Abstract
In this study we have investigated a new class of cationic lipids – “bolaamphiphiles” or “bolas” for their ability to efficiently deliver small interfering RNAs (siRNAs) to cancer cells. The bolas of this study consist of a hydrophobic chain with one or more positively charged head groups at each end. Recently, we reported that micelles of the bolas GLH-19 and GLH-20 (derived from vernonia oil) efficiently deliver siRNAs, while having relatively low toxicities in vitro and in vivo. Our previous studies validated that; bolaamphiphiles can be designed to vary the magnitude of siRNA shielding, its delivery, and its subsequent release. To further understand the structural features of bolas critical for siRNAs delivery, new structurally related bolas (GLH-58 and GLH-60) were designed and synthesized from jojoba oil. Both bolas have similar hydrophobic domains and contain either one, in GLH-58, or two, in GLH-60 positively charged head groups at each end of the hydrophobic core. We have computationally predicted and experimentally validated that GLH-58 formed more stable nano sized micelles than GLH-60 and performed significantly better in comparison to GLH-60 for siRNA delivery. GLH-58/siRNA complexes demonstrated better efficiency in silencing the expression of the GFP gene in human breast cancer cells at concentrations of 5 μg/mL, well below the toxic dose. Moreover, delivery of multiple different siRNAs targeting the HIV genome and further inhibition of virus production was demonstrated.
Keywords: olaamphiphiles, molecular dynamics simulations, cryo-EM, FRET, siRNA drug delivery, RNAi induced gene silencing
Graphical abstract
1. Introduction
Fire and Mello, showed that double-stranded RNAs mediated RNA interference (RNAi) based on an RNA sequence specific protein assisted process that can target and induce silencing of complementary mRNA expression [1]. It was also shown that relatively long double-stranded RNA molecules (dsRNAs) which are processed into small interfering RNAs (siRNAs) by an enzyme dicer can elicit RNA interference (RNAi) in mammalian cells [2]. Typically, dsRNAs are cleaved by dicer to form siRNA duplexes (21–23 nucleotides), which together with argonaute form RNA-induced silencing complexes (RISC). The so-called “sense” strand is removed by the argonaute protein and the “antisense” strand remains associated with the argonaute protein. Thereafter, the antisense binds to the target mRNA and its cleavage is catalyzed by argonaute to knock down the expression of the specific target gene [2]. siRNA-based strategies have become an attractive approach for therapeutic targeting and inhibition of gene expression responsible for pathological disorders [3]. Recently, attempts have been made to develop siRNA into anti-cancer therapeutics, as in, for example, gynecologic, liver, lung and prostate cancers [3].
However, siRNA-based therapeutic approaches face a number of technical challenges and the success of this technology is subject to efficient and suitable delivery tools [3, 4]. Before siRNA reaches its target site, it encounters a number of impediments that block the assembly of the RISC machinery. After entering the blood circulation, siRNAs are subject to renal excretion due to their small size and highly anionic character, which disfavors their interactions and uptake by cells [3, 4]. Moreover, non-modified “naked” siRNA is vulnerable to degradation by endogenous nucleases and this also decreases its chance to be taken up by cells. Therefore, in order to achieve successful siRNA-based therapy, a delivery system that will protect the siRNA from nucleases and will extend its pharmacokinetic profile should be used to achieve efficient gene silencing.
Viral and non-viral vectors have been used to deliver nucleic acids into cells. Viral vectors, although efficient in delivering nucleic acids, have several drawbacks such as immunogenicity, inflammatory reactions and problems associated with scale-up. These limitations of the viral vectors focused the research on non-viral vectors, particularly on nanoparticles. A wide variety of nanoparticles composed of different materials including polymers, lipids, and inorganic materials have been developed for the delivery of nucleic acids [5, 6]. The characteristics of nanoparticles that make them attractive for the delivery of nucleic acids are their small dimensions that allow maximal uptake into cells and the possibility of targeting the nanoparticles to specific cells in which the nucleic acids exert their action. However, nanoparticles also have limitations with respect to the delivery of nucleic acids. Nanoparticles for the delivery of nucleic acids have to form a stable complex with the nucleic acid in order to protect it from degradation extracellularly, then, it should arrive to the cell of interest, get internalized and escape the endosome to reach its target site within the cell. The most popular nucleic acid carriers are lipid based nanoparticles such as lipoplex and cationic liposomes [7]. Yet, most of these lipid-based carriers have drawbacks that hamper their use in vivo. The most serious problem is toxicity [8], since in order to achieve good complexation with the negatively charged nucleic acids, the lipid carrier should be cationic and cationic lipids have been shown to be toxic [9]. Another problem is stability as nanoparticles made of cationic lipids show very short survival in blood circulation [10] and their head groups may be sensitive to relatively low pHs [11]. To increase the stability of cationic liposomes attempts are made to modify the lipids such incorporating polyether branches [10] or coating the liposomes with polyethylene glycol moieties [12], which may interfere with cellular uptake and targeting.
We have recently described cationic bolaamphiphiles for targeted drug delivery [13–18] that show low toxicity with high in vitro and in vivo stability [17]. These cationic bolalipids showed good transfection efficiency of siRNA [19], RNA/DNA hybrids [20] and RNA nanoparticles [21] in vitro and in vivo with superior features compared to nanoparticles made of amphoteric lipids with one head groups such as phospholipids, DOTAP and other lipids used to make carriers for the delivery of nucleic acids. The basic structure of one major class of bolas (GLH-19 and GLH-20) that we studied, consists of only one hydrophobic alkyl chain in contrast to phospholipids that typically contain two long fatty acyl chains. The ends of the alkyl chains of the bolas that we investigate are covalently linked to one or more positively charged acetyl choline head groups (ACh-HG) that bind with the negatively charged nucleic acids.
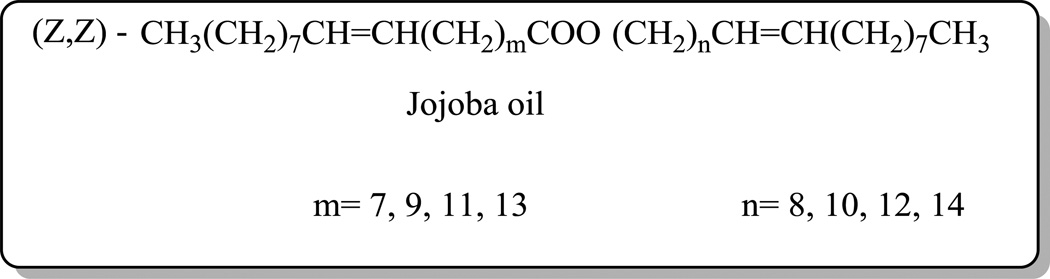
Our group has shown that the positively charged ACh-HG of bolas GLH-19 and GLH20 play a major role in complexation with siRNAs [19] and thus, it was hypothesised that an increase in the number of head groups might increase the binding affinity of siRNAs to the bolas and improve their delivery into cells. Therefore, we synthesized bolas bearing more than two ACh-HG and investigated their interactions with siRNAs and their silencing abilities. This work describes two novel bolas, GLH-58 and −60, that are synthesized from jojoba oil shown in Figure 1 and that differ only in the number of ACh-HGs. GLH-58 carries two positively charged ACh-HGs, one at each end, and GLH-60 contains four positively charged ACh-HG, two at each end of the chain (Figure 2). GLH-58 and GLH-60 were investigated computationally and the computational data were validated experimentally for their capability to interact, deliver siRNAs into cancer cells and silence a specific gene in these cells.
Figure 1.
