Abstract
In this Communication, we describe an enzyme-responsive, paclitaxel-loaded nanoparticle and assess its safety and efficacy in vivo in a human fibrosarcoma murine xenograft. The material was generated via graft-through block copolymerization of norbornene monomers with hydrophilic targeting peptides together with hydrophobic paclitaxel prodrugs. This work represents a proof-of-concept study demonstrating the utility of enzyme-responsive nanoscale drug carriers capable of targeted accumulation and retention in tumor tissue in response to overexpressed endogenous enzymes. Critically, we observed low systemic toxicity in healthy mice following intravenous administration, with the maximum tolerated dose (MTD) exceeding 240 mg/kg with respect to paclitaxel. Furthermore, we observed efficacy against tumorigenesis paralleling that of paclitaxel at equivalent intravenous dosing, and near complete tumor growth suppression when administered intratumorally. This work represents a significant departure from traditional targeted drug delivery systems and presents a new avenue of exploration for nanomedicine.
Keywords: drug delivery, enzyme-responsive, nanomaterials, self assembly, stimuli responsive
Graphical abstract
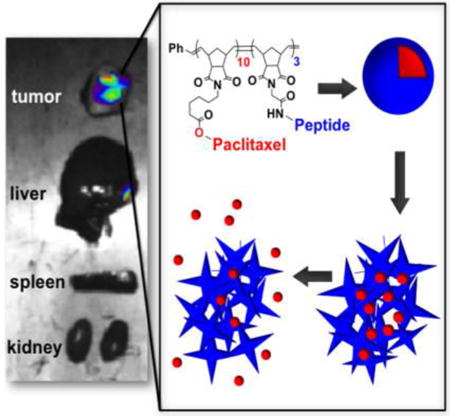
The goal of nanomedicine is to treat disease through selective accumulation of therapeutics in diseased tissue. Nanoparticles offer the potential to package large quantities of drug cargo per carrier entity, to be decorated with targeting moieties in a multivalent fashion, and to have the potential to decrease off-target effects associated with conventional treatment regimes, while simultaneously increasing efficacy.[1] With respect to cancer therapy, the enhanced permeability and retention (EPR) effect has been implicated, albeit somewhat controversially,[2] as a mechanism by which nanomaterials accumulate in the fenestrated vasculature of tumor tissue. However, among other factors, this effect is limited to diseases that undergo rapid angiogenesis in their pathology.[3] Furthermore, the EPR effect is a passive mechanism of accumulation. To achieve active targeting, nanoparticle drug carriers have utilized receptor-mediated endocytosis[4] and hence, rely on the overexpression of surface receptors on disease-associated cells. Therefore, researchers have focused on a recurring set of ligand-receptor combinations, including RGD with integrin αvβ3,[5] NGR with aminopeptidase N,[6] and folic acid with the folate receptor.[7]
We envisioned a different targeting method, wherein an enzymatic signal endogenous to tumor tissue directs a build-up of material selectively at the tumor site.[8, 9-11] Specifically, we aimed to utilize matrix metalloproteinases (MMPs), overexpressed in an array of cancer types and present as catalytic, extracellular or membrane-bound tumor markers.[12] In this strategy, nanoparticles have shells decorated with peptides containing a substrate for MMPs. Upon exposure to the enzyme, the materials undergo a nano- to microscale change in size, coupled with a change in morphology[10]. In this way, the tumor guides the accumulation process through MMP expression patterns resulting in active accumulation through catalytic amplification. To date, we have demonstrated this targeting method for the accumulation of fluorescent probes with the aim of developing approaches for guided surgery[13] and for diagnostic purposes.[9-11]
Given our experience with targeting fluorescent probes, we hypothesized that an MMP-targeted nanoparticle platform could be employed as a tool for the delivery of chemotherapeutics (Figure 1). Towards this end, we generated micellar nanoparticles through the direct diblock copolymerization of a novel paclitaxel conjugate with a MMP substrate (Figure 1A). Both functional monomers were synthesized as norbornene analogues amenable to ring opening metathesis polymerization (ROMP),[14, 15, 16] utilizing a highly functional group tolerant Ru-based initiator[15, 17, 18] capable of producing polymers with low dispersity in a highly reproducible manner.[18] The resulting amphiphilic block copolymers assemble into micellar nanoparticles with a surface comprised of shell of MMP-substrates and a hydrophobic paclitaxel core (Figure 1B). Notably, the drug is polymerized directly and is covalently bound via a biodegradable ester linkage. Upon exposure to MMP, the peptide shell is cleaved and the nanoparticles undergo a drastic change in morphology from discrete, spherical micelles 20 nm in diameter to form micron-scale assemblies visualized by transmission electron microscopy (Figure 1C-D).[10, 11] This transition amounts to a tumor-guided implantation of the polymer-bound drug conjugate via intravenous (IV) injection.
