Abstract
Bioorthogonal reactions that are fast and reversible under physiologic conditions are in high demand for biological applications. Herein, we show that an ortho boronic acid substituent makes aryl ketones to rapidly conjugate with α-nucleophiles at neutral pH. Specifically, 2-acetylphenylboronic acid and derivatives were found to conjugate with phenylhydrazine with rate constants of 102 to 103 M−1 s−1, comparable to the fastest bioorthogonal conjugations known to date. 11B-NMR analysis reveals varied extent of iminoboronate formation of the conjugates, in which the imine nitrogen forms a dative bond with boron. The iminoboronate formation activates the imines for hydrolysis and exchange, rendering these oxime/hydrazone conjugations reversible and dynamic under physiologic conditions. The fast and dynamic nature of the iminoboronate chemistry should find wide applications in biology.
Keywords: iminoboronate, oxime, hydrazone, dynamic combinatorial chemistry, α-nucleophiles
Bioorthogonal reactions prove to be powerful tools for the interrogation of complex biological systems.[1] Among the earliest known bioorthogonal reactions, the hydrazone/oxime chemistry has found limited applications due to the slow kinetics under physiologic conditions. Elegant work from several groups has identified aniline-based catalysts that greatly accelerate the hydrazone and oxime conjugation chemistry.[2] However, the high concentrations (up to 100 mM) of catalysts needed are less ideal for broad applications in biology.
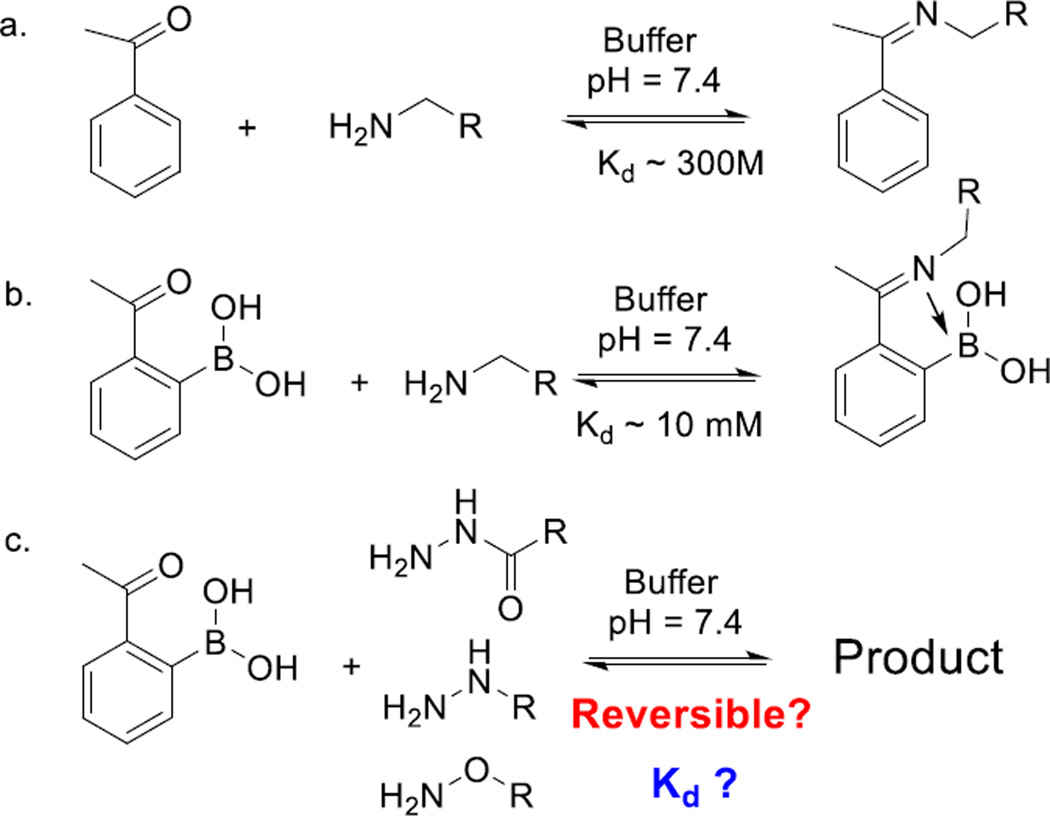
Iminoboronate refers to structures where a dative bond being formed between an imine nitrogen and an electron-deficient boron. We[3] and others[4] have shown that, in comparison to simple imines,[5] iminoboronates display much improved thermodynamic stability and allows labeling of biological amines at low millimolar concentrations (Scheme 1a, b). In this contribution, we extend our investigation of the iminoboronate chemistry to α-nucleophiles (Scheme 1c). Our results show that aryl ketones with an ortho-boronic acid rapidly conjugate with α-nucleophiles. Further, due to the iminoboronate formation, the reactions are reversible and the conjugates display varied thermodynamic stability depending on the nucleophile. Although a number of oxime/hydrazone structures with adjacent boronic acids or esters exist in literature,[6] the thermodynamic and kinetic details of these conjugation reactions have not been previously elucidated, save one recent report by Gillingham and coworkers.[7]
Scheme 1.
