Abstract
Over 800 mutations in the ABCA4 gene cause autosomal recessive Stargardt disease. Due to extensive genetic heterogeneity, observed variant-associated phenotypes can manifest tremendous variability of expression. Furthermore, the high carrier frequency of pathogenic ABCA4 alleles in the general population (~1:20) often results in pseudo-dominant inheritance patterns further complicating the diagnosis and characterization of affected individuals. This study describes a genotype/phenotype analysis of an unusual family with multiple macular disease phenotypes spanning across two generations and segregating four distinct ABCA4 mutant alleles. Complete sequencing of ABCA4 discovered two known missense mutations, p.C54Y and p.G1961E. Array comparative genomic hybridization revealed a large novel deletion combined with a small insertion, c.6148-698_c.6670del/insTGTGCACCTCCCTAG, and complete sequencing of the entire ABCA4 genomic locus uncovered a new deep intronic variant, c.302+68C>T. Patients with the p.G1961E mutation had the mildest, confined maculopathy phenotype with peripheral flecks while those with all other mutant allele combinations exhibited a more advanced stage of generalized retinal and choriocapillaris atrophy. This family epitomizes the clinical and genetic complexity of ABCA4-associated diseases. It contained variants from all classes of mutations, in the coding region, deep intronic, both single nucleotide variants (SNV) and copy number variants (CNV) that accounted for varying phenotypes segregating in an apparent dominant fashion. Unequivocally defining disease-associated alleles in the ABCA4 locus requires a multifaceted approach that includes advanced mutation detection methods and a thorough analysis of clinical phenotypes.
Keywords: ABCA4, Stargardt disease, pseudo-dominance, copy number variation, deletion-insertion, intron, mutations
Introduction
Mutations in the ABCA4 gene are responsible for a wide range of autosomal recessive retinal dystrophy phenotypes, from Stargardt disease (STGD1; OMIM #248200) (Allikmets et al. 1997) to cone-rod dystrophy (CRD) (Cremers et al. 1998; Maugeri et al. 2000) and, in some advanced cases, generalized choriocapillaris dystrophy (GCCD) (Bertelsen et al. 2014) and pan-retinal dystrophies with extensive pigment migration resembling retinitis pigmentosa (RP) (Cremers et al. 1998; Martinez-Mir et al. 1998; Shroyer et al. 2001). STGD1 is a predominantly juvenile-onset macular dystrophy associated with central visual impairment, progressive bilateral atrophy of the retinal pigment epithelium, and the accumulation of yellow, pisciform flecks, defined as lipofuscin deposits, in or around the macula or posterior pole of the retina.
Complete sequencing of the ABCA4 coding and adjacent intronic sequences in patients with STGD1 usually identifies the expected two disease-associated alleles in 65–70% of patients, one mutation in ~15–20%, and no mutations in the remaining ~15% (Zernant et al. 2011). Clinically diagnosed cases of STGD1 with no detected ABCA4 mutations most often represent phenocopies (Zernant et al. 2011; Zernant et al. 2014), whereby mutations in other gene(s) manifest a STGD1-like phenotype (Riveiro-Alvarez et al. 2015; Tsang et al. 2014; Yamamoto et al. 2014). However, in STGD1 cases with one ABCA4 mutant allele, the second usually resides in the ABCA4 locus. Such non-coding alleles in ABCA4 belong to two main classes of mutations: 1) copy number variants (CNV), i.e., large deletions or insertions of one exon or more which elude detection by PCR-based sequencing techniques, and 2) deep intronic variants >10bp away of exons, i.e., outside of splice sites. While the ABCA4 gene and locus show extensive variability in single nucleotide mutation variants (SNV), large CNVs are rare in the ABCA4 gene (<1% of all disease-associated alleles); there have been only a few reports describing these (Stenirri et al. 2006; Yatsenko et al. 2003; Zernant et al. 2011; Zernant et al. 2014). Recently, many deep intronic disease-associated variants which may affect splicing or other regulatory functions have been described (Braun et al. 2013; Zernant et al. 2014). However, these are also individually very rare and often difficult to unequivocally associate with the disease due to inaccessibility of ABCA4 RNA (the gene/protein is expressed only in photoreceptors) and lack of a direct functional assay (Zernant et al. 2014). All of the above makes the molecular analysis of the ABCA4 locus in STGD1 patients very challenging. Moreover, although all ABCA4-associated diseases are recessive, the high carrier frequency of pathogenic ABCA4 alleles in the general population (1:20) (Maugeri et al. 1999; Yatsenko et al. 2001; Zernant et al. 2011) can often result in a ‘dominant-looking’ pattern of inheritance in families.
Here we describe a genotype/phenotype analysis of a large family presenting with four distinct macular disease phenotypes spanning across two successive generations. Four distinct ABCA4 mutant alleles were identified, two of which are novel, from three classes of mutations, which account for two of the four phenotypes and the seemingly pseudo-dominant inheritance pattern. This study demonstrates the genetic and clinical complexity of ABCA4-associated disorders and highlights the extensive molecular analyses required to unequivocally solve such challenging cases involving multiple mutations and phenotypes in a single family.
Materials and Methods
Patients and Clinical Evaluation
Ten members of a two-generation family were consented and enrolled under the protocol IRB-AAAI9906, approved by the Institutional Review Board at Columbia University and adhering to tenets of the Declaration of Helsinki. Each affected and unaffected relative underwent a complete ophthalmic examination by a retina specialist (S.H.T. and S.B.) which included fundus autofluorescence (AF) images obtained using a confocal scanning-laser ophthalmoscope (cSLO, Heidelberg Retina Angiograph 2, Heidelberg Engineering, Dossenheim, Germany) by illuminating the fundus with argon laser light (488 nm) and viewing the resultant fluorescence through a band pass filter with a short wavelength cut-off at 495 nm. Simultaneous AF and spectral domain-optical coherence tomography (SD-OCT) images were acquired using a Spectralis HRA+OCT (Heidelberg Engineering, Heidelberg, Germany). Color fundus photography was acquired using a FF 450plus Fundus Camera (Carl Zeiss Meditec AG, Jena, Germany).
