Abstract
DNA polymerase ζ (pol ζ) is exceptionally important for maintaining genome stability. Inactivation of the Rev3l gene encoding the polymerase catalytic subunit causes a high frequency of chromosomal breaks, followed by lethality in mouse embryos and in primary cells. Yet it is not known whether the DNA polymerase activity of pol ζ is specifically essential, as the large REV3L protein also serves as a multiprotein scaffold for translesion DNA synthesis via multiple conserved structural domains. We report that Rev3l cDNA rescues the genomic instability and DNA damage sensitivity of Rev3l-null immortalized mouse fibroblast cell lines. A cDNA harboring mutations of conserved catalytic aspartate residues in the polymerase domain of REV3L could not rescue these phenotypes. To investigate the role of REV3L DNA polymerase activity in vivo, a Rev3l knock-in mouse was constructed with this polymerase-inactivating alteration. No homozygous mutant mice were produced, with lethality occurring during embryogenesis. Primary fibroblasts from mutant embryos showed growth defects, elevated DNA double-strand breaks and cisplatin sensitivity similar to Rev3l-null fibroblasts. We tested whether the severe Rev3l-/- phenotypes could be rescued by deletion of DNA polymerase η, as has been reported with chicken DT40 cells. However, Rev3l-/- Polh-/- mice were inviable, and derived primary fibroblasts were as sensitive to DNA damage as Rev3l-/- Polh+/+ fibroblasts. Therefore, the functions of REV3L in maintaining cell viability, embryonic viability and genomic stability are directly dependent on its polymerase activity, and cannot be ameliorated by an additional deletion of pol η. These results validate and encourage the approach of targeting the DNA polymerase activity of pol ζ to sensitize tumors to DNA damaging agents.
Author Summary
Translesion synthesis allows DNA replication to occur in the presence of damaged DNA. This process is mediated by low-fidelity DNA polymerases (such as pol ζ or pol η) that maintain genomic stability. The action of these polymerases is crucial to limit cancer. In mice, complete deletion of DNA pol ζ leads to embryonic lethality, and conditional deletion enhances tumorigenesis. Pol ζ is a large protein with many domains that interact with other essential proteins and maintain the structural integrity of pol ζ. It is not known if the polymerase activity of pol ζ mediates its essential activities. Using a cell culture complementation system and in vivo knock-in mice, our work shows that pol ζ–mediated maintenance of genomic stability in the presence of DNA damage is absolutely dependent on its DNA polymerase activity. Others have demonstrated in chicken cells that co-deletion of pol ζ and pol η rescues the pol ζ-dependent phenotypes, but our work in mice and in mouse cell culture does not support that conclusion. These results demonstrate the physiological importance of pol ζ polymerase activity, and show that employing small-molecule inhibitors of the polymerase reaction is a valid strategy for sensitizing tumor cells to chemotherapeutic agents.
Introduction
In eukaryotes, DNA polymerase ζ (pol ζ) is critical for the tolerance of many types of DNA replication blocks, by playing a central role in translesion DNA synthesis (TLS). Primary replicative DNA polymerases (pol δ or pol ε) are stalled when they encounter many types of template DNA adducts or DNA sequences forming stable secondary structures. Such stalled replication forks are prone to formation of a dangerous DNA double-strand break. The process of TLS helps avoid catastrophes by using a lower fidelity DNA polymerase (such as pol ζ or pol η), to incorporate nucleotides across from a lesion. TLS may occur either in S phase during primary DNA replication or in G2 phase during post-replication DNA synthesis. In yeast and in mammalian cells, pol ζ is important for this process, but it leads to endogenous and DNA damage-induced point mutations because of errors introduced during TLS [1–5]. Elimination of the pol ζ catalytic subunit Rev3l in mice leads to death during embryogenesis (reviewed in [6]). Primary cells in culture also cannot survive in the absence of Rev3l, because chromosomal DNA breaks quickly accumulate [7, 8]. Circumvention of damage-dependent checkpoints by SV40 large T antigen (TAg) immortalization of cells or by Tp53 knockout allows Rev3l-deficient cell lines to grow, but the cells continue to display gross chromosomal instability and DNA damage sensitivity [8–10]. Mice conditionally deleting Rev3l in a fraction of hematopoietic cells or in basal skin keratinocytes are viable, but exhibit enhanced tumor incidence, as a consequence of the chromosomal instability of Rev3l-null cells [7, 11]. The hypersensitivity of REV3L-defective cells to some clinically-used DNA damaging agents indicates that REV3L is a possible target for enhancing the sensitivity of tumors to chemotherapeutic agents [12].
Although the consequences of pol ζ disruption are dramatic, it is not clear that these arise from a specific DNA polymerase defect. In mammalian cells, REV3L is a large protein (>3000 amino acids), with multiple functional domains. The DNA polymerase domain occupies only the last third of the protein (Fig 1A). The structural integrity of REV3L may be required in DNA processing complexes and for protein-protein interactions necessary to maintain cell viability and DNA integrity. Indeed, REV3L serves as a multi-DNA polymerase scaffold. The central region harbors two adjacent binding domains for REV7 (gene name MAD2L2). REV7 is necessary for pol ζ activity in vitro and serves an important function as a bridge protein for interaction with the REV1 protein [13–15]. REV1 in turn interacts with Y-family DNA polymerases that insert bases opposite sites of DNA damage and work in tandem with pol ζ [16–18]. REV7 also has other cellular functions in chromatin assembly and structure [19–21]. An N-terminal region of REV3 is conserved with yeast homologs [22]. At the C-terminus of REV3L [23], an Fe-S cluster is present that binds two other subunits of the pol ζ enzyme, POLD2 and POLD3. Both of these proteins also serve as subunits of the replicative DNA polymerase δ [23–26]. More recently, a conserved positively charged domain in the central region has been recognized as necessary for the efficient polymerase function of the recombinant protein [24]. Another domain in the central region has strong homology to the KIAA2022 gene (S1 Fig).
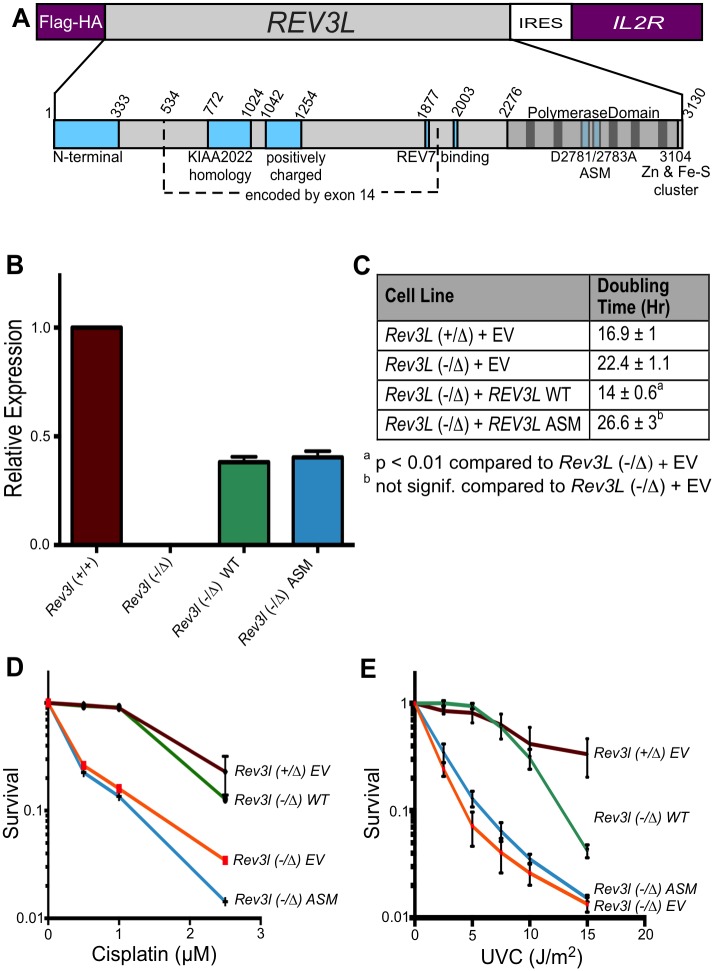
Fig 1. Expression of human REV3L complements Rev3l-deficient MEFs.
(A) Top, the human REV3L gene was cloned into a pOZ vector for expression in mammalian cells with an N-terminal FLAG-HA epitope tag. The vector also expresses the interleukin 2 receptor (IL2R) gene via an internal ribosomal entry site (IRES). Below, domains in mammalian REV3L protein. Indicated here are the N-terminal domain, positively-charged domain, two REV7-binding domains, the KIAA2022 homology domain (see S1 Fig), and the C-terminal Fe-S cluster for interaction with other subunits. Vertical bars in the polymerase domain represent highly conserved motifs. The location of the D2781A/D2783A active site mutations (ASM) is shown. (B) Expression of REV3L in MEF cell lines. A set of primers and a Taqman probe were used that recognizes both human and mouse Rev3l, but does not amplify knockout transcript. Functional mouse Rev3l is expressed in Rev3l+/+ but not Rev3l-/Δ cells. Wild-type or ASM recombinant REV3L mRNA was expressed in immortalized Rev3l-/Δ MEFs at about half of the endogenous level. (C) Doubling time (in hr) of MEFs harboring empty vector (EV) Rev3l+/Δ, Rev3l-/Δ, and Rev3l-/Δ MEFs expressing wild-type or ASM recombinant REV3L. (D) Survival of MEFs harboring empty vector (EV): Rev3l+/Δ (maroon), Rev3l-/Δ (orange); and Rev3l-/Δ MEFs expressing wild-type (green) or ASM (blue) recombinant REV3L. ATP content was measured 48 hr after addition of cisplatin. (E) Survival of these cell lines 48 hr after ultraviolet C (UVC) radiation as measured by ATP content. Data represent mean ± SEM.
