Abstract
Early genome-wide association (GWA) studies on Parkinson’s disease (PD) have not been able to yield conclusive, replicable signals of association, perhaps due to limited sample size. We aimed to investigate whether association signals derived from the meta-analysis of the first two GWA investigations might be replicable in different populations. We examined six single-nucleotide polymorphisms (SNPs) (rs1000291, rs1865997, rs2241743, rs2282048, rs2313982, and rs3018626) that had reached nominal significance with at least two of three different strategies proposed in a previous analysis of the original GWA studies. Investigators from the “Genetic Epidemiology of Parkinson’s Disease” (GEOPD) consortium were invited to join in this study. Ten teams contributed replication data from 3,458 PD cases and 3,719 controls. The data from the two previously published GWAs (599 PD cases, 592 controls and 443 sibling pairs) were considered as well. All data were synthesized using both fixed and random effects models. The summary allelic odds ratios were ranging from 0.97 to 1.09 by random effects, when all data were included. The summary estimates of the replication data sets (excluding the original GWA data) were very close to 1.00 (range 0.98–1.09) and none of the effects were nominally statistically significant. The replication data sets had significantly different results than the GWA data. Our data do not support evidence that any of these six SNPs reflect susceptibility markers for PD. Much stronger signals of statistical significance in GWA platforms are needed to have substantial chances of replication. Specifically in PD genetics, this would require much larger GWA studies and perhaps novel analytical techniques.
Keywords: Parkinson’s disease, meta-analysis, genome-wide association
INTRODUCTION
Genome-wide screening for genetic associations is a promising approach for the exploration of the relationship between the genetic determinants and common complex diseases [McCarthy et al., 2008]. Meta-analysis is a technique that entails the combination of different studies or data sets. Recently, it has become a powerful tool for the synthesis of different genome-wide association (GWA) studies for the same phenotype [Evangelou et al., 2007; Baum et al., 2008; Zeggini et al., 2008]. The first attempt to synthesize results from GWA studies has been performed for the genetics of Parkinson’s disease (PD) [Evangelou et al., 2007]. The effect estimates from a two-tier design study (LEAPS) [Maraganore et al., 2005] and from a single-tier design (NINDS) [Fung et al., 2006] study were synthesized to identify potentially significant signals. Given the limited sample sizes of both combined GWA data sets, the attained statistical significance for the summary results were not impressive. Nevertheless, some tentative signals were noted and six single-nucleotide polymorphisms (SNPs) were found to be nominally significant at the P <0.05 level even by random effects calculations with at least two of the three meta-analytic strategies that were investigated.
The replication of the findings from GWAs and the detection of loci with realistic effect sizes require the use of large sample sizes [Burton et al., 2008]. In order to assess if the signals from the meta-analysis reflect true findings, we co-ordinated an independent large-scale multicenter international replication effort. Our study incorporated also data from the previous published GWAs [Maraganore et al., 2005; Fung et al., 2006] along with the replication data. Meta-analytical techniques were applied for the combination of the available data.
METHODS
Study Population
Investigators consisting the “Genetic Epidemiology of Parkinson’s Disease” (GEOPD) consortium were invited to join in this study. Ten teams participated in the replication study, contributing with individual-level data on PD cases and PD-free controls. Data from the two already published GWAs were also available for comparison with the replication data and for inclusion in the overall analysis.
Polymorphisms
The following six polymorphisms were evaluated: rs1000291, rs1865997, rs2241743, rs2282048, rs2313982, and rs3018626. These SNPs had been found to have nominally statistical significant association even by random effects calculations, in at least two of the three strategies used for the combination of data from the first two published GWA studies: enhancement of first-stage data, enhancement of replication data, and joint analysis of all available data sets [Evangelou et al., 2007]. Three of the six had nominally significant results with all three strategies and no or minimal heterogeneity (I2 0–15%) across the combined data sets. The summary data from the combined GWA data sets for these six polymorphisms in the joint analysis varied from 0.76 to 0.81 (for protective effects) and 1.25 to 1.65 (for susceptibility effects) by fixed effects and from 0.76 to 0.80 and 1.25 to 1.67 by random effects analyses. P-values ranged from 0.0001 to 0.004 by fixed effects and from 0.0001 to 0.038 by random effects.
Genotyping of DNA samples was undertaken either on-site or through commercial contract. Methods that were used was fluorescence polarization single-base extension (Wirdefeldt), allelic discrimination (real-time PCR) (Tan), mass spectrometry (van Broeckhoven), TaqMan assay (Hadjigeorgiou, Ferrarese, Annesi), Illumina genotyping chips (Illumina Humanhap550) (Sharma), Sequenom Platform (Mellick), direct sequencing (Brice), and Illumina Golden Gate assay (Elbaz).