Full-field electroretinograms (ERG) were acquired with the Diagnosys Espion Electrophysiology System (Diagnosys LLC, Littleton, MA, USA). For all recordings, the pupils were maximally dilated before full-field ERG testing using guttate Tropicamide (1%) and Phenylephrine Hydrochloride (2.5%); and the corneas were anesthetized with guttate Proparacaine 0.5%. Silver impregnated fiber electrodes (DTL; Diagnosys LLC, Littleton, MA) were used with a ground electrode on the forehead. Full-field ERGs to test generalized retinal function were performed using extended testing protocols incorporating the International Society for Clinical Electrophysiology of Vision standard.(McCulloch et al. 2015)
Sequencing
Patient samples were analyzed by the Illumina Truseq Custom Amplicon target enrichment strategy followed by sequencing on Illumina MiSeq platform (Illumina, San Diego, CA). For sequencing of the ABCA4 gene (coding region) 58 amplicons of 425bp were designed, resulting in 11,258 bp cumulative target with 100% coverage. For sequencing of the entire ABCA4 locus the genomic region 94,456,700- 94,591,600 on chromosome 1 was targeted, including the ABCA4 locus, and 4895bp of 5′UTR, and 1694bp of 3′UTR sequences. The genomic region was divided into 9 targets, with seven 500bp and one 1100bp gap introduced by DNA repeats. The cumulative target included 130,319 bp and was covered at 94% with 421 amplicons of 425bp each. The next-generation sequencing reads were analyzed with the NextGENe variant discovery software (SoftGenetics, State College, PA). The reads were aligned against the targeted region in the haploid reference genome GRCh37/hg19. For a variant to be called, we required the read containing the variant to match 85% to the aligned position, the variant to be covered by at least 10 reads and the variant to be present in 20% of all reads aligned to the given position. We also used the overall confidence score of 10 of the NextGENe software and the previously determined ABCA4 coding variants as controls for further filtering criteria.
Analysis of the ABCA4 variants
All variants and their allele frequencies were compared to the 1000 Genomes database (Genomes Project et al. 2012), and to the Exome Variant Server (EVS) dataset, NHLBI Exome Sequencing Project, Seattle, WA, USA (http://snp.gs.washington.edu/EVS/; accessed April 2015). New variants that were not recorded in these databases were further analyzed by a combination of predictive in silico methods and statistical analyses. The possible effect of the two new non-coding ABCA4 variants on splicing was assessed using 5 different algorithms (SpliceSiteFinder, MaxEntScan, NNSPLICE, GeneSplicer, Human Splicing Finder) via Alamut software (http://www.interactive-biosoftware.com). In order to assess the regulatory potential of the new ABCA4 intronic variants we compared their chromosome coordinates against the predicted regulatory regions from two ENCODE datasets: 1) Combined DNaseI hypersensitivity clusters from 125 cell types (“Digital DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE” http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=366969521&c=chr1&g=wgEncodeRegDnaseClusteredV2, filename: wgEncodeRegDnaseClusteredV2.bed.gz); and 2) ChIP-seq clustered regions for 161 transcription factors in 91 cell types (“Transcription Factor ChIP-seq V4 (161 factors) with Factorbook motifs from ENCODE” (ENCODE project; Date submitted 2015-04-21, http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=366969521&c=chr1&g=wgEncodeRegTfbsClusteredV3, filename: wgEncodeRegTfbsClusteredV3.bed.gz). Evolutionary conservation of the variants was noted via UCSC Genome Browser (http://genome.ucsc.edu). The Combined Annotation Dependent Depletion (CADD) algorithm (http://cadd.gs.washington.edu/score) was used to estimate combined predicted general deleteriousness of every variant. The variants’ segregation with the disease was analyzed by Sanger sequencing.
Array-Comparative Genome Hybridization (aCGH)
Custom CGH (Agilent Technologies) arrays, in an 8X60K format, were designed with high-density probes tiling the critical genetic locus of ABCA4-associated diseases. The details of the array design have been described previously.(Zernant et al. 2014)
Array-CGH analysis was performed on DNA from patients I-3, II-1 and II-3, for each of whom only one mutation had been found by sequencing of the entire ABCA4 coding region. A DNA sample with a known, previously reported 1030 bp heterozygous deletion of exon 18 in ABCA4, was used as positive control (Yatsenko et al. 2003). Experimental procedures for aCGH were performed as described previously (Gonzaga-Jauregui et al. 2010) with minor modifications. Agilent Genomic Workbench version 7.0 software (Agilent Technologies) was used for data analysis. CNVs were called by using the ADM-2 algorithm with a threshold of 4.0. PCR validations were performed for the plausible true-positive CNVs after being filtered against several criteria.
TaKaRa LA Taq (Clontech) was used for the PCR amplifications. Sanger sequencing was performed for the PCR products. DNA sequences were compared to the reference genome (hg19) to map the breakpoint junctions. PCR primers used for breakpoint mapping: ABCA4-bkpt-F, 5′-ACCCCAATAAACAGAGGGCAAGAGTT-3′; ABCA4-bkpt-R, 5′-TTTAGGAGTGAAGGGCTGTGATGAGT-3′. A PCR genotyping assay using the same pair of breakpoint junction specific primers was performed for the family members with available DNA samples.
