techniques.
We performed this study to determine whether quantitative imaging features from pretreatment PET can enhance overall survival risk stratification beyond what can be achieved with conventional prognostic factors in patients with stage III NSCLC.
Materials and Methods
Patients
The institutional review board approved this retrospective chart review study and waived the requirement to obtain informed consent. We complied with all Health Insurance Portability and Accountability Act regulations. The medical records of patients with stage III NSCLC treated definitively with external-beam radiation therapy between January 2008 and January 2013 were retrospectively reviewed by D.V.F. (5 years of experience). These dates were chosen for two reasons: (a) to ensure that patients’ PET scans underwent three-dimensional reconstruction, which our institution implemented in 2008, and (b) to ensure that patients had a minimum potential follow-up of 1 year at the time of analysis. We excluded 30 patients because they had undergone treatment for another solid tumor less than 5 years earlier, had multiple primary lesions, or had primary lesions with tumor volumes of less than 5 mL as measured with PET. Those with primary tumor volumes less than 5 mL were excluded because we have observed in preliminary work that quantitative features extracted from these lesions yield less reproducible and consistent results compared with those in larger tumors. This yielded 195 patients for analysis (mean age, 65 years). The conventional prognostic factors and treatment characteristics are listed in Table 1. There was no significant difference between the median ages of men (median age, 65 years; age range, 40–88 years) and women (median age, 67 years; age range, 38–82 years) by using a Wilcoxon rank sum test (P = .23). The median follow-up for all living patients was 37 months (range, 3–70 months). Three patients were lost to follow-up before 1 year. Overall survival was measured as the time from the initiation of treatment until death in months. Treatment initiation was defined as the first cycle of chemotherapy for patients receiving induction chemotherapy or the first day of radiation treatment for patients receiving radiation therapy upfront. Patients not experiencing an event were censored at the last known follow-up date.
Table 1.
Patient Conventional Prognostic Factors and Treatment Characteristics
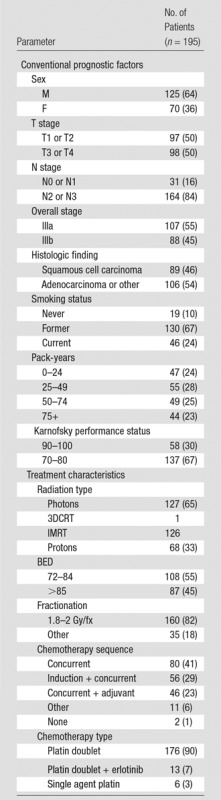
Note.—The mean patient age was 66 years. Numbers in parentheses are percentages. BED = biological equivalent dose, fx = fraction, IMRT = intensity-modulated radiation therapy, 3DCRT = three-dimensional conformal radiation therapy.
Imaging
All patients underwent PET/CT before initiation of treatment. PET/CT scans were obtained with either a Discovery RX or STE scanner (GE Healthcare, Waukesha, Wis). All images were reconstructed with three-dimensional ordered subset expectation maximization by using two iterations, 20–21 subsets, and a 6-mm postprocessing Gaussian blurring filter. All images were composed of 128 × 128 pixels with voxel dimensions of 5.47 × 5.47 × 3.27 mm. Patients fasted for at least 6 hours before administration of an average injected dose of 381 Mbq (range, 255–540 Mbq). The average duration from injection to scanning was 78 minutes (range, 50–124 minutes). Low-dose unenhanced CT was performed for attenuation correction by using 120 kVp, automated milliampere modulation, a pitch of 1.35, and 3.75-mm-thick sections.
Conventional Prognostic Factors
We obtained the patient’s T stage (T1 or T2 vs T3 or T4), N stage (N0 or N1 vs N2 or N3), overall stage (IIIa vs IIIb), age, sex, histologic findings (squamous cell carcinoma vs other), Karnofsky performance status (score, 90–100 vs <90), smoking status (current, former, never), estimated pack-years, use of induction chemotherapy, and gross tumor volume from the medical records. These factors were included because they have all been suggested to be prognostic in stage III NSCLC (13). All TNM staging was performed according to the 7th edition of the American Joint Committee on Cancer Staging Manual (14). Gross tumor volume consisted of both the primary and nodal disease as defined by the treating radiation oncologist for definitive radiation therapy. The gross tumor volume was transformed before modeling by using the logarithm to the base.