A provocative hypothesis has been put forward to explain the severe genotoxic effects of Rev3l deletion [27]. It was suggested that these are the consequence of the function of a second DNA polymerase, pol η (gene Polh). As in mammalian cells, chicken DT40 cells with a disruption of pol ζ exhibit growth defects, chromosomal aberrations and DNA damage sensitivity [27]. Remarkably, it was reported that co-disruption of Polh and Rev3l corrects all of these phenotypes in DT40. The suggested interpretation was that pol η and pol ζ always work together in bypass of DNA damage, and that a toxic intermediate is formed by pol η that cannot be resolved in the absence of pol ζ. It is clearly important to determine, in mammalian cells, whether the genome instability caused by pol ζ disruption is dependent on pol η.
Here we describe experiments with knockout cells and a specific knock-in mouse model to test whether the catalytic activity of pol ζ is responsible for the phenotypes observed in pol ζ knockout mutants. We describe complementation of Rev3l-deficient mouse embryonic fibroblasts (MEFs) by expression of full-length human wild-type REV3L, and show that DNA polymerase-defective mutant REV3L cDNA is unable to complement cell survival or increased levels of DNA breaks. Using a Rev3l polymerase-dead knock-in mouse model, we show that specific disruption of the polymerase activity prevents the completion of embryogenesis. Finally, we tested whether pol ζ defects can be rescued by ablation of pol η function.
Results
Expression of REV3L cDNA in mouse embryonic fibroblasts rescues slow growth
Rev3l deletion in mouse cell lines is associated with an elevated baseline level of DNA breaks and an increased sensitivity to DNA damaging agents such as cisplatin and UV radiation [3, 8–10]. We wanted to test definitively whether these phenotypes are caused by the disruption of Rev3l. A pOZ expression vector harboring an IL2R selectable marker (Fig 1A) [28] was used to express human REV3L cDNA in Rev3l-deficient MEFs [8]. Cells were selected for IL2R expression by repeated cycles of magnetic bead sorting and clonal populations were isolated. The integrity of the expression vector was confirmed by PCR-based detection, and cells were assayed for expression of REV3L mRNA by real-time RT-PCR. Human REV3L was expressed in the Rev3l-deficient MEFs at about one-half of the normal endogenous level (Fig 1B). Mouse cells expressing one or two alleles of Rev3l have indistinguishable low levels of spontaneous senescence, apoptosis, and chromosome aberrations [8] and there is no haploinsufficiency apparent regarding embryonic or adult viability in mice [7].
We expressed both wild-type REV3L (WT), as well as REV3L with a dual point mutation (ASM: D2781A; D2783A) in residues essential for divalent metal binding in conserved DNA polymerase motif I. Equivalent changes in all other tested DNA polymerases inactivate Mg2+ coordination in the active site, and eliminate enzymatic activity [29, 30].
We tested the growth of Rev3l-proficient and deficient cells expressing an empty vector (EV), as well as deficient cells expressing WT and ASM REV3L cDNA. Rev3l-deficient cells experienced S-phase associated delay and mitotic failure, leading to a population doubling time that was longer than Rev3l-proficient populations [8]. REV3L re-expression in the deficient cell lines significantly decreased their doubling time to a level similar to Rev3l-/+ cells, whereas expression of the polymerase-inactive mutant had no effect (Fig 1C).
Rescue of DNA damage sensitivity and DNA breaks by REV3L expression
Deletion of Rev3l causes sensitivity to DNA damaging agents [8–10]. To determine whether REV3L expression could rescue this phenotype, cells were exposed to cisplatin or UVC radiation and cell survival was measured. Rev3l-deficient cells displayed the expected sensitivity to these damaging agents when compared to the Rev3l-proficient cells (Fig 1D and 1E). Assays were repeated with multiple clones for each genotype. Expression of wild-type REV3L rescued the sensitivity to all three DNA damaging agents, but expression of ASM REV3L did not.
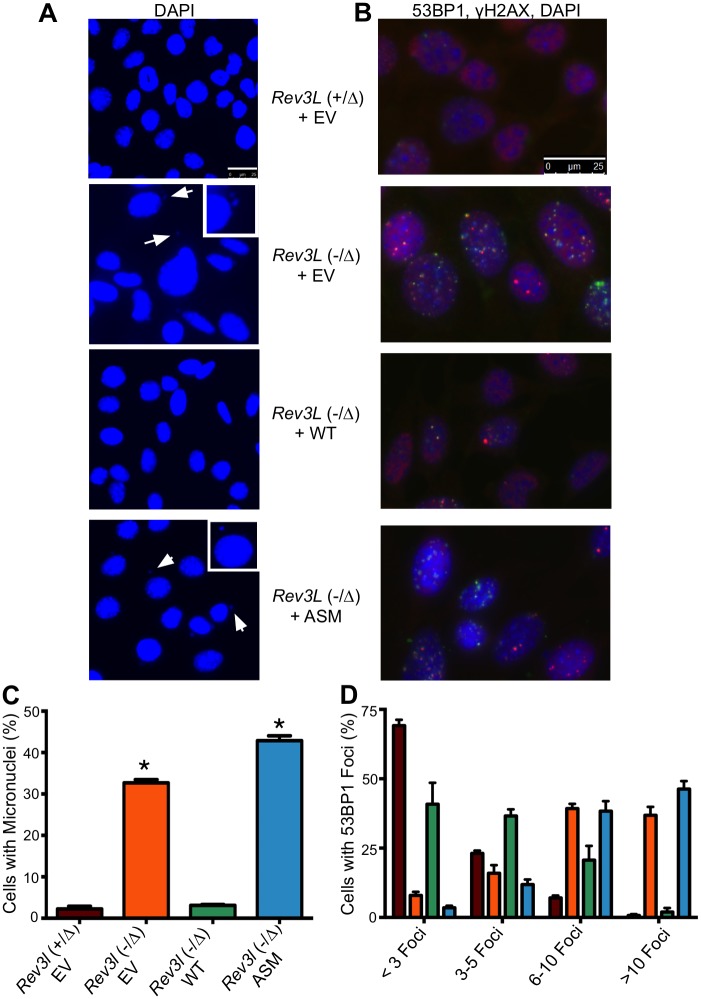
Rev3l-deficient cells manifest an increased formation of DNA breaks in the absence of exogenous DNA damage. We measured a 10 to 20-fold increase in cellular micronuclei (Fig 2A and 2C) in Rev3l-defective cells, with 30–40% of all cells displaying micronuclei. The Rev3l defect was also accompanied by an increased frequency of DNA breaks as quantified by 53BP1 foci per cell, with a pronounced shift in distribution towards larger numbers of foci per cell (Fig 2B and 2D). Expression of wild-type REV3L in Rev3l-deficient MEFs rescued both of these phenotypes, but expression of ASM REV3L did not. The frequency of sister chromatid exchange (SCE) was not decreased in Rev3l-deficient cells (Table 1), indicating that this mitotic recombination event is not impaired by a REV3L defect. These experiments demonstrate that sensitivity to DNA damaging agents and the presence of DNA breaks in Rev3l-deficient cells is caused by the absence of the REV3L protein, and REV3L polymerase activity is required for prevention of these phenotypes.
Fig 2. Spontaneous DNA double-strand break formation is reduced in Rev3l complemented MEFs.
(A) DAPI staining of empty vector (EV)-expressing Rev3l+/Δ, Rev3l-/Δ, as well as Rev3l-/Δ MEFs expressing wild-type or ASM recombinant REV3L; arrows indicate micronuclei, with an enlarged example in the inset. (B) Merged immunofluorescence staining of the same MEFs as in (A) with DAPI (blue), 53BP1 (red) and γ-H2AX (green); foci indicate areas of DNA double-strand breaks. (C) Quantification of percent of nuclei that have associated micronuclei. (D) Quantification of cells with fewer than 3, 3 to 5, 6 to 10 or greater than 10 53BP1 foci (as measured using CellProfiler). The bars are color-coded exactly as in Part C to indicate the genotype of the MEFs (*) p < 0.01. Data represent mean ± SEM.
Table 1. Sister Chromatid Exchange (SCE) frequency in immortalized MEF cell lines.
| Cell line | Rev3l status | % Av SCE/chrom ± s.d. | Av no. chrom ± s.d. |
|---|---|---|---|
| Rev3l+/Δ TAg | +/- | 15.9 ± 7.9 | 72 ± 15 |
| Rev3l-/Δ TAg | -/- | 27.3 ± 9.3 | 105 ± 29 |
| Rev3l+/+ Tp53-/- | +/+ | 15.5 ± 6.2 | 80.9 ± 7 |
| Rev3l+/- Tp53-/- | +/- | 23.9 ± 9.0 | 51.7 ± 11 |
SCE frequencies are given as the average number per chromosome (total SCE observed / total chromsomes counted). For each cell line, 30–35 metaphases were scored. All cell lines were polyploid, as is commonly observed following immortalization. One MEF cell line pair was derived from p53-defective embryos and were described by Wittschieben et al. [9]; these are the Rev3l+/+ Tp53-/- B2 cell line and the Rev3l+/- Tp53-/- B4-9 cell line. The second MEF cell line pair was derived by T-antigen immortalization of primary cells as described [8], these were Rev3l+/Δ 1(+)cl2 TG1Het1 and Rev3l-/Δ 5(-)cl7 TG2Het5. No reduction of SCE frequency per chromosome was found in Rev3l-defective cells.
We also investigated two reported human REV3L knockout lines designated 332 and 504, derived from the Burkitt lymphoma cell line BL2 [31]. However, Rev3l mRNA is still transcribed in the 332 and 504 subclones, the subclones were no more sensitive to cisplatin than the parental BL2, there was no significant increase in spontaneous double-strand break incidence in the subclones, and no complementation of the mild UV sensitivity was observed with Rev3L cDNA (S2 Fig). These results and uncertainties regarding the targeting strategy (S3 Fig) indicate that the BL2 subclones may not be pol ζ defective.