Results
Patients and disease phenotypes
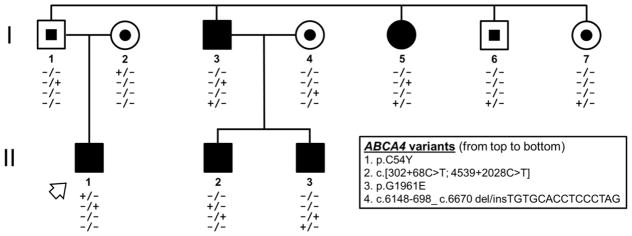
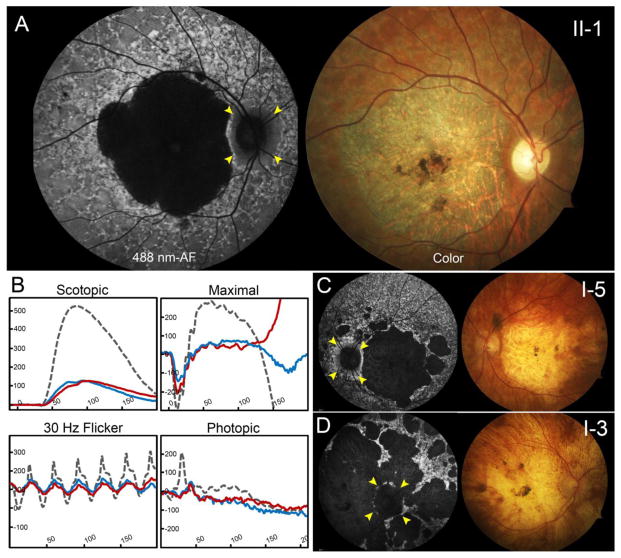
Ten members of a two-generation family presented to the clinic with a history of dominant retinal disease (Figure 1, pedigree). A summary of demographic and genetic characteristics is provided in Table 1 (below). Fundus examinations were abnormal in five members of the family exhibiting retinal phenotypes within the spectrum described for ABCA4-associated disease. The proband (II-1), a 43-year-old man of German descent, and his paternal aunt (I-4) and uncle (I-3) reported experiencing symptoms of uncorrectable vision loss within the first decade of life. At the time of examination each were found to have poor best-corrected visual acuities (BCVA) ranging from 20/400 to hand motion and peripheral light perception with eccentric peripheral fixation. Funduscopy and autofluorescence imaging in the proband revealed a large area of well-delineated chorioretinal atrophy and central pigment clumping within the vascular arcades. The macular lesions in each eye were circumscribed by a dense border of hyperautofluorescence adjacent to a reticular pattern of granular flecks in the periphery. The peripapillary region around the optic nerve was notably spared of any apparent disease-associated changes (Figure 2A, yellow arrowheads). Full-field electroretinogram (ERG) testing in the proband revealed significant reductions in both rod and cone function that is indicative of advanced-stage, generalized photoreceptor loss throughout the retina. (Figure 2B). Atrophic chorioretinal lesions in the proband’s aunt and uncle were comparatively more advanced extending further into the posterior pole of the retina. Retinal vessels were visibly attenuated and residual areas of fundus tissue (RPE) around the numerous lesions were speckled with densely resorbed, hyperautofluorescent flecks. The peripapillary regions in both were affected with flecks or partially spared, respectively. (Figure 2C and 2D, yellow arrowheads).
Figure 1.
Pedigree of the family illustrating the segregation of four disease-causing ABCA4 alleles with two Stargardt disease (STGD1) phenotypes of varying severity. Filled circles and squares denote affected males and females, respectively, and centrally filled shapes denote heterozygous carriers. ABCA4 variants, and their order on the chromosome, are listed in the key. The symbol (+) denotes the presence of a disease-causing ABCA4 variant while (−) represents the wild-type allele. The mother of the proband (I-2, arrowhead) was affected with pattern macular dystrophy which was phenotypically distinct from the other affected individuals and the father (I-1) was affected with chronic central serous chorioretinopathy.
Table 1.
Demographic and Genetic Characteristics of Individuals within the Family
| Subject | Age (y) | Relationship to proband | Gender | Diagnosis | ABCA4 Mutation | |
|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | |||||
| I-1 | 75 | Father | M | CSC/AMD | c.[302+68C>T;4539+2028C>T] | |
| I-2 | 75 | Mother | F | PD | p.C54Y | |
| I-3 | 69 | Uncle (P) | M | STGD1 | c.6148-698_c.6670del/insTGTGCACCTCCCTAG | c.[302+68C>T;4539+2028C>T] |
| I-4 | 68 | Aunt | F | p.G1961E | ||
| I-5 | 67 | Aunt (P) | F | STGD1 | c.6148-698_c.6670del/insTGTGCACCTCCCTAG | c.[302+68C>T;4539+2028C>T] |
| I-6 | 63 | Uncle (P) | M | c.6148-698_c.6670del/insTGTGCACCTCCCTAG | ||
| I-7 | 59 | Aunt | F | c.6148-698_c.6670del/insTGTGCACCTCCCTAG | ||
| II-1 | 43 | Proband | M | STGD1 | p.C54Y | c.[302+68C>T;4539+2028C>T] |
| II-2 | 40 | Cousin | M | STGD1 | c.[302+68C>T;4539+2028C>T] | p.G1961E |
| II-3 | 37 | Cousin | M | STGD1 | c.6148-698_c.6670del/insTGTGCACCTCCCTAG | p.G1961E |
Abbreviations: y, years; (P), paternal lineage; M, male; F, female; CSC, central serous chorioretinopathy; AMD, age-related macular degeneration; PD, pattern dystrophy; STGD1, Stargardt disease
Figure 2.
Advanced ABCA4 phenotype of the proband, paternal aunt and uncles. (A) The proband, a 43-year-old man of German descent harboring the p.C54Y and c.[302+68C>T;4539+2028C>T] variants, presented with large well-delineated lesions of chorioretinal atrophy and central pigment clumping in both eyes on autofluorescence imaging and color fundus photographs. Surrounding areas of dense hyperautofluorescence preceding a reticular pattern of granular flecks were found extended throughout the periphery. (B) Full-field electroretinogram testing in the proband (II-2) revealed significantly reduced amplitudes in rod and cone responses in the right (blue trace) and left (red trace) eyes when compared to an age-matched control (dotted gray trace). The paternal aunt (c.[302+68C>T;4539+2028C>T]; c.6148-698_c.6670del/insTGTGCACCTCCCTAG) presented with similarly large area of chorioretinal atrophy while the paternal uncle (D) with the same compound heterozygous genotype exhibited generalized atrophy of the posterior pole. Various degrees of sparing of the peripapillary region in each patient are marked with yellow arrowheads.