For teams where the genotyping call rate was <95%, we requested a second effort to genotype the missing samples, so that the 95% threshold could be exceeded for completeness of genotyping information. We also used an exact test to examine whether there is a deviation from the Hardy–Weinberg equilibrium (HWE) among controls in each team. Deviation for HWE was deemed significant for P-value <0.05. In the presence of significant deviation from HWE, the specific teams were also asked to consider re-genotyping their samples. We specified upfront that we would perform for each SNP sensitivity analyses that would exclude data from teams where the missing rate was >5% or there was nominally significant deviation from HWE in the controls.
Data Synthesis
The natural log odds ratio and their respective standard errors were calculated for each evaluated SNP within each team. For consistency, we computed all the effect estimates based on the major versus minor allele contrast. Assignment of minor allele was based on the allele frequencies of the control samples in the NINDS GWA study. Then the log ORs were synthesized using both fixed [Lau et al., 1997] and random effects models [DerSimonian and Laird, 1986]. In fixed effects models, it is assumed that the risk of the alleles is always the same across the comparisons. In random effects models, the risk is varying around an overall average. Thus, random effects models can be considered more conservative. In absence of heterogeneity, summary effects obtained from fixed and random effects models coincide. We tested the presence of between-study heterogeneity using Cochran’s Q test and we calculated the extent of inconsistency using I2, which ranges from 0% to 100% [Higgins and Thompson, 2002; Higgins et al., 2003]. We also calculated 95% confidence intervals for I2 [Ioannidis et al., 2007].
In the main analysis, all the effect estimates from the replication effort and the published GWA studies were considered. Another analysis was also undertaken where summary effects were provided separately for replication and already published data. Even for genuine associations, it is expected that effect sizes may be inflated in the discovery (GWA) phase [Zollner and Pritchard, 2007] [Ioannidis, 2008b] and this may result in heterogeneity in the data when both the GWA and replication data are considered.
As above, in a sensitivity analysis we excluded studies that deviated from HWE and studies that had genotyping call rate < 95% in cases or in controls groups only.
We calculated the Bayes factor for nominally significant associations (uncorrected for multiple comparisons), considering a spike and smear prior [Ioannidis, 2008a] and assuming that average effect sizes for associations in PD may reflect odds ratios of 1.3. Bayes factor evaluates if a nominally statistically significant result increases the credibility of a postulated association.
The six evaluated SNPs were very common with minor allele frequencies ranging from 22.7% to 49.3% in the combined GWA data sets, with one exception (rs2313982) where the minor allele frequency was only 9.4%. For the lowest minor allele frequency among them, odds ratios of 1.25 would require almost 6,800 cases and controls to have 80% power to replicate at alpha =0.05, and about 10,450 cases and controls to have 80% power to replicate at alpha =0.05/6 =0.0083, assuming 1:1 allocation and homogeneous effects. For minor allele frequencies of 22.7%, the respective numbers are 3,400 and 5,240 cases and controls for odds ratios of 1.25 for P =0.05 and 0.0083, respectively. For odds ratios of 0.8, the respective numbers are 3,840 cases and controls for P =0.05 and 5,920 cases and controls for P =0.0083.
All analyses were performed with Intercooled Stata 10 (College Station, TX).
RESULTS
Characteristics of Participating Studies
Ten replicating teams contributed 3,458 cases and 3,719 controls. Characteristics of these 10 study populations are shown in Table I. The previously published GWAs included (a) 443 sibling pairs (family-based design) (cases were enrolled at the Department of Neurology of the Mayo Clinic in Rochester, MN, and their siblings were contacted, if permitted, in order to exclude Parkinsonism via a validated tool) and 332 matched case-unrelated control pairs (LEAPS, two-tier design) [Maraganore et al., 2005] and (b) 267 publicly available samples from a cohort of PD patients and 270 neurologically normal controls (NINDS, single-tier design) with publicly available samples [Fung et al., 2006].
TABLE I.