Specific inactivation of Rev3l DNA polymerase activity causes embryonic lethality
To determine the in vivo consequence of specifically inactivating the DNA polymerase function of Rev3l, a genetically engineered mouse was constructed to express an ASM knock-in allele from the endogenous promoter (Fig 3A). Variant lox sites [32] were used to control knock-in of the Rev3l allele. The mice were crossed to CMV-Cre, producing a constitutive ASM allele (abbreviated the “M” allele for the mice here), in a pure C57BL/6J background. All steps of genomic engineering were extensively monitored by Southern blotting analysis (Fig 3B), PCR analysis and DNA sequencing.
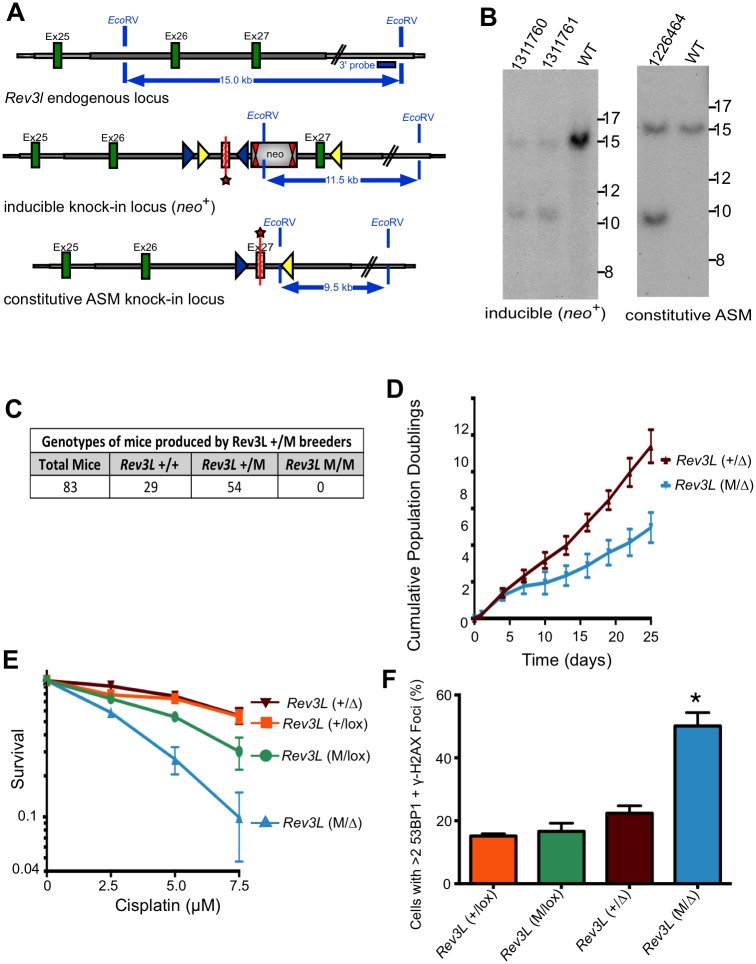
Fig 3. Knock-in mice and MEFs expressing active site mutant Rev3l have knockout phenotypes.
(A) Diagram of the mouse Rev3l ASM knock-in allele, with the wild-type (WT) locus shown at the top. Green rectangles indicate Rev3l coding sequences and the gray line represents chromosomal sequence. In the middle diagram, FRT sites are represented by double red triangles, loxP sites by blue triangles and lox511 sites by yellow triangles. The targeted exon 27 (starred) carries D2773A and D2775A point mutations and is inserted in an inverted orientation between wild-type exons 26 and 27. Splicing donor and acceptor sites flanking wild-type and ASM exon 27 are kept intact. The knock-in was produced by a Cre-dependent genetic switch. First, the neomycin positive selection cassette (neo) was excised by breeding with C57BL/6 Flp deleter mice. A subsequent cross with Cre-expressing mice led to excision of the wild-type exon 27 and inversion of ASM mutant exon 27 into the functional orientation. In the constitutive ASM knock-in locus shown in the lower diagram, the D2773A/D2775A Rev3l gene is expressed under the control of the endogenous Rev3l promoter and wild-type Rev3l exon 27 is absent from the locus. Heterozygous ASM knock-in mice (Rev3L+/M) were then used for breeding. (B) Example of Southern blot analysis of (left) the inducible knock-in locus (neo+) and (right) the constitutive ASM knock-in locus. Genomic DNA of the tested animals was compared with C57BL/6 wild-type genomic DNA (WT). EcoRV digested DNA was blotted on a nylon membrane and hybridized with the external 3’ probe with the position shown at the top of part A. Restriction fragments of 15 kb, 11.5 kb and 9.5 kb were observed for the wild-type, inducible knock-in locus (neo+) and constitutive ASM knock-in locus, respectively. Genomic DNA was further analyzed extensively and confirmed by specific PCR assays and complete DNA sequencing as described in the Materials and Methods. (C) Genotypes of mouse pups produced by breeding parental Rev3l+/M mice. (D) Growth of Rev3l+/Δ and Rev3lM/Δ cells. These cells were produced by addition of AdCre to Rev3lM/lox or Rev3l+/lox MEFs, deleting the floxed allele of Rev3l. (E) Survival of Rev3l+/lox, Rev3lM/lox, Rev3l+/Δ and Rev3lM/Δ primary MEFs 120 hr after addition of cisplatin, as measured by ATP content. (F) The MEFs as in (E) were stained with DAPI, and for 53BP1 and γ-H2AX by immunofluorescence as in Fig 2 to detect foci of DNA double-strand breaks. The quantification shows the percentage of cells with >2 53BP1 and γ-H2AX foci in Rev3l+/lox, Rev3lM/lox, Rev3l+/Δ and Rev3lM/Δ primary MEFs 9 days after AdCre treatment. (*) p < 0.01. Data represent mean ± SEM.
Heterozygote mutant Rev3l+/M mice were viable and fertile, demonstrating that the mutant allele does not have dominant-negative activity affecting viability. Heterozygous mutant Rev3l+/M mice were bred and pups genotyped. No homozygous mutant animals were identified at weaning (Fig 3C). In addition, 48 embryos from 6 pregnancies were isolated between 8.5 and 10.5 dpc. Rev3lM/M embryos were rare at the earlier timepoints, and by 10.5 dpc only a few very small Rev3lM/M embryos were identifiable. The severely impaired development of homozygous Rev3l ASM embryos mirrors the lethality of the Rev3l null allele on a C57BL/6 background [33].
Growth defects and accumulation of DNA strand breaks in Rev3l ASM cell lines
Due to the early embryonic lethality in Rev3lM/M embryos, we were never successful in deriving MEFs from them. To circumvent this problem we crossed Rev3l+/M mice with Rev3l-/lox mice. This mating produced embryos for derivation of viable Rev3lM/lox MEFs. The floxed (lox) allele of Rev3l is functional, but becomes a knockout allele (termed the Δ allele) after action of the Cre recombinase. We expressed Cre recombinase in the cells to yield Rev3lM/Δ MEFs. The mice also harbored the mT/mG transgene to monitor Cre activity. This mT/mG transgene constitutively expresses red fluorescent protein (RFP). When Cre is active, the RFP gene is removed and green fluorescent protein (GFP) is expressed [34]. This allows GFP to be used for flow sorting and as a marker of cells in which Cre recombinase has been expressed.
Cre was introduced via an adenovirus vector into primary MEFs [8] to compare Rev3lM/Δ MEFs with Rev3lM/+ MEFs (retaining a wild-type allele of Rev3l). We measured cell growth, cisplatin sensitivity and DNA double-strand breaks in GFP-positive cells. ASM MEFs had a growth defect compared to wild-type allele-containing MEFs (Fig 3D) and eventually failed to thrive. ASM MEFs were hypersensitive to cisplatin, compared to control MEFs (Fig 3E). Additionally, there was a two to three-fold increase in the number of ASM MEFs containing 53BP1 and γ-H2AX foci (a measure of DNA breaks) compared to controls at 9 days after Cre recombinase expression (Fig 3F). These phenotypes are similar to those seen in Rev3l null primary MEFs [8] (and compare Figs 3F and 4B). This result demonstrates that the DNA polymerase activity of REV3L is specifically required to allow for cell proliferation, to protect genome stability and to moderate cisplatin sensitivity.
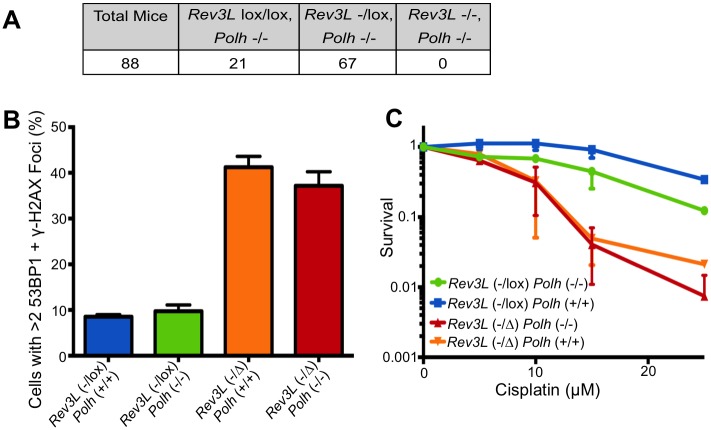
Fig 4. Deletion of Polh does not ameliorate phenotypes caused by knockout of Rev3l.