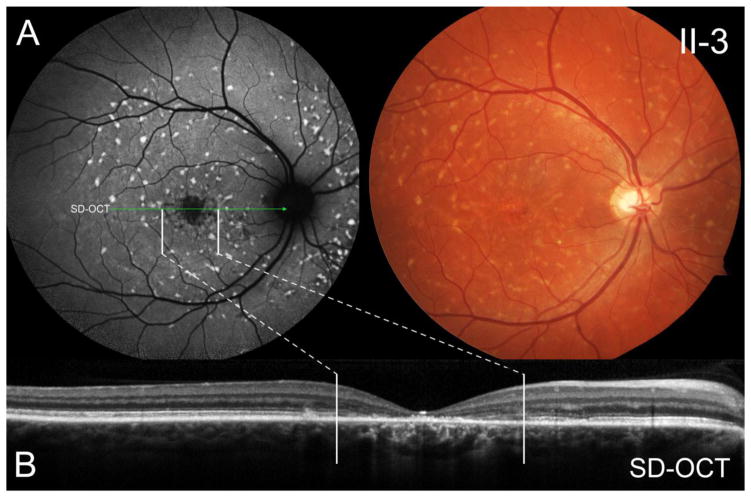
The paternal cousins of the proband (sons of the uncle) presented with a comparatively milder ABCA4 phenotype. Aged 36 years (II-3) and 39 years (II-2) at the time of examination, both reported an onset of symptoms within the third decade of life signifying a shorter disease duration as compared to the other affected relatives described above. Their phenotypes were similar to one another; measured BCVA ranged from 20/100 to 20/150 in both eyes. A lesion of mottled atrophy and photoreceptor loss was confined to the central macula in each eye (Figure 3A and 3B) with a scattered, confluent pattern of subretinal pisciform flecks centrifugally distributed throughout the mid-periphery. Sparing of the peripapillary region was also apparent in both cases. A comparative summary of ABCA4-associated phenotypic features in these affected individuals is provided in Table 2.
Figure 3.
The paternal cousin (36-year-old son of the paternal uncle) harboring the p.G1961E allele and the c.6148-698_c.6670del/insTGTGCACCTCCCTAG variant presented at a comparatively milder disease stage. (A) Autofluorescence imaging revealed a localized lesion of retinal pigment epithelium mottling and photoreceptor loss in the central macula (B) as correlated with spectral domain-optical coherence tomography. The lesion is surrounded by a confluent pattern of round, pisciform flecks centrifugally-distributed across the midperiphery of the retina.
Table 2.
Summary of ABCA4-Associated Phenotypes in Affected Individuals
| Subject | Onset (y) | Duration (y) | BCVA | Geographic atrophy | Extent of atrophy | Fleck Distribution | Peripapillary sparing | ABCA4 Mutation | ||
|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | Allele 1 | Allele 2 | |||||||
| I-3 | 5 | 67 | CF | CF | Posterior pole | Choriocapillaris | Resorbed | Partial | del/insa | intronicb |
| I-5 | 5 | 62 | CF | 20/400 | Extra-macular | Choriocapillaris | Resorbed | Partial | del/insa | intronicb |
| II-1 | 9 | 34 | 20/400 | 20/200 | Macular | Choriocapillaris | Resorbed/Reticular | Spared | p.C54Y | intronicb |
| II-2 | 30 | 10 | 20/100 | 20/100 | n/a | Outer retina | Scattered | Spared | intronicb | p.G1961E |
| II-3 | 30 | 7 | 20/150 | 20/150 | n/a | Outer retina | Scattered | Spared | del/insa | p.G1961E |
c.6148-698_ c.6670del/insTGTGCACCTCCCTAG;
c.[302+68C>T;4539+2028C>T]
Abbreviations: y, years; BCVA, best-corrected visual acuity; OD, right eye; OS, left eye; CF, counting; n/a, not applicable
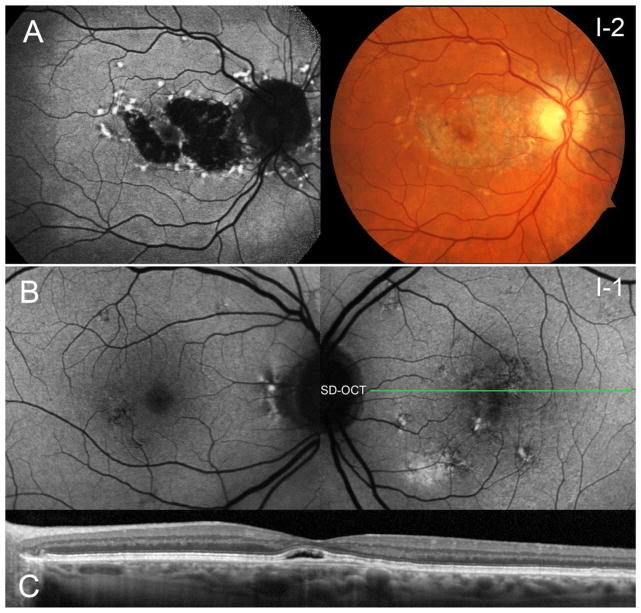
Remarkably, both parents of the proband were also affected with macular diseases; however, with phenotypes usually not associated with ABCA4 mutations. The mother (I-2) reported a late onset of visual symptoms and had measured BCVA’s of 20/20 and 20/30 in the right and left eyes, respectively. A confined area of central atrophy surrounded by large amorphous hyperautofluorescent flecks was found in each eye. The fovea was spared of disease-associated changes in both eyes which likely accounted for her preserved visual acuity and stable fixation. It was noted that atrophy from the macular lesion extended to the peripapillary region around the optic disc (Figure 4A), resulting in a phenotype described as ‘pattern dystrophy’. The proband’s father (I-1), aged 75 years, also presented with a recent onset of macular findings consistent with non-exudative central serous chorioretinopathy in the left eye (Figure 4B); SD-OCT revealed a minor pigment epithelial detachment in this eye (Figure 4C). Minor pigmentary changes and drusenoid deposits were found in both eyes which are likely attributable to the onset of dry age-related macular degeneration (AMD).
Figure 4.