Chemical structure of jojoba oil
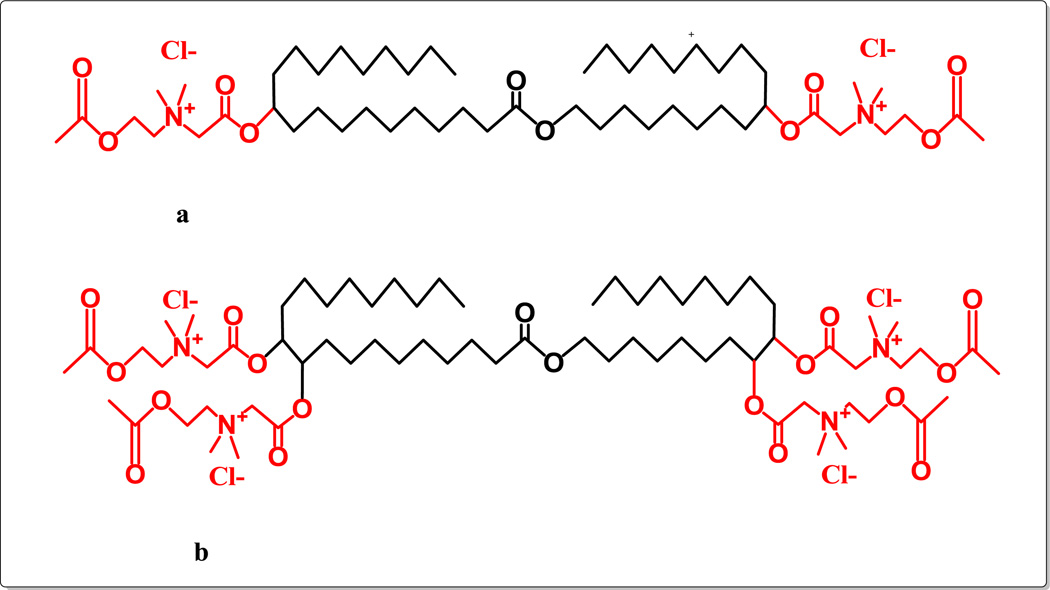
Figure 2.
Chemical structure of bolaamphiphilic compounds with positively charged head groups as ACh synthesized from jojoba oil: (a) GLH-58 (MW 983.45), a bolaamphiphile with two ACh head groups; (b) GLH-60 (MW 1359.87), a bolaamphiphile with four ACh head groups.
2. Material and methods
Standard chemicals were purchased from Sigma-Aldrich Chemical Co. Israel. Jojoba oil was obtained from Kibbutz Hatzerim, Israel. Epoxy groups were determined by potentiometric titration [22]. FT-IR analysis was carried out on a Nicolet spectrometer. 1H and 13C NMR (500 MHz) spectra were recorded on Brucker WP-500 SY spectrometers, in CDCl3 with TMS as the internal standard or d6 DMSO solutions. HPLC analysis was carried out on a C18RP column with an evaporative light scattering detector (evaporation temperature 46°C; mobile phase methanol/methanol: acetone: hexane (2:1:1) (70/ 30, v/v); flow rate 0.5 mL/min). MS analysis was carried out on a Waters Micromass Q-TOF Premier Mass spectrophotometer (Waters-Micromass, Milford, MA, USA). MALDI was carried out on a MS Spectrophotometer MALDI-TOF Reflex-IV (Broker Bremen Germany).
Nucleic acid sequences and fluorescently labeled nucleic acid sequences for making duplexes were obtained from Integrated DNA Technologies, Inc (Coralville, Iowa, USA), MISSION siRNA universal negative control was purchased from Sigma-Aldrich (St. Louis, MO). The Human breast cancer cell line MDA-MB-231, 293T cells, and Hela cells were procured from ATCC (Manassas, VA, USA). The green fluorescent protein expressing MDA-MB-231/GFP cells were obtained from Cell Biolabs Inc. (San Diego, CA, USA). Nuclease/protease-free water was purchased from Quality Biological Inc. (Gaithersburg, MD), Cell titer blue reagent and RQ1 RNase free DNase (Promega, Madison, WI, USA). Cell culture reagents and media were purchased from Gibco (Invitrogen, NY, USA).
2.1. Syntheses of the bolaamphiphile compounds
Bolaamphiphiles GLH-58 and GLH-60 were synthesized by using jojoba oil and the products obtained from each step of synthesis to get to the final structures of both the bolas were characterized by FT-IR, NMR techniques. The details of the syntheses methods are given in Supporting Information.
2.2. Nucleic acid duplex assembly
All sequences used in this project are listed below. Dicer substrate of RNA (DS RNAs), fluorescent duplexes of Alexa-488 labeled RNA/DNA hybrids and Alexa-546 labeled RNA/DNA hybrids designed against green fluorescent protein (GFP) and DS RNAs designed against multiple sites of HIV-1 were used in this study. The individual sense and antisense strands of all the duplexes were heated at 90°C for 1 min in assembly buffer (50 mM NaCl and 2 mM Mg2+) and then annealed at room temperature before further use [20, 21].
RNA sequences used in this project
Dicer Substrate RNAs (DS RNA) designed against GFP [23]
DS RNA duplex:
Sense: 5’ – pACCCUGAAGUUCAUCUGCACCACCG
Antisense: 5’ – CGGUGGUGCAGAUGAACUUCAGGGUCA
DS RNAs selected against multiple sites of HIV-1 [42,43] and used in mixture:
Targeting PBS-Matrix:
Antisense: 5’- GACGGACUCGCACCCAUCUCUCUCCUU
Sense: 5’-pGGAGAGAGAUGGGUGCGAGUUCGUC
Targeting Envelope/gp120:
Antisense: 5’- GGACAAUUGGAGAAGUGAAUUAUAUU
Sense: 5’-pUAUAAUUCACUUCUCCAAUUGUCC
Targeting Capsid:
Antisense: 5’- CCUGGAAUGCUGUCAUCAUUUCUUCUU
Sense: 5’-pGAAGAAAUGAUGACAGCAUUUCAGG
Targeting Reverse Transcriptase:
Antisense: 5’- AUUUAUCUACUUGUUCAUUUCCUCCA
Sense: 5’-pGAGGAAAUGAACAAGUAGAUAAAU
Targeting Protease:
Antisense: 5’- CUUCUAAUACUGUAUCAUCUGCUCCU
Sense: 5’-pGAGCAGAUGAUACAGUAUUAGAAGA
Targeting Nef:
Antisense: 5’- GGAGGAAAUUAGCCCUUCCAGUCCCUU
Sense: 5’-pGGGACUGGAAGGGCUAAUUUUCUCC
5’ side of sense strand is phosphorylated.
Fluorescently labeled DNA sequences
DNA antisense_Alexa488: 5’ – CGGTGGTGCAGATGAACTTCAGGGTCAtt/3AlexF488N/
DNA sense_Alexa546: 5’ – /5AlexF546N/aaTGACCCTGAAGTTCATCTGCACCACCG
DNA sense_Iowa Black FQ: 5’ -/5IAbFQ/ ACCCTGAAGTTCATCTGCACCACCG
13-mer and 12mer RNA duplexes used for MD simulations
13-mer
5’- AUCUGCACCACCG
5’- CGGUGGUGCAGAU
12-mer
5’- ACCCUGAAGUUC
5’- GAACUUCAGGGUCA
2.3. Formation of Bola/duplex complexes
Bolas GLH-58 and −60 pre-dissolved in chloroform were dried with a stream of nitrogen to make a thin film and desiccated overnight under vacuum. The bolas were then resuspended at 1 mg/mL in nuclease/protease-free water while vortexing. Bola suspensions were used either alone or in complexation with nucleic acid duplexes after 30 min of incubation before each experiment (see sections below). The specific conditions for various experiments are described below and in respective figure legends. The concentrations mentioned for bolas and nucleic acids in the manuscript are final concentrations.
2.4. Fluorescent anisotropy/polarization measurements
To determine the binding affinities of GLH-58 and −60 with fluorescently labeled nucleic acids, fluorescent anisotropy/polarization measurements were conducted using Tecan Infinite M1000 (Tecan, USA). Bolas at different concentrations (1- 20 μg/mL) were incubated with fluorescent Alexa-488 labeled RNA/DNA hybrid duplexes (200 nM) designed against GFP (λex 460 nm/λem 520 nm). Changes in fluorescent (anisotropy/polarization) values of fluorescent RNA/DNA hybrid duplexes were used to study binding with bolas. The anisotropy data was fitted using the Hill equation below:
Where R is the anisotropy, R0 is the anisotropy of the hybrid in solution, ΔRmax is the difference in anisotropy of the hybrid within the micellar complex versus in solution, [GLH] is the concentration of the bolaamphiphile, Kd is the dissociation constant and n is the Hill coefficient. The fitting was performed using a least square approach in Sigmaplot.