Figure 1.
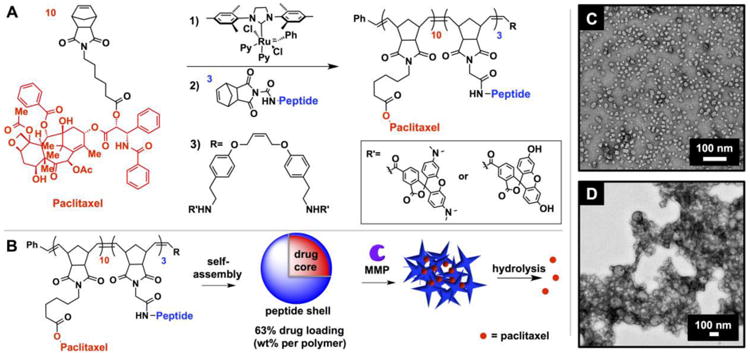
Synthesis of drug-loaded, enzyme responsive micellar nanoparticles. A) Polymerization scheme of enzyme-responsive, paclitaxel-conjugated diblock copolymers. B) Nanoparticles assemble, and subsequently change morphology in response to MMP. C) TEM image (dry state, negative uranyl acetate stain) of NPL before and D) after exposure to MMP, demonstrating enzyme-induced morphology change.
We utilized paclitaxel (PTX) in the hydrophobic block of the copolymer and as the therapeutic moiety in this motif, as it is a potent microtubule-stabilizing agent[19] and standard component of chemotherapy regimes for many malignant and metastatic cancers. The free 2′-hydroxyl group of PTX is absolutely required for its antitumor activity[20], but is available for conjugation via a biodegradable ester formed with a carboxylic acid-functionalized norbornene (PTX-norbornenyl ester). This ensures PTX is completely inactivated, and thus is delivered as a prodrug prior to hydrolysis from the polymer scaffold. The peptide sequence GPLGLAGGERDG was employed as the hydrophilic moiety and MMP recognition sequence. The sequence was amenable to graft-through polymerization affording precise control of the polymer chemistry and subsequent enzymatic response.[16, 21]
Uniform nanoparticles with high drug loading (63% by weight per polymer) spontaneously assembled upon dialysis of the copolymers initially dissolved in DMSO against aqueous solution. Two analogous systems whose hydrophilic blocks were comprised of either all L- or all D-amino acid peptides, were generated to afford enzyme-responsive or nonresponsive, negative control nanoparticles respectively. Additionally, both systems were split into two batches during polymerization of the second block, and terminated with either fluorescein or rhodamine, which form a FRET pair when formulated into a single particle[9, 10] to enable tracking of these materials in vivo. With both responsive (NPL) and nonresponsive (NPD) nanoparticles in hand, we confirmed the ability of these materials to respond to MMP and aggregate in vitro (Figure S1-S3). In summary, catalytic exposure of NPL to MMP-12 for 4 hours led to the aggregated material. Conversely, NPD showed no change in structure when exposed to the same conditions. On the basis of these observations, we hypothesized that NPL would collect within tumor tissue upon IV injection, or be retained following intratumoral (IT) injection. This would lead, in turn, to release of PTX within the tumor tissue achieving a measurable therapeutic dose via hydrolysis induced by the tumor microenvironment. By contrast, NPD would not be retained, but rather clear from the tumor tissue, before PTX hydrolysis and release could lead to a therapeutic dose.