Macular findings in the mother and father of the proband who each carry a single ABCA4 variant, p.C54Y and c.[302+68C>T;4539+2028C>T], respectively. (A) The mother presented with a confined lesion of atrophy and large hyperfluorescent flecks sparing the fovea in each eye. Notably, atrophy extends into the peripapillary region of the optic nerve in both eyes. (B) The father exhibited a recent onset of central serous chorioretinopathy in the left eye with fluid and pigment epithelial detachment over the fovea in the left eye. Pigmentary changes noted in the macula are likely attributed to early, dry age-related macular degeneration.
Discovery of new disease-associated variants by next generation sequencing
Sequencing of the ABCA4 gene and the entire genomic locus in patients II-1 and II-3, at an average depth of coverage of 100X, identified the disease-associated missense variants, p.C54Y and p.G1961E, respectively. Both of these variants are known to cause STGD1, the p.G1961E being the most frequent disease-associated allele (Burke et al. 2012). Interestingly, both of these mutations originated from carrier mothers (I-2, I-4) of the two STGD1 patients (Figure 1). Locus sequencing in II-1 detected a new c.302+68C>T variant and a previously described c.4539+2028C>T allele (Braun et al. 2013; Zernant et al. 2014). Both of these variants were on the same chromosome as a complex allele. This haplotype was on the other allele from the p.C54Y variant in patient II-1, and was detected also in 5/350 other STGD1 cases, all of whom had a second ABCA4 mutation.
According to in silico analyses, neither c.302+68C>T nor c.4539+2028C>T was predicted to have any effect on splicing, either involving existing cryptic splice sites or creating new sites. This was also confirmed by the RNA analysis from leukocytes via an ‘illegitimate transcript’ experimental approach (data not shown); however, this method has been shown to have limited value for the analysis of the ABCA4 gene, which is expressed only in photoreceptors, often producing results that can lead to erroneous interpretations (Albert 2015). Comparing these two positions and flanking sequences in primates reveals that c.302+68C>T lies in a relatively less conserved area than c.4539+2028C>T. Both variants were not detected in the 1000 Genomes Project and segregated with the disease in this large family (and in 5 other STGD1 patients), making them very likely candidates for intronic ABCA4 mutations.
Analysis of regulatory sequences
To assess the potential functional effects of the two newly described ABCA4 intronic variants on putative regulatory regions we compared their location against the chromosome coordinates of the DNaseI hypersensitivity and transcription factor binding clusters from ENCODE. The c.4539+2028C>T variant maps to a region of weak DNaseI hypersensitivity (score 192; scale from 0–1000), while the c.302+68C>T does not overlap with DNaseI hypersensitivity nor transcription factor binding consensus sequences.
The new ABCA4 intronic variants were also subjected to the Combined Annotation Dependent Depletion (CADD) algorithm (http://cadd.gs.washington.edu/score). The CADD algorithm combines a diverse array of annotations into one metric (C score) for each variant, which correlates with allelic diversity, pathogenicity, and experimentally measured regulatory effects (Kircher et al. 2014); a score >10 indicates that the variant is among the top 10% of most deleterious substitutions in the human genome. C scores for c.302+68C>T and c.4539+2028C>T were similar, 3.15 and 3.5, respectively, suggesting no significant deleterious effect for either of these variants.
Analysis of the copy number variants by aCGH arrays
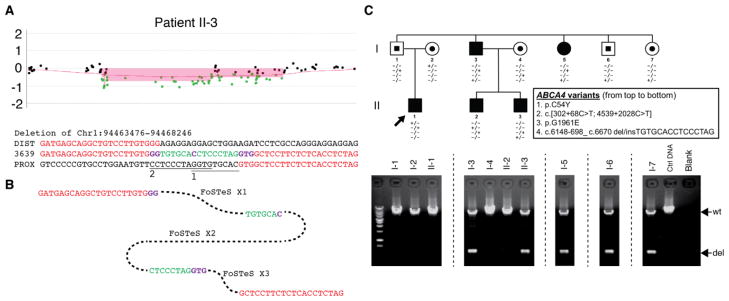
Copy number variations in ABCA4, specifically in the form of large deletions encompassing one or more exons, have been shown to account for a small portion of Stargardt disease alleles (Yatsenko et al. 2003). While these reports have been rare, we screened the patients I-3, II-1 and II-3 on the using custom designed high density array CGH, which revealed a ~5 kb heterozygous deletion at the ABCA4 locus (Figure 5A) in patients I-3 and II-3. Breakpoint junction sequencing mapped the precise genomic interval (Chr1: 94463476- 94468246), revealing a 4771 bp deletion (Figure 5B). A short inserted DNA fragment (TGTGCACCTCCCTAG) was revealed at the deletion breakpoint junction, indicating a deletion/insertion haplotype. The insertion could be split into two halves; both were potentially from regions close to the proximal end of the deletion (Figure 5B). Microhomologies were identified at the breakpoint junctions (Figure 5B). These suggest fork stalling and template switching/microhomology-mediated break-induced replication (FoSTeS/MMBIR) as the potential CNV generating mechanism via three template switches (Hastings et al. 2009; Lee et al. 2007; Zhang et al. 2009).
Figure 5.
The deletion/insertion variant in the ABCA4 locus in the family. A. aCGH log2 ratio plot showing the heterozygous deletion identified in patient II-3. The breakpoint junction sequences alignment underneath the plot reveals two inserted DNA fragments (green) conjugated by microhomologies (purple). The potential origins of the inserted DNA fragments are marked with underlines labeled with ”1” and “2”, respectively. DIST/PROX, human genome reference sequences at the distal/proximal end of the junction. B. Potential molecular mechanism explaining the deletion etiology. FoSTeS, fork stalling and template switching. C. Pedigree of the family identified with the deletion/insertion variant allele (c.6148-698_c.6670del/insTGTGCACCTCCCTAG). The gel pictures underneath the pedigree demonstrate the segregation of the deletion allele with three other potential disease-causing alleles identified in this family. wt, wild type allele; del, deletion/insertion variant allele.