Characteristics of the Participating Teams Evaluating the Replication of the Six SNPs
| Team | Location | Cases (N) | Controls (N) | Age at onset, cases mean (SD) | Age at study, cases mean (SD) | Age at study, controls mean (SD) | Gender, females (%) | Familial PDa (%) | Diagnostic criteria | Genotyping methods |
|---|---|---|---|---|---|---|---|---|---|---|
| Annesi | Italy | 200 | 200 | 61.2 (10.5) | 67.0 (9.2) | 44.7 (15.5) | 49.25 | NA | UKPDBB | TaqMan assay |
| Brice | France | 291 | 250 | 47.6 (10.1) | 57.8 (11.5) | 57.8 (11.9) | 40.7 | 0 | At least two of the Parkinsonian triad of signs (bradykinesia, rigidity, rest tremor), at least 30% improvement after L-dopa therapy and absence of other PD case among first-degree relatives | Direct sequencing |
| Elbaz | France | 209 | 501 | 63.5 (7.3) | 67.0 (7.0) | 66.9 (7.2) | 41.55 | 6.5 | Bower | Illumina Golden Gate assay |
| Ferrarese | Italy | 100 | 100 | 58.8 (8.2) | 65.5 (7.0) | 65.0 (6.9) | 44.5 | NA | Gelb | TaqMan assay |
| Hadjigeorgiou | Greece | 300 | 300 | 64.4 (9.7) | 69.5 (9.4) | 70.0 (8.4) | 42.64 | 0 | Bower | TaqMan assay |
| Mellick | Australia | 1,013 | 681 | 59.4 (11.5) | 72.3 (10.7) | 71.2 (10.9) | 46.93 | 24.6 | UKPDBB | Sequenom Platform |
| Sharma | Germany | 742 | 944 | NA | NA | NA | NA | NA | UKPDBB | Illumina Humanhap 550 |
| Tan | Singapore | 203 | 201 | 61.5 (10.0) | 66.3 (10.0) | 53.5 (11.0)) | 48.76 | 5.9 | UKPDBB | Allelic discrimination (real-time PCR) |
| Van Broeckhoven | Belgium | 305 | 364 | NA | 60.3 (11.3) | 60.8 (14.4) | 51.7 | NA | Pals/Engelborghs | Mass spectrometry |
| Wirdefeldt | Sweden | 95 | 179 | 65.9 (10.9) | 75.7 (8.6) | 74.9 (9.1) | 48.18 | 25.2 | Gelb | Fluorescence polarization single-base extension |
UKPDBB, United Kingdom Parkinson’s Disease Brain Bank.
Proportion of PD cases that have family history of PD.
Analysis Including All Data
Allele frequencies per team and per SNP of interest are shown in Table II. The meta-analysis of these data did not reveal any nominally statistically significant associations by random effects with the exception of rs3018626. The summary OR for this SNP was 1.09 (95% CI 1.01–1.17) with P =0.037 uncorrected for multiple comparisons (Table II). With fixed effects analysis the summary estimate for the specific SNP was also nominally statistically significant (P =0.032 uncorrected) and low heterogeneity was observed (I2 =6%) (Table II).
TABLE II.
Allele Counts for Each of the Six SNPs in Each of the Replication Teams
| Team | Cases, N |
Controls, N |
Cases (major allele/minor allele)
|
Controls (major allele/minor allele)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1000291, A/G |
rs1865997, A/G |
rs2241743, T/C |
rs2282048, A/G |
rs2313982, C/T |
rs3018626, G/T |
rs1000291, A/G |
rs1865997, A/G |
rs2241743, T/C |
rs2282048, A/G |
rs2313982 C/T |
rs3018626, G/T |
|||
| Annesi | 200 | 200 | 216/184 | 253/147 | 237/163 | 219/181 | 376/24 | 308/92 | 205/195 | 243/157 | 259/141 | 205/195 | 365/35 | 314/86 |
| 0.54/0.46 | 0.63/0.37 | 0.59/0.41 | 0.55/0.45 | 0.94/0.06 | 0.77/0.23 | 0.51/0.49 | 0.61/0.39 | 0.65/0.35 | 0.51/0.49 | 0.91/0.09 | 0.79/0.21 | |||
| Brice | 291 | 250 | 295/287 | 372/210 | 339/239 | 239/237 | 521/59 | 444/134 | 252/248 | 308/192 | 298/202 | 305/195 | 463/37 | 367/133 |