(A) Genotypes of mouse pups produced by breeding parental Rev3l-/lox Polh-/—mice. (B) MEFs with the indicated genotypes were stained with DAPI, and for 53BP1 and γ-H2AX by immunofluorescence as in Fig 2 to detect foci of DNA double-strand breaks. The quantification shows the percentage of cells with >2 53BP1 and γ-H2AX foci in Rev3l-/lox Polh+/+, Rev3l-/lox Polh-/-, Rev3l-/Δ Polh+/+ and Rev3l-/Δ Polh-/- MEFs 9 days after AdCre treatment. (C) Survival of primary MEFs as in part B, 120 hr after addition of cisplatin, as measured by ATP content. For panels B& C, Data represent mean ± SEM.
Deletion of DNA polymerase eta does not rescue Rev3l-deficient phenotypes in mice
We wanted to determine in mammalian cells whether the DNA damage sensitivity and genome instability caused by pol ζ disruption is dependent on pol η, as has been reported for the DT40 cell line [27]. We crossed parental mice with the genotypes Rev3l-/lox Polh-/-, and investigated the genotypes of the pups. In the Polh-/- background, no Rev3l-/- mice were born (Fig 4A), consistent with the complete lethality of the Rev3l-/- genotype in a Polh+/+ background [6]. We attempted to produce Rev3l-/- Polh-/- MEFs from mouse embryos, but were unable to obtain sufficient material to produce viable MEFs because of the early death during embryogenesis. Instead we derived primary MEFs from viable Rev3l-/lox Polh-/- embryos. Following introduction of Cre via an adenovirus, Rev3l-/Δ Polh-/- cells were produced. These Rev3l-defective primary MEFs had an elevated level of DNA breaks that was indistinguishable from Rev3l-/Δ Polh+/+ cells (Fig 4B). Consistent with published results [35] the pol η defect in Rev3l-/lox Polh-/- MEFs conferred enhanced sensitivity to cisplatin (by comparison with Rev3l-/lox Polh+/+ MEFs) (Fig 4C). A Rev3l defect independently enhanced cisplatin sensitivity, and the sensitivity of the Rev3l-/Δ Polh-/- and the Rev3l-/Δ Polh+/+ MEFs was similar. Therefore, deletion of pol η does not rescue the cell and organismal defects caused by loss of pol ζ, showing that the absence of pol ζ does not create a pol η-dependent toxic intermediate in mouse cells.
Discussion
The polymerase activity of pol ζ is essential for embryonic development and for limiting genome damage
A major objective of this study was to determine whether the catalytic activity of pol ζ is responsible for the severe consequences observed in pol ζ mutant mouse cells. These include hypersensitivity to DNA damaging agents, a greatly increased generation of double-strand breaks in unchallenged cells, a slower growth rate, and a required role for pol ζ in embryonic viability. The impetus for this question is the existence of numerous other functional domains within the catalytic subunit of REV3L. These include a conserved N-terminal domain, two REV7 binding domains [14, 19], and a C-terminal Fe-S cluster that interacts with the POLD2 subunit and is necessary for in vitro activity. In addition, the central region contains a conserved positively charged domain [24] that likely promotes protein-protein and protein-DNA interactions, and a KIAA2022 homology domain, described in detail here for the first time (S1 Fig). The presence of all of these domains introduces the possibility that the essential functions of REV3L could be structural, rather than directly related to the DNA polymerase activity itself. A catalytically deficient but otherwise intact REV3L may have been able to specifically interact with protein partners and DNA substrates, allowing viability of cells and mice.
There is ample precedent for such a situation. One example is the mammalian ERCC2/XPD gene. Complete disruption of XPD is incompatible with viability [36]. However, an amino acid substitution that inactivates the catalytic helicase activity of XPD specifically compromises nucleotide excision repair capacity, but allows cellular viability [37]. This is because the presence of XPD as a subunit of transcription factor TFIIH is necessary for the integrity of that complex, even though XPD activity itself is unnecessary for transcription [38, 39]. Another example is provided by the REV1 protein. REV1 has a DNA polymerase domain that can catalyze dCMP incorporation in DNA. Cells lacking REV1 are hypersensitive to UV radiation, but this DNA damage tolerance activity does not require the polymerase catalytic domain of REV1. Instead, the damage tolerance activity is conferred by a protein-protein interaction domain at the C-terminus of REV1 that interacts with REV7 in pol ζ and with Y family DNA polymerases [40]. Recently, a non-catalytic role has been reported for human DNA pol κ in protection against oxidative stresses [41].
Here, we analyzed the consequence of a homozygous mutation of the Rev3l DNA polymerase active site. No viable homozygous mice were produced, and the corresponding embryos died early in embryogenesis, as with a complete knockout allele. To investigate cell-autonomous consequences of the specific polymerase alteration, we derived primary MEFs that carried one null Rev3l allele, and one active site mutant allele. The growth defects, DNA break formation and cisplatin sensitivity of these cells were similar to cells harboring two null alleles [8]. These results show that the DNA polymerase activity of REV3L is essential for all functions so far measured in mice and in cells.
Rescue of phenotypes by expression of the wild-type REV3L gene
Loss of Rev3l causes chromosomal instability in cells. This complicates studies of the consequences of Rev3l deficiency, as genomic alterations may accumulate during each cell cycle and lead to new phenotypes. A rigorous way to determine which phenotypes are directly caused by Rev3l loss is to complement the cells by expression of Rev3l cDNA. Here we utilized a complementation system for REV3L in mammalian cells, allowing definitive testing of whether phenotypes seen in Rev3l-deleted cells are due to Rev3l-deletion [19, 42]. Our results with specific mutant cDNAs establish that the polymerase activity of REV3L is specifically essential for preserving genome integrity and protecting against DNA damage. It is of course possible that other domains within REV3L also have critical functions for viability or genome integrity, and this complementation system will allow investigation of that possibility. For example, we recently demonstrated that the REV7-binding domains of REV3L are essential for pol ζ function [19].
We also attempted complementation of Rev3l-deficient phenotypes using human BL2 cell lines, reported to carry disruptions of REV3L [31]. It is notable that there were no major differences in phenotypes between the wild-type BL2 cells and the nominal 332 and 504 REV3L mutants. In contrast to the marked phenotypes found with Rev3l-deficient MEFs, the BL2 lines exhibited no statistically significant differences in cell doubling times, micronuclei formation or double-strand break formation as assessed by 53BP1 foci per cell. A modest sensitivity of 332 and 504 cells to UVC radiation and cisplatin was not rescued by complementation with REV3L. The limited sensitivity of 332 and 504 cells to a variety of DNA damaging agents has been noted [43–45]. Others have also reported no significant differences in spontaneous DNA breaks in 332 and 504 cells compared to wild-type BL2 cells [43]. In a study with wild-type BL2 cell extracts and extracts from the nominal REV3L-deficient cells [46], it was concluded that REV3L does not contribute to acetylaminofluorine-induced frameshift mutagenesis. This should probably be re-examined with a different REV3L-defective cell system. It is possible that the modest increased sensitivity of the BL2 subclones to UV radiation and cisplatin [43] might be due to inadvertent disruption of an unrelated gene by the targeting strategy, as may have occurred with BL2 cells deleted for pol ι [47–49]. Our data indicate that the 332 and 504 cell lines may not be truly (or only) REV3L-deficient, and are not well-suited for studies of REV3L function.
Cooperation of pol η and pol ζ in conferring genome protection
The DT40 chicken cell line has been widely used to examine the consequences of DNA repair defects, because it is amenable to genetic manipulation by homologous recombination. Some characteristics of Rev3l-deficient DT40 cells are similar to Rev3l-deficient mouse cells, including elevated levels of spontaneous DNA breaks and sensitivity to DNA damaging agents. Intriguingly, it was reported that deletion of polh (pol η) could rescue the severe phenotypes of Rev3l-deficient DT40 cells [27]. This led to the model that the major defects in Rev3l –deficient cells are a consequence of a polh-dependent toxic intermediate. To test this model in mammalian cells, we investigated whether Rev3l-/- Polh-/- mice could be generated. We found that embryonic lethality of this double mutant was complete and similar in timing to Rev3l-/- Polh+/+ mice. Moreover, Rev3l-/Δ Polh-/- MEFs showed levels of DNA breaks and cisplatin sensitivity analogous to that seen with Rev3l deletion in the presence of pol η. The Rev3l-/lox Polh-/- MEFs were more sensitive to cisplatin than the Rev3l-/lox Polh+/+ MEFs, consistent with the cisplatin sensitivity of human polh-defective cells [35]. Notably, the pol η defective and pol η pol ζ double mutant MEFs had similar sensitivities to cisplatin. This epistatic interaction suggests that these two proteins act in the same pathway to mediate resistance to cisplatin. In fact both polymerases can cooperate to bypass a cisplatin-DNA adduct [24]. In summary, the severe phenotypes caused by Rev3l deletion cannot be rescued in murine cells by concurrent deletion of pol η. This is consistent with results found in the yeast S. cerevisiae, where a Rev3 Rad30 (pol ζ pol η) mutant is more sensitive to ultraviolet radiation than a single Rev3 mutant [50, 51]. Although the absence of pol η causes sensitivity to some DNA damaging agents, it is not specifically toxic in the absence of pol ζ. In the absence of pol ζ, it is possible that TLS does not occur at all, and that other modes of replication fork rescue are relied upon, which leads to a higher prevalence of DNA double-strand breaks [1]. The genetic interaction between pol η and pol ζ reported for chicken DT40 cells might reflect a peculiarity of that cell line. DT40 cells harbor mutations in TP53, and no poli gene has been found in the chicken genome. but it seems unlikely that either gene is relevant in this context. Previously reported Tp53-/- Rev3l-/- MEFs are also pol i deficient (an allele from the 129 ES cell background), and show major genome instability and DNA damage sensitivity [9]. Polh poli double mutant mice are apparently normal with no deficits in development. A poli defect does not exacerbate the UV radiation sensitivity of polh-defective mouse cells, indicating that pol ι does not have a significant backup function protecting against lethality in the absence of pol η [52].