The deletion/insertion spanned from the intron upstream of exon 45 to exon 48. The exons 45–47 were completely deleted, while exon 48 was partially deleted. Integrating the insertion, the variant may be represented as c.6148-698_c.6670del/insTGTGCACCTCCCTAG. Genotyping analysis of c.6148-698_c.6670del/insTGTGCACCTCCCTAG revealed its presence in unaffected individuals I-6 and I-7, who were carriers of this variant, and in affected individuals I-3 and I-5 in combination with a second variant consisting of a complex deep intronic mutant allele c.[302+68C>T;4539+2028C>T] (Figure 5C).
Discussion
The ABCA4 gene has been the subject of intensive genetic research since it was first described as the causal gene for STGD1 18 years ago (Allikmets et al. 1997). Similarly, phenotypes caused by ABCA4 mutant alleles have also been extensively studied and found to be notable for a high degree of variability of expression (Fujinami et al. 2014; Fujinami et al. 2015; Mullins et al. 2012; Noupuu et al. 2014; Zahid et al. 2013). Genetic analyses are challenging due to the size of the gene and the extensive genetic heterogeneity. Moreover, the gene is expressed only in rod and cone photoreceptors and, consequently, it is impossible to obtain photoreceptor samples in vivo from patients for RNA analyses. With the lack of patient RNA for direct structural and expression studies, the analysis is limited to the assessment of variant frequencies in STGD1 patients and in the matched general populations, to in silico suggestions by predictive software programs, and to segregation analyses in families. The latter approach is often complicated due to the variants in ABCA4 coding and non-coding regions being extremely rare, most often represented in singleton cases and/or in nuclear families where the segregation analysis has limited power.
In the current study we analyzed the genotypes and phenotypes in a large two-generation family segregating two distinct forms of STGD1 in a pseudo-dominant inheritance pattern. The disease in its variable expression was due to different combinations of 4 ABCA4 mutant alleles 2 well known missense mutations and two new variant alleles, a large deletion/insertion (including exons 45–48) and a complex allele containing 2 deep intronic variants. Segregation of the 4 mutant alleles explained the seemingly dominant (in fact, pseudo-dominant) inheritance pattern and variable expression of the disease. The complex allele with the two deep intronic variants (c.[302+68C>T;4539+2028C>T]) appeared to impart an aggressive, early-onset phenotype in patients II-1, I-3 and I-4 (Figure 2). The deletion variant (c.6148-698_ c.6670del/insTGTGCACCTCCCTAG) is also likely a null allele since it causes a shift of the reading frame and is predicted to result in protein truncation (Figure 5); however, II-2 and II-3 presented with comparatively milder disease phenotypes (Figure 3) despite harboring each of the intronic and deletion variants, respectively. This is likely due to the p.G1961E mutation shared by II-2 and II-3 on the opposite, maternal allele, which has been previously associated with a late-onset, milder disease phenotype characterized by more localized disease confined to the central macula (Burke et al. 2012; Cella et al. 2009). This apparent resistance to disease severity conferred by p.G1961E is clearly exemplified in this family; this could potentially be attributed to G1961E representing a hypomorphic allele, although its precise mechanism remains to be elucidated (Allikmets 2000; Lewis et al. 1999).
While neither of the two deep intronic variants c.302+68C>T and c.4539+2028C>T, which compose a complex allele, had an effect on splicing or regulatory elements as determined by in silico analyses, the complex allele segregated with the disease. Moreover, this complex allele has been also detected as the second ABCA4 disease-associated allele in five more cases from our large (>700 cases) STGD1 cohort at Columbia University (data not shown). In addition, there is precedent at the ABCA4 locus for complex alleles to have more severe functional consequences than the individual constituent variants and also have more severe clinical phenotypic consequences (Shroyer et al. 2001). The assessment of a potential effect on splicing using prediction programs is correct in only 70–80% of variants examined (Liu et al. 2009). The same is true for predicting the effect of intronic variants on regulatory elements affecting gene expression, such as transcription factor binding sites, enhancers, promoters, etc. (Hardison and Taylor 2012). A good example of a disease-associated intronic variant in the ABCA4 locus which, until most recently, had no proven functional effect is the c.5461-10T>C variant in intron 38, which is one of the most frequently observed variants in STGD1 patients (Zernant et al. 2011). This variant always segregates with the disease phenotype in families, is very rare in the general population (<0.001) and was shown not to affect splicing using the ‘minigene’ approach (Rivera et al. 2000). Therefore it had been assumed that this variant is not a pathogenic allele, but in LD with an unknown ABCA4 variant. However, it was most recently shown by obtaining RNA from iPS-derived photoreceptor progenitor cells from patients carrying this allele, that the variant causes skipping of exons 39 and 40 in the ABCA4 gene (Albert 2015). Taken together, these data strongly suggest pathogenic consequences for the deep intronic complex allele. The unequivocal proof will be obtained by analyzing RNA derived from patients’ iPS cells, as described (Albert 2015).
The two additional macular phenotypes found in patients I-1 and I-2 (Figure 4), which were chronic central serous chorioretinopathy (CSC) and pattern dystrophy (PD), respectively, initially provided yet another challenge in assessing the disease entity in the family. While we were not able to determine the exact genetic cause of the phenotypes in these patients, we speculate that the heterozygous ABCA4 variants were not causal in these cases. The genetic cause of CSC is largely unknown. Variants in the CDH5 gene, currently the only gene reliably associated with the CSC phenotype (Schubert et al. 2014), were not detected in patient I-1. PD, as presented in patient I-2, refers to a group of slowly progressive macular diseases that manifest with minimal diminution of visual acuity and photoreceptor atrophy in older individuals (Marmor and McNamara 1996; Watzke et al. 1982). Many PD cases have been attributed to mutations in the PRPH2/RDS (Boon et al. 2007; Francis et al. 2005). The genetic cause of the PD phenotype can be different and it is often difficult to diagnose based on phenotype alone. However, while the patient carried one ABCA4 allele, p.C54Y, and exhibited certain phenotypic characteristics similar to STGD1, the distinct lack of peripapillary sparing (a pathognomonic characteristic of STGD1 (Cideciyan et al. 2005)) is clinically contraindicative of ABCA4-associated disease. No other potentially disease-associated variants in either the ABCA4 or in PRPH2 genes were detected in this patient.