2.5. Nuclease degradation
The ability of bolas to protect nucleic acid duplexes against degradation by nucleases was assessed by Förster resonance energy transfer (FRET) based experiments following published protocol [19]. Quenched DNA duplexes designed against GFP in which the 3’ end of the antisense strand was labeled with Alexa-488 (fluorescent) and the 5’ end of the sense strand was labeled with Iowa Black FQ (fluorescence quencher) were used. DNA duplexes alone or complexed with bolas were treated according to the manufacturer’s protocol by RQ1 RNase free DNase and degradation of the duplexes was monitored following dequenching of Alexa-488 (λex 460 nm, λem 520 nm). The excitation slit width was kept at 2 nm and the dequenching was recorded at 30 sec intervals and the measurements were performed using Fluoromax-3 fluorimeter (Jobin-Yon, Horiba Scientific, Edison, NJ).
2.6. Cell culture studies
Human breast cancer cell lines MDA-MB-231 and MDA-MB-231/GFP that stably express Green Fluorescent Protein (GFP) were maintained in a Dulbecco’s modified Eagle’s medium (DMEM) 10 % (v/v) heat-inactivated FBS (fetal bovine serum), 100 i.u./mL penicillin and 100 μg/mL streptomycin (serum containing media) under a humidified 5% CO2 incubator at 37°C. The cells were then used according to the requirements of the experiments.
2.7. Cell viability assay
MDA-MB-231 cells were seeded in 96 well plates at a density of 30,000 cells/well in serum containing media 16–24 hours prior to experiments. Bolas (GLH-58 and GLH-60) suspensions at different concentrations (1–100 μg/mL) with and without DS RNAs designed against GFP were added to the cells in triplicate and incubations were continued for 24 h at 37°C. At the end of the incubations, cell titer blue reagent (Promega, Madison, WI) was added to each well according to the manufacturer’s protocol and the cells were further incubated for 4 hours at 37°C. The fluorescence of the resofurin (converted from resazurin by viable cells) was measured at λex 560 nm and λem 590 nm with an auto cut-off in a fluorescent ELISA plate reader (SpectraMAX2, Molecular Devices, Sunnyvale, CA).
2.8. Transfection and silencing measurements by flow cytometry
100,000 cells per well of MDA-MB-231 and 30,000 cells/well of MDA-MB-231/GFP were plated in serum containing media in two separate 12 well plates one day prior to uptake and silencing experiments. On the day of transfection, the serum-containing media from the cells was replaced with serum free media containing different concentrations of bolas complexed with either Alexa-488 RNA/DNA hybrid duplexes or Dicer substrates RNAs (DS RNAs) designed against GFP. The cells were then incubated for 4h at 37°C. After 4h, the serum free media was replaced with serum containing media. Efficiency of uptake of RNA/DNA hybrid duplexes (27 bps hybrid duplex) and the extent of GFP silencing caused by DS RNA (25 bps RNA duplex) were statistically analyzed 1 day and 3 days after the transfections respectively by fluorescence-activated cell sorting (FACS) using Cell Quest software and fluorescent microscopy (described below).
2.9. Fluorescent microscopy
Cells were imaged 72 hours (three days later) after the transfection with a Nikon 200 TE inverted microscope (Melville, NJ) to evaluate the silencing efficiency. PanFluor 20X, ELWD, NA=0.45 objective and a Nikon B-2E/C, 465–495/505/515–555 cube for GFP imaging (Chroma Technology Corp., Rockingham, VT) was used. Meta Morph software (Universal Imaging Co., Downingtown, PA) was used to image the samples.
2.10. Endosomal colocalization experiments
The fluorescently labeled early endosomal (EEA1) marker was used for colocalization experiments with fluorescently labeled Alexa-546 RNA/DNA hybrids designed against GFP in MDA-MB-231 cells. 30,000 cells per quadrant of a culture dish were seeded one day before the transfection. The next day, cells were transfected in serum free media with fluorescently labeled hybrids associated with bolas. After 4h of incubation, the cells were washed three times with 1% BSA in 1X PBS and then fixed with 4% paraformaldehyde for 20 min. The cells were then washed again three times with 1% BSA in 1X PBS and permeabilized with 0.2% Triton X-100 for 10 min. After washing the cells three times, the cells were treated with primary antibodies against EEA1 (Cell signaling, Danvers, MA) in 1% BSA in 1X PBS for 1h. The cells were then washed three times and were stained with secondary antibody labeled with Alexa-488. Thereafter, the cells were fixed again with 4% paraformaldehyde and were imaged and analyzed for endosomal colocalization using a LSM 710 confocal microscope (Carl Zeiss, Munchen, Germany) with a 63X, 1.4 NA magnification lens. All images were taken with a pinhole adjusted to 1 airy unit.
2.11. Transfection of DS RNAs designed against HIV
The day before the transfection 50,000 293T cells were plated in 6 well plates. 30 min before the transfection, the growth media was removed and replaced with 500 μL of OPTI-MEM (serum free media). Two simultaneous transfections took place: (i) Cells were transfected with 2 μg HIV-RFP and 0.5 μg VSV-g plasmids using HilyMax transfection reagent (Dojindo Molecular Technologies) according to the Hilymax protocol, (ii) Two different concentrations (1x, 2x) of GLH-58 and GLH-60 were incubated for 30 min with 0.5 μM of DS RNAs mixture against HIV RNA. As controls extra reactions of bolas without HIV DS RNAs were included. Both reactions were added to the cells and the OPTI-MEM media was changed to DMEM with 10% FBS (serum containing media) after 4h of incubation at 37°C. Virus containing supernatants were collected after 48h and filtered. Equal volumes of these supernatants were used for infections of HeLa cells. Two days after infection, the percentage of infected cells was assessed using FACS.
2.12. Dynamic Light Scattering (DLS) experiments
Nanoparticles from the bolas GLH-58 and GLH-60 (made from different concentrations of the bolas, ranging from 1–20 μg/mL, were analyzed for their hydrodynamic size before and after complexation with 50 nM DS RNAs designed against GFP in nuclease-free water at 25°C using Zetasizer Nano ZS (Malvern Instruments Ltd, Westborough, MA) equipped with a 633 nm laser and a back-scattering detector [24]. The Stokes-Einstein equation was used to measure the hydrodynamic size of the samples. Each sample was run at least 3 times and the representative graph for each sample was selected and presented.
2.13. Cryo-EM experiments
Quantifoil Copper 200 mesh R 3.5/1 grids were washed overnight with acetone. To prepare a frozen, hydrated grid, 2.5 μL of sample of bolas alone or bola/DS RNA (designed against GFP) was applied to the grid, blotted, and plunged into liquid ethane using Vitrobot III (FEI, Hillsboro, OR). Images were collected at liquid nitrogen temperature (∼100 K) on a JEM-2010F (JEOL, Tokyo, Japan) transmission electron cryo-microscope equipped with a field emission gun. JEM-2010F was operating at 200 kV and was equipped with a Gatancryo-holder (model 626) (Gatan, Pleasanton, CA). Images were recorded on DE-12, a 12.6 megapixel (3,072 × 4,096) Direct Detection Device sensor (Direct Electron LP, San Diego, CA). Samples were imaged at 13,900X effective magnifications targeted at 3–4 μm under focus. We used a total specimen exposure for each image of 30 e-/Å2 second.
2.14. In silico studies of bolas (MD simulation)
Explicit solvent molecular dynamics (MD) simulations were used to study the formation of bola micelles and bola interactions with RNA. Spherical aggregate structures were constructed for GLH-58 and GLH-60 which were modeled as micelles and thus are referred to in this paper as micelles. In addition, the self-aggregation of each bola was also tested, starting with randomly distributed individual bolas. The aggregation of bolas on RNA surfaces was simulated with two halves of an siRNA, a 12-mer duplex with a two-nucleotide overhang on one strand (illustrated in all MD results figures) and a 13-mer RNA duplex (results not illustrated). Two protocols, one using RNA with individual bolas at the start, and the second using RNA with bola clusters (micelles) were employed (described in the Supporting Information). In the simulations of the RNA with bola clusters (micelles), equimolar MD conditions using 14 bolas of each kind, as well as simulations with 19 GLH-58 bolas were employed, the second case bringing the mass concentrations to comparable levels (16.6 mg/mL for 19 GLH-58s and 16.9 mg/mL for 14 GLH-60s). In these simulations the final states of pure bola aggregation MD runs were used to select cluster sets maintaining the mean cluster sizes observed for each bola (4 for GLH-58 and 2 for GLH-60). Auxiliary runs with RNA surrounded by randomly placed individual bolas were also performed for comparison purposes. Additional information on the MD protocols and sequences of two RNAs is provided in the Supporting Information.
