Osteoarthritis of the knee is a major cause of pain, disability and the use of healthcare resources among middle-aged and older people.1 Although osteoarthritis is multifactorial, it is known to have a significant genetic contribution and a number of studies have attempted to dissect such a contribution (see Valdes and Spector2 for review).
The GDF5 gene encodes the growth differentiation factor 5, a bone morphogenetic protein involved in joint formation, expressed in different joint structures, which has been shown to ameliorate tendon, ligament and bone healing after trauma in mice.3,4
A promoter polymorphism (rs143383) in GDF5 has been found to be strongly associated with both hip and knee osteoarthritis in Asian individuals,4 and is the most widely replicated genetic association with knee osteoarthritis, although much less so for hip and hand osteoarthritis.5 This variant is functional, with the lower gene expression variant having increased genetic risk.4
A large-scale meta-analysis reported the association of the major (T) allele with knee osteoarthritis achieved OR 1.15 p=9.7×10−7 and achieved p=9×10−5 (OR 1.13, 95% CI 1.06 to 1.20) when Asian subjects were excluded.5
The genome-wide statistical significance level of p<5×10−8 is increasingly seen as the threshold at which genetic associations are considered credible.6 The aim of our study was to prove that common genetic variation in the GDF5 gene is important in knee osteoarthritis beyond reasonable doubt.
We genotyped 3303 controls and 2235 knee osteoarthritis cases from the UK, Estonia and The Netherlands, added published data from the Chingford Study (259 cases and 509 controls),7 and combined with published effect size estimates from the recent large-scale meta-analysis5 using both fixed and random effects models as described in Evangelou et al.5 A full detailed description of each study cohort on recruitment, radiographic and clinical assessment is found in Hofman et al8 and Valdes et al.9 The studies were approved by the relevant ethics committees and informed consent was obtained from all study participants. Genotypes were subtracted from the genome-wide association dataset of the Rotterdam Study III with methods described previously.10 DNA from UK and Estonian study participants was genotyped by Kbioscience (Hertfordshire UK) using methods described elsewhere.7 The total, including previously reported data, is 10 103 controls and 6861 knee osteoarthritis cases of European descent and 1844 controls and 718 cases of Asian descent, which has 86% statistical power to detect the association as genome-wide significant with an OR of 1.15 or higher for the T allele. The descriptive characteristics of new samples studied are shown in table 1 along with the OR for the T allele and CI. The individual study effect sizes are shown in figure 1.
Table 1.
Descriptive statistics of study samples included, number of cases and controls for new and published studies and OR for the GDF5 rs143383 T allele
| New data
|
Published data
|
|||||
|---|---|---|---|---|---|---|
| Rotterdam study III | Estonian knee osteoarthritis study | Hertfordshire cohort plus Nottingham cases | Genetics of osteoarthritis and lifestyle | Chingford study | Ten studies from large-scale meta-analysis (*) | |
| GDF5 association data from | This study | This study | This study | This study | 7 | 5 |
| Country of origin | Netherlands | Estonia | UK | UK | UK | Europe, China, Japan |
| Ethnicity | Caucasian | Caucasian | Caucasian | Caucasian | Caucasian | Asian and Caucasian |
| No of knee osteoarthritis cases/controls | 151/1582 | 65/427 | 1141/536 | 867/758 | 259/509 | 5085/8135 |
| Knee osteoarthritis definition | TF K/L ≥2 | TF K/L ≥2 | TF or PF K/L ≥2 or TKR | TKR | TF K/L ≥2 | Mixed |
| Women % | 56 | 69 | 52 | 49 | 100 | NA |
| BMI mean (SD) | 27.7 (4.7) | 28.1 (5.4) | 28.2 (5.1) | 29.3 (5.3) | 26.7 (4.7) | NA |
| Age mean (SD) | 56.0 (5.5) | 47.1 (6.4) | 67.2 (7.4) | 66.5 (7.9) | 64.1 (6.0) | NA |
| rs143383 T allele % in knee osteoarthritis/controls | 64.6/61.0% | 67.7/60.3% | 63.3/62.9% | 68.5/62.1% | 67.6/59.5% | NA |
| OR (95% CI) | 1.16 (0.91 to 1.49) | 1.38 (0.93 to 2.04) | 1.02 (0.87 to 1.1.8) | 1.32 (1.14 to 1.53) | 1.42 (1.13 to 1.77) | 1.15 (1.09 to 1.22) |
Data for all studies, the summary statistics reported for each of the individual studies were used separately and are shown in figure 1.
BMI, body mass index; K/L, Kellgren–Lawrence radiographic grade; PF, patellofemoral; TF, tibiofemoral; TKR, total knee replacement; NA, not applicable because it refers to 10 different studies.
Figure 1.