Comparative overview of imine chemistry. a) Imine formation of a primary amine with acetophenone. b) Iminoboronate formation of a primary amine with 2-acetylphenylboronic acid (2-APBA) showing much improved thermodynamic propensity for conjugation. c) Conjugation of 2-APBA with α-ncuelophiles, which is the focus of this contribution.
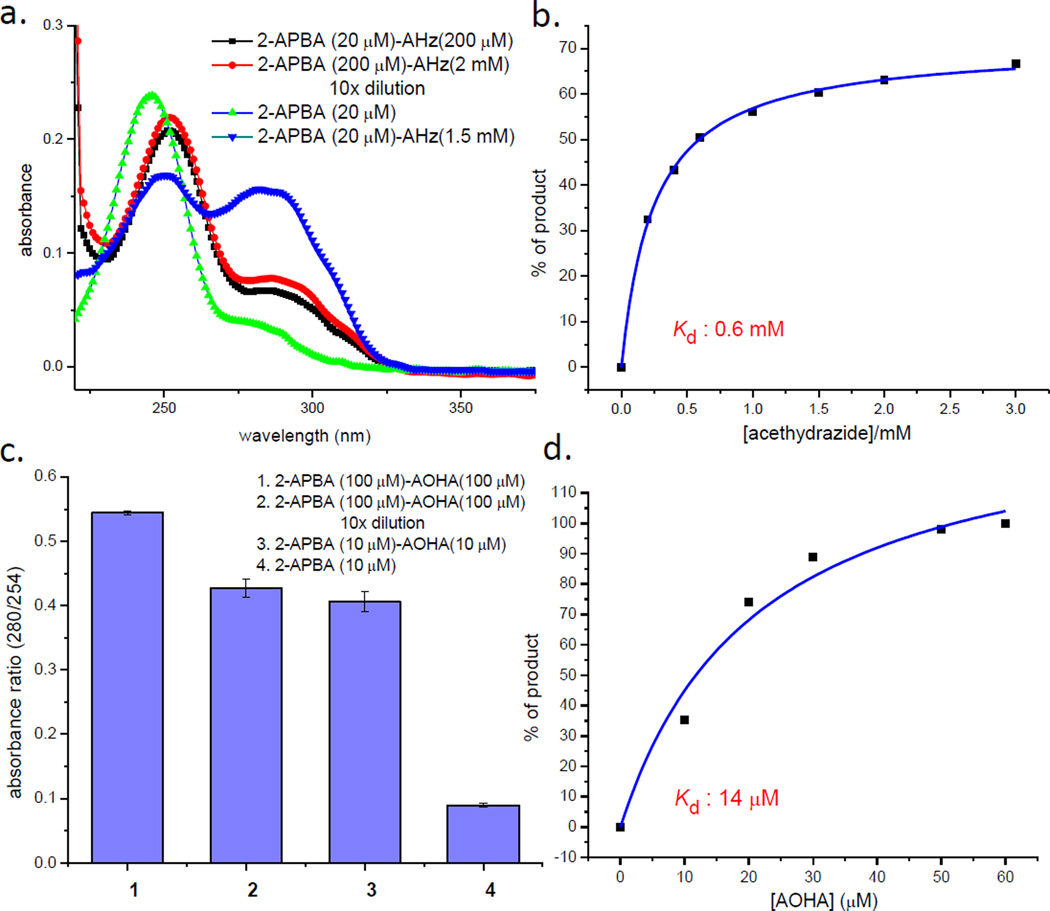
To study the iminoboronate chemistry of α-nucleophiles, we chose 2-acetylphenylboronic acid (2-APBA) as a model compound due to its commercial availability, and also due to its stability in comparison to the corresponding aldehydes. The conjugation of 2-APBA to various nucleophiles was monitored via UV-vis spectroscopy, NMR spectroscopy and mass spectrometry. 2-APBA alone in a neutral buffer displays an absorption peak at 254 nm. Mixing with an excess amount of acethydrazide gives a new peak at 290 nm (Figure 1a) and formation of the acylhydrazone is corroborated by the NMR and mass-spec characterizations (Figure S1). This conjugation reaction reaches equilibrium extremely fast (< 10 s) under the experimental conditions, which makes it difficult to measure the exact reaction rate. However, our data suggest that the reaction rate should be on the order of 103 M−1 s−1 or higher. The fast conjugation contrasts sharply to previous reports where engineered proteins presenting m- or p-acetyl phenylalanines were found to conjugate with a hydrazide only with extended incubation (>12 hrs).[8] To assess the reversibility of the reaction, we diluted a mixture of 2-APBA and acethydrazide (200 µM and 2.0 mM respectively) by 10 folds. The diluted sample gives essentially identical UV-vis spectrum as the one prepared by directly mixing 20 µM 2-APBA and 0.20 mM acethydrazide (Figure 1a), demonstrating the rapid reversibility of the reaction. The binding affinity of 2-APBA to