Due to the high carrier frequency of disease-causing ABCA4 alleles in the general population (1:20), pseudo-dominant inheritance in STGD1 has been described (Cremers et al. 1998; Lewis et al. 1999; Yatsenko et al. 2001). However, similar families mostly occur in isolated populations with elevated occurrence of consanguineous marriages, and contain no more than 3 ABCA4 mutations (Cremers et al. 1998; Yatsenko et al. 2001). The family reported here that segregates four pathogenic ABCA4 variant alleles is: i) derived from an outbred population and, ii) harbors two known ABCA4 missense mutations which were introduced into the family by marriage, which is a very rare event (1:400).
In summary, the analysis of the entire ABCA4 locus in a large family with multiple members presenting four very different phenotypes in a seemingly dominant fashion revealed four different ABCA4 variants belonging to 3 separate classes of mutations segregating with the disease and accounting for at least 2 out of 4 phenotypes. This family epitomizes the extremely complex mutational spectrum underlying ABCA4-associated diseases and suggests a thorough analysis of all cases where the phenotype falls into the spectrum of ABCA4-associated diseases. Genetic analysis should include not only complete sequencing of the gene and adjacent canonical splice junctions, but also the variation in the entire genomic locus, including copy number variant analysis.
Acknowledgments
This work was supported, in part, by grants from the National Eye Institute/NIH EY021163, EY019861, EY021237 and EY019007 (Core Support for Vision Research); National Human Genome Research Institute/NIH HG0065342; Robert L. Burch III Fund, Columbia University, New York, NY, New York Community Trust Fredrick J. and Theresa Dow Wallace Fund, Columbia University, New York, NY; Foundation Fighting Blindness (Owings Mills, Maryland), and unrestricted funds from Research to Prevent Blindness (New York, NY) to the Department of Ophthalmology, Columbia University.
Footnotes
Competing interests: None declared.
References
- Albert SSR, Bax N, Roosing S, van den Born L, den Engelsman-van Dijk A, Ramlal A, Stone E, Hoyng C, Cremers F. Towards the identification of deep-intronic ABCA4 mutations in Stargardt patients by using induced pluripotent stem cell-derived photoreceptor progenitor cells. Association for Research in Vision and Ophthalmology 2015 [Google Scholar]
- Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet. 2000;67:487–91. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–46. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- Bertelsen M, Zernant J, Larsen M, Duno M, Allikmets R, Rosenberg T. Generalized choriocapillaris dystrophy, a distinct phenotype in the spectrum of ABCA4-associated retinopathies. Invest Ophthalmol Vis Sci. 2014;55:2766–76. doi: 10.1167/iovs.13-13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon CJ, van Schooneveld MJ, den Hollander AI, van Lith-Verhoeven JJ, Zonneveld-Vrieling MN, Theelen T, Cremers FP, Hoyng CB, Klevering BJ. Mutations in the peripherin/RDS gene are an important cause of multifocal pattern dystrophy simulating STGD1/fundus flavimaculatus. Br J Ophthalmol. 2007;91:1504–11. doi: 10.1136/bjo.2007.115659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun TA, Mullins RF, Wagner AH, Andorf JL, Johnston RM, Bakall BB, Deluca AP, Fishman GA, Lam BL, Weleber RG, Cideciyan AV, Jacobson SG, Sheffield VC, Tucker BA, Stone EM. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–45. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TR, Fishman GA, Zernant J, Schubert C, Tsang SH, Smith RT, Ayyagari R, Koenekoop RK, Umfress A, Ciccarelli ML, Baldi A, Iannaccone A, Cremers FP, Klaver CC, Allikmets R. Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53:4458–67. doi: 10.1167/iovs.11-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella W, Greenstein VC, Zernant-Rajang J, Smith TR, Barile G, Allikmets R, Tsang SH. G1961E mutant allele in the Stargardt disease gene ABCA4 causes bull’s eye maculopathy. Exp Eye Res. 2009;89:16–24. doi: 10.1016/j.exer.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Swider M, Aleman TS, Sumaroka A, Schwartz SB, Roman MI, Milam AH, Bennett J, Stone EM, Jacobson SG. ABCA4-associated retinal degenerations spare structure and function of the human parapapillary retina. Invest Ophthalmol Vis Sci. 2005;46:4739–46. doi: 10.1167/iovs.05-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Deutman AF, Hoyng CB. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum Mol Genet. 1998;7:355–62. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- Francis PJ, Schultz DW, Gregory AM, Schain MB, Barra R, Majewski J, Ott J, Acott T, Weleber RG, Klein ML. Genetic and phenotypic heterogeneity in pattern dystrophy. Br J Ophthalmol. 2005;89:1115–9. doi: 10.1136/bjo.2004.062695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinami K, Singh R, Carroll J, Zernant J, Allikmets R, Michaelides M, Moore AT. Fine central macular dots associated with childhood-onset Stargardt Disease. Acta Ophthalmol. 2014;92:e157–9. doi: 10.1111/aos.12259. [DOI] [PubMed] [Google Scholar]
- Fujinami K, Zernant J, Chana RK, Wright GA, Tsunoda K, Ozawa Y, Tsubota K, Robson AG, Holder GE, Allikmets R, Michaelides M, Moore AT. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology. 2015;122:326–34. doi: 10.1016/j.ophtha.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Zhang F, Towne CF, Batish SD, Lupski JR. GJB1/Connexin 32 whole gene deletions in patients with X-linked Charcot-Marie-Tooth disease. Neurogenetics. 2010;11:465–70. doi: 10.1007/s10048-010-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13:469–83. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lewis RA, Shroyer NF, Singh N, Allikmets R, Hutchinson A, Li Y, Lupski JR, Leppert M, Dean M. Genotype/Phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet. 1999;64:422–34. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Li CG, Zhou SF. Prediction of deleterious functional effects of non-synonymous single nucleotide polymorphisms in human nuclear receptor genes using a bioinformatics approach. Drug Metab Lett. 2009;3:242–86. doi: 10.2174/187231209790218145. [DOI] [PubMed] [Google Scholar]