| 0.51/0.49 | 0.64/0.36 | 0.58/0.42 | 0.59/0.41 | 0.90/0.10 | 0.77/0.13 | 0.505/0.495 | 0.62/0.38 | 0.60/0.40 | 0.61/0.39 | 0.93/0.07 | 0.73/0.27 | |||
| Elbaz | 209 | 501 | 219/197 | 248/166 | 256/156 | 258/140 | 390/24 | 317/99 | 543/453 | 615/377 | 604/390 | 582/362 | 934/64 | 780/216 |
| 0.53/0.47 | 0.60/0.40 | 0.62/0.38 | 0.65/0.35 | 0.94/0.06 | 0.76/0.24 | 0.55/0.45 | 0.62/0.38 | 0.61/0.39 | 0.62/0.38 | 0.94/0.06 | 0.78/0.22 | |||
| Ferrarese | 100 | 100 | 105/95 | 113/87 | 124/76 | 108/92 | 190/10 | 158/42 | 123/77 | 116/82 | 116/84 | 93/107 | 187/13 | 152/48 |
| 0.53/0.47 | 0.57/0.43 | 0.62/0.38 | 0.54/0.46 | 0.95/0.05 | 0.79/21 | 0.62/0.38 | 0.59/0.41 | 0.58/0.42 | 0.47/0.53 | 0.94/0.06 | 0.76/0.24 | |||
| Hadjigeorgiou | 300 | 300 | 287/289 | 387/183 | 336/232 | 307/273 | 546/36 | 441/135 | 333/239 | 354/238 | 345/239 | 293/307 | 570/26 | 451/145 |
| 0.50/0.50 | 0.68/0.32 | 0.59/0.41 | 0.53/0.47 | 0.94/0.06 | 0.77/0.23 | 0.58/0.42 | 0.60/0.40 | 0.59/0.41 | 0.49/0.51 | 0.96/0.04 | 0.76/0.24 | |||
| Mellick | 1,013 | 681 | 1,018/972 | 1,129/871 | 1,090/890 | 1,236/768 | 1,869/147 | 1,489/469 | 723/613 | 793/553 | 751/577 | 822/528 | 1,263/95 | 996/324 |
| 0.51/0.49 | 0.56/0.44 | 0.55/0.45 | 0.62/0.38 | 0.93/0.07 | 0.76/0.24 | 0.54/0.46 | 0.59/0.41 | 0.57/0.43 | 0.61/0.39 | 0.93/0.07 | 0.75/0.25 | |||
| Sharma | 742 | 944 | 778/706 | NA | NA | NA | 1,372/112 | 1,159/323 | 1,026/862 | NA | NA | NA | 1,727/159 | 1,456/430 |
| 0.52/0.48 | 0.92/0.08 | 0.78/0.22 | 0.54/0.46 | 0.92/0.08 | 0.77/0.23 | |||||||||
| Tan | 203 | 201 | 86/318 | 251/147 | 154/252 | 154/248 | 384/20 | 337/65 | 108/294 | 225/163 | 138/262 | 129/265 | 379/23 | 333/65 |
| 0.21/0.79 | 0.63/0.37 | 0.38/0.62 | 0.38/0.62 | 0.95/0.05 | 0.84/0.16 | 0.27/0.73 | 0.58/0.42 | 0.35/0.65 | 0.33/0.67 | 0.94/0.06 | 0.84/0.16 | |||
| Van Broeckhoven | 305 | 364 | 317/283 | 367/237 | 358/242 | 391/211 | 549/49 | 462/128 | 343/337 | 385/305 | 403/283 | 415/273 | 625/63 | 542/158 |
| 0.53/0.47 | 0.61/0.39 | 0.60/0.40 | 0.65/0.35 | 0.92/0.08 | 0.78/0.22 | 0.505/0.495 | 0.56/0.44 | 0.59/0.41 | 0.60/0.40 | 0.91/0.09 | 0.77/0.23 | |||
| Wirdefeldt | 95 | 179 | 107/83 | 114/76 | 112/78 | 105/83 | 165/25 | NA | 198/158 | 207/147 | 199/157 | 221/131 | 318/38 | NA |
| 0.56/0.44 | 0.60/0.40 | 0.59/0.41 | 0.56/0.44 | 0.87/0.13 | 0.56/0.44 | 0.58/0.42 | 0.56/0.44 | 0.63/0.37 | 0.89/0.11 | |||||
| Total | 3,458 | 3,719 | ||||||||||||
NA, not available.
Some genotypes may be missing in some cases and controls groups.
In two cases, one study deviated from HWE equilibrium in the control group (Brice for rs3018982 and Annesi for rs2282048 [P <0.0001 and 0.02, respectively]). Moreover, in six studies even though the overall genotyping call rate was >95%, more than 5% missing genotypes existed in the control group. We performed a sensitivity analysis excluding these studies and the results were essentially unaltered (Table III). For rs2313982, a nominally statistically significant association was detected (OR =1.15) (95% CI 1.02–1.31) (P =0.026, uncorrected for multiple corrections), but moderate heterogeneity was observed. The observed association was not nominally significant with random effects analysis (Table II).
TABLE III.