All MD simulations in our study were performed using the Amber 12 package [25]. The general AMBER force field (GAFF) was used for both GLH-58 and −60, and the ff10 force field was used for the RNA duplexes. In the MD simulations of the bola aggregates’ formation, Na+ and Cl− ion pairs were added in excess of the bola-neutralizing Cl− ions for an effective 0.15 M salt concentration. In the MD simulations of bola/RNA complexes, only the ions needed to neutralize the net charge of the whole system (RNA and bolas) were added to reduce the ionic interference with the interactions between bola head groups and RNA phosphate groups. The following ion parameters were used: Na+ radius 1.868 Å and a well depth 0.00277 kcal/mol; and Cl- radius 1.948 Å and a well depth 0.265 kcal/mol. The particle mesh Ewald summation (PME) [26] was used to calculate the electrostatic interactions. Minimization of the system was done using harmonic constraints on the RNAs and bolas.
After minimization, the system was heated to 300 K while initially holding the solute with a 200 kcal/(mol) harmonic restraint. Restraints were slowly released as the system equilibrated in 16 stages for a total duration of 0.4 ns. In order to remove the fastest hydrogen vibrations and to allow longer simulation time steps, SHAKE was applied to all hydrogen atoms. A constant temperature of 300 K was maintained using a weak-coupling algorithm while the pressure was maintained at 1.0 Pa. All production simulations were performed with a 2 fs time step. Analyses of the MD simulations were performed with the ptraj and cpptraj modules of the Amber package.
3. Results
3.1. Syntheses of bolaamphiphiles
In previous studies we described the synthesis of novel bolaamphiphiles containing acetylcholine head groups (ACh-HG) namely GLH-19 and −20 [14, 17, 27]. These bolas interact with siRNAs to form particles that are internalized by cells and silence genes following their internalization both in vitro and in vivo [19, 20]. GLH-19 and GLH-20 were synthesized from vernolic acid by a multi-step synthesis. This route involves first the synthesis of the skeleton of the bola lipid (i.e. the hydrophobic part) and then the addition of the ACh-HG to the bolaamphiphilic skeleton. Hence the overall bola synthesis using the vegetable oils is a complex multi-step process.
In the present work, we used jojoba oil (Figure 1) as the starting material to simplify the synthesis of the new bolas, GLH-58 and −60, that contain two and four ACh-HG, respectively. Contrary to the triglyceride vegetable oils, jojoba oil is a liquid wax with a 40–42 carbon atom chain composed mainly of straight chain monoesters of C20 and C22 monounsaturated acids and alcohols. Jojoba oil constitutes a unique starting material for the syntheses of bolaamphiphiles as its chemical structure provides a hydrophobic skeleton of 40–44 carbon atoms and the ACh-HG can be conjugated directly to the jojoba oil, which is used as the bolaamphiphilic skeleton.
The two double bonds on either side of the jojoba’s aliphatic chain are used to attach the head groups. To attach the ACh-HG to the jojoba skeleton, two strategies were explored: (a) direct addition of haloacetic acid to the double bond followed by quaternization of the head group, and (b) epoxidation of the double bonds and opening the epoxy group; e.g. esterification of the hydroxyl groups formed with a haloacetic acid followed by quaternization of the tertiary amine to give the desired bolalipids (Figure 2). The chemical synthesis of new bolas is detailed in Supporting Information.
3.2. Experimental characterization of bolaamphiphiles
3.2.1. Fluorescent anisotropy/polarization
Fluorescent anisotropy/polarization measurements were performed to investigate the relative binding affinities between GLH-58 and −60 bolas and nucleic acids (Figure 3a). Bolas GLH-58 and −60 at different concentrations ranging from 1 to 20 μg/mL were incubated with fixed amounts of Alexa 488 fluorescently labeled RNA/DNA hybrids designed against GFP (at 200 nM). After incubation, the samples were loaded into separate wells of a 96-well plate and were read at λex 470 nm and λem 517 nm of Alexa 488. The increment in the anisotropy values of fluorescent RNA/DNA hybrids on increasing concentrations of GLH-58 and −60 showed that both bolas were binding to duplexes. GLH-60 showed higher binding affinity to the RNA/DNA duplexes in comparison to GLH-58. GLH-60 started binding at lower molar concentrations than GLH-58. At higher bola concentrations (∼20 μg/mL), the anisotropy values for the bolas upon complexing with R/DNA hybrids seemed to reach saturation (Figure 3a). The anisotropy values for GLH-58 and 60 were fitted as described in Methods section. The fitting of the anisotropy data with the Hill equation is represented in figure 3a (dotted lines). The 4 individual experiments yielded an average Kd of 12.6 μM for GLH-58 and 6.39 μM for GLH-60 which would indicate about 125 GLH-58 and 64 GLH-60 per hybrid molecule. A Hill coefficient above 3 in both cases indicated a cooperative binding.
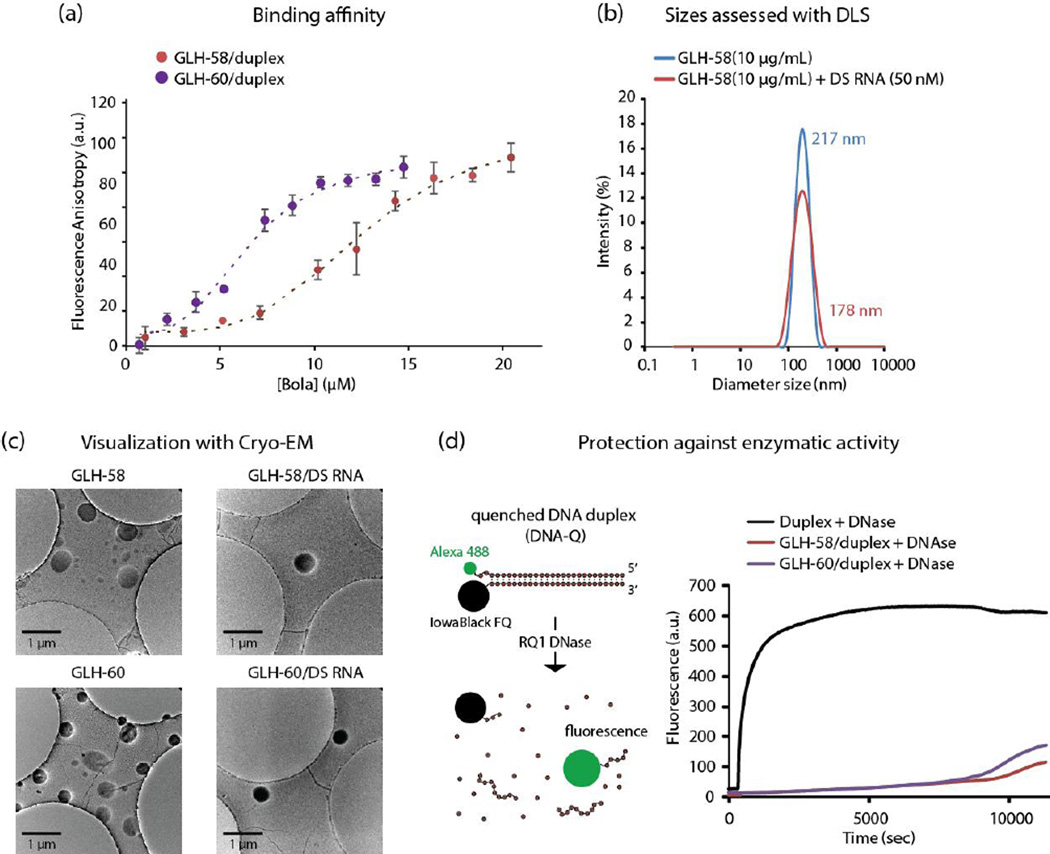
Figure 3.