We examined the safety and efficacy of PTX-loaded NPs in three proof-of-concept in vivo studies (Figure 2); 1) maximum tolerated dose (MTD) following IV administration 2) efficacy post-IT injection and 3) efficacy post-IV injection. For these studies, employed an HT-1080 fibrosarcoma xenograft cancer model known to overexpress MMPs[22] and to rapidly proliferate in a predictable manner after subcutaneous implantation. All animal procedures were approved by the University of California, San Diego's institutional animal care and use committee (IACUC).
Figure 2.
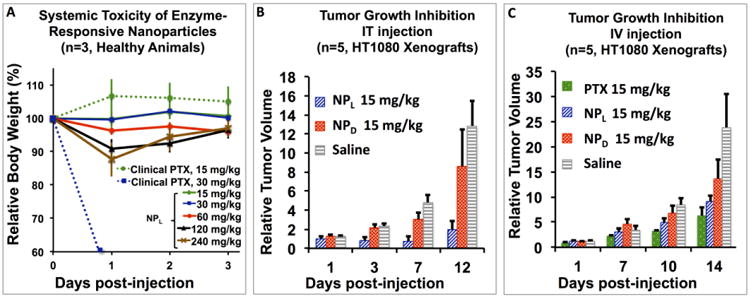
A) Maximum Tolerated Dose (MTD) of IV injection of NPL and clinically formulated PTX. Note: LD50 of clinical PTX is 30 mg/kg and MTD is 15 mg/kg. B) Comparison of NPL to NPD following IT injection. NPL effectively inhibits tumor growth up to 12 days post-injection, whereas NPD has no observable effect. Note: clinical paclitaxel cannot be IT injected without severe adverse effects (ulceration). C) Comparison of NPL and NPD vs. clinical paclitaxel following IV injection.
To examine the safety of our system, an MTD was determined in healthy nu/nu mice. In animal models, toxicity was secondarily measured as a function of animal body weight,[23] with lethality and/or weight loss of greater than 20% suggestive of severe adverse reactions. The MTD in mice of clinically formulated PTX as a suspension in 1:1 Cremophor EL (polyoxyethylated castor oil) to ethanol has been previously established as being in a range between 10-30 mg/kg.[24] In our hands the clinical formulation had a MTD of 15 mg/kg when administered via single tail-vein IV injection. Conversely, we were able to administer NPL via tail vein IV at a dose of 240 mg/kg. Therefore, NPL exhibited a MTD 16 times greater than PTX without overt clinical toxicity, except for a 10% weight loss at 1 day with slow recovery over the next 3 days (Figure 2A). This suggests our enzyme-responsive materials are safely administered, even at exceptionally high doses. To examine efficacy, NPL was tested against NPD, clinically formulated PTX, and saline in a series of IV and IT studies, with all injection concentrations standardized to the equivalent of a 15 mg/kg dose of PTX. In brief, tumor xenografts of HT-1080 were established by inoculating each mouse subcutaneously with ∼106 cells. Drug treatments were initiated once the tumors reached approximately 50 mm[3] in size. Tumor growth was assessed by daily measurement of tumor diameter through B-Mode Ultrasound (US) (Figure S4).[25]
To confirm that morphology change is necessary to retain our materials and further, to determine whether this accumulation event leads to a release of drug cargo at the tumor site, we conducted an efficacy study in which the effect on tumor growth of NPL was compared to that of both NPD and saline (negative control) following IT injection. Live-animal fluorescence imaging (Figure S5) was used to monitor the retention of our materials post-injection as a function of FRET (Förster Resonance Energy Transfer) signal with the eXplore Optix preclinical scanner (λex= 470 nm and λem= 590 nm). Briefly, PTX-nanoparticles contained both fluorescein- and rhodamine-labeled polymers, as these molecules form a FRET pair. We monitored the presence of a viable FRET signal by exciting the donor at 470 nm and monitoring the emission of the acceptor at 590 nm. FRET is only manifest when donor and acceptor molecules are within the Förster radius, as they are in these materials. The use of a FRET signal, rather than a single-dye system, eliminates much of the background signal caused by autofluorescence. As shown in Figure 3, FRET is observable up to 5 days following IV injection of NPL, suggesting that these materials are accumulating and being retained over a long time-scale. Importantly, FRET is only observed for the first 5 hours following IV injection of NPD, indicative of rapid clearance of the material, presumably due to the lack of MMP-induced morphology change. Excitingly, we observed superior tumor growth suppression by NPL up to 12 days post-injection, and in fact, one animal in the cohort experienced complete remission beyond two months post-treatment (Figure 2B). Conversely, there is no observable difference between NPD and saline throughout the duration of the study. These results provide evidence that morphology change is required for the function of these materials.