acethydrazide was assessed by using UV-vis (Figure S2) and 1H-NMR spectroscopy (Figure 1b), which yielded comparable dissociation constants in the sub-millimolar range (Kd: ~0.6 mM). In comparison to primary amines, which typically conjugate with 2-APBA at low millimolar concentrations,[3] acethydrazide displays a higher propensity for conjugation with 2-APBA. Similar behavior was observed for benzhydrazide, for which a Kd value of 0.5 mM was obtained (Figure S3).
Figure 1.
Conjugation of 2-APBA to hydrazides and oxyamines. a) UV-vis characterization of the 2-APBA-acethydrazide conjugation demonstrating its quick reversibility; AHz: acethydrazide. b) Titration curve of the acylhydrazone formation generated via 1H-NMR. c) UV-vis characterization of the oxime formation of 2-APBA demonstrating its quick reversibility. d) Titration curve of the oxime formation of 2-APBA generated via UV-vis measurements.
2-APBA was found to rapidly conjugate with an alkoxyamine (6-aminoxy hexanoic acid, AOHA) to form oximes as well. NMR and mass-spec characterization reveals that, when AOHA is in excess, 2-APBA is completely converted into the oxime conjugate (Figure S4a–c). A dilution experiment shows that the oxime formation is rapidly reversible (Figure 1c), suggesting that the boronic acid activates the oxime for hydrolysis. The quick hydrolysis was also clearly seen during HPLC analysis of the oxime, in which 2-APBA was regenerated (Figure S4f). The fast reversibility differs dramatically from oximes without such boronic acid substituent, which typically display hydrolysis rates of 10−6 s−1 at neutral pH.[9] Binding curves of 2-APBA and AOHA were generated by monitoring the changes of UV-vis absorption (Figure 1d and Figure S4d). Data fitting yields a Kd of ~14 µM, showcasing a more favorable equilibrium for the formation of oximes than acylhydrazones.