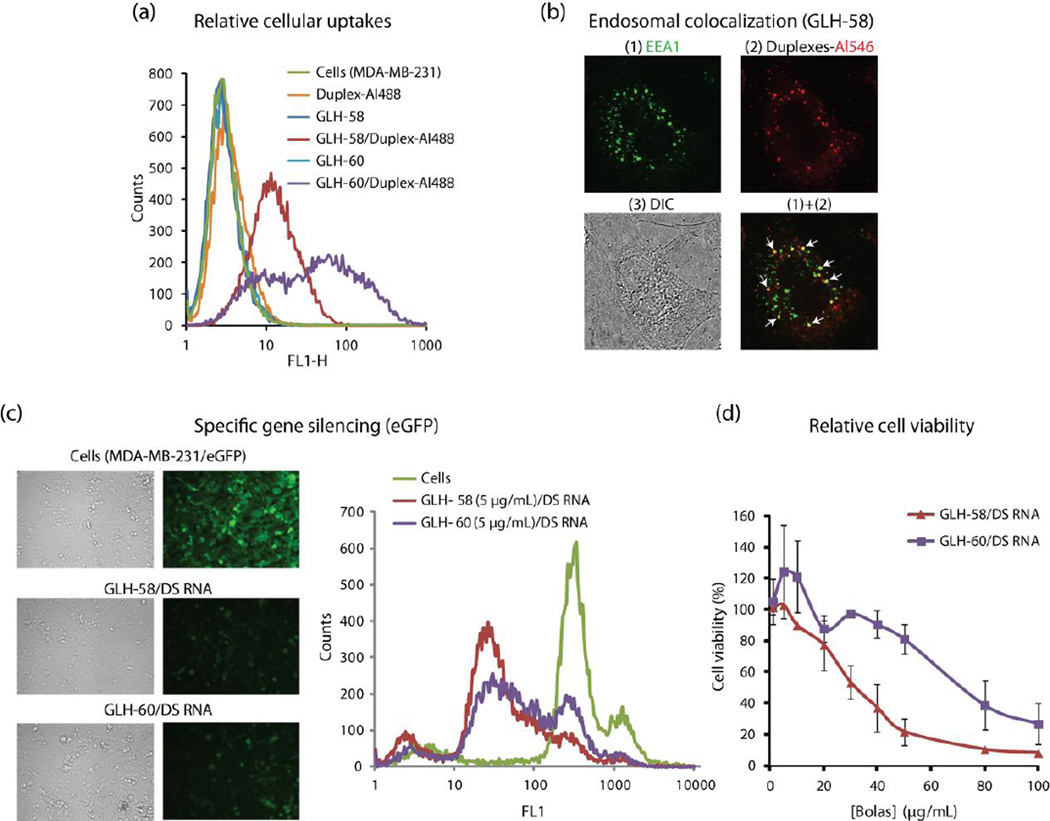
Biophysical characterization of bola/nucleic acid complexes. The nucleic acids used to obtain the above results were designed against GFP. (a) Binding affinity of GLH-58 and GLH-60 to Alexa-488 labeled RNA/DNA hybrid duplexes. The fitting (dashed lines) is described in Methods. (b) Size analysis graphs for bola micelles alone and bola/DS RNA complexes, (c) Cryo-EM images of the GLH-58 and GLH-60 micelles alone and in complexation with DS RNAs, (d) Ability of 5 ug /mL of GLH-58 and GLH-60 to protect quenched DNA duplexes (50 nM) upon DNase digestion. All the concentrations mentioned are final.
3.2.2. Size analysis with dynamic light scattering (DLS)
Bolas GLH-58 and −60 at different concentrations were analyzed for their sizes alone and in complexation with 50 nM DS RNA designed against GFP using DLS. Results shown in Figure 3b demonstrate that GLH-58 form stable nano sized micelles with an average diameter of about 200 nm at 1, 5, 10 and 20 μg/mL concentrations alone and with DS RNA (data shown only for 10 μg/mL). Although the average peak intensity of the bolas did not change upon addition of DS RNA, size analysis of the resulting complexes exhibited relatively wider size distribution and an average diameter of 178 nm as compared to the bolas alone (217 nm). This slight change is also reflected in the cryo-EM images. GLH-60 could bind DS RNAs but could not form homogeneous micelles (data not shown).
3.2.3. Cryo-EM imaging
We further studied the individual bolas and the bolas complexed with DS RNA designed against GFP using cryogenic electron microscopy (cryo-EM). The cryo-EM images presented in Figure 3c showed that a majority of GLH-58 and −60 nanoaggregates (or micelles) were attracted to the hydrophobic carbon surface. It is also possible that the micelles stick to the carbon surface and get flattened. Therefore, the micelles’ diameter was increased to around 400 nm on the carbon surface compared to 200 nm, the size in solution as observed by DLS. Similar transitions from surface-attached liposomes to bilayer patches on solid supports were previously observed by atomic force microscopy [28, 29] and electron cryo microscopy [30]. Electron microscopy images in our study showed light and dark spherical aggregates without DS RNA (left column of the Cryo-EM micrographs, Figure 3c). The light particles were mostly observed in the middle of the carbon surface and seemed to form a flat non-spherical layer. The darker ones were mostly on the edge of the surface. When the micelles were complexed with DS RNA, only dark circles that did not collapse were observed (micrographs of the right column, Figure 3c).
3.2.4. Nuclease degradation assays
We examined the ability of bolas to protect the nucleic acids from degradation by nucleases (Figure 3d). For this purpose, due to cost effectiveness, a DNA duplex (rather than RNA) designed against GFP was subjected to treatment with DNase, as a model system. The DNA duplexes contained sequences that were identical to the DS RNAs (designed against GFP) used for transfection experiments in breast cancer cells (MDA-MB-231). The 3’-end of the DNA strand corresponding to the DS RNA antisense strand was labeled with Alexa 488 and the 5’-end of the DNA corresponding to the DS RNA sense strand with the Iowa Black fluorescence quencher [19]. Due to the proximity of the Alexa 488 strand and the fluorescence quencher, the DNA duplexes could not fluoresce until fully digested by the DNase. GLH-58 and −60 bolas complexed with 50 nM of fluorescently quenched DNA duplexes were incubated and equilibrated for 2 min at 37°C in the fluorimeter and then DNase was added to induce degradation of the duplexes in the form of dequenching of Alexa 488 at λex 460 nm and λem 520 nm. From the results of this experiment we concluded that both bolas offered similar protection capability to DNA duplexes over a relatively long time period (Figure 3d).
3.2.5. Cellular uptake experiments
Bolas GLH-58 and −60 were then investigated for their nucleic acid delivery efficiency. To measure cellular uptake of the bola/nucleic acid complexes, we used RNA/DNA hybrid duplexes (designed against GFP) containing a DNA labeled fluorophore. These hybrids contain RNA sense strands which are duplexed with antisense DNA strands where the 3’-end was labeled with the Alexa 488 fluorophore. Bolas at different concentrations were complexed with fluorescent RNA/DNA duplexes and then added to human breast cancer cells MDA-MB-231. The day after transfection, efficiencies were analyzed by fluorescence-activated cell sorting (FACS). Significant cellular uptake of GLH-58/duplex and GLH-60/duplex complexes was observed at bola concentrations of 1,5 and 10 μg/mL in comparison to control samples. GLH-60 at 5 μg/mL and 10 μg/mL showed significantly higher uptake of duplexes than GLH-58 (Figure 4a and supporting Figure S7).
Figure 4.
Cellular characterization of bola/nucleic acid complexes in human breast cancer cells (MDA-MB-231) (a) Uptake of Alexa-488 labeled RNA/DNA hybrid duplexes (50 nM) mediated by GLH-58 and GLH-60 bolas (at 10 ug/ml), (b) Endosomal co-localization of Alexa-546 labeled RNA/DNA hybrid duplexes delivered by GLH-58, (c) Silencing of GFP mediated by the release of DS RNA designed against GFP by GLH-58 and GLH-60 in GFP expressing MDA-MB-231 cells, (d) Effect of addition of GLH-58/DS RNA and GLH-60/DS RNA complexes on viability of cells. All the concentrations mentioned are final.
3.2.6. Endosomal co-localization
The co-localization of the fluorescently labeled Alexa-546 RNA/DNA hybrid duplexes designed against GFP in the endosomes of breast cancer cells (MDA-MB-231) delivered by GLH-58 and −60 was confirmed by co-staining with the early endosome (EEA1) marker using confocal microscopy. (Figure 4b, co-localization for GLH-60 is similar and therefore, not shown).
3.2.7. Gene silencing efficiency
To examine the release of the siRNAs from the bola/DS RNA complexes for silencing a particular gene (in our case GFP), DS RNA duplexes containing a sequence to knock down the GFP gene were designed. For this purpose, bola/DS RNA complexes at different concentrations of bolas complexed with a fixed concentration of DS RNA (50 nM final) were transfected into human breast cancer cells stably expressing GFP. Three days later, the decrease in the fluorescence, a measurement for silencing activity of GFP expression was evaluated with fluorescence microscopy and FACS shown in Figure 4c. We observed significant silencing activity of GFP by GLH-58/DS RNA complexes at lower doses compared to GLH-60/DS RNA complexes (Figure 4c). Based on the data collected from three independent experiments, GFP silencing activity was 82.15 ± 3.57 and 75.72 ± 5.58 % for GLH-58 and GLH-60 respectively when 5 μg/mL lipid concentrations were used.