Implications for cancer therapy
Our results with the Rev3l knock-in polymerase mutant mouse are relevant to development of REV3L as a target for chemotherapy. Suppression of REV3L sensitizes cancer cells to cisplatin in mouse model systems, and can limit chemo-resistance [12, 53] because loss of pol ζ diminishes point mutagenesis [2–5]. These studies used siRNA knockdown of REV3L to demonstrate this effect, but future use of small molecule DNA polymerase inhibitors may be more clinically feasible. Until now it has not been known whether inhibition of the catalytic activity of REV3L mimics the cytotoxic effects of a knockdown of the entire gene. Our work demonstrates that loss of REV3L catalytic activity is equivalent, in the assays used here, to gene knockout. This validates and encourages strategies to directly inhibit pol ζ DNA polymerase activity.
Materials and Methods
Cell lines
Rev3l-deficient TAg-immortalized MEFs were derived as in Lange et al [8]. Briefly, MEFs were made from mouse embryos with the genotypes mT/mG+/- Rev3l-/lox or mT/mG+/- Rev3l+/lox, where “lox” represents a functional allele flanked by loxP sites. The mT/mG transgene constitutively expresses RFP, until Cre recombinase activity removes the RFP and allows expression of GFP [34]. The strain background of the mice used to derive these alleles was mixed C57BL/6 and 129. We genotyped DNA polymerase iota (pol ι) in these cell lines, because 129 mice carry a mutant allele of pol ι [54]. All cell lines were heterozygous for this mutation, and so can be considered pol ι proficient. These cell lines were immortalized with SV40 large T-antigen, and then treated with adenovirus Cre (AdCre) to delete the floxed allele of Rev3l (and generate the knockout Δ allele). The cells were subcloned and selected for GFP positivity and for complete deletion of the floxed Rev3l allele. They were grown as in Lange et al [8], in an atmosphere containing 2% O2. The primary MEFs were also derived and cultured in 2% O2 as in Lange et al [8]. They were made from mouse embryos with the genotypes mT/mG+/- Rev3lM/lox or mT/mG+/- Rev3l+/lox, as well as from Rev3l-/lox Polh-/- or Rev3l-/lox Polh+/+ embryos. The loxP-flanked allele of the Rev3l gene was deleted using AdCre adfection, and the deletion efficiency was measured as described [8]. For all cell lines, cell number was counted at each passage, and was used to calculate population doublings and doubling time.
The BL2 parental cell line and subclones 332 and 504 [31] were kindly provided by Claude-Agnés Reynaud (Institut Gustave Roussy, Villejuif, France). Genomic DNA samples from the three cell lines were compared using short tandem repeat (STR) fingerprinting by the Cell Line Identification Core at MD Anderson. All yielded identical profiles of the 16 standard STR markers, confirming the relationship of the three cell lines.
Expression of human REV3L in cell lines
The human REV3L full-length cDNA was acquired in the pUC19M1 vector from Zhigang Wang [55]. The following modifications were made to the REV3L cDNA: a C-terminal Flag tag was added and the 5’-UTR was eliminated and replaced with an optimized mammalian Kozak sequence. This cDNA was cloned into the pTSIGN vector, which contains an EF1α promoter and an internal ribosomal entry site (IRES) fused to a neomycin-eGFP reporter. The active site mutation (residues D2781A/D2783A in human REV3L) was introduced into this pTSIGN-REV3L vector using PCR primers containing the REV3L mutations, and then the mutated PCR fragment was ligated into the REV3L-vector, replacing the wild-type sequence. The full-length human REV3L gene was PCR amplified from the pTSIGN-REV3L and pTSIGN-REV3L-ASM vectors and was cloned into the pETDuet-1 vector (Novagen). The REV3L gene was removed from the REV3L-pETDuet-1 vectors using XhoI/NotI digestion, and the resulting fragments were inserted into the pOZN vector (contains a Flag-HA tag on the N-terminal side of the inserted gene [28]). For the pCDH vector, the XhoI/NotI fragment from the full-length REV3L-pETDuet-1 vector was inserted into the pCDH-EF1α-Flag-HA-MCS-IRES-Puro vector (System Biosciences). All vectors were completely sequenced to verify the integrity of the REV3L gene and the plasmid backbone. Full-length Flag-HA tagged REV3L can be expressed from this cDNA [19].
The pOZN-REV3L or pCDH-REV3L vectors were transfected into HEK-293T cells using lipofectamine 2000 (Life Technologies), together with the retroviral packaging vectors psPAX2 (plasmid 12260, Addgene) and pMD2.G (plasmid 12259, Addgene). 48 hr later, the media (containing pOZ or pCDH lentivirus) was collected. It was filtered, and polybrene was added to 4 μg/mL. This media was added to plates of immortalized MEFs (pOZ) or flasks of BL2 cells (pCDH). 48 hr later, the cells began selection for puromycin expression (pCDH, 10 day incubation), or for IL2R expression (pOZ). The latter required incubation of the infected cells with IL2R-antibody conjugated magnetic beads followed by washing of the beads (as in [56, 57]; IL2R antibody from Millipore, 05–170). This was repeated 5 times. The population was then sorted for single-cells, and clones were selected and verified. The cells were confirmed to contain both the N and C-terminal portions of the REV3L expression construct using the following PCR primers: NFwd: 5’ TAC ACA GTC CTG CTG ACC AC 3’, NRev: 5’ GAG GTA AGG AAA GAT GCC ATG TAG 3’, CFwd: 5’ ACC TAA CTC AGC ATG GCA TCT G 3’, CRev: 5’ CGG AAT TGA TCC GCT AGA G 3’ (at an annealing temperature of 50°C).
Expression of the recombinant human REV3L was confirmed using a human-specific Taqman assay (Life Technologies) at the exon 14–15 boundary: Ex14Fwd: 5’ CAC CTG GCC TTA GCC CAT TAT 3’, Ex15Rev: 5’ CTC TTC TAA GAG TGT CAG TAT TAC TTC CTT TC 3’ Probe: FAM-MGB-5’ CAA CAG AAC CAA AAA CA 3’. In order to compare the recombinant expression to that of endogenous mouse Rev3l, we designed a set of primers and a probe that would recognize both human and mouse Rev3l, and would not amplify any knockout transcript. The primers/probe were at the exon 26/27 boundary: Ex26Fwd: 5’ GTG AAT GAT ACC AAG AAA TGG GG 3’; Ex27Rev: 5’ GTG AAT GAT ACC AAG AAA TGG GG 3’; Probe: FAM-MGB-5’ TAC TGA CAG TAT GTT TGT 3’. An additional gene expression analysis was completed on the hREV3L-expressing BL2 cells in order to distinguish the endogenous REV3L transcript (which was expressed at approximately equal levels in the REV3L knockout and wild-type BL2 cells) from the exogenously expressed REV3L. We used primers and a probe that crossed the FLAG tag on the exogenous gene: FlagFwd: 5’–GTCTTTGTTTCGTTTTCTGTTCTG C– 3’; FlagRev: 5’–GCTTGTCATCGTCGTCCTTG– 3’; Probe: FAM-MGB-5’–GCT GTG ACC GGC GCC TAC TCT AG– 3’. Gene expression (with mouse or human GAPDH as an expression control) was measured on an Applied Biosystems 7900HT Fast Real-Time PCR System.
Mice
Ethics statement
All animal work in this study was done according to The University of Texas, MD Anderson Cancer Center Institutional Animal Care and Use Committee guidelines, and approved by the MD Anderson Animal Care and Use Committee (IACUC).
Construction of the targeting vector
The targeting vector construction and the FlEx strategy [32] (Cre-dependent genetic switch approach) were designed and performed by genOway (Lyon, France). The Rev3l targeting vector was constructed from C57BL/6 mouse strain genomic DNA with a long (5.7 kb) homology arm upstream of exon 27, and a short (1.5 kb) homology arm downstream of exon 27. A D2773A/D2775A Rev3l mutant exon 27 and a Neo cassette (selection marker flanked by FRT sites for use in Flp-mediated excision) were inserted into intron 26. The targeting vector also incorporated a diphtheria toxin negative selection cassette. The action of Cre recombinase switched the allele to express mutant REV3L via the use of a combination of specifically oriented loxP and lox511 sites flanking the exons, as in Fig 4A.
Screening of Rev3lD2773A/D2775A-Neo targeted ES cell clones
Linearized targeting vector was transfected into C57BL/6 ES cells (genOway, Lyon, France) according to genOway's electroporation procedures (i.e. 5 x 106 ES cells with 40 μg of linearized plasmid, 260 V, 500 μF). Positive selection started 48 hr after electroporation in medium containing 200 μg/ml of G418 (150 μg/ml of active component, Life Technologies, Inc.). Rev3l resistant clones were isolated and amplified in 96-well plates. Duplicates of 96-well plates were made. The set of plates containing ES cell clones amplified on gelatin were genotyped by both PCR and Southern blot analysis.
For PCR analysis, one primer pair was designed to amplify sequences spanning the 3’ homology region. This primer pair was designed to specifically amplify the targeted locus:
Forward (Neo cassette): 5’-ATGCTCCAGACTGCCTTGGGAAAAG-3'
Reverse: 5'-CTGGGGTGCTACTGTTCTTGTTAGAGTGC-3'
A second PCR was designed to confirm the integration of the FlEx cassette (mutant exon 27 and loxP/lox511 sites):
Forward: 5’- GCCAAAGAGACATGCAGTGAGAAGAGTACC-3'
Reverse: 5'- TGAGTGGGCTTGCAGAAGTCAGCA-3'
PCR products were then sequenced in order to validate the presence of all FlEx elements. The targeted locus was confirmed by Southern blotting using internal and external probes for both 3’ and 5’ ends (Fig 3). Eight clones were identified as correctly targeted at the Rev3l locus.