Quantitative Analysis
The patient’s primary and nodal tumor volumes were delineated by using the PETedge feature from MIM software (version 6.2; MIM Software, Cleveland, Ohio). This method was chosen because it was found in a review by Werner-Wasik et al (15) to be the most accurate and consistent technique for target volume contouring for lung cancer lesions on PET scans. In addition, the PETedge algorithm is semiautomated and thus capable of higher throughput than manual contouring. Images and their corresponding contours were imported into our in-house quantitative image software (IBEX), which was designed by using a commercial software package (Matlab, version 8.1.0; MathWorks, Natick, Mass) (16). IBEX extracts quantitative image features from delineated regions of interest. We used diagnostic radiologists’ notes from the medical record along with contours from the radiation treatment plan to determine the location of the primary tumor and nodal disease.
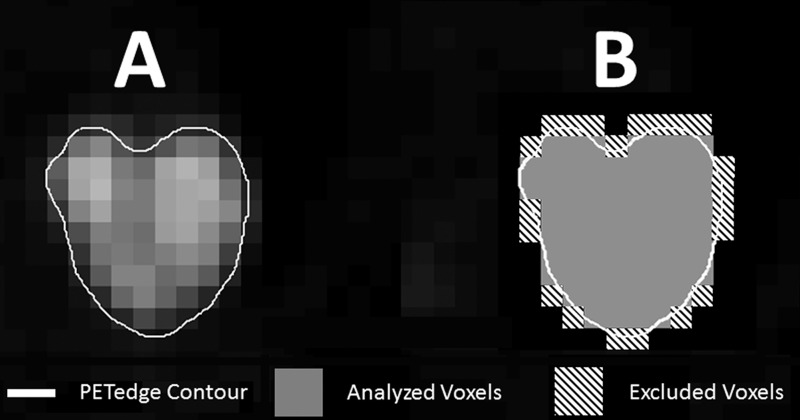
Because the resolution of tumor contours was on a 512 × 512 image grid and our images were only 128 × 128, we designed a resampling algorithm that measured what percentage of the image voxel was included in the tumor contour. This was used to determine which voxels should be analyzed and which should be excluded (Fig 1, B). All voxels with less than 50% of their volume within the contour were excluded. This algorithm was needed because IBEX is written such that any voxel containing any portion of the contour, no matter how small, is included in the binary mask used to determine which voxels should be used for analysis.
Figure 1:
Illustration of voxel exclusion based on percentage of voxel within the PETedge contour. A, Original contour. B, Illustration of analyzed and excluded voxels.
The contours were delineated (by D.V.F., with 5 years of thoracic contouring experience with CT and PET) and used to calculate eight histogram features, four co-occurrence matrix features, and four shape or volume features (Fig 2) (17). Surface area and convex hull volume were transformed by using the logarithm to the base 2 before model development. For the co-occurrence matrix features, images were first scaled to the number of gray levels between the minimum and maximum of the tumor SUV by using the minimum and maximum as the gray level limits. This effectively rounded the SUVs within the contour to the nearest whole number and subtracted the minimum SUV. For example, a lesion with a minimum SUV of 3.2 and a maximum SUV of 17.8 would first be scaled to be comprised of values ranging from 3 to 18 and then the minimum value (ie, 3) subtracted, resulting in values from 0 to 15. This allowed for the analyses to have a finite number of gray levels and ensured that the new scaled values had a consistent relationship to the underlying SUVs (ie, a difference of one between scaled values represented an SUV change of one). By subtracting the minimum SUV, the co-occurrence matrix features were calculated by using variability in uptake regardless of underlying amplitude. Other metrics (eg, maximum and mean SUV) were used as metrics to quantify amplitude of uptake. The co-occurrence matrix features were computed by generating the co-occurrence matrix for each of the four unique two-dimensional directions (up, up-right, right, down-right) across the entire tumor in a one-voxel neighborhood. These four directions are unique in that their reflections would generate the same information. Once calculated, these matrices are added together, made symmetric across the diagonal, and normalized to sum to one. We chose to use only the two-dimensional directions rather than each three-dimensional direction to both represent the entire tumor and prevent the anisotropic voxel sizes from having an effect (ie, adjacent voxels in the x-y plane have a larger distance than voxels between image sections).