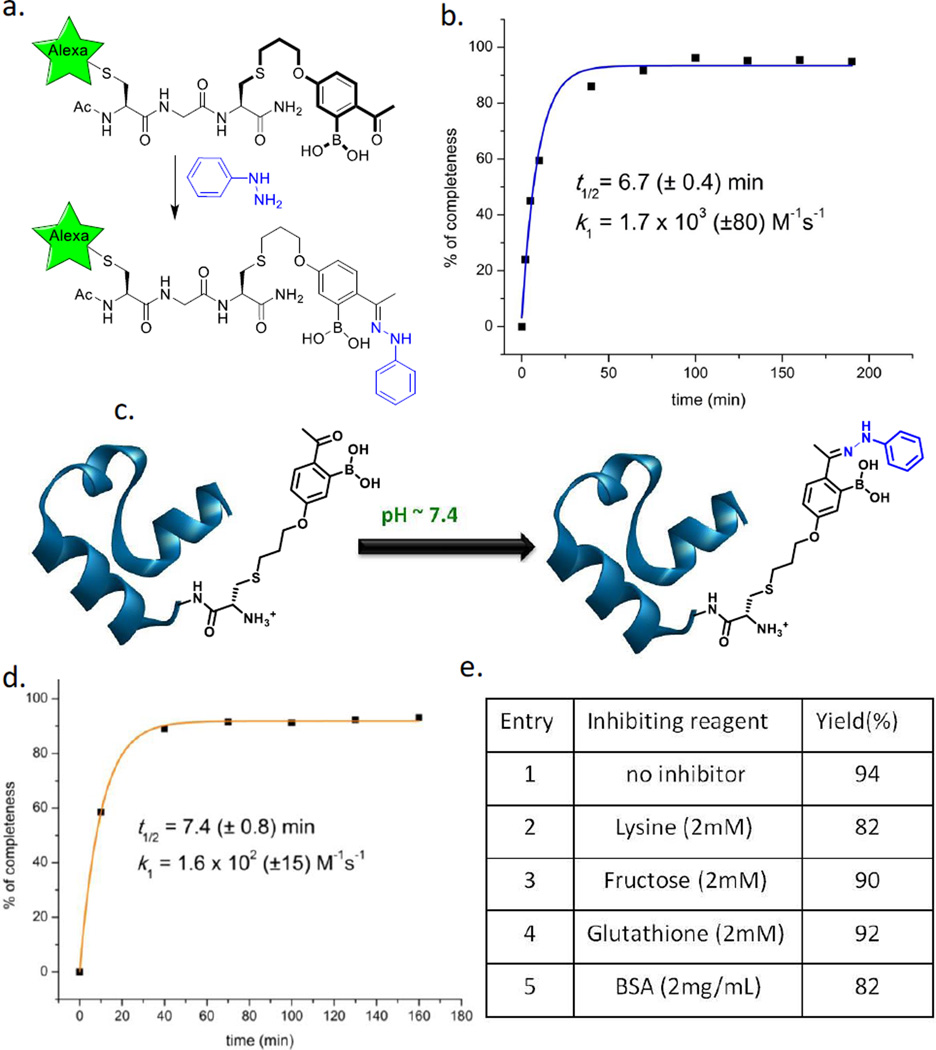
Similar to the oxime formation, 2-APBA readily conjugates with phenylhydrazine (Phzn) to give a hydrazone as the single product (Figure S5). The hydrazone formation proceeds at even lower concentrations of the reactants: 2-APBA was close to be fully converted to the hydrazone even with the reactants at 10 µM (Figure S5b). To analyze the reaction at lower concentrations, we incorporated the 2-APBA structure into a fluorophore labeled peptide by using an unnatural amino acid (dubbed AB1) that displays the 2-APBA structure as its side chain (Figure 2a). The synthesis of AB1 was recently reported by our group in the application of iminoboronate chemistry for bacterial imaging.[3] A short peptide CGAB1 with the sequence of Ac-C*G(AB1)-NH2 was synthesized, where C* denotes a cysteine labeled with Alexa Fluor 488 (AF488, Figure 2a). The conjugation reactions, with 0.2 µM peptide and Phzn over a range of concentrations, were analyzed by using LC-MS. With 1 µM Phzn, CGAB1 was over 90% labeled (Figure S6), demonstrating the high propensity of 2-APBA to conjugate with Phzn to form hydrazones. The peptide labeling was half completed within 7 min with 1 µM Phzn (Figure 2b). Fitting the kinetic data yielded a rate constant (k1) of 1.7 × 103 (±80) M−1 s−1, which compares well to the fastest ligation reactions known in a neutral buffer.[10] Data fitting also revealed a rate constant (k−1) of 1.2 × 10−4 s−1 for hydrolysis, which is ~ 100 times faster than structurally similar hydrazones without the boronic acid.[2c] In terms of the thermodynamic equilibrium, the hydrazone formation gives a Kd value of 0.07 µM (Kd = k−1/k1), which is much more favorable than the formation of oximes and acylhydrazones.
Figure 2.
Fast hydrazone formation of 2-APBA. a) Illustration of peptide (CGAB1) labeling via hydrazone conjugation; the side chain structure of AB1 is highlighted in bold. b) Kinetic profile of peptide labeling with Phzn. c) Illustration of VHP35 labeling via the hydrazone chemistry. d) Kinetic characterization of VHP35 labeling with Phzn. e) Inhibition of VHP35 labeling by abundant biomolecules; the results are tabulated by showing % of labelled proteins.