Generation of mosaic mice and breeding scheme
Clones were microinjected into albino C57BL/6 blastocysts, and gave rise to male mosaics with a significant ES cell contribution (as determined by a black coat color). Mice were bred to C57BL/6 mice expressing the Flp recombinase to remove the Neo cassette (Rev3lD2773A/D2775A-flox mice), which resulted in the inducible allele gene architecture. These mice were then crossed with CMV-Cre mice to remove the endogenous exon 27 and allow expression of the mutant exon 27 cassette, resulting in the constitutive ASM allele (Rev3lD2773A/D2775A mice) (Fig 3A).
Genotyping of the Rev3lD2773A/D2775A-flox inducible mouse line
The following genotyping primers were used to genotype the inducible Rev3lD2773A/D2775A-flox allele:
Forward: 5’- GCCAAAGAGACATGCAGTGAGAAGAGTACC-3'
Reverse: 5'- TGAGTGGGCTTGCAGAAGTCAGCA-3'
No amplification is expected for the wild-type allele, 1680-bp for the Rev3lD2773A/D2775A-flox allele, 3353-bp for the Rev3lD2773A/D2775A-Neo allele. Animals were then validated by Southern blot analysis using a 3’ external probe: the wild-type allele gives rise to a 15 kb signal while the Rev3lD2773A/D2775A-flox allele gives rise to a 10.4 kb signal, and the Rev3lD2773A/D2775A-Neo allele is expected to give a signal at 11.5 kb.
Genotyping of the Rev3lD2773A/D2775A constitutive mouse line
The following genotyping primers were used to genotype the induced Rev3lD2773A/D2775A mice:
Forward: 5’- GCCAAAGAGACATGCAGTGAGAAGAGTACC-3'
Reverse: 5'- TGAGTGGGCTTGCAGAAGTCAGCA-3'
No amplification is expected for the wild-type allele, 1680-bp for the Rev3lD2773A/D2775A-flox allele, 1131 bp for the Rev3lD2773A/D2775A allele.
Animals were then validated by Southern blot analysis using a 3’ external probe: the wild-type allele gives rise to a 15 kb signal, the Rev3lD2773A/D2775A allele gives rise to a 9.5 kb signal, and the Rev3lD2773A/D2775A-Neo allele is expected to give a signal at 11.5 kb (Fig 3).
The presence of the mutant allele in the mice can be confirmed by PCR using the following primers: ASMWTFwd: 5’ TTG GGG CAT TGG TTT ACA GGT GGG 3’ and ASMWTRev: 5’ GCT GCT GAT ACT ACT ACT ACC ACC ACC ACT ACC 3’. Using these primers, the wild-type allele produces a 236 bp product, and the mutant allele a 345 bp product (at an annealing temperature of 65°C). Heterozygous mutant mice (which were maintained as pure C57BL/6 mice) were crossed in an attempt to observed homozygous mutant progeny. When this was unsuccessful, embryos were isolated at days 8.5–12.5 dpc. No viable embryos that were homozygous mutant were identified. To produce Rev3l mutant/floxed mice, Rev3l+/M mice were crossed to mT/mG+/+, Rev3l-/lox mice (the latter were on a C57BL6/N; 129 strain).
To investigate the phenotypes of animals and cell lines that were knockout for both Rev3l and DNA polymerase η, we obtained mice from P. Gearhart with the genotype Rev3l-/lox Polh-/- (Rev3L heterozygous and Polh knockout, as in [58]). These mice were in a mixed C57BL/6 and 129X1 background (formerly termed 129/SvJ) background and all had a pure white color. SNP analysis by the MD Anderson Genetics Services Core showed that the mice analyzed were homozygous for 129X1 alleles throughout the region of chromosome 7 flanking the albino Tyr locus. Because the 129Sv strain contains a nonsense mutation in pol ι [54] we genotyped pol ι in these Rev3l-/lox, Polh-/- mice and determined that they harbored the Poli+/+ C57BL/6 allele (homozygous for wild-type pol ι). After crossing Rev3l-/lox Polh-/- mice, the resulting progeny were genotyped as in Lange et al [8], and Saribasak et al [58]. Embryos were also isolated at e9.5 and e10.5 dpc, but no viable Rev3l-/-, Polh-/- embryos were identified. To study the phenotypes of Rev3l and pol η deficient cell lines, we crossed Rev3l-/lox, Polh-/- mice and isolated MEFs with this genotype.
DNA damage sensitivity
To test sensitivity to chemical DNA damaging agents, the immortalized MEFs or BL2 cells were plated into white 96-well plates (immortalized MEFs– 5,000 cells/well; BL2 cells– 10,000 cells/well). The following day, various concentrations of cisplatin (Sigma) or bleomycin (Sigma) were added to the wells, and the cells were incubated for 48 hr. Then the cells were lysed, a reagent was added that emits light in the presence of ATP (ATPLite One Step, Perkin Elmer), and luminescence was measured using a plate reader (Biotek Synergy II). The luminescence measurement was normalized to undamaged control. To test cisplatin sensitivity in Rev3l-deleting primary MEFs, 1 day after deleting the Rev3l floxed allele with AdCre, the cells were plated into white 96-well plates (10,000 cells/well). On day 3, cisplatin at various concentrations was added, and the cells were incubated for 5 days. Then ATP content was measured by luminescence, as above.
To test sensitivity of immortalized MEFs or BL2 cells to UVC radiation, 3 x 105 cells were pelleted and resuspended in 300 μL of phosphate-buffered saline. Three 100 μL drops were placed into the middle of a plastic dish and 10 μL aliquots from each were plated into 100 μL of growth media in a white 96-well plate after 0, 2.5, 5, 7.5, 10, 15 or 20 J/m2 UVC radiation at a fluence of 0.4 J/m2 s-1. 48 hr after irradiation, ATP content was measured as above.
Immunofluorescence
To measure the formation of DNA double-strand breaks, immortalized MEFs were plated in an 8-well chamber slide. The following day they were fixed and stained for DAPI, 53BP1 and γ-H2AX, as in Lange et al [8]. BL2 cells were applied to microscope slides using a Cytospin (Thermo Scientific), and then fixed and stained as with the MEFs. Immunofluorescence images were photographed through a Leica DMI6000B microscope. Micronuclei were counted based on small, separate DAPI foci associated with DAPI-stained nuclei. 53BP1 foci per cell were counted using the CellProfiler program 1 [59] with a threshold correction factor of 1.7. To measure DNA double-strand breaks in the primary MEFs, cells were plated into 8-well chamber slides 7 days after deletion of the Rev3l floxed allele using AdCre. 48 hr later they were fixed and stained for DAPI, 53BP1 and γ-H2AX as above. Photographs of the immunofluorescence were taken on the Leica microscope, and cells containing double-strand breaks were scored as those with 3 or more 53BP1 + γ-H2AX foci.
Chromosomal analysis
Assessment of the Rev3L-/Δ and Rev3L+/Δ cell lines for sister chromatid exchanges (SCEs) was as described [57]. BrdU (10 μM) was added to growing TAg-immortalized MEFs for a period of two cell cycles, followed by a 4 hr incubation with 0.02 μg/mL colcemid. The cells were then harvested and incubated with hypotonic solution (0.075 M KCl) for 10 min at 37°C. Then 50 μL of fresh fixative solution (3:1 methanol:acetic acid) was added and the cells were pelleted at 1000 rpm for 10 min. After aspiration of the supernatant, 5 mL of fresh 4°C Carnoy’s fixative (6:3:1 ethanol: chloroform: glacial acetic acid) was added dropwise to the pellet and the cells were incubated at 4°C for 30 min followed by centrifugation for 10 min at 1000 rpm at 4°C. The supernatant was aspirated, and this process was repeated. 1 mL of Carnoy’s fixative was added to the final cell pellet and the cells were dropped onto clean microscope slides in a humid environment to favor chromosome spreading. The slides were stained for 45 min in 0.5X SSC buffer containing 2 μg/mL Hoechst 33258 for 45 min, and then were washed twice in SSC buffer for 5 min each. The slides were then immersed in 0.5X SSC buffer and exposed to UVA light (350 nm wavelength, 15 W) at a distance of 10 cm for 1 hr. Then the slides were incubated for 1 hr in fresh 0.5X SSC buffer at 60°C, and were stained for 15 min with 3% Giemsa dye in Sorenson’s buffer (Sigma, diluted 1:15 in 0.025 M KH2PO4 pH 6.8). The chromosome spreads were viewed at 600X magnification under oil. Thirty to thirty-five chromosome spreads were counted for each genotype, and both total chromosome number and number of SCEs was assessed.
Statistical analysis
Analysis of cells with associated micronuclei or > 2 53BP1 + γ-H2AX foci was done using one-way ANOVA with Tukey’s multiple comparisons test (P < 0.05). Statistical analysis of cell survival after cisplatin or UVC treatment, or of cell growth, was done using linear regression analysis, and the lines were compared based on equality of slope and intercepts.
Supporting Information
Mammalian REV3L is about twice as long as S. cerevisiae Rev3, owing largely to a central domain of 1500 amino acids. A Blast search using the central domain detects similarity to the KIAA2022 gene on the human X chromosome, as we noted previously [60]. The KIAA2022 protein is predicted to have 1516 amino acids, largely encoded by exon 3 of the four exon gene [61]. KIAA2022 is disrupted in a family with X-linked mental retardation [61]. Most of the central domain of mammalian REV3L is encoded by a single large exon (Exon 14, 4162 bp in human REV3L), encoding 1386 amino acids. A Clustal X alignment, using default parameters, was generated by aligning this region of the indicated REV3L orthologs with the 1458 residue human KIAA2022 exon 3-encoded region from the same species. Most homology between the KIAA2022 and REV3L gene products is confined to a region of about 250 amino acids, with the three regions of highest similarity shown here. The third region includes a proposed nuclear localization signal [62]. Part of KIAA2022 appears to have been retrotransposed to the genome of a multicellular eukaryote ancestor of the REV3L gene. This is an example of the frequent gene traffic between the mammalian X chromosome and autosomes observed during evolution [63].