Figure 3.
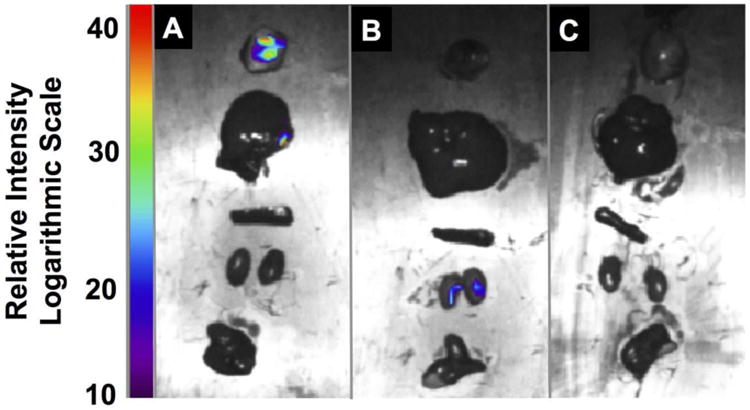
Ex vivo tissue analysis. Fluorescence imaging of A) NPL B) NPD and C) saline cohorts at 14 days post-IV injection. Organs were imaged immediately after excision, and include tumor, liver, spleen, kidneys, heart, and lung, from top to bottom in each panel.
Further evidence of efficacy was elucidated through a preliminary IV study. The effect on tumorigenesis of NPL was compared to that of clinically formulated PTX (positive control) and of saline (negative control), following a single tail vein IV injection (Figure 2C). In the literature, it is accepted that in vivo tumor growth follows an exponential curve until it reaches a lethal tumor volume of 109 cells (1 cubic centimeter).[26] After 10 days post-injection, mice in the saline cohort experienced rapid proliferation until reaching nearly lethal tumor volume within 14 days. By contrast, NPL successfully suppressed tumor growth for up to two weeks post-treatment, and in fact, paralleled that of PTX, within standard error, throughout the duration of the study. This, coupled with the MTD data, suggests that at equivalent doses, enzyme-responsive nanoparticle scaffolds have potentially very low toxicity for equivalent efficacy.
In conjunction with the IV efficacy study, the targeting capabilities of our materials following IV injection were analyzed via live animal fluorescence imaging to monitor for FRET signal generation at the tumor site. Indeed, a FRET signal is generated at the tumor within 3 hours post-injection, and remains observable for up to 3 days (see SI). Furthermore, ex vivo tissue analysis of animals sacrificed at 14 days post-injection shows the highest fluorescence signal intensity in the excised tumors, with fluorescence observed to a lesser extent in the liver, spleen, and kidneys (Figure 3, Figure S6). This suggests that a mode of clearance of our system is through the reticuloendothelial system (RES).[27] However, the limited toxicity established in the MTD study suggests that although RES-associated organs may sequester these materials, they are not being processed to release their payloads at off-target sites at a rate high enough to achieve toxic doses in the animals. Full pharmacokinetic analysis is underway to further elucidate the underlying mechanisms of these findings.