Summary Odds Ratios and 95% Confidence Intervals for the Six SNPs Including Both the Original GWA and Replication Data
| dbSNP id | No. studies | No.a cases/controls | Fixed effects OR (95% CI) |
P-value | Random effects OR (95% CI) |
P-value | I2 (%) (95% CI) |
|---|---|---|---|---|---|---|---|
| rs1000291 | |||||||
| Overall | 13 | 4,020/4,257 | 1.03 (0.97–1.10) | NS | 1.05 (0.94–1.18) | NS | 67 (40–81) |
| rs1865997 | |||||||
| Overall | 12 | 3,278/3,322 | 0.98 (0.92–1.05) | NS | 0.99 (0.87–1.12) | NS | 67 (39–82) |
| Sensitivity analysis | 11 | 2,976/2,977 | 0.96 (0.90–1.03) | NS | 0.97 (0.85–1.10) | NS | 65 (34–82) |
| rs2241743 | |||||||
| Overall | 12 | 3,266/3,316 | 1.05 (0.98–1.12) | NS | 1.06 (0.97–1.16) | NS | 32 (0–66) |
| Sensitivity analysis | 10 | 2,682/2,681 | 1.06 (0.98–1.14) | NS | 1.08 (0.97–1.20) | NS | 44 (0–73) |
| rs2282048 | |||||||
| Overall | 12 | 3,224/2,714 | 0.99 (0.93–1.07) | NS | 0.98 (0.86–1.13) | NS | 71 (48–84) |
| Sensitivity analysis | 9 | 2,524/2,290 | 0.94 (0.87–1.02) | NS | 0.93 (0.79–1.09) | NS | 74 (50–87) |
| rs2313982 | |||||||
| Overall | 13 | 4,301/4,552 | 1.11 (0.99–1.25) | NS | 1.13 (0.95–1.34) | NS | 49 (4–73) |
| Sensitivity analysis | 11 | 3,712/3,952 | 1.15 (1.02–1.31) | 0.026 | 1.18 (0.98–1.43) | NS | 48 (0–74) |
| rs3018626 | |||||||
| Overall | 12 | 3,900/4,090 | 1.08 (1.01–1.17) | 0.032 | 1.09 (1.01–1.17) | 0.037 | 6 (0–61) |
CI, confidence interval; NS, not nominally significant (P >0.05); OR, odds ratio.
Sensitivity analysis: analysis excluding studies with >5% missing genotype rate or significant deviation from Hardy–Weinberg equilibrium.
Four hundred forty-nine sibling pairs were evaluated in all analyses.
None of the aforementioned nominally significant associations revealed with either main or sensitivity analyses would remain significant after adjusting even for six tested SNPs.
Analysis Including Only Replication Studies
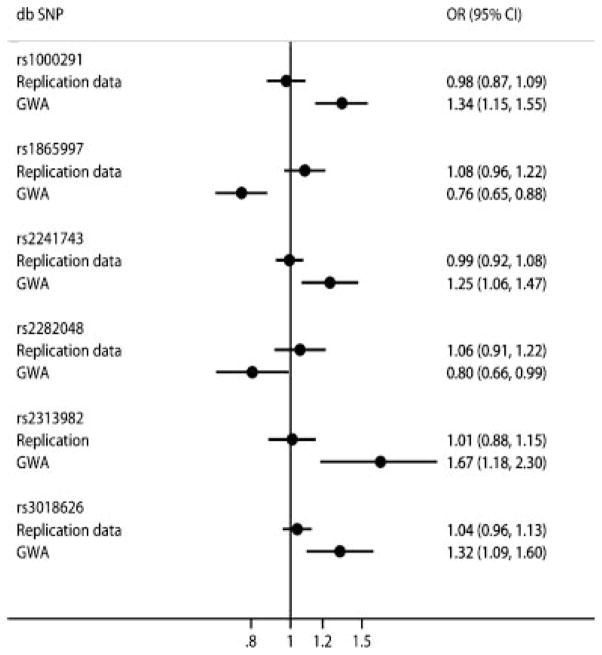
All summary effects obtained from the replication effort were very close to unity (Fig. 1) with estimated summary ORs ranging from 0.98 to 1.09 with random effects analyses. None of the effects were nominally statistically significant. Also, no heterogeneity was observed in three SNPs (rs2241743, rs2313982, and rs3018626 with I2 ranging from 0% to 6%), whereas moderate to large heterogeneity was observed for the other three SNPs under study (rs1000291, rs1865997, and rs2241743) with I2 ranging from 49% to 67%.
FIG. 1.
Forest plot of subgroup summary effects estimates presenting ORs and 95% CIs computed by random effects for the replication data and for the original meta-analysis of GWA data.
The summary effects obtained from the replication data were statistically significantly different from the results of the synthesis of the GWA data. Four out of six SNPs (rs1000291, rs1865997, rs2241743, and rs2282048) had summary effects that were even in the opposite direction compared to summary effects obtained from the original GWA data.