(TIFF)
Two reported human REV3L knockout lines designated 332 and 504 were derived from the Burkitt lymphoma cell line BL2 [31]. We expressed human REV3L in these cell lines using a pCDH vector system with puromycin as a selectable marker (B-cells express IL2R, preventing the use of pOZ vectors [64]). We have successfully expressed REV3L in human cells using pCDH [19]. (A) Expression of recombinant Flag-tagged empty vector in BL2 WT cells or recombinant Flag-tagged REV3L in 332 cells. The 332 and 504 cells still expressed detectable levels of endogenous REV3L cDNA, but expression of exogenous recombinant REV3L could be achieved above the endogenous level. Expression of exogenous REV3L cDNA was confirmed using primers specific to that cDNA. (B) Doubling time (in hr) of BL2 WT, REV3L-deficient 332 and 504 cells, and 332 cells expressing exogenous REV3L. Expression of wild type REV3L cDNA did not alter the doubling time of these cells. (C) Survival of the same cell lines as in (B) as measured by ATP concentration 48 hr after addition of cisplatin. The 332 and 504 cell lines were not more sensitive to cisplatin than the parental cells. (D) Survival of the cell lines as measured by ATP concentration 48 hr after ultraviolet C (UVC) radiation. Both lines appeared moderately UVC radiation-sensitive, but REV3L re-expression did not rescue this sensitivity. (E) Quantification of percent of BL2 cell nuclei with associated micronuclei. (F) Quantification of cells with fewer than 3, 3 to 5, 6 to 10 or greater than ten 53BP1 foci (as measured using CellProfiler). There was no significant elevation in the spontaneous incidence of DNA double-strand breaks in 332 or 504 cells (compare S2E and S2F Fig with Fig 2C and 2D). Data represent mean ± SEM.
(TIFF)
The major transcript variants of human REV3L are shown. Transcript variant 1 (NM_002912) encodes DNA polymerase ζ catalytic subunit isoform a, with 33 exons translating to 3130 aa. Transcript variant 2 (NM_001286431) encodes DNA polymerase ζ catalytic subunit isoform b, with 35 exons translating to 3052 aa. The two additional exons in transcript variant 2 are indicated (yellow). A short upstream ORF is also present in transcript variant 2. Gueranger et al., who made the 332 and 504 cell lines, designed a targeting strategy with the intention of removing the exon encoding the initiating ATG codon for REV3L, and a downstream exon [31]. In retrospect, this targeting was designed for transcript variant 2, and affects exons 3 and 4 of the major REV3L transcript variant 1. The bottom part of the figure shows the location of the primer pair given in [31] to assess targeting of the neo marker intended to disrupt exon 5. The primer 3'-KO-zeta-2 for this analysis is located 100 kb away from exons 5, a distance not compatible with PCR genotyping. We note also that both primers used to test absence of Rev3l mRNA by Gueranger et al. are located in the deleted region, and therefore no PCR product would be generated in cells even if REV3L mRNA were present.
(TIFF)
Acknowledgments
We thank David Trono for performing the SCE analysis and J. Michael Liskay (Oregon Health Sciences University) for archiving some of the p53-defective MEFs. We appreciate discussions and comments on the manuscript from Amanda McCullough and Sara Martin. We are grateful for the help provided by University of Texas MD Anderson Cancer Center core facilities (Genetic Services Core, Characterized Cell Line Core, Research Support and Animal Facility) and Department of Epigenetics & Molecular Carcinogenesis core facilities (Molecular Biology Core, Flow Cytometry and Imaging Core).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institutes of Health grants CA132840 and CA097175 (RDW) www.nih.gov and by the Grady F. Saunders Ph.D. Distinguished Research Professorship from MD Anderson Cancer Center (RDW) www.mdanderson.org. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. 10.1038/nrc2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen X, Jun S, O'Neal LE, Sonoda E, Bemark M, Sale JE, et al. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA). J Biol Chem. 2006;281(20):13869–72. . [DOI] [PubMed] [Google Scholar]
- 3.Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, et al. Polζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206(2):477–90. Epub 2009/02/11. 10.1084/jem.20080669 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto K, Cho Y, Yang IY, Akagi J, Ohashi E, Tateishi S, et al. The vital role of polymerase ζ and REV1 in mutagenic, but not correct, DNA synthesis across benzo[a]pyrene-dG and recruitment of polymerase ζ by REV1 to replication-stalled site. J Biol Chem. 2012;287(12):9613–22. 10.1074/jbc.M111.331728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–93. Epub 2009/01/21. 10.1038/emboj.2008.281 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan GN, Wittschieben JP, Wittschieben BØ, Wood RD. DNA polymerase ζ (polζ) in higher eukaryotes. Cell Res. 2008;18(1):174–83. . [DOI] [PubMed] [Google Scholar]
- 7.Lange SS, Bedford E, Reh S, Wittschieben JP, Carbajal S, Kusewitt DF, et al. Dual role for mammalian DNA polymerase ζ in maintaining genome stability and proliferative responses. Proc Natl Acad Sci USA. 2013;110(8):E687–E96. 10.1073/pnas.1217425110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange SS, Wittschieben JP, Wood RD. DNA polymerase ζ is required for proliferation of normal mammalian cells. Nucleic Acids Res. 2012;40(10):4473–82. Epub 2012/02/10. 10.1093/nar/gks054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittschieben JP, Gollin SM, Reshmi SC, Wood RD. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 2006;66(1):134–42. . [DOI] [PubMed] [Google Scholar]
- 10.Zander L, Bemark M. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair (Amst). 2004;3(7):743–52. . [DOI] [PubMed] [Google Scholar]
- 11.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, et al. Lack of DNA polymerase θ (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172(2):165–74. Epub 2009/07/28. 10.1667/RR1598.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Xie K, Zhang XQ, Pridgen EM, Park GY, Cui DS, et al. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc Natl Acad Sci USA. 2013;110(46):18638–43. 10.1073/pnas.1303958110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J Biol Chem. 2012;287(40):33847–52. 10.1074/jbc.M112.396838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanafusa T, Habu T, Tomida J, Ohashi E, Murakumo Y, Ohmori H. Overlapping in short motif sequences for binding to human REV7 and MAD2 proteins. Genes Cells. 2010. 10.1111/j.1365-2443.2009.01380.x . [DOI] [PubMed] [Google Scholar]
- 15.Wojtaszek J, Liu J, D'Souza S, Wang S, Xue Y, Walker GC, et al. Multifaceted recognition of vertebrate Rev1 by translesion polymerases ζ and κ. J Biol Chem. 2012;287(31):26400–8. Epub 2012/06/16. 10.1074/jbc.M112.380998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22(24):6621–30. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, D'Andrea AD, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J Biol Chem. 2012;287(40):33836–46. Epub 2012/08/04. 10.1074/jbc.M112.394841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pozhidaeva A, Pustovalova Y, D'Souza S, Bezsonova I, Walker GC, Korzhnev DM. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochemistry. 2012;51(27):5506–20. 10.1021/bi300566z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomida J, Takata K, Lange SS, Schibler AC, Yousefzadeh MJ, Bhetawal S, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 2015;43(2):1000–11. 10.1093/nar/gku1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe N, Mii S, Asai N, Asai M, Niimi K, Ushida K, et al. The Rev7 Subunit of DNA Polymerase ζ Is Essential for Primordial Germ Cell Maintenance in the Mouse. The Journal of biological chemistry. 2013. Epub 2013/03/07. 10.1074/jbc.M112.421966 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirouz M, Pilarski S, Kessel M. A critical function of Mad2l2 in primordial germ cell development of mice. PLoS Genet. 2013;9(8):e1003712 10.1371/journal.pgen.1003712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci USA. 1998;95(12):6876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8(1):125–32. Epub 2011/11/29. 10.1038/nchembio.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111(8):2954–9. 10.1073/pnas.1324001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci USA. 2012. Epub 2012/06/20. 10.1073/pnas.1206052109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranovskiy AG, Lada AG, Siebler H, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerases δ and ζ switching by sharing the accessory subunits of DNA polymerase δ. J Biol Chem. 2012;287(21):17281–7. Epub 2012/04/03. 10.1074/jbc.M112.351122 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirota K, Sonoda E, Kawamoto T, Motegi A, Masutani C, Hanaoka F, et al. Simultaneous Disruption of Two DNA Polymerases, Pol η and Pol ζ, in Avian DT40 Cells Unmasks the Role of Pol η in Cellular Response to Various DNA Lesions. PLoS Genet. 2010;6(10):e1001151 10.1371/journal.pgen.1001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–44. Epub 2004/01/10. 10.1016/S0076-6879(03)70037-8 . [DOI] [PubMed] [Google Scholar]
- 29.Copeland WC, Wang TS. Mutational analysis of the human DNA polymerase α. The most conserved region in α-like DNA polymerases is involved in metal-specific catalysis. J Biol Chem. 1993;268(15):11028–40. . [PubMed] [Google Scholar]
- 30.Polesky AH, Steitz TA, Grindley ND, Joyce CM. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990;265(24):14579–91. . [PubMed] [Google Scholar]
- 31.Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, et al. Role of DNA polymerases η, ι and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst). 2008;7(9):1551–62. Epub 2008/07/01. 10.1016/j.dnarep.2008.05.012 . [DOI] [PubMed] [Google Scholar]
- 32.Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21(5):562–5. 10.1038/nbt811 . [DOI] [PubMed] [Google Scholar]
- 33.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Current biology. 2000;10(19):1221–4. Epub 2000/10/26. . [DOI] [PubMed] [Google Scholar]