Figure 2:
Extracted quantitative PET features. * = Features were calculated for primary disease, nodal disease, and total disease (primary plus nodal). † = Feature was only calculated for nodal disease and total disease (primary plus nodal).
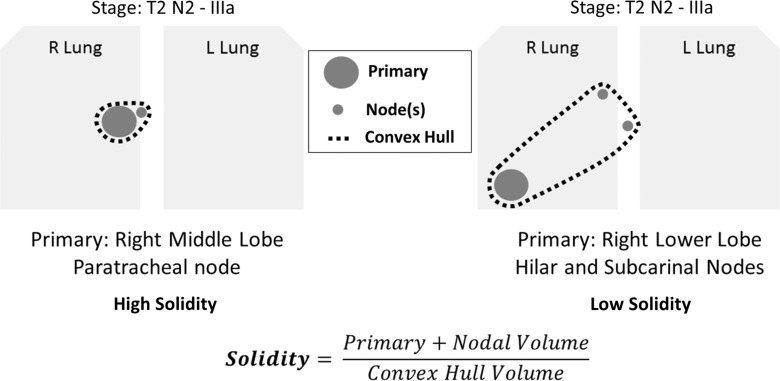
The peak SUV was defined as the average of the 3 × 3-pixel neighborhood surrounding the maximum SUV (18). The convex hull volume was defined as the volume of the smallest convex shape that could encompass the disease being examined as defined by the PETedge tool. Solidity was defined as the disease volume divided by the convex hull volume; therefore, a lower value for solidity was indicative of the disease being more dispersed (Fig 3).
Figure 3:
Illustration of solidity metric.
Statistical Analysis
All statistical analyses were performed in R 3.0.2 with the following R packages: survival v (version 2.37-4), penalized (version 0.9-42), and survcomp v (version 1.10.0) (R Foundation, Vienna, Austria). Eleven conventional prognostic factors and 27 quantitative imaging features (features calculated for primary, nodal, and total disease were defined as separate features) extracted from each patient’s pretreatment PET scan were entered into a penalized multivariate Cox proportional hazards model. Development of cross-validated predictions by using conventional prognostic factors alone and conventional prognostic factors plus quantitative imaging features was performed separately. This modeling framework simultaneously carries out covariate selection alongside model development. The penalization is directed by the L1 penalty parameter, which balances model fit and model complexity. The penalty parameter is determined by maximizing the cross-validated likelihood. The R penalized package standardizes all covariates by their unit central L2 norm before penalization to minimize the influence of covariates that are off vastly different scales. The model coefficients are subsequently rescaled to reflect their original magnitudes.