Bayes Factor and Credibility of the Association
We calculated the Bayes factors for the association between rs3018626 and PD that was nominally statistically significant when all data were included. For an expected OR =1.3, the −log10 Bayes factor is 0.01. This means that the probability of the association being true does not increase given the observed data, compared with what we thought before obtaining these data.
DISCUSSION
Our analysis of six SNPs suggested by a meta-analysis of two GWA data sets does not show any convincing associations with PD. None of the SNPs under study reached even nominal significance in the replication data sets and inclusion of all data (including the GWA discovery data) yielded only one SNP with a P-value of 0.037, which should be dismissed given the extreme multiplicity of the agnostic discovery approach in the GWA setting. The Bayes factor for the combined data was not supportive of an association being present.
The collaborative analysis has sufficient power to detect modestly small associations for five of the six tested SNPs. However, this would not be the case for very subtle associations. For example, effects in the range of odds ratios of <1.12 may have been missed, and experience from meta-analysis of GWA in type 2 diabetes (T2D) suggests that such effects cannot be excluded [Zeggini et al., 2008]. On the other hand, we managed to assemble one of the largest studies in the history of PD research. Perusal of very subtle effects, if any such exist, would really require full exploitation of all available samples from multiple consortia [Seminara et al., 2007]. Moreover, if diagnostic criteria and patient characteristics create genuine heterogeneity in the genetic effects, replication would be difficult or even impossible even with extremely large sample sizes [Mooneshinghe et al., 2008]. Power may also be diluted by genotyping errors and unfortunately we were not able to perform central genotyping or central assessment/quality control for the assessment of genotyping errors. Nevertheless, while genuine heterogeneity and genotyping errors can never be totally excluded, we doubt that they primarily explain the non-replication in the observed results.
The most likely explanation for our findings is that none of these six SNPs has a genuine association with PD risk. Even when combined, the two GWAs remain underpowered to reveal efficiently true markers with modest effects. Moreover, testing only six SNPs is unlikely to have hit genuine markers for PD susceptibility. Due to resource constrains, we could not evaluate a wider range of SNPs that might have been more informative. It is expected that genuine associations are not on the very top of the list in the data that arise from GWA studies, regardless of the selection analytical strategy [McCarthy et al., 2008]. Moreover, we should acknowledge that one SNP (rs2313982) has been further evaluated with published data before, and this information is widely available at www.pdgene.org. This database provides a comprehensive assessment of all prior genetic association studies in PD. Analysis of this SNP shows nominally statistically significant random effects odds ratios in the PDGene meta-analyses after exclusion of the initial study (P =0.012). However, both main and sensitivity analyses by random effects did not show any statistical significant results in our sample. An informal meta-analysis of previous data (excluding the first data) plus our new data (excluding previously published information) by random effects would give an odds ratio of 1.08 (95% CI 1.01–1.17), which is not nominally significant (P =0.055, no between-study heterogeneity, I2 =0, 95% CI 0–45%).
Given our current experience, and the recurrent inability to replicate top hits derived from modest size GWA data sets [Elbaz et al., 2006], we believe that a more extensive list of candidate markers for replication should await the accumulation of much larger sample sizes from GWA platforms. Additional GWA studies are currently being completed on PD and their data would be very useful to combine with the existing information. Given that different platforms are used in these GWA investigations, imputation of polymorphisms that are not directly typed [Marchini et al., 2007] can enhance also the genome coverage compared with the first meta-analysis of GWA data that was limited only to common, overlapping polymorphisms across Perlegen and Illumina platforms [Evangelou et al., 2007]. Concurrently, novel analytical approaches, such as pathways analyses [Lesnick et al., 2007; Wang et al., 2007; Li et al., 2008; Srinivasan et al., 2008], and extension to other kinds of genetic variation, such as copy number variation or rare variants [Jakobsson et al., 2008] may be helpful in determining whether we can expand our knowledge base about genetic determinants of PD.
Acknowledgments
Grant sponsor: Inserm; Grant sponsor: MSA; Grant sponsor: Agence Nationale de la Recherche, Agence Française de Sécurité Sanitaire de l’Environnement et du Travail, France Parkinson; Grant sponsor: FIRB 2003 GENOPOLIS Project; Grant number: RBLA038RMA_003; Grant sponsor: National Institutes of Health (NIH); Grant number: 2R01 ES10751; Grant number: ES10758; Grant number: AG 08724; Grant sponsor: Michael J. Fox Grants (Linked Efforts to Accelerate Parkinson’s Solutions, and Edmond J. Safra Global Genetics Consortia); Grant sponsor: The Swedish Medical Research Council, the Swedish Society of Medicine and the Parkinson Foundation in Sweden; Grant sponsor: VIB Genetic Service Facility; Grant sponsor: The Biobank of the Institute Born-Bunge; Grant sponsor: Fund for Scientific Research Flanders, the Institute for Science and Technology – Flanders (IWT-V); Grant sponsor: Foundation for Alzheimer Research (SAO/FRMA); Grant sponsor: Interuniversity Attraction Poles Program P6/43 of the Belgian Science Policy Office, Belgium.