To further explore the potential of the hydrazone chemistry for biological applications, we have incorporated AB1 into villin headpiece subdomain (VHP35), a small model protein that displays a variety of amino acids.[11] The mutant protein VHP35-AB1 was treated with Phzn and the labeling kinetics was analyzed via LC-MS (Figure 2c and Figure S7). Curve fitting yielded a rate constant of 1.6 × 102 (±15) M−1 s−1 (Figure 2d), which is still comparable to the fastest hydrazone conjugation reactions where aniline catalysts are used.[2c, 2e] The labeling of VHP35 appears to be slower than that of CGAB1, likely due to AB1 being trapped by the protein via hydrophobic effect or conjugation with lysines. We further assessed the inhibitory effect of protein labeling by lysine (2 mM), fructose (2 mM), glutathione (2 mM) and bovine serum albumin (BSA, 2mg/mL) respectively (Figure 2e). The results showed essentially no inhibitory effect for fructose and glutathione. The presence of lysine or BSA only elicited a marginal reduction (<10%) of protein labeling by Phzn as well (Figure S8).
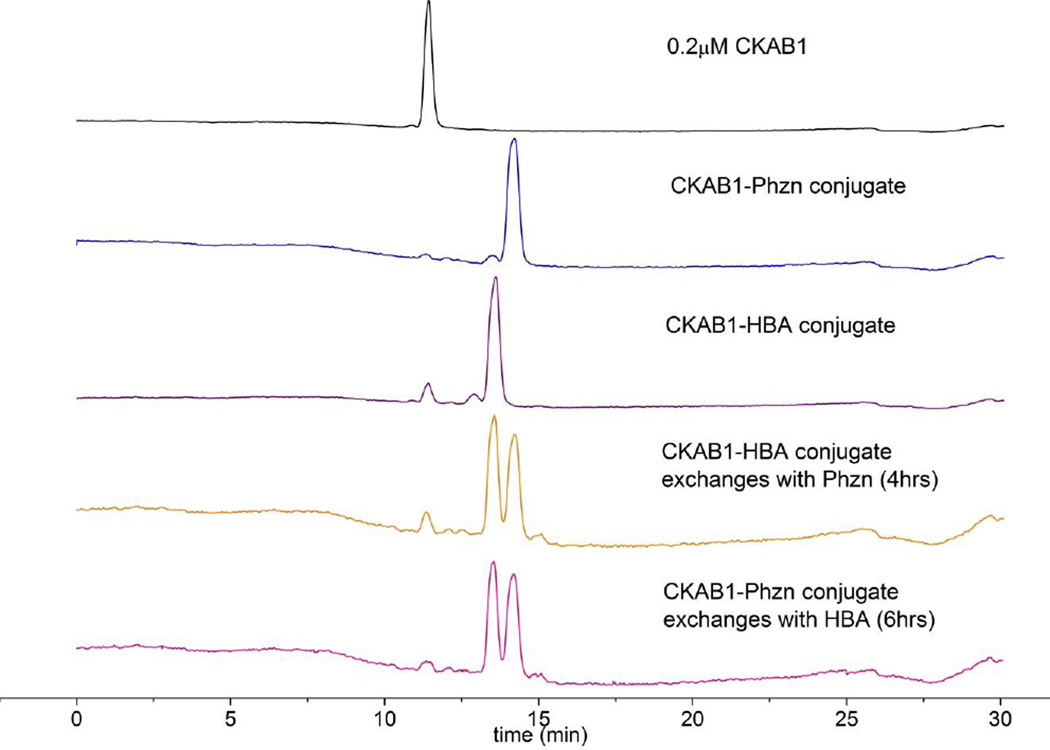
Fast and reversible conjugation chemistry is highly desirable for dynamic combinational chemistry (DCC), in which virtual libraries can be created via the conjugation and exchange of various building blocks.[12] Typical hydrazone and oxime structures are not suitable for DCC due to the lack of exchange a neutral pH,[13] a problem that the chemistry of 2-APBA should be able to circumvent. Towards this end, we examined the hydrazone-hydrazine exchange by using a peptide (CKAB1, Figure S10) that can form hydrazones with Phzn or 4-hydrazinobenzoic acid (HBA) respectively. Titrating HBA (1 equivalent) to a preformed peptide-Phzn conjugate elicits reequilibration over several hours to give the two hydrazones at ~1:1 ratio (Figure 3). The reverse titration gives the same products, indicating that the hydrazone formation of 2-APBA is under thermodynamic control. For comparison, no exchange was observed at all when acetophenone was used instead of 2-APBA as a building block (Figure S11). The contrasting results highlight the catalytic role of the boronic acid moiety in the exchange reactions.