To adjust for the bias associated with training and testing a model on the same internal dataset, we predominantly used methods suggested by Simon et al (19) to generate cross-validated Kaplan-Meier curves. This method allows a reasonable estimate for out-of-sample performance of our models while using only an internal dataset (20). Cross-validated Kaplan-Meier curves are generated by using model predictions for patients that are derived from models developed without the patients’ inclusion in model training. When performing leave-one-out cross-validation, a patient is left out of model development and a prediction for this patient is generating by using the remaining cohort. The patient that is left out is changed and this process repeated such that each observation in the sample has a prediction from when it was not involved in model development. These predictions (we used the linear predictor generated during each fold of cross-validation) are used to stratify patients into risk groups. The linear predictor is defined as the sum of each of the model coefficient times the corresponding covariate value of that specific patient. Therefore, the higher the linear predictor the higher the predicted risk. The generated linear predictors for each patient from the fold in which they were left out of model development were categorized into one of five groups by using k-means clustering. Rather than splitting data by the median or quartiles, the k-means clustering considers the underlying distribution of patient predictions and therefore generates groups that are more homogeneous in their predictions. We believe this approach allows the resulting Kaplan-Meier curves to better illustrate the calibration between actual and predicted survival. The Harrell concordance index was calculated by using all 195 patients and subsequently only for patient pairs with survival differences of at least 6, 12, 18, and 24 months. The use of survival differences other than zero was done to allow for clinical assessment of what survival difference is “meaningful.” For instance, we would expect a model’s ability to differentiate between two patients whose survivals are vastly different to be superior to its ability to discriminate between two patients whose survivals are only separated by a few months. A likelihood ratio test was performed to assess the significance of adding the quantitative imaging features that were selected during cross-validation (solidity and co-occurrence matrix energy) to the model developed with use of conventional prognostic factors alone. To generate an overall model for conventional prognostic factors and the combination of conventional prognostic factors and quantitative imaging features, we fit a Cox proportional hazards models with covariates that were included in more than 50% of the 195 folds.
Results
The patient conventional prognostic factors and characteristics of patient treatment are listed in Table 1.
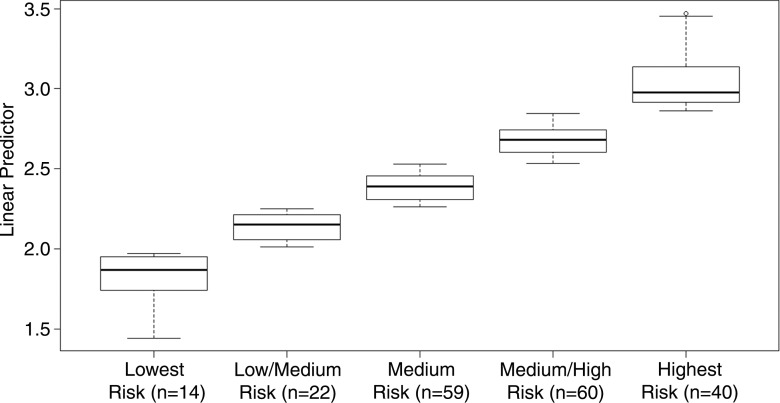
A k-means clustering illustrating the divisions of the linear predictors into lowest- to highest-risk groups by using both conventional prognostic factors and quantitative imaging features is illustrated in Figure 4. Kaplan-Meier plots comparing conventional prognostic factors models with conventional prognostic factors and quantitative imaging features models are shown in Figure 5, A and B. Figure 5, C, compares the concordance index between models with and models without quantitative imaging features, with patient survival differences ranging from 0 to 24 months.
Figure 4:
Box-and-whisker plot shows k-means clustering of linear predictor from cross-validation. Whiskers are maximum of 1.5 × interquartile range.
Figure 5:
A, Graph of Kaplan-Meier risk groups based on linear predictors from conventional prognostic factors. B, Graph of Kaplan-Meier risk groups based on linear predictors from conventional prognostic factors and quantitative imaging features. C, Concordance index (C-Index) values from both models at various time points. CPFs = conventional prognostic factors, CV = cross-validation, QIFs = quantitative imaging features.
According to the log-rank statistic, the separation between curves is much better defined in the model with quantitative imaging features than in the model with conventional prognostic factors alone (P = .18 vs P = .0001, respectively). Furthermore, the concordance index at all patient survival differences is higher in models generated with quantitative imaging features than with models generated by using conventional prognostic factors alone. The addition of solidity and co-occurrence matrix energy to the conventional prognostic factors alone model yielded a significant improvement with the likelihood ratio test (P = .007). Models that used factors that were selected in more than 50% of the cross-validation folds are shown in Table 2.
Table 2.