Dr. Eng-King Tan thanks the National Medical and Biomedical Research Councils, and the Duke-NUS Graduate Medical School, Singapore Millennium Foundation. B.M. holds a PhD fellowship of the IWT-V.
Australia: G.T. Sutherland Eskitis Institute for Cell and Molecular Therapies, School of Biomolecular Physical Sciences, Griffith University, Nathan, QLD, G.A. Siebert Eskitis Institute for Cell and Molecular Therapies, School of Biomolecular Physical Sciences, Griffith University, Nathan, QLD. Belgium: Jessie Theuns, PhD Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; Laboratory of Neurogenetics, Institute Born-Bunge; University of Antwerp, Antwerpen, David Crosiers Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; Laboratory of Neurobiology, Institute Born-Bunge; University of Antwerp and University Hospital Antwerp, Antwerpen, Barbara Pickut, MD Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen, Philippe Pals, MD, PhD Laboratory of Neurobiology, Institute Born-Bunge and University Hospital Antwerp; Antwerpen, Sebastiaan Engelborghs, MD, PhD Laboratory of Neurochemistry and Behavior, Institute Born-Bunge; University of Antwerp and Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen Belgium, Karen Nuytemans Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; Laboratory of Neurogenetics, Institute Born-Bunge; and University of Antwerp, Antwerpen, Peter P. De Deyn, MD, PhD Laboratory of Neurochemistry and Behavior, Institute Born-Bunge; University of Antwerp; and Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen, Patrick Cras, MD, PhD Laboratory of Neurobiology, Institute Born-Bunge; University of Antwerp; and University Hospital Antwerp, Antwerpen. France: From the French Parkinsons Disease Genetics Study Group: Agid Y, Bonnet A-M, Borg M, Brice A, Broussolle E, Damier P, Destée A, Dürr A, Durif F, Lesage S, Lohmann E, Pollak P, Rascol O, Tison F, Tranchant C, Viallet F, Vidailhet M. Also, Christophe Tzourio Inserm U708, Paris, Philippe Amouyel Inserm U744, Lille, Marie-Anne Loriot Inserm UMRS775, Paris. Germany: Prof. Thomas Gasser Department of Neurology, University Hospital Tuebingen, Olaf Riess, PhD Department of Neurology, University Hospital Tuebingen, Daniela Berg, MD Department of Neurology, University Hospital Tuebingen, Claudia Schulte, MSc Department of Neurology, University Hospital Tuebingen. Also, Christine Klein, PhD Department of Neurology, University of Lübeck, Ana Djarmati, PhD Department of Neurology, University of Lübeck, Katja Lohmann, PhD Department of Neurology, University of Lübeck. Greece: Georgia Xiromerisiou, MD Institute of Biomedical Research Technology, CERETETH, Larissa, Efthimios Dardiotis, MD Faculty of Medicine, Department of Neurology, University of Thessaly; and Institute of Biomedical Research Technology, CERETETH, Larissa, Persa Kountra, MD Faculty of Medicine, Department of Neurology, University of Thessaly. Japan: Nobutaka Hattori, MD, PhD Department of Neurology, Juntendo University School of Medicine, Tokyo, Hiroyuki Tomiyama, MD, PhD Department of Neurology, Juntendo University School of Medicine, Tokyo, Manabu Funayama, PhD Research Institute for Diseases of Old Age, Juntendo University School of Medicine, Tokyo, Hiroyo Yoshino, BS Research Institute for Diseases of Old Age, Juntendo University School of Medicine, Tokyo, Yuanzhe Li, MD, PhD Department of Neurology, Juntendo University School of Medicine, Tokyo. Italy: Enza Maria Valente, MD, PhD Mendel Institute, Casa Sollievo della Sofferenza Hospital, Rome, Alessandro Ferraris, MD, Mendel Institute, Casa Sollievo della Sofferenza Hospital, Rome, Anna Rita Bentivoglio, MD, PhD Institute of Neurology, Catholic University, Rome, Tamara Ialongo, MD, PhD Institute of Neurology, Catholic University, Rome, Chiara Riva, PhD Department of Neuroscience, University of Milano-Bicocca, Monza, Barbara Corradi, PhD Department of Paediatrics, University of Milano-Bicocca, Monza. Poland: Grzegorz Opala, MD, PhD Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice, Barbara Jasinska-Myga, MD, PhD Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice, Gabriela Klodowska-Duda, MD, PhD Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice, Magdalena Boczarska-Jedynak, MD, PhD Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice. Sweden: Andrea Carmine Belin, PhD Department of