Together, the foregoing results demonstrate that this novel, innovative class of nanoscale carrier is capable of transporting small molecule chemotherapeutics specifically and selectively to the disease site while limiting off-target toxicity. A distinct advantage of this system is that therapeutic moieties are incorporated into the nanoparticle scaffold via labile covalent bonds enabling high drug-loading, highly reproducible synthesis, and no observable release of material until accumulation occurs at the tumor site. Furthermore, these systems are potentially generalizable, as any therapeutic capable of conjugation to a norbornene handle can be incorporated into the center of the nanoparticle scaffold. Future studies will center on the optimization of this system, and include investigation of higher PTX doses, exploration of the effect of surface chemistry on RES uptake, and tuning the biodegradation of the drug-to-polymer bond via incorporation of linkages sensitive to other stimuli present in tumor tissue, such as lowered pH and oxidative stress. Finally, we note the promising effects observed for IT administration. Although IV administration is certainly the gold standard for development of chemotherapeutics, there are several instances in which IT administration is used clinically, and is highly efficacious against primary and metastatic disease,[28] thus this route may prove a powerful translational tool. In summary, the system introduced here constitutes a new paradigm in the design of drug-carrying nanomaterials: the use of switchable morphology to guide in vivo accumulation for enhanced safety and efficacy.
Supplementary Material
Acknowledgments
The authors acknowledge support for this work from the NIH (NIBIB - 1R01EB011633). C.E.C. thanks the Cancer Researchers in Nanotechnology Program for fellowship support. The authors also thank the UCSD Chemistry and Biochemistry Molecular MS Facility for high-resolution MS analysis, as well as Christian Nilewski and K.C. Nicolau for helpful input in the synthetic approach to the paclitaxel monomer.
Footnotes
Supporting Information: Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Cassandra E. Callmann, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA
Christopher V. Barback, Department of Radiology, University of California, San Diego, La Jolla, CA 92093, USA
Matthew P. Thompson, Dr., Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA
David J. Hall, Department of Radiology, University of California, San Diego, La Jolla, CA 92093, USA
Robert F. Mattrey, Department of Radiology, University of California, San Diego, La Jolla, CA 92093, USA
Nathan C. Gianneschi, Email: ngianneschi@ucsd.edu, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA.
References
- 1.a) Davis ME, Chen Z, Shin DM. Nat Rev Drug Discov. 2008;7:771. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]; b) Blum AP, Kammeyer JK, Rush AM, Callmann CE, Hahn ME, Gianneschi NC. J Am Chem Soc. 2015;137:2140. doi: 10.1021/ja510147n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Bae YH, Park K. J Controlled Release. 2011;153:198. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kobayashi H, Watanabe R, Choyke PL. Theranostics. 2014;4:81. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Jain RK, Stylianopoulos T. Nat Rev Clin Oncol. 2010;7:653. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J Controlled Release. 2000;65:271. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 4.Danhier F, Feron O, Préat J V. J Controlled Release. 2010;148:135. doi: 10.1016/j.jconrel.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 5.a) Arap W, Pasqualini R, Ruoslahti E. Science. 