Comparison of Model with Conventional Prognostic Factors Alone and Model with Conventional Prognostic Factors Plus Quantitative Imaging Features
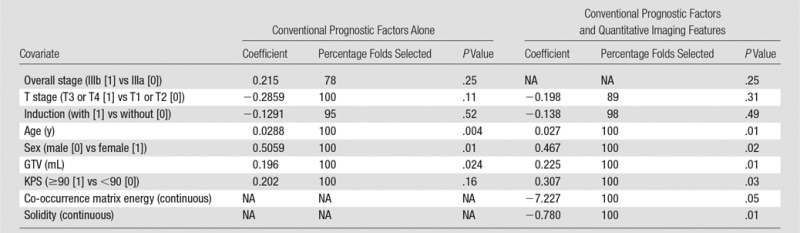
Note.—GTV = gross tumor volume, KPS = Karnofsky performance status, NA = not applicable.
Discussion
We were able to demonstrate that the incorporation of pretreatment PET quantitative imaging features alongside conventional prognostic factors in survival models enabled improved model fit and better stratification of patients in terms of overall survival compared with models that used conventional prognostic factors alone. The use of cross-validation allows the use of all data in both training and testing and thus is more efficient than splitting data into independent test and validation sets. The results from cross-validation should more aptly reflect how the model would perform in an independent cohort comprised of similar patients.
Recent data have suggested that quantification of intratumoral heterogeneity may yield information that could improve prediction of response and/or prognosis in patients with NSCLC (9–11). It is hypothesized that tumor heterogeneity in fluorodeoxyglucose PET tracer uptake may reflect underlying tumor biology such as hypoxia, angiogenesis, and necrosis (21). Therefore, these methods could be used to identify tumors that are predisposed to aggressive behavior. In NSCLC specifically, preliminary data exist that suggest a relationship between quantitative imaging features and patient outcome (9,11). However, these studies do not sufficiently adjust for conventional prognostic factors when assessing the significance of new quantitative imaging features. This study is unique in that substantial effort was made to generate multivariate models that implement both quantitative imaging features and conventional prognostic factors to assess the added benefit of quantitative imaging features to models that use conventional prognostic factors. Furthermore, models frequently use cohorts comprised of patients with varying stages of disease, whereas our cohort is comprised solely of patients with stage III NSCLC. Models capable of stratifying patients with a homogeneous disease stage may be more clinically useful because different stages of disease frequently dictate different treatment courses. Furthermore, solidity and co-occurrence matrix energy were consistently selected during cross-validation; conventional PET metrics such as mean and maximum SUV and metabolic tumor volume were not selected with nearly the same frequency. This suggests that our quantitative imaging features that examine spatial heterogeneity of uptake may be more useful than conventional PET metrics when adjusting for conventional prognostic factors. Solidity in our study quantifies how dispersed the primary and nodal disease are in a local region context (all patients with stage III NSCLC). Co-occurrence matrix energy quantifies the uniformity of the SUVs within the primary tumor while taking into account the spatial orientation of the voxels. The co-occurrence matrix energy metric is calculated by determining the probabilities for different voxel-adjacent voxel pairs within the tumor, squaring these values, and summing them together. Therefore, a completely uniform tumor would have a co-occurrence matrix energy of 1 whereas a heterogeneous tumor where few adjacent voxels have the same SUV would have a co-occurrence matrix energy value that is very small.
The use of quantitative imaging features from routinely obtained images has the potential to provide beneficial information to clinicians and their patients without any added expense or radiation exposure. Pretreatment risk stratification could enable clinicians to deliver more patient-specific treatment tailored to their individual risk. Particularly in our cohort of patients with advanced NSCLC, accurate predictions might aid in determining the appropriate level of treatment aggressiveness and maintaining as much of a patient’s quality of life as possible. In addition, more accurate models could ensure more balanced and/or appropriate treatment arms in prospective trials.
If the use of quantitative imaging features are ever going to develop outside the realm of academic research and into the clinic, large-scale validated studies that examine uniformly calculated metrics are needed. IBEX provides a framework that researchers across institutions can use to compare metrics that are consistently defined and calculated.