Neuroscience, Karolinska Institutet, Stockholm, Prof. Lars Olson Department of Neuroscience, Karolinska Institutet, Stockholm, Dagmar Galter, PhD Department of Neuroscience, Karolinska Institutet, Stockholm, Marie Westerlund, PhD Department of Neuroscience, Karolinska Institutet, Stockholm, Olof Sydow, PhD Department of Clinical Neuroscience, Karolinska University Hospital, Stockholm. Also Christer Nilsson, MD, PhD Department of Geriatric Psychiatry, Lund University, Andreas Puschmann, MD Department of Neurology, Lund University Hospital, Department of Geriatric Psychiatry, Lund University. USA: Demetrius M. Maraganore, MD Department of Neurology, Mayo Clinic, Rochester, MN, J. Eric Ahlskog PhD, MD Department of Neurology, Mayo Clinic, Rochester, MN, USA, Mariza de Andrade, PhD Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, Timothy G. Lesnick, MS Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, and Walter A. Rocca, MD, MPH Departments of Neurology and Health Sciences Research, Mayo Clinic, Rochester, MN, USA. Also, Harvey Checkoway, PhD Department of Environmental and Occupational Health Sciences, University of Washington.
References
- Baum AE, Hamshere M, Green E, Cichon S, Rietschel M, Noethen MM, Craddock N, McMahon FJ. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol Psychiatry. 2008;13(5):466–467. doi: 10.1038/mp.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Hansell AL, Fortier I, Manolio TA, Khoury MJ, Little J, Elliott P. Size matters: Just how big is BIG? Quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2008;38(1):263–273. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Elbaz A, Nelson LM, Payami H, Ioannidis JP, Fiske BK, Annesi G, Carmine Belin A, Factor SA, Ferrarese C, Hadjigeorgiou GM, et al. Lack of replication of thirteen single-nucleotide polymorphisms implicated in Parkinson’s disease: A large-scale international study. Lancet Neurol. 2006;5(11):917–923. doi: 10.1016/S1474-4422(06)70579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Maraganore DM, Ioannidis JP. Meta-analysis in genome-wide association datasets: Strategies and application in Parkinson disease. PLoS ONE. 2007;2(2):e196. doi: 10.1371/journal.pone.0000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung HC, Scholz S, Matarin M, Simon-Sanchez J, Hernandez D, Britton A, Gibbs JR, Langefeld C, Stiegert ML, Schymick J, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Calibration of credibility of agnostic genome-wide associations. Am J Med Genet Part B. 2008a;147B(6):964–972. doi: 10.1002/ajmg.b.30721. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008b;19(5):640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. Br Med J. 2007;335(7626):914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- Lesnick TG, Papapetropoulos S, Mash DC, Ffrench-Mullen J, Shehadeh L, de Andrade M, Henley JR, Rocca WA, Ahlskog JE, Maraganore DM. A genomic pathway approach to a complex disease: Axon guidance and Parkinson disease. PLoS Genet. 2007;3(6):e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rowland C, Xiromerisiou G, Lagier RJ, Schrodi SJ, Dradiotis E, Ross D, Bui N, Catanese J, Aggelakis K, et al. Neither replication nor simulation supports a role for the axon guidance pathway in the genetics of Parkinson’s disease. PLoS ONE. 2008;3(7):e2707. doi: 10.1371/journal.pone.0002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci USA. 2008;105(2):617–622. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara D, Khoury MJ, O’Brien TR, Manolio T, Gwinn ML, Little J, Higgins JP, Bernstein JL, Boffetta P, Bondy M, et al. The emergence of networks in human genome epidemiology: Challenges and opportunities. Epidemiology. 2007;18(1):1–8. doi: 10.1097/01.ede.0000249540.17855.b7. [DOI] [PubMed] [Google Scholar]
- Srinivasan BS, Doostzadeh J, Absalan F, Mohandessi S, Jalili R, Bigdeli S, Wang J, Mahadevan J, Lee CL, Davis RW, et al. Whole genome survey of coding SNPs reveals a reproducible pathway determinant of Parkinson disease. Hum Mutat. 2008;30(2):228–238. doi: 10.1002/humu.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-Based Approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner S, Pritchard JK. Overcoming the winner’s curse: Estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80(4):605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]