Figure 3.
Hydrazone-hydrazine exchange under physiologic conditions enabled by boronic acid activation. The same products are obtained with different preformed hydrazones, indicating the hydrazone-hydrazine exchange is under thermodynamic control. The equilibrium is reached over several hours under the experimental conditions.
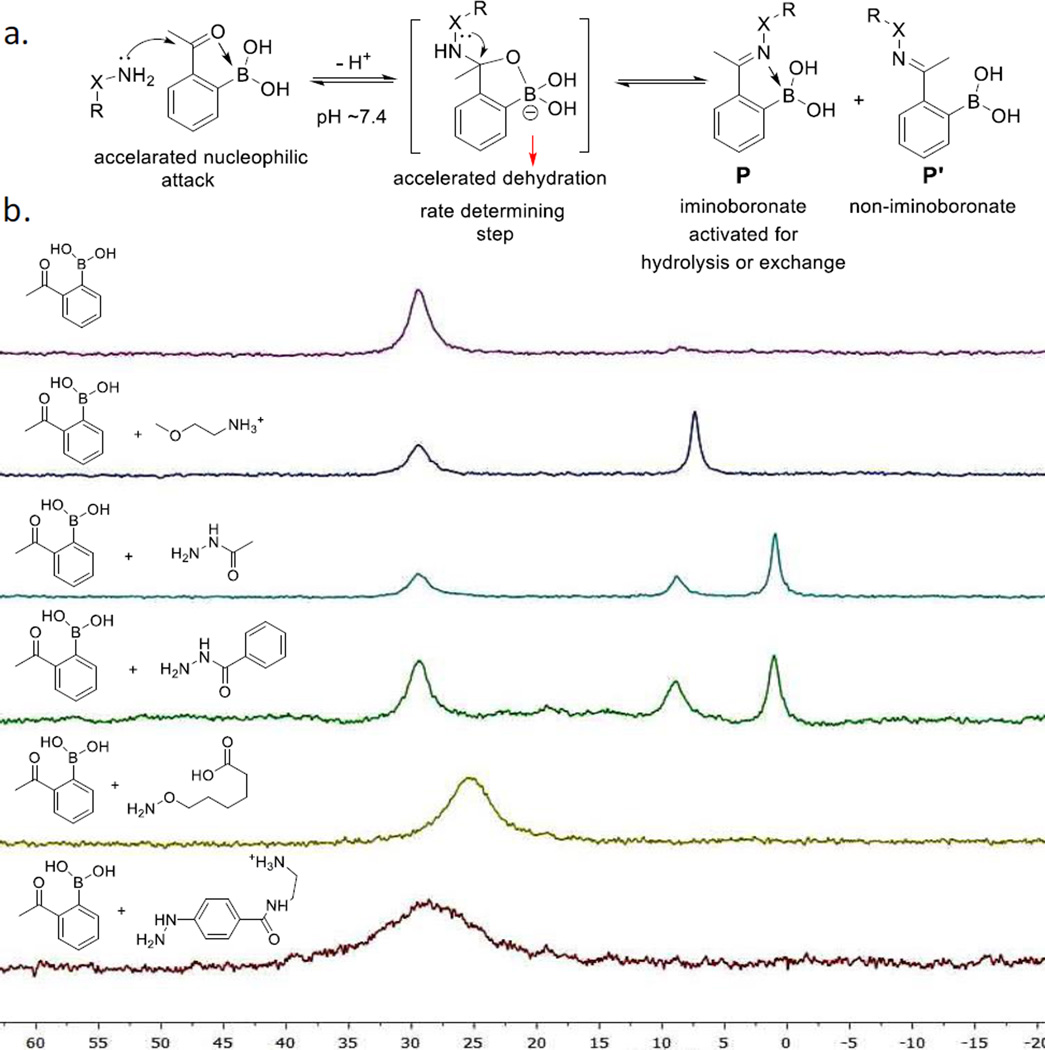
Based on the aforementioned reaction profiles, we propose a plausible mechanism to realize the fast kinetics and quick reversibility of the oxime/hydrazone formation at biological pH (Figure 4a). We anticipate that the ortho-boronic acid group greatly favors the dehydration step, which is believed to be the rate determining step for imine formation. This is in line with the recent work by Kool and coworkers, where adjacent acidic groups accelerate hydrazone formation.[14] As illustrated in Figure 4a, rotation along the C-C bond (highlighted in bold) of the hydrozone/oxime product resuts in a pair of conformational isomers (P and P’), one of which allows formation of iminoboronates with a dative bond formed between the imine nitrogen and the boron. Kinetically, iminoboronate formation activates the imine for nucleophilic attack by water and other nucleophiles, leading to rapid hydrolysis or exchange. To support this mechanism, we recorded the 11B-NMR spectra of the 2-APBA conjugates with various nucleophiles (Figure 4b). 2-APBA alone displays a peak around 30 ppm. Mixing 2-APBA with a primary amine results in a new peak at 5 ppm, which is characteristic of anionic boron as in iminoboronates. Acylhydrazone formation of 2-APBA affords two new peaks (~8 and 0 ppm), indicating formation of two stable products both presenting an iminoboronate. In other words, for acylhydrazone formation, the iminoboronate conjugate P consists of two isomers, the existence of which is consistent with the 1H-NMR data (Figure S1 and S3). Possible structures of such isomers are illustrated in Figure S1. In contrast, mixing 2-APBA with AOHA gives a single broad peak at 25 ppm. The high chemical shift value indicates the non-iminoboronate isomer (P’) being the predominant species. However, the upfield shift and peak broadening suggest quick exchange between P’ and P. Formation of the iminoboronate conformer P, although only to a small extent, activates the oxime for quick hydrolysis. The conjugate of 2-APBA and Phzn also exhibits a broad peak, with marginal upfield shift in comparison to 2-APBA, consistent with the improved stability of the hydrazone in comparison to the oxime.