1998;279:377. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]; b Horton MA. Int J Biochem Cell Biol. 1997;29:721. doi: 10.1016/s1357-2725(96)00155-0. [DOI] [PubMed] [Google Scholar]; c) Jin H, Varner J. Br J Cancer. 2004;90:561. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Temming K, Schiffelers RM, Molema G, Kok RJ. Drug Resist Updates. 2005;8:381. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]; e) Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]; f) Lawler J, Weinstein R, Hynes RO. J Cell Biol. 1988;107:2351. doi: 10.1083/jcb.107.6.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Cancer Res. 2000;60:722. [PMC free article] [PubMed] [Google Scholar]; b) Curnis F, Sacchi A, Borgna L, Magni F, Gasparri A, Corti A. Nat Biotech. 2000;18:1185. doi: 10.1038/81183. [DOI] [PubMed] [Google Scholar]; c) Garde SV, Forté AJ, Ge M, Lepekhin EA, Panchal CJ, Rabbani SA, Wu JJ. Anti-Cancer Drugs. 2007;18:1189. doi: 10.1097/CAD.0b013e3282a213ce. [DOI] [PubMed] [Google Scholar]; d) Pastorino F, Brignole C, Marimpietri D, Cilli M, Gambini C, Ribatti D, Longhi R, Allen TM, Corti A, Ponzoni M. Cancer Res. 2003;63:7400. [PubMed] [Google Scholar]
- 7.a) Low PS, Antony AC. Adv Drug Delivery Rev. 2004;56:1055. doi: 10.1016/j.addr.2004.02.003. [DOI] [PubMed] [Google Scholar]; b) Stella B, Arpicco S, Peracchia MT, Desmaële D, Hoebeke J, Renoir M, D'Angelo J, Cattel L, Couvreur P. J Pharm Sci. 2000;89:1452. doi: 10.1002/1520-6017(200011)89:11<1452::aid-jps8>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]; c) Leamon CP, Reddy JA. Adv Drug Delivery Rev. 2004;56:1127. doi: 10.1016/j.addr.2004.01.008. [DOI] [PubMed] [Google Scholar]; d) Lee RJ, Low PS. Biochim Biophys Acta, Biomembr. 1995;1233:134. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]; e) Leamon C, Low P. J Drug Targeting. 1994;2:101. doi: 10.3109/10611869409015898. [DOI] [PubMed] [Google Scholar]
- 8.a) Galande AK, Hilderbrand SA, Weissleder R, Tung CH. J Med Chem. 2006;49:4715. doi: 10.1021/jm051001a. [DOI] [PubMed] [Google Scholar]; b) Nguyen QT, Olson ES, Aguilera TA, Jiang T, Scadeng M, Ellies LG, Tsien RY. Proc Nat Ac Soc. 2010;107:4317. doi: 10.1073/pnas.0910261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien MP, Thompson MP, Lin EC, Gianneschi NC. Chem Sci. 2012;3:2690. doi: 10.1039/C2SC20165H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien MP, Thompson MP, Barback CV, Ku TH, Hall DJ, Gianneschi NC. Adv Mat. 2013;25:3599. doi: 10.1002/adma.201300823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien MP, Carlini AS, Hu D, Barback CV, Rush AM, Hall DJ, Orr G, Gianneschi NC. J Am Chem Soc. 2013;135:18710. doi: 10.1021/ja408182p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Kessenbrock K, Plaks V, Werb Z. Cell. In: Olson ES, Jiang T, Aguilera TA, Nguyen QT, Ellies LG, Scadeng M, Tsien RY, editors. Proc Nat Ac Soc. Vol. 141. Vol. 107. 2010. 2010. p. 52.p. 4311. [Google Scholar]; b) Bremer C, Bredow S, Mahmood U, Weissleder R, Tung CH. Radiology. 2001;221:523. doi: 10.1148/radiol.2212010368. [DOI] [PubMed] [Google Scholar]; c) Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc Nat Ac Soc. 2004;101:17867. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metildi C, Felsen C, Savariar E, Nguyen Q, Kaushal S, Hoffman R, Tsien R, Bouvet M. Ann Surg Oncol. 2014;1 doi: 10.1245/s10434-014-4144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Bielawski CW, Grubbs RH. Angew Chem, Int Ed. 2000;39:2903. doi: 10.1002/1521-3773(20000818)39:16<2903::aid-anie2903>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]; b) Trnka TM, Grubbs RH. Acc Chem Res. 2001;34:18. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]; c) Slugovc C. Macromol Rapid Commun. 2004;25:1283. [Google Scholar]; d) Sanford MS, Love JA, Grubbs RH. Organometallics. 2001;20:5314. [Google Scholar]; e) Sanford MS, Love JA, Grubbs RH. J Am Chem Soc. 2001;123:6543. doi: 10.1021/ja010624k. [DOI] [PubMed] [Google Scholar]; f) Grubbs RH. Tetrahedron. 2004;60:7117. [Google Scholar]