Figure 4c shows a representative micrograph of GFP expression in the absence or presence of bola/DS RNA incubations at a concentration of 5 μg/mL bolas. The left panel of Figure 4c also shows a representative data set of FACS analysis at 5 μg/mL indicating superior GFP silencing when bola GLH-58 was used as the carrier. To further substantiate this observation, we performed a dose response curve with varying concentrations of bola lipids complexed with 10 nM of DS RNA. (Figure S8). These data clearly indicate the superior activity of GLH-58 at lower bola concentrations.
To rule out the possibility of non-specific effects of bolas/siRNA complexes on GFP silencing, we used a negative control RNA and silencing experiments were performed as above. The data is shown in supporting information (Figure S8). These results clearly demonstrate that GFP expression remains unaffected when scrambled RNA/bola compelxes (both GLH-58 and 60) were used for transfection. This data further substantiates the specificity of gene silencing upon transfection with bola lipids complexes with DS RNA.
3.2.8. Cell viability assay
The effect of increasing concentrations (1–100 μg/mL) of GLH-58 and −60 complexed with DS RNA designed against GFP (at 50 nM) was investigated for the viability of breast cancer cells (Figure 4d). At lower concentrations of bolas (up to 10 μg/mL) both the bola/DS RNA complexes showed no cytotoxicity. However, at concentrations equal to or greater than 20 μg/mL GLH-58/DS RNA complexes were more cytotoxic than the GLH-60/DS RNA complexes. GLH-58/DS RNA complexes showed 20% cell viability at 50 μg/mL and almost complete killing of cells at 80 μg/mL of GLH-58 whereas 25% cell viability was observed at the highest concentration of 100 μg/mL of GLH-60 for GLH-60/DS RNA complexes shown in Figure 4d. Thus the concentration that were shown to result in gene silencing were significantly below the toxic levels.
3.2.9. Transfection of DS RNAs designed against HIV
293T cells were transfected with HIV plasmid together with VSV-g. Simultaneously, the same cells were transfected with a mixture of DS RNAs against HIV [34] using two different concentrations of bolas (see materials and methods). As can be observed from the supporting information Figure S9, DS RNAs transfected into the cells with GLH-58 and 60 significantly inhibit the production of HIV (lines 1–4). This implies an efficient delivery of siRNAs into 293T cells by bolas. Furthermore bolas without the DS RNAs did not affect the production of HIV (lines 8–10) thus, underlying the minimum toxicity of these two compounds to 293T cells even at higher concentration.
3.3. MD simulations
3.3.1. Micelle Formation
The aggregate structures formed by GLH-58 and GLH-60 were simulated using the micellar structures as the major nanostructure. The examination of micelle formation of GLH-58 and −60 using molecular dynamics (MD) simulations showed that GLH-58 formed a stable micelle while GLH-60 did not. Micelle formation was explored with two different initial configurations of each bola. In the first configuration, a spherically shaped micelle was constructed from 20 pre-folded bolas, with the hydrophobic chains placed inside the micelle and the positively charged head groups placed on the micelle surface. Thus, the GLH-58 micelle had 40 head groups on its surface, while GLH-60 had 80 head groups on its surface. During 80 ns long MD simulations GLH-58 maintained the micelle configuration while the initial micelle configuration of GLH-60 began to break apart within 5 ns and split into 7 clusters at the end of 80 ns-long simulation (Figure S11).
We observed analogous results in the second configuration simulating self-aggregation of randomly placed 24 individual bolas. After approximately 60 ns of simulation, GLH-58 aggregated into six clusters with a mean size of 4, while the GLH-60 bolas formed 11 clusters with a mean size of 2 per cluster (Figure S12). In a low density variant of this configuration only 14 bolas were used in the beginning, and MD yielded similar results in 25 ns simulations (results not shown). Thus, the results of three different MD simulation protocols indicated that GLH-58 could form and maintain larger stable micelles, while GLH-60 was able to form only small clusters and not able to maintain a large micelle due to the strong electrostatic repulsion of its ACh-HGs. The results of these MD simulations agree well with the experimental DLS results, which showed that GLH-58 formed an aggregate structure categorized as a stable micelle, while GLH-60 did not.
3.3.2. Aggregation of bolas on RNA surfaces
MD simulations were also used to study the interactions between RNA and each bola, and they showed that a larger number of GLH-58 bolas accumulated on the RNA than GLH-60. For these MD simulations we used selected clusters from the self-aggregation bola runs described in the previous paragraph. We used the mean cluster sizes from those runs for a total of 14 GLH-58 and −60 bolas in equimolar runs and 19 GLH-58 and 14 GLH-60 for the comparable mass concentrations of 16.6 mg/mL for GLH-58 and 16.9 mg/mL for GLH 60 in another set of runs. These bola clusters were placed in a solvent box with a 12-mer RNA duplex with a two-nucleotide overhang.
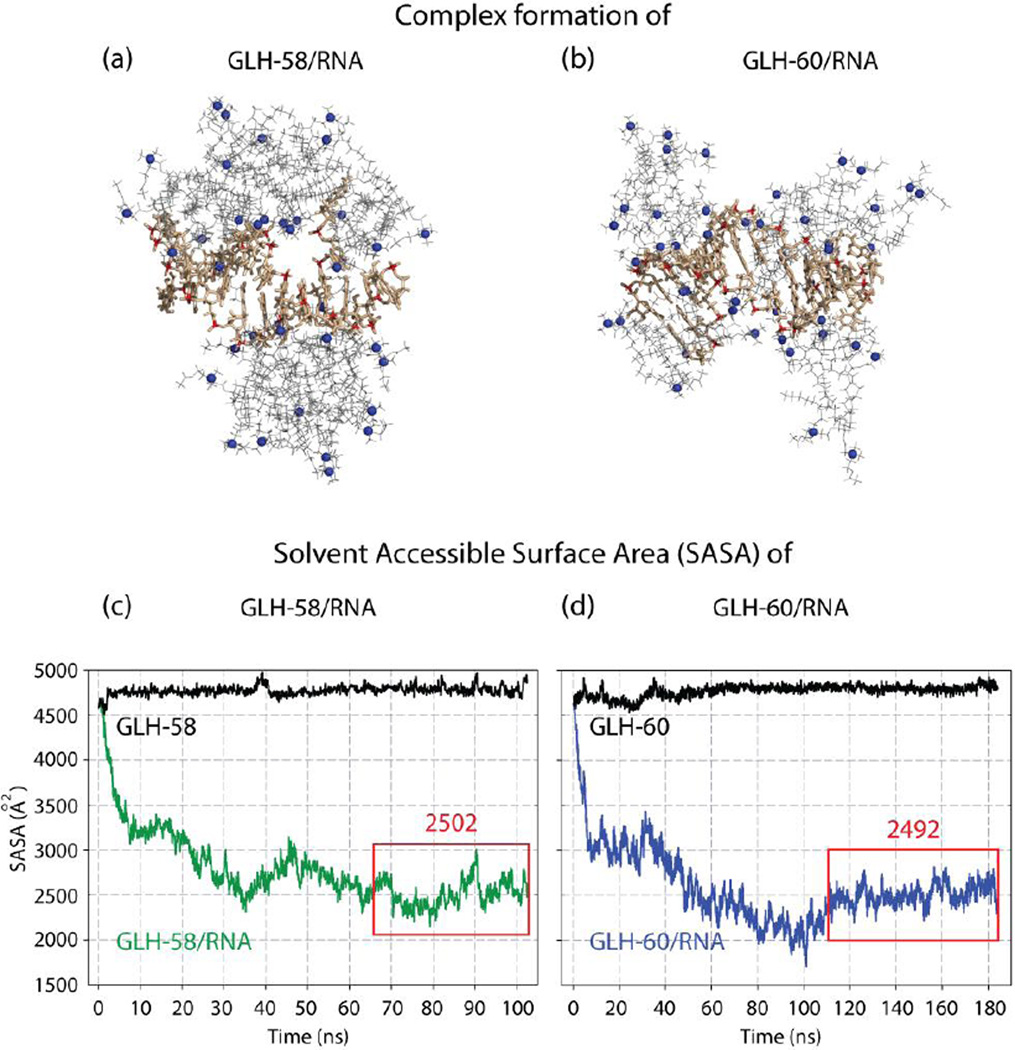
The interactions between the bolas and the RNA were first evaluated based on the number of bola head groups which were associated with the RNA within a short range (less than 6 Å). In the GLH-58/RNA complex, 14 positively charged bola head groups were located approximately 5 Å from the atoms of the RNA’s backbone phosphate groups at the end of the run, with an additional three (for a total of 17) placed in the proximity of other RNA atoms (Figure 5a). In the GLH-60/RNA complex a maximum of 17 positively charged GLH-60 head groups were located mostly within 5 Å of the phosphate groups, with an additional 9 (for a total of 26) being in the proximity of other RNA atoms at the 100 ns mark in the MD simulation.