Models that include quantitative imaging features have been shown to have significant potential; however, a few limitations should be noted. First, most of the evidence for the association of quantitative imaging features (including the evidence in this study) comes from retrospective reviews and not from prospective assessment. In addition, to generate sizable cohorts, several studies have used patient data acquired on a variety of scanners that implement various and/or outdated reconstruction parameters (eg, differing voxel sizes or use of two-dimensional reconstruction). Recently, the literature has shown that these differences have a significant impact on the reproducibility of the extracted quantitative imaging features (22–25). Leijenaar et al (24) analyzed the reproducibility of quantitative imaging features by using test-retest scans and specifically found that the co-occurrence matrix energy feature had an intraclass correlation coefficient of 0.96. This suggests that this feature may be “portable” enough to have a broad implementation. Tumor delineation on PET images is also less than straightforward. Many delineation methods exist, such as manual contouring, value thresholding, percentage thresholding, and a variety of other semiautomated techniques.
In our study, we used a semiautomated gradient technique because a review by Werner-Wasik et al (15) found this method to be the most robust in terms of accuracy and consistency for NSCLC tumors. Variations in tumor delineation could easily influence the extracted quantitative imaging features. Another issue to be cognizant of is the issue of tumor volume. A volume threshold below which quantitative features cannot be accurately and/or reproducibility measured is an area that requires further investigation. A couple of publications have examined this issue, but a consensus on size limitation has not been reached to ensure adequate sampling (22,26). We have done preliminary work examining this issue and found that feature reproducibility with features from 5-mL tumors (approximately 90 voxels) was similar to that with much larger lesions.
Our work has several limitations. First, our data are retrospective and results are derived only from a single institution cohort and are thus hypothesis generating. Proper validation with use of a separate cohort of patients is needed. Second, we originally considered 27 distinct quantitative imaging features and did not perform our analysis solely by using the two quantitative imaging features found to have an association. This may have led to overly optimistic results and P values. However, the use of cross-validation for simultaneous multivariate selection of these features should provide a better assessment of predictive model fit than resubstitution statistics.
Accurate knowledge of a patient’s prognosis is a valuable tool in medicine, particularly in oncology. We demonstrated that quantitative imaging features extracted from pretreatment PET images could enhance stratification of patients on the basis of overall survival compared with conventional prognostic factors. Appropriate use of these models could aid the treating clinicians and the patients themselves.
Advance in Knowledge
■ Fluorodeoxyglucose positron emission tomography (PET)–based quantitative image features improved the fit of our developed survival model in patients with stage III non–small cell lung cancer compared with the use of only conventional prognostic factors (P = .001).
Implication for Patient Care
■ This work provides preliminary evidence that quantitative image features from fluorodeoxyglucose PET may have the potential to improve prognosis assessment for patients and their caregivers.
Received December 18, 2014; revision requested February 13, 2015; revision received April 6; accepted April 29; final version accepted May 8.
Supported in part by the American Legion Auxiliary, American Association of Physicists in Medicine Graduate Fellowship, and University of Texas Graduate School of Biomedical Sciences at Houston.
Funding: This research was supported by the National Institutes of Health (grant CA 16672).
Disclosures of Conflicts of Interest: D.V.F. disclosed no relevant relationships. O.M. disclosed no relevant relationships. L.Z. disclosed no relevant relationships. X.F. disclosed no relevant relationships. S.Z. disclosed no relevant relationships. G.I. disclosed no relevant relationships. Z.L. disclosed no relevant relationships. L.E.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: received grants from NCI, Varian Medical Systems, and Elekta Medical Systems. Other relationships: disclosed no relevant relationships.