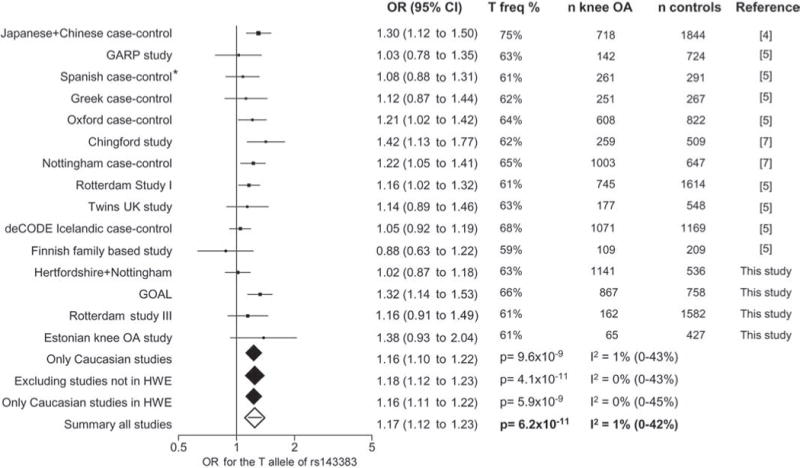
Forest plot of study-specific estimates and fixed-effects summary OR estimates and 95% CI for the association between the rs143383 polymorphism of the GDF5 gene and knee osteoarthritis (OA) in current and previously published studies. The summary statistics and corresponding p values for all studies and for sensitivity analyses excluding studies out of Hardy–Weinberg equilibrium (HWE; indicated by *) and/or Asian studies are shown. The between-study heterogeneity I2 and the respective 95% CI are also included.
The results of the combined meta-analysis show that the T allele of GDF5 rs143833 is associated with a 17% increased risk of knee osteoarthritis (OR 1.17, 95% CI 1.12 to 1.23). When all data were analysed the genetic association reached genome-wide significance with p=6.2×10−11 in all samples and with p=8.3×10−9 in European descent samples alone (figure 1). There was no significant between-study heterogeneity. The p value was even smaller when one study violating Hardy–Weinberg equilibrium5 was excluded (OR 1.18, 95% CI 1.12 to 1.23, p=4.1×10−11). Stratification according to gender did not reveal differences in the effect size (OR 1.14, 95% CI 1.05 to 1.23, p=1.6×10−3 in men vs OR 1.19, 95% CI 1.10 to 1.27, p=3.7×10−6 in women by random effects).
We have shown that the association between a functional promoter single nucleotide polymorphism in the GDF5 gene and knee osteoarthritis achieves genome-wide statistical significance. The association is consistently replicated and no significant heterogeneity is detected between studies, further strengthening the robustness of GDF5 as a risk factor for knee osteoarthritis.
Acknowledgments
The authors thank Marijn Verkerk and Karol Estrada for their help creating the genome-wide association database for RS III and Dieuwke Schiphof for scoring the knee radiographs of RSIII. The authors thank all the study participants and the participating general practitioners and pharmacists contributing to all the study cohorts.
Funding This work was supported by EC framework 7 programme grant 200800 TREAT-osteoarthritis, Arthritis Research UK (ARUK), the Medical Research Council (UK), the Oxford NIHR Musculoskeletal Biomedical Research Unit, the Estonian Science Foundation grant no 5308, the Estonian Ministry of Social Affairs grants no 9.6-4/2035 and 12.1-5/597 and by AstraZeneca, Macclesfield, UK. The generation and management of genome-wide association genotype data for the Rotterdam Study is supported by The Netherlands Organisation of Scientific Research NWO Investments (no 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2) and The Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) (project no 050-060-810).
Footnotes
Competing interests RM is an Astra Zeneca plc employee and owns Astra Zeneca stock. All other authors declare no competing interests.
Patient consent Obtained.
Ethics approval This study was conducted with the approval of the the Nottingham case–control and the GOAL study protocols were approved by the Nottingham City Hospital and North Nottinghamshire ethical committees. The Hertfordshire Cohort Study was approved by the East and North Hertfordshire ethical committees. The medical ethics committee of Erasmus University Medical School approved the Rotterdam study III. The Ethics Committee of the University of Tartu approved the Estonian knee osteoarthritis study.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet. 2005;365:965–73. doi: 10.1016/S0140-6736(05)71086-2. [DOI] [PubMed] [Google Scholar]
- 2.Valdes AM, Spector TD. The clinical relevance of genetic susceptibility to osteoarthritis. Best Pract Res Clin Rheumatol. 2010;24:3–14. doi: 10.1016/j.berh.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Mikic B. Multiple effects of GDF-5 deficiency on skeletal tissues: implications for therapeutic bioengineering. Ann Biomed Eng. 2004;32:466–76. doi: 10.1023/b:abme.0000017549.57126.51. [DOI] [PubMed] [Google Scholar]
- 4.Miyamoto Y, Mabuchi A, Shi D, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–33. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 5.Evangelou E, Chapman K, Meulenbelt I, et al. Large-scale analysis of association between GDF5 and FRZB variants and osteoarthritis of the hip, knee, and hand. Arthritis Rheum. 2009;60:1710–21. doi: 10.1002/art.24524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evangelou E, Maraganore DM, Annesi G, et al. Non-replication of association for six polymorphisms from meta-analysis of genome-wide association studies of Parkinson’s disease: large-scale collaborative study. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:220–8. doi: 10.1002/ajmg.b.30980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valdes AM, Spector TD, Doherty S, et al. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Ann Rheum Dis. 2009;68:1916–20. doi: 10.1136/ard.2008.102236. [DOI] [PubMed] [Google Scholar]
- 8.Hofman A, Breteler MM, van Duijn CM, et al. The Rotterdam Study: 2010 objectives and design update. Eur J Epidemiol. 2009;24:553–72. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdes AM, Arden NK, Tamm A, et al. A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis. Osteoarthr Cartil. 2010;18:699–704. doi: 10.1016/j.joca.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Kerkhof HJ, Lories RJ, Meulenbelt I, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62:499–510. doi: 10.1002/art.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]