Figure 5.
Complex formation of (a) GLH-58/RNA and (b) GLH-60/RNA in Molecular Dynamics simulations; bola hydrophobic skeleton (gray) its head groups (blue spheres). RNA (tan) its P atoms of the backbone phosphate groups (red). Solvent Accessible Surface Area (SASA) plots for the Molecular Dynamics (MD) simulations of (c) 19 GLH-58 and (d) 14 GLH-60 bolas complexed with an RNA. Plots shown in black correspond to the SASA of the RNA alone. Plots shown in color indicate SASA variations of the RNA surface exposed to the solvent in the bola/RNA complex. Red boxes indicate time intervals of stable complex states over which the median SASA values shown above them were calculated. 12 mer RNA used in these studies is one-half the DS RNA designed against GFP.
However, this result for GLH-60 coincides in time with the Solvent Accessible Surface Area (SASA) value being below the final stable median (discussed in the following paragraph). For a time point with the median SASA value, a total of 24 head groups were present in the proximity of the RNA, with 14 head groups near the RNA phosphates and additional 10 near other RNA atoms (Figure 5b). Importantly, the majority of GLH-60 bolas had two or more positively charged ACh-HG per bola in the proximity of the RNA phosphate groups, while it was only one ACh-HG per bola for the GLH-58. In addition, monitoring of hydrogen bonds between the bolas and the RNA indicated more of them for the GLH-60 (8 at the 100 ns point and 9 at the 183 ns point) than for the GLH-58 (only 2). These results indicate that GLH-60 can bind more tightly to the RNA than GLH-58. Consequently, GLH-58 can release RNA more easily than GLH-60, ultimately allowing for more efficient silencing. These results are consistant with the experimentally obtained binding affinity values for the two bolas.
Equimolar runs indicated that all the GLH-58 bolas (14 bolas in 3 clusters) associate with the RNA (Figure 5c, supporting Figure S13), and the auxiliary MD simulations with individual bolas surrounding the RNA indicated that more than 14 bolas could interact with the RNA and further lower the SASA results of the exposed RNA in the bola/RNA complexes. Therefore, we added another bola cluster to the GLH-58/RNA MD simulations for a total of 19 bolas in 4 clusters. This allowed the ratio between the two types of bolas complexed with the RNA to reach comparable mass concentrations (see Supporting Information). In the approximately 100 ns long MD simulations all of the 19 GLH-58 bolas associated with the RNA, while only 11 of the 14 GLH-60 bolas aggregated on the RNA within 35 ns and no more association was observed for the rest of the MD simulation, which had to be extended to 184 ns to reach a stable conformation, based on the SASA output (Figure 5d). In addition, we observed that of the 19 GLH-58 bolas, 15 were in direct contact with the RNA, while the remaining 4 added a “second layer” interacting only with other bolas. For GLH-60, the numbers were 10 and 1, respectively. These differences, however, did not affect the SASA we measured for both complexes. Despite the different dynamics of RNA/bola interactions both bolas yielded comparable levels of surface protection in the final states, with no statistically significant differences between them (P=0.439). The median, 25 percentile and 75 percentile values for the RNA surface area left exposed to solvent (measured in Å2) in the complexes with GLH-58 and GLH-60 were (2501.8, 2407.0, 2615.0) and (2492.3, 2421.9, 2577.4), respectively (Figure 5c–d). The SASA results appear to agree well with the experimental data for the protection against enzymatic activity (Figure 3d). The propensity to form multiple layers of bolas around the RNA by the GLH-58 may indicate a potential for extended protection by a thicker, multi-layer bola structure, if local excess of GLH-58 is available. Altogether, the MD simulation results agree well with the results of the protection and siRNA release experiments.
4. Discussion
To better understand bola lipids as efficient delivery agents for nucleic acids, the present study focuses on the characterization of newly designed bolas, GLH-58 and GLH-60, for their interaction with nucleic acids and their siRNA delivery efficiency particulary in the context of silencing green fluorescent protein (GFP) gene. These bolas contain similar hydrophobic domains as described previously but vary in the number of positively charged head groups; one positively charged head group at each end of the hydrophobic skeleton in the case of GLH-58 and two positively charged head groups at each end of the hydrophobic skeleton in the case of GLH-60. Our biophysical studies demonstrated that GLH-58 formed more stable micelles as compared to GLH-60. We have substantial experimental and MD simulation results to prove this. In addition, GLH-58 micelles were a superior delivery agent for silencing the gene expression of GFP. GLH-58 was able to silence the GFP expression even at the lower dose of 1 μg/mL whereas GLH-60 could not. This could be explained by the difference between the binding affinity GLH-58 and GLH-60 with nucleic acids.
Fluorescent anisotropy studies showed that GLH-60 had higher binding affinity than GLH-58 . The differences in binding affinities are in agreement with our equimolar computational analysis in which GLH-60 bolas associated with the RNA with more of their head groups than did the GLH-58 bolas. In addition more hydrogen bond interactions were observed for the GLH-60. Approximately a third fewer GLH-60 bolas (10 compared with 15 GLH-58) were predicted to make direct contact with the RNA while yielding comparable surface protection (as measured by SASA). Both the experimental and the computational results indicated that GLH-60 molecules bind more tightly to RNA molecules than GLH-58 and thereby may hamper the release of nucleic acids. In contrast, the uptake of fluorescent RNA/DNA hybrid duplexes by GLH-60 had an increased dose dependency whereas the uptake of duplexes by GLH-58 did not show such a trend with an increase in dose and no significant difference in uptake was observed at all the doses tested. The difference in the amount of uptake and the silencing efficiency by both the bolas can only be speculated to be related to the increase in release of the nucleic acids by GLH-58 inside the cell. Thus, a sufficient amount of siRNA molecules were not available at the lower concentration of 5 μg/mL to cause the silencing effect due to strong binding of DS RNA with GLH-60, whereas due to lower binding affinity of DS RNA with GLH-58, this bola showed significant silencing activity at lower concentrations compared to GLH-60. The results of this study suggest that our bolaamphiphile carrier is at least as effective as other lipid-based carriers, but the higher stability of bolaamphiphiles of the type described here [17, 31] as compared to amphiphiles with one hydrophilic head group, such as those found in lipoplexes, and the lower toxicity of these types of bolaamphiphiles [17] compared to other cationic lipids used for the delivery of nucleic acids [8], indicate the superiority of bolaamphiphiles over other amphiphiles in delivering nucleic acids.
5. Conclusion
Two types of bolaamphiphiles with different head groups were synthesized from natural jojoba oil and compared experimentally. These bolas can form positively charged micelles in aqueous solutions with very low cytotoxicities. The current work presents the chemical synthesis with in silico and in vitro studies of interactions between polycationic micelles and functional siRNAs. We confirmed the formation of stable complexes, which protect nucleic acids from degradation. The work is also supported by extensive cell culture experments. Bolas are shown to effectively deliver and release siRNAs into cancer cells causing significant silencing of a target gene.
The results of our experimental studies are well supported by computational molecular dynamics (MD) simulations and we surmise that among both bolas (GLH-58 and −60) tested, GLH-58 with one positively charged ACh-HG at each end of the hydrophobic domain formed more stable nano sized micelles, had lower binding affinity to nucleic acids and were superior delivery agents for the delivery of siRNAs compared to GLH-60 with two positively charged ACh-HGs at each end of the hydrophobic domain. GLH-58 was able to release siRNAs more efficiently compared to GLH-60 so that significant silencing of the GFP expression was observed at a concentration as low as 5 μg/mL. Moreover, these bolas were used to deliver various thereapeutic siRNAs to HIV infected human cells and can also be a carrier for various functional RNA nanoparticles (characterized previously [32–34]) thus, becoming instrumental in the emerging field of therapeutic RNA nanotechnology [20, 21, 35–41].