- 34.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. Epub 2007/09/18. 10.1002/dvg.20335 . [DOI] [PubMed] [Google Scholar]
- 35.Albertella MR, Green CM, Lehmann AR, O'Connor MJ. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65(21):9799–806. . [DOI] [PubMed] [Google Scholar]
- 36.de Boer J, Donker I, de Wit J, Hoeijmakers JH, Weeda G. Disruption of the mouse xeroderma pigmentosum group D DNA repair/basal transcription gene results in preimplantation lethality. Cancer Res. 1998;58(1):89–94. . [PubMed] [Google Scholar]
- 37.Andressoo JO, Mitchell JR, de Wit J, Hoogstraten D, Volker M, Toussaint W, et al. An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell. 2006;10(2):121–32. . [DOI] [PubMed] [Google Scholar]
- 38.Winkler GS, Araújo SJ, Fiedler U, Vermeulen W, Coin F, Egly J-M, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–66. [DOI] [PubMed] [Google Scholar]
- 39.Kuper J, Braun C, Elias A, Michels G, Sauer F, Schmitt DR, et al. In TFIIH, XPD Helicase Is Exclusively Devoted to DNA Repair. PLoS Biol. 2014;12(9):e1001954 10.1371/journal.pbio.1001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33(4):1280–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanemaru Y, Suzuki T, Niimi N, Gruz P, Matsumoto K, Adachi N, et al. Catalytic and non-catalytic roles of DNA polymerase kappa in the protection of human cells against genotoxic stresses. Environ Mol Mutagen. 2015;56(8):650–62. 10.1002/em.21961 . [DOI] [PubMed] [Google Scholar]
- 42.Takezawa J, Aiba N, Kajiwara K, Yamada K. Caffeine abolishes the ultraviolet-induced REV3 translesion replication pathway in mouse cells. Int J Mol Sci. 2011;12(12):8513–29. Epub 2012/01/25. 10.3390/ijms12128513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, et al. Differential roles for DNA polymerases η, ζ, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30(5):1217–30. Epub 2009/12/24. 10.1128/MCB.00993-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, et al. REV1 and polymerase ζ facilitate homologous recombination repair. Nucleic Acids Res. 2012;40(2):682–91. Epub 2011/09/20. 10.1093/nar/gkr769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S, Shah NA, Joiner AM, Roberts KH, Canman CE. DNA polymerase ζ is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol Pharmacol. 2012;81(6):778–87. 10.1124/mol.111.076828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janel-Bintz R, Wagner J, Haracska L, Mah-Becherel MC, Bichara M, Fuchs RP, et al. Evidence for a Rad18-independent frameshift mutagenesis pathway in human cell-free extracts. PLoS One. 2012;7(4):e36004 10.1371/journal.pone.0036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud CA, Weill JC. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase ι. Nature. 2002;419(6910):944–7. . [DOI] [PubMed] [Google Scholar]
- 48.Shimizu T, Azuma T, Ishiguro M, Kanjo N, Yamada S, Ohmori H. Normal immunoglobulin gene somatic hypermutation in Pol κ Pol ι double-deficient mice. Immunol Lett. 2005;98(2):259–64. 10.1016/j.imlet.2004.11.022 . [DOI] [PubMed] [Google Scholar]
- 49.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204(1):17–23. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol ζ- and Pol η-dependent frameshift mutagenesis. Genetics. 2006;172(3):1487–98. 10.1534/genetics.105.052480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatterjee N, Pabla R, Siede W. Role of polymerase η in mitochondrial mutagenesis of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2013;431(2):270–3. 10.1016/j.bbrc.2012.12.119 . [DOI] [PubMed] [Google Scholar]
- 52.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, et al. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol Cell Biol. 2006;26(20):7696–706. 10.1128/MCB.01076-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, et al. Suppression of Rev3, the catalytic subunit of Pol ζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA. 2010;107(48):20786–91. Epub 2010/11/12. 10.1073/pnas.1011409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, et al. 129-derived strains of mice are deficient in DNA polymerase ι and have normal immunoglobulin hypermutation. J Exp Med. 2003;198(4):635–43. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin W, Wu X, Wang Z. A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat Res. 1999;433(2):89–98. . [DOI] [PubMed] [Google Scholar]
- 56.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15(11):1138–46. 10.1038/nsmb.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takata K, Reh S, Tomida J, Person MD, Wood RD. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nature Communications. 2013;4 10.1038/ncomms3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saribasak H, Maul RW, Cao Z, Yang WW, Schenten D, Kracker S, et al. DNA polymerase ζ generates tandem mutations in immunoglobulin variable regions. The Journal of experimental medicine. 2012;209(6):1075–81. Epub 2012/05/23. 10.1084/jem.20112234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–9. [DOI] [PubMed] [Google Scholar]
- 61.Cantagrel V, Lossi AM, Boulanger S, Depetris D, Mattei MG, Gecz J, et al. Disruption of a new X linked gene highly expressed in brain in a family with two mentally retarded males. J Med Genet. 2004;41(10):736–42. 10.1136/jmg.2004.021626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh B, Li X, Owens KM, Vanniarajan A, Liang P, Singh KK. Human REV3 DNA Polymerase Zeta Localizes to Mitochondria and Protects the Mitochondrial Genome. PLoS One. 2015;10(10):e0140409 10.1371/journal.pone.0140409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303(5657):537–40. 10.1126/science.1090042 . [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Ma A, Young F, Alt FW. IL-2 receptor alpha chain expression during early B lymphocyte differentiation. Int Immunol. 1994;6(8):1265–8. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mammalian REV3L is about twice as long as S. cerevisiae Rev3, owing largely to a central domain of 1500 amino acids. A Blast search using the central domain detects similarity to the KIAA2022 gene on the human X chromosome, as we noted previously [60]. The KIAA2022 protein is predicted to have 1516 amino acids, largely encoded by exon 3 of the four exon gene [61]. KIAA2022 is disrupted in a family with X-linked mental retardation [61]. Most of the central domain of mammalian REV3L is encoded by a single large exon (Exon 14, 4162 bp in human REV3L), encoding 1386 amino acids. A Clustal X alignment, using default parameters, was generated by aligning this region of the indicated REV3L orthologs with the 1458 residue human KIAA2022 exon 3-encoded region from the same species. Most homology between the KIAA2022 and REV3L gene products is confined to a region of about 250 amino acids, with the three regions of highest similarity shown here. The third region includes a proposed nuclear localization signal [62]. Part of KIAA2022 appears to have been retrotransposed to the genome of a multicellular eukaryote ancestor of the REV3L gene. This is an example of the frequent gene traffic between the mammalian X chromosome and autosomes observed during evolution [63].
(TIFF)
Two reported human REV3L knockout lines designated 332 and 504 were derived from the Burkitt lymphoma cell line BL2 [31]. We expressed human REV3L in these cell lines using a pCDH vector system with puromycin as a selectable marker (B-cells express IL2R, preventing the use of pOZ vectors [64]). We have successfully expressed REV3L in human cells using pCDH [19]. (A) Expression of recombinant Flag-tagged empty vector in BL2 WT cells or recombinant Flag-tagged REV3L in 332 cells. The 332 and 504 cells still expressed detectable levels of endogenous REV3L cDNA, but expression of exogenous recombinant REV3L could be achieved above the endogenous level. Expression of exogenous REV3L cDNA was confirmed using primers specific to that cDNA. (B) Doubling time (in hr) of BL2 WT, REV3L-deficient 332 and 504 cells, and 332 cells expressing exogenous REV3L. Expression of wild type REV3L cDNA did not alter the doubling time of these cells. (C) Survival of the same cell lines as in (B) as measured by ATP concentration 48 hr after addition of cisplatin. The 332 and 504 cell lines were not more sensitive to cisplatin than the parental cells. (D) Survival of the cell lines as measured by ATP concentration 48 hr after ultraviolet C (UVC) radiation. Both lines appeared moderately UVC radiation-sensitive, but REV3L re-expression did not rescue this sensitivity. (E) Quantification of percent of BL2 cell nuclei with associated micronuclei. (F) Quantification of cells with fewer than 3, 3 to 5, 6 to 10 or greater than ten 53BP1 foci (as measured using CellProfiler). There was no significant elevation in the spontaneous incidence of DNA double-strand breaks in 332 or 504 cells (compare S2E and S2F Fig with Fig 2C and 2D). Data represent mean ± SEM.
(TIFF)
The major transcript variants of human REV3L are shown. Transcript variant 1 (NM_002912) encodes DNA polymerase ζ catalytic subunit isoform a, with 33 exons translating to 3130 aa. Transcript variant 2 (NM_001286431) encodes DNA polymerase ζ catalytic subunit isoform b, with 35 exons translating to 3052 aa. The two additional exons in transcript variant 2 are indicated (yellow). A short upstream ORF is also present in transcript variant 2. Gueranger et al., who made the 332 and 504 cell lines, designed a targeting strategy with the intention of removing the exon encoding the initiating ATG codon for REV3L, and a downstream exon [31]. In retrospect, this targeting was designed for transcript variant 2, and affects exons 3 and 4 of the major REV3L transcript variant 1. The bottom part of the figure shows the location of the primer pair given in [31] to assess targeting of the neo marker intended to disrupt exon 5. The primer 3'-KO-zeta-2 for this analysis is located 100 kb away from exons 5, a distance not compatible with PCR genotyping. We note also that both primers used to test absence of Rev3l mRNA by Gueranger et al. are located in the deleted region, and therefore no PCR product would be generated in cells even if REV3L mRNA were present.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.