- 15.Scholl M, Ding S, Lee CW, Grubbs RH. Org Lett. 1999;1:953. doi: 10.1021/ol990909q. [DOI] [PubMed] [Google Scholar]
- 16.Kammeyer JK, Blum AP, Adamiak L, Hahn ME, Gianneschi NC. Polym Chem. 2013;4:3929. doi: 10.1039/C3PY00526G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Biagini SCG, Gareth Davies R, North M, Gibson VC, Giles MR, Marshall EL, Robson DA. Chem Comm. 1999;235 [Google Scholar]; b) Maynard HD, Okada SY, Grubbs RH. Macromolecules. 2000;33:6239. [Google Scholar]
- 18.a) Leitgeb A, Wappel J, Slugovc C. Polymer. 2010;51:2927. [Google Scholar]; b) Sutthasupa S, Shiotsuki M, Sanda F. Polym J. 2010;42:905. [Google Scholar]
- 19.a) Schiff PB, Fant J, Horwitz SB. Nature. 1979;277:665. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]; b) Band Horwitz S. Trends Pharmacol Sci. 1992;13:134. doi: 10.1016/0165-6147(92)90048-b. [DOI] [PubMed] [Google Scholar]
- 20.Rowinsky EK, Donehower RC. N Engl J Med. 1995;332:1004. doi: 10.1056/NEJM199504133321507. [DOI] [PubMed] [Google Scholar]
- 21.Blum AP, Kammeyer JK, Yin J, Crystal DT, Rush AM, Gilson MK, Gianneschi NC. J Am Chem Soc. 2014;136:15422. doi: 10.1021/ja5088216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon SO, Kim MM, Chung AS. J Biol Chem. 2001;276:20085. doi: 10.1074/jbc.M101143200. [DOI] [PubMed] [Google Scholar]
- 23.a) Cao S, Black JD, Troutt AB, Rustum YM. Cancer Res. 1998;58:3270. [PubMed] [Google Scholar]; b) Cao S, Rustum YM. Cancer Res. 2000;60:3717. [PubMed] [Google Scholar]
- 24.a) Huang GS, Lopez-Barcons L, Freeze BS, Smith AB, Goldberg GL, Horwitz SB, McDaid HM. Clin Cancer Res. 2006;12:298. doi: 10.1158/1078-0432.CCR-05-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yamori T, Sato S, Chikazawa H, Kadota T. Jpn J Cancer Res. 1997;88:1205. doi: 10.1111/j.1349-7006.1997.tb00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Trail PA, Willner D, Bianchi AB, Henderson AJ, TrailSmith MD, Girit E, Lasch S, Hellström I, Hellström KE. Clin Cancer Res. 1999;5:3632. [PubMed] [Google Scholar]
- 25.Ayers GD, McKinley ET, Zhao P, Fritz JM, Metry RE, Deal BC, Adlerz KM, Coffey RJ, Manning HC. J U M. 2010;29:891. doi: 10.7863/jum.2010.29.6.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a) Simpson-Herren L, Lloyd HH. Cancer Chemother Rep, Part 1. 1970;54:143. [PubMed] [Google Scholar]; b) Yorke ED, Fuks Z, Norton L, Whitmore W, Ling CC. Cancer Res. 1993;53:2987. [PubMed] [Google Scholar]
- 27.Gref R, Domb A, Quellec P, Blunk T, Müller RH, Verbavatz JM, Langer R. Adv Drug Delivery Rev. 1995;16:215. doi: 10.1016/0169-409X(95)00026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Goldberg EP, Hadba AR, Almond BA, Marotta JS. J Pharm Pharmacol. 2002;54:159. doi: 10.1211/0022357021778268. [DOI] [PubMed] [Google Scholar]; b) Walter KA, Tamargo RJ, Olivi A, Burger PC, Brem H. Neurosurgery. 1995;37:1129. [PubMed] [Google Scholar]; c) Tong Y, Song W, Crystal RG. Cancer Res. 2001;61:7530. [PubMed] [Google Scholar]; d) Lammers T, Peschke P, Kühnlein R, Subr V, Ulbrich K, Huber P, Hennink W, Stormy G. NEOPFL. 2006;8:788. doi: 10.1593/neo.06436. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Celikoglu F, Celikoglu SI, Goldberg EP. Lung Cancer. 2008;61:1. doi: 10.1016/j.lungcan.2008.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