Figure 4.
Mechanistic understanding of the iminoboronate chemistry. a) Proposed mechanism for the conjugation chemistry between 2-APBA and α-nucleophiles. b) 11B-NMR spectra of various imine products of 2-APBA (2-APBA: 4mM; nucleophiles: 6 mM). The upfield shift of the peaks indicates iminoboronate formation, which activates the imine for hydrolysis. The extent of iminoboronate formation inversely correlates with the stability of the conjugates. A cationic derivative of Phzn was used for better solubility.
It is interesting to note that the oxime conjugation of 2-APBA is quickly reversible in our study, while the recent report from the Gillingham group describes rather stable oxime formation by 2-formylphenyl boronic acids (2-FPBA).[7] The contrasting results can be rationalized by the extent of iminoboronate formation as revealed by 11B-NMR (Figure S13). Specifically, the oxime formation of 2-FPBA causes essentially no upfield shift of the boron peak, indicating minimal existence of the 2-FPBA oxime as iminoboronates.
To summarize, this contribution describes the effect of an ortho-boronic acid moiety on the oxime/hydrazone formation of aryl ketones. Our results show that the boronic acid moiety greatly accelerates the conjugation of 2-APBA to various α-nucleophiles at neutral pH. Importantly, the oxime/hydrazone conjugates also display faster rates of hydrolysis and dynamic exchange due to formation of iminoboronates. In comparison to the recent report by Gillingham and co-workers,[7] which describes rapid oxime formation facilitated by boronic acids, our work herein shows that the boronic acid-accelerated conjugation also applies to hydrazone formation. Furthermore, our contribution examines the reverse reaction and highlights the dynamic nature of the oxime/hydrazone conjugates that can form iminoboronates. We anticipate these dynamic conjugation reactions to find many applications in the context of dynamic combinatorial chemistry in biological settings. Of equal importance, we note that 2-APBA displays nanomolar potency for conjugation with phenylhydrazines (Kd: 0.07 µM). The high potency, together with the fast kinetics, makes the hydrazone chemistry of 2-APBA highly attractive for labeling biomolecules as well.
Experimental Section
Experimental Details are provided in the Supporting Information.
Supplementary Material
Acknowledgements
We acknowledge the National Institutes of Health (GM102735) for financial support.