- Marmor MF, McNamara JA. Pattern dystrophy of the retinal pigment epithelium and geographic atrophy of the macula. Am J Ophthalmol. 1996;122:382–92. doi: 10.1016/s0002-9394(14)72065-3. [DOI] [PubMed] [Google Scholar]
- Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzalez-Duarte R, Balcells S. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998;18:11–2. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67:960–6. doi: 10.1086/303079. S0002-9297(07)63287-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugeri A, van Driel MA, van de Pol DJ, Klevering BJ, van Haren FJ, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Dahl N, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. The 2588G-->C Mutation in the ABCR Gene Is a Mild Frequent Founder Mutation in the Western European Population and Allows the Classification of ABCR Mutations in Patients with Stargardt Disease. Am J Hum Genet. 1999;64:1024–1035. doi: 10.1086/302323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch DL, Marmor MF, Brigell MG, Hamilton R, Holder GE, Tzekov R, Bach M. ISCEV Standard for full-field clinical electroretinography (2015 update) Doc Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Kuehn MH, Radu RA, Enriquez GS, East JS, Schindler EI, Travis GH, Stone EM. Autosomal recessive retinitis pigmentosa due to ABCA4 mutations: clinical, pathologic, and molecular characterization. Invest Ophthalmol Vis Sci. 2012;53:1883–94. doi: 10.1167/iovs.12-9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noupuu K, Lee W, Zernant J, Tsang SH, Allikmets R. Structural and genetic assessment of the ABCA4-associated optical gap phenotype. Invest Ophthalmol Vis Sci. 2014;55:7217–26. doi: 10.1167/iovs.14-14674. iovs.14-14674 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveiro-Alvarez R, Xie YA, Lopez-Martinez MA, Gambin T, Perez-Carro R, Avila-Fernandez A, Lopez-Molina MI, Zernant J, Jhangiani S, Muzny D, Yuan B, Boerwinkle E, Gibbs R, Lupski JR, Ayuso C, Allikmets R. New Mutations in the RAB28 Gene in 2 Spanish Families With Cone-Rod Dystrophy. JAMA Ophthalmol. 2015;133:133–9. doi: 10.1001/jamaophthalmol.2014.4266. 1918815 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, Weber BH. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet. 2000;67:800–13. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C, Pryds A, Zeng S, Xie Y, Freund KB, Spaide RF, Merriam JC, Barbazetto I, Slakter JS, Chang S, Munch IC, Drack AV, Hernandez J, Yzer S, Merriam JE, Linneberg A, Larsen M, Yannuzzi LA, Mullins RF, Allikmets R. Cadherin 5 is regulated by corticosteroids and associated with central serous chorioretinopathy. Hum Mutat. 2014;35:859–67. doi: 10.1002/humu.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Lupski JR. Null missense ABCR (ABCA4) mutations in a family with stargardt disease and retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2001;42:2757–61. [PubMed] [Google Scholar]
- Stenirri S, Battistella S, Fermo I, Manitto MP, Martina E, Brancato R, Ferrari M, Cremonesi L. De novo deletion removes a conserved motif in the C-terminus of ABCA4 and results in cone-rod dystrophy. Clin Chem Lab Med. 2006;44:533–7. doi: 10.1515/CCLM.2006.116. [DOI] [PubMed] [Google Scholar]
- Tsang SH, Burke T, Oll M, Yzer S, Lee W, Xie YA, Allikmets R. Whole exome sequencing identifies CRB1 defect in an unusual maculopathy phenotype. Ophthalmology. 2014;121:1773–82. doi: 10.1016/j.ophtha.2014.03.010. S0161-6420(14)00234-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzke RC, Folk JC, Lang RM. Pattern dystrophy of the retinal pigment epithelium. Ophthalmology. 1982;89:1400–6. doi: 10.1016/s0161-6420(82)34632-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, Zhang K, Bayat V, David G, Li T, Chen K, Gala U, Harel T, Pehlivan D, Penney S, Vissers LE, de Ligt J, Jhangiani SN, Xie Y, Tsang SH, Parman Y, Sivaci M, Battaloglu E, Muzny D, Wan YW, Liu Z, Lin-Moore AT, Clark RD, Curry CJ, Link N, Schulze KL, Boerwinkle E, Dobyns WB, Allikmets R, Gibbs RA, Chen R, Lupski JR, Wangler MF, Bellen HJ. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–14. doi: 10.1016/j.cell.2014.09.002. S0092-8674(14)01113-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. Late-onset Stargardt disease is associated with missense mutations that map outside known functional regions of ABCR (ABCA4) Hum Genet. 2001;108:346–55. doi: 10.1007/s004390100493. [DOI] [PubMed] [Google Scholar]
- Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. An ABCA4 genomic deletion in patients with Stargardt disease. Hum Mutat. 2003;21:636–44. doi: 10.1002/humu.10219. [DOI] [PubMed] [Google Scholar]
- Zahid S, Jayasundera T, Rhoades W, Branham K, Khan N, Niziol LM, Musch DC, Heckenlively JR. Clinical phenotypes and prognostic full-field electroretinographic findings in Stargardt disease. Am J Ophthalmol. 2013;155:465–473. e3. doi: 10.1016/j.ajo.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Schubert C, Im KM, Burke T, Brown CM, Fishman GA, Tsang SH, Gouras P, Dean M, Allikmets R. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci. 2011;52:8479–87. doi: 10.1167/iovs.11-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Xie YA, Ayuso C, Riveiro-Alvarez R, Lopez-Martinez MA, Simonelli F, Testa F, Gorin MB, Strom SP, Bertelsen M, Rosenberg T, Boone PM, Yuan B, Ayyagari R, Nagy PL, Tsang SH, Gouras P, Collison FT, Lupski JR, Fishman GA, Allikmets R. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet. 2014;23:6797–806. doi: 10.1093/hmg/ddu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]