Supplementary Material
Acknowledgements
This work is dedicated to Sarina Grinberg who recently passed away. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD and the National Cancer Institute’s Advanced Biomedical Computing Center (ABCC) of the Frederick National Laboratory, Frederick. We thank Dr. Bindu Lakshman and Dr. Andrew Stephen, Protein Chemistry Laboratory, Cancer Research Technology Program, Leidos Biomedical Inc. Frederick, MD for their help in using the fluorescent anisotropy technique. This publication has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract HHSN 261200800001E to WKK. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nature Materials. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsabahy M, Nazarali A, Foldvari M. Non-viral nucleic acid delivery: key challenges and future directions. Curr Drug Deliv. 2011;8(3):235–244. doi: 10.2174/156720111795256174. [DOI] [PubMed] [Google Scholar]
- 6.Sunshine JC, Bishop CJ, Green JJ. Advances in polymeric and inorganic vectors for nonviral nucleic acid delivery. Ther Deliv. 2011;2(4):493–521. doi: 10.4155/tde.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pensado A, Seijo B, Sanchez A. Current strategies for DNA therapy based on lipid nanocarriers. Expert Opin Drug Deliv. 2014;11(11):1721–1731. doi: 10.1517/17425247.2014.935337. [DOI] [PubMed] [Google Scholar]
- 8.Kuo WT, Huang HY, Huang YY. Intracellular trafficking, metabolism and toxicity of current gene carriers. Curr Drug Metab. 2009;10(8):885–894. doi: 10.2174/138920009790274504. [DOI] [PubMed] [Google Scholar]
- 9.Khatri N, Baradia D, Vhora I, Rathi M, Misra A. Development and characterization of siRNA lipoplexes: Effect of different lipids, in vitro evaluation in cancerous cell lines and in vivo toxicity study. AAPS PharmSciTech. 2014;15(6):1630–1643. doi: 10.1208/s12249-014-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garinot M, Mignet N, Largeau C, Seguin J, Scherman D, Bessodes M. Amphiphilic polyether branched molecules to increase the circulation time of cationic particles. Bioorg Med Chem. 2007;15(9):3176–3186. doi: 10.1016/j.bmc.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 11.Alhakamy NA, Elandaloussi I, Ghazvini S, Berkland CJ, Dhar P. Effect of Lipid Headgroup Charge and pH on the Stability and Membrane Insertion Potential of Calcium Condensed Gene Complexes. Langmuir. 2015;31(14):4232–4245. doi: 10.1021/la504970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Liu R, Shi Y, Zhang Z, Zhang X. Zwitterionic poly(carboxybetaine)-based cationic liposomes for effective delivery of small interfering RNA therapeutics without accelerated blood clearance phenomenon. Theranostics. 2015;5(6):583–596. doi: 10.7150/thno.11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dakwar GR, Abu Hammad I, Popov M, Linder C, Grinberg S, Heldman E, Stepensky D. Delivery of proteins to the brain by bolaamphiphilic nano-sized vesicles. J Control Release. 2012;160(2):315–321. doi: 10.1016/j.jconrel.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 14.Grinberg S, Kolot V, Linder C, Shaubi E, Kas’yanov V, Deckelbaum RJ, Heldman E. Synthesis of novel cationic bolaamphiphiles from vernonia oil and their aggregated structures. Chem Phys Lipids. 2008;153(2):85–97. doi: 10.1016/j.chemphyslip.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Grinberg S, Linder C, Heldman E. Progress in lipid-based nanoparticles for cancer therapy. Crit Rev Oncog. 2014;19(3–4):247–260. doi: 10.1615/critrevoncog.2014011815. [DOI] [PubMed] [Google Scholar]
- 16.Popov M, Grinberg S, Linder C, Waner T, Levi-Hevroni B, Deckelbaum RJ, Heldman E. Site-directed decapsulation of bolaamphiphilic vesicles with enzymatic cleavable surface groups. J Control Release. 2012;160(2):306–314. doi: 10.1016/j.jconrel.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Popov M, Linder C, Deckelbaum RJ, Grinberg S, Hansen IH, Shaubi E, Waner T, Heldman E. Cationic vesicles from novel bolaamphiphilic compounds. J Liposome Res. 2010;20(2):147–159. doi: 10.3109/08982100903218900. [DOI] [PubMed] [Google Scholar]
- 18.Puri A, Loomis K, Smith B, Lee JH, Yavlovich A, Heldman E, Blumenthal R. Lipid-Based Nanoparticles as Pharmaceutical Drug Carriers: From Concepts to Clinic. Critical Reviews in Therapeutic Drug Carrier Systems. 2009;26(6):523–580. doi: 10.1615/critrevtherdrugcarriersyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, Grinberg S, Linder C, Blumenthal RP, Shapiro BA. In Silico, In Vitro, and In Vivo Studies Indicate the Potential Use of Bolaamphiphiles for Therapeutic siRNAs Delivery. Mol Ther Nucleic Acids. 2013;2:80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, Heldman E, Jaeger L, Blumenthal R, Shapiro BA. Activation of different split functionalities on re-association of RNA-DNA hybrids. Nature nanotechnology. 2013;8(4):296–304. doi: 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA. Multifunctional RNA nanoparticles. Nano letters. 2014;14(10):5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jay RR. Direct Titration of Epoxy Compounds + Aziridines. Analytical Chemistry. 1964;36(3):667. [Google Scholar]
- 23.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic acids research. 2005;33(13):4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta K, Jang H, Harlen K, Puri A, Nussinov R, Schneider JP, Blumenthal R. Mechanism of membrane permeation induced by synthetic beta-hairpin peptides. Biophysical journal. 2009;105(9):2093–2103. doi: 10.1016/j.bpj.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case DA, Darden TA, Cheatham TE, 3rd, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER12. San Francisco: University of California; 2012. [Google Scholar]
- 26.Essmann U, Perera L, Berkowitz ML, Darden TA, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 27.Stern A, Guidotti M, Shaubi E, Popov M, Linder C, Heldman E, Grinberg S. Steric environment around acetylcholine head groups of bolaamphiphilic nanovesicles influences the release rate of encapsulated compounds. Int J Nanomedicine. 2014;9:561–574. doi: 10.2147/IJN.S53563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jass J, Tjarnhage T, Puu G. From liposomes to supported, planar bilayer structures on hydrophilic and hydrophobic surfaces: an atomic force microscopy study. Biophysical journal. 2000;79(6):3153–3163. doi: 10.1016/S0006-3495(00)76549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reviakine I, Brisson A. Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir. 2000;16(4):1806–1815. [Google Scholar]
- 30.Mornet S, Lambert O, Duguet E, Brisson A. The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy. Nano letters. 2005;5(2):281–285. doi: 10.1021/nl048153y. [DOI] [PubMed] [Google Scholar]
- 31.Hutter T, Linder C, Heldman E, Grinberg S. Interfacial and self-assembly properties of bolaamphiphilic compounds derived from a multifunctional oil. Journal of Colloid and Interface Science. 2012;365(1):53–62. doi: 10.1016/j.jcis.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 32.Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc. 2011;6(12):2022–2034. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, Shapiro BA. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano letters. 2012;12(10):5192–5195. doi: 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano letters. 2011;11(2):878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, Isambert H, Guo P, Rossi JJ, Tenenbaum SA, Shapiro BA. A boost for the emerging field of RNA nanotechnology. ACS Nano. 2011;5(5):3405–3418. doi: 10.1021/nn200989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo P. The emerging field of RNA nanotechnology. Nature nanotechnology. 2010;5(12):833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao C, Li X, Tian C, Jiang W, Wang G, Mao C. Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nature communications. 2014;5:3890. doi: 10.1038/ncomms4890. [DOI] [PubMed] [Google Scholar]
- 38.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano letters. 2005;5(9):1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nature nanotechnology. 2011;6(10):658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Advanced drug delivery reviews. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA. In silico design and enzymatic synthesis of functional RNA nanoparticles. Accounts of chemical research. 2014;47(6):1731–1741. doi: 10.1021/ar400329z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Low JT, Knoepfel SA, Watts JM, ter Brake O, Berkhout B, Weeks KM. Shapedirected discovery of potent ShRNA inhibitors of Hiv-1. Mol. Ther. 2012;20:820–828. doi: 10.1038/mt.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ter Brake O, t Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.