References
- 1.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 2.a) Dirksen A, Dirksen S, Hackeng TM, Dawson PE. J. Am. Chem. Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]; b) Dirksen A, Hackeng TM, Dawson PE. Angew. Chem. Int. Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; c) Dirksen A, Dawson PE. Bioconjug. Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Crisalli P, Kool ET. J. Org. Chem. 2013;78:1184–1189. doi: 10.1021/jo302746p. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Larsen D, Pittelkow M, Karmakar S, Kool ET. Org. Lett. 2015;17:274–277. doi: 10.1021/ol503372j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay A, McCarthy KA, Kelly MA, Gao J. Nat. Commun. 2015;6 doi: 10.1038/ncomms7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Hutin M, Bernardinelli G, Nitschke JR. Chem. Eur. –J. 2008;14:4585–4593. doi: 10.1002/chem.200800074. [DOI] [PubMed] [Google Scholar]; b) Galbraith E, Kelly AM, Fossey JS, Kociok-Kohn G, Davidson MG, Bull SD, James TD. New J. Chem. 2009;33:181–185. [Google Scholar]; c) Gutierrez-Moreno NJ, Medrano F, Yatsimirsky AK. Org. Biomol. Chem. 2012;10:6960–6972. doi: 10.1039/c2ob26290h. [DOI] [PubMed] [Google Scholar]; d) Cal PM, Vicente JB, Pires E, Coelho AV, Veiros LF, Cordeiro C, Gois PM. J. Am. Chem. Soc. 2012;134:10299–10305. doi: 10.1021/ja303436y. [DOI] [PubMed] [Google Scholar]
- 5.Crugeiras J, Rios A, Riveiros E, Amyes TL, Richard JP. J. Am. Chem. Soc. 2008;130:2041–2050. doi: 10.1021/ja078006c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Tschampel P, Snyder HR. J. Org. Chem. 1964;29:2168–2172. [Google Scholar]; b) Dunn HE, Catlin JC, Snyder HR. J. Org. Chem. 1968;33:4483–4486. [Google Scholar]; c) Mears RJ, Sailes HE, Watts JP, Whiting A. J. Chem. Soc. Perkin Trans. 1. 2000:3250–3263. [Google Scholar]; d) Tickell DA, Mahon MF, Bull SD, James TD. Org. Lett. 2013;15:860–863. doi: 10.1021/ol303566k. [DOI] [PubMed] [Google Scholar]; e) Xu SY, Sun XL, Ge HB, Arrowsmith RL, Fossey JS, Pascu SI, Jiang YB, James TD. Org. Biomol. Chem. 2015;13:4143–4148. doi: 10.1039/c4ob02267j. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt P, Stress C, Gillingham D. Chem. Sci. 2015;6:3329–3333. doi: 10.1039/c5sc00921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Wang L, Zhang Z, Brock A, Schultz PG. Proc. Natl. Acad. Sci. USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang Z, Smith BA, Wang L, Brock A, Cho C, Schultz PG. Biochemistry. 2003;42:6735–6746. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- 9.Kalia J, Raines RT. Angew. Chem. Int. Ed. 2008;47:7523–7526. doi: 10.1002/anie.200802651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjug. Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Song W, Wang Y, Qu J, Madden MM, Lin Q. Angew. Chem. Int. Ed. 2008;47:2832–2835. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]; d) Schmidt P, Zhou L, Tishinov K, Zimmermann K, Gillingham D. Angew. Chem. Int. Ed. 2014;53:10928–10931. doi: 10.1002/anie.201406132. [DOI] [PubMed] [Google Scholar]
- 11.Chiu TK, Kubelka J, Herbst-Irmer R, Eaton WA, Hofrichter J, Davies DR. Proc. Natl. Acad. Sci. USA. 2005;102:7517–7522. doi: 10.1073/pnas.0502495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Huc I, Lehn JM. Proc. Natl. Acad. Sci. USA. 1997;94:2106–2110. doi: 10.1073/pnas.94.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Herrmann A. Org. Biomol. Chem. 2009;7:3195–3204. doi: 10.1039/b908098h. [DOI] [PubMed] [Google Scholar]
- 13.a) Bunyapaiboonsri T, Ramstrom H, Ramstrom O, Haiech J, Lehn JM. J. Med. Chem. 2003;46:5803–5811. doi: 10.1021/jm030917j. [DOI] [PubMed] [Google Scholar]; b) Nguyen R, Huc I. Chem. Commun. 2003:942–943. doi: 10.1039/b211645f. [DOI] [PubMed] [Google Scholar]
- 14.a) Crisalli P, Kool ET. Org. Lett. 2013;15:1646–1649. doi: 10.1021/ol400427x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kool ET, Park DH, Crisalli P. J. Am. Chem. Soc. 2013;135:17663–17666. doi: 10.1021/ja407407h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.