Abbreviations:
- NSCLC
- non–small cell lung cancer
- SUV
- standardized uptake value
References
- 1.Dehing-Oberije C, Yu S, De Ruysscher D, et al. Development and external validation of prognostic model for 2-year survival of non–small-cell lung cancer patients treated with chemoradiotherapy. Int J Radiat Oncol Biol Phys 2009;74(2):355–362. [DOI] [PubMed] [Google Scholar]
- 2.MacManus M, Nestle U, Rosenzweig KE, et al. Use of PET and PET/CT for radiation therapy planning: IAEA expert report 2006–2007. Radiother Oncol 2009;91(1):85–94. [DOI] [PubMed] [Google Scholar]
- 3.Berghmans T, Dusart M, Paesmans M, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non–small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol 2008;3(1):6–12. [DOI] [PubMed] [Google Scholar]
- 4.Machtay M, Duan F, Siegel BA, et al. Prediction of survival by [18F]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: results of the ACRIN 6668/RTOG 0235 trial. J Clin Oncol 2013;31(30):3823–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Im HJ, Pak K, Cheon GJ, et al. Prognostic value of volumetric parameters of (18)F-FDG PET in non–small-cell lung cancer: a meta-analysis. Eur J Nucl Med Mol Imaging 2015;42(2):241–251. [DOI] [PubMed] [Google Scholar]
- 6.Al-Jahdali H, Khan AN, Loutfi S, Al-Harbi AS. Guidelines for the role of FDG-PET/CT in lung cancer management. J Infect Public Health 2012;5(Suppl 1):S35–S40. [DOI] [PubMed] [Google Scholar]
- 7.de Geus-Oei LF, van der Heijden HF, Corstens FH, Oyen WJ. Predictive and prognostic value of FDG-PET in nonsmall-cell lung cancer: a systematic review. Cancer 2007;110(8):1654–1664. [DOI] [PubMed] [Google Scholar]
- 8.Fried DV, Tucker SL, Zhou S, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non–small cell lung cancer. Int J Radiat Oncol Biol Phys 2014;90(4):834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang SR, Song HC, Byun BH, et al. Intratumoral metabolic heterogeneity for prediction of disease progression after concurrent chemoradiotherapy in patients with inoperable stage III non–small-cell lung cancer. Nucl Med Mol Imaging 2014;48(1):16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tixier F, Hatt M, Valla C, et al. Visual versus quantitative assessment of intratumor 18F-FDG PET uptake heterogeneity: prognostic value in non–small cell lung cancer. J Nucl Med 2014;55(8):1235–1241. [DOI] [PubMed] [Google Scholar]
- 11.Cook GJ, Yip C, Siddique M, et al. Are pretreatment 18F-FDG PET tumor textural features in non–small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med 2013;54(1):19–26. [DOI] [PubMed] [Google Scholar]
- 12.van Gómez López O, García Vicente AM, Honguero Martínez AF, et al. Heterogeneity in [18F]fluorodeoxyglucose positron emission tomography/computed tomography of non–small cell lung carcinoma and its relationship to metabolic parameters and pathologic staging. Mol Imaging 2014;13(0):1–12. [DOI] [PubMed] [Google Scholar]
- 13.Berghmans T, Paesmans M, Sculier JP. Prognostic factors in stage III non–small cell lung cancer: a review of conventional, metabolic and new biological variables. Ther Adv Med Oncol 2011;3(3):127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egner JR. AJCC cancer staging manual. JAMA 2010;304(15):1726–1727. [Google Scholar]
- 15.Werner-Wasik M, Nelson AD, Choi W, et al. What is the best way to contour lung tumors on PET scans? Multiobserver validation of a gradient-based method using a NSCLC digital PET phantom. Int J Radiat Oncol Biol Phys 2012;82(3):1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42(3):1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;SMC-3(6):610–621. [Google Scholar]
- 18.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med 2009;50(Suppl 1):122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform 2011;12(3):203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fushiki T. Estimation of prediction error by using K-fold cross-validation. Stat Comput 2011;21(2):137–146. [Google Scholar]
- 21.Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non–small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266(1):326–336. [DOI] [PubMed] [Google Scholar]
- 22.Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med 2014;55(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010;49(7):1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leijenaar RT, Carvalho S, Velazquez ER, et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 2013;52(7):1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med 2012;53(5):693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatt M, Cheze-le Rest C, van Baardwijk A, Lambin P, Pradier O, Visvikis D. Impact of tumor size and tracer uptake heterogeneity in (18)F-FDG PET and CT non–small cell lung cancer tumor delineation. J Nucl Med 2011;52(11):1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]