Abstract
Biological molecular processes are often studied in model systems, which simplifies their inherent complexity but may cause investigators to lose sight of the effects of the molecular environment. Information obtained in this way must therefore be validated by experiments in the cell. NMR has been used to study biological cells since the early days of its development. The first NMR structural studies of a protein inside a cell (by solution-state NMR) and of a membrane protein (by solid-state NMR) were published in 2001 and 2011, respectively. More recently, dynamic nuclear polarization, which has been used to enhance the signal in solid-state NMR, has also been applied to the study of frozen cells. Much progress has been made in the past 5 years, and in this review we take stock of this new technique, which is particularly appropriate for the study of biological membranes.
Main Text
NMR is a noninvasive technique that can tackle individual molecules as well as whole animals. For NMR, three types of samples can qualify as in cell: living cells (in vivo NMR), intact cells, and intact cellular compartments such as organelles and cellular envelopes (in situ NMR). Biological cells were studied by NMR as early as 1955 (1), and 15 years later the focus shifted from looking at water molecules to examining lipids (2) and small metabolites (3). In the 2000s, larger soluble proteins within bacterial cells were studied by NMR (4), and in-cell NMR became a field in and of itself, with investigators identifying common obstacles and finding ways to circumvent them. In particular, a meaningful in-cell experiment requires a stable environment to avoid the degradation (e.g., molecular oxidation, hydrolysis, or proteolysis by proteases) that can occur in such samples in a matter of hours or days. Several approaches have been suggested to prolong sample survival (5, 6), but the state of the sample should be regularly checked independently by biochemical methods, such as electrophoresis or activity assays. The first decade of in-cell solution-state NMR was reviewed in 2011 (7).
The weak signal/noise ratio provided by molecules in cells is due not only to their low abundance but also to environmental heterogeneity and the slow molecular tumbling of molecules that are either too large or are interacting with other molecules. It has been shown that the rotational correlation time (the time it takes for a molecule to rotate one radian) of proteins inside a cell is longer because of increased viscosity, but in most cases it is increased by only a factor of 2 (8), which should not be a major obstacle for solution-state NMR. Nevertheless, the molecular rotational correlation time can become prohibitively long for specific cellular or membrane interactions, in which case solid-state NMR and magic-angle spinning (MAS) may be the best option (9). In addition, MAS also averages out sample heterogeneity and magnetic susceptibility anisotropies (10), thereby improving the spectral resolution.
Solid-state NMR has been applied to a variety of biomolecules that are naturally solid, ranging from human biopsies (10) to amyloid fibrils (11), mussel byssus (12), whole nematodes (13), and whole living flies (14), but it has also proved useful for studying membrane-bound molecules, which become solid because of the boundary imposed by the membrane. Such molecules include lipids, membrane proteins, and components of the cell wall and extracellular matrix that can also be studied separately or reconstituted into model systems (15). In 2011, solid-state NMR joined solution-state NMR in the quest to determine the structure of proteins within cells (16). The concomitant development of dynamic nuclear polarization (DNP) for increased sensitivity (17) has made solid-state NMR an important player in this field, and over the past 5 years investigators have made much progress in optimizing sample preparation protocols and NMR and DNP conditions, and gathering important preliminary data.
In this review, we assess the state of the art of in-cell solid-state NMR and discuss the achievements that have been made in studying cell-envelope molecules such as lipids, membrane proteins, and lipopolysaccharides, as well as cell wall and biofilm extracellular matrix components, within their natural environment.
Sample preparation
As indicated above, in-cell NMR suffers from a major limitation: the short period of time before the sample irreversibly degrades. With some rare exceptions, in-cell solid-state NMR is often performed at low temperature to preserve the integrity of the membrane, and in certain cases, cellular samples are even frozen or lyophilized, which makes it impossible to probe dynamic phenomena. Although it is debatable whether results obtained in this way qualify as in cell, the important factor is that the studied molecules are never taken out of their native environment. Sample spinning has also been criticized as being too violent for cells because of its centrifugal and dehydrating effects, but spinning rates and times can be adjusted to preserve cellular integrity and even cell survival (14, 18, 19, 20).
In addition, in-cell NMR faces the same obstacles as spectroscopy in general: sensitivity and resolution. Although in-cell solid-state NMR has benefited from recent developments such as MAS for high-resolution solid-state NMR and DNP for signal enhancement, the application of in-cell NMR to proteins also relies on one’s ability to overexpress these biomolecules to ensure that they are 1) concentrated enough to be rapidly visible by NMR (on the order of 100 μM) and 2) more abundant than other molecules that may obscure them. Protein overexpression in bacteria has been improved throughout the years and is now very common, although it remains a challenge in the case of eukaryotic protein expression (21).
One can also achieve increased sensitivity to reduce the experimental time and improve the cell survival rate by using isotopic labeling. It is now quite simple to achieve uniform isotopic labeling, using mostly ammonium chloride for nitrogen labeling and glucose, glycerol, or sodium carbonate for carbon labeling. Carbon background labeling is often detrimental, and specific labeling for carbon may be preferable. For proteins, this can be done by using other labeled precursors (e.g., amino acids or ketoacids to label only the methyl groups), which reduces the background but presents other problems, such as metabolic scrambling. Investigators in this area of research are actively searching for alternatives, and even more specific labeling can be achieved by using auxotrophic bacterial strains that force bacteria to use the incorporated labeled molecule for a specific metabolic pathway (22). Although labeling is often unnecessary for lipids, researchers have used deuterated fatty acids to label lipids and observe them inside bacterial cell membranes (23).
Lipids and lipopolysaccharides
Pioneering work in solid-state NMR, before the advent of MAS, relied on low-resolution NMR spectra for protons and carbons, and wide-line NMR for phosphorous and deuterium (24). Lipids constitute approximately half of the membrane weight, and phospholipids are abundant in most membrane cells (except for some plants), making 31P, an abundant and sensitive isotope, a nucleus of choice for in-cell NMR. In addition, rather than simply assigning signals, wide-line 31P NMR provides structural and dynamic information about the lipid headgroups, such as phase changes, polymorphism, and changes in the order parameters. Therefore, 31P NMR has been used to study a variety of cells ranging from rabbit muscle cells (25) to bacteria (26, 27). Surprisingly, although MAS also improves the resolution in 31P NMR and theoretically allows the separation of lipids according to their headgroups, 31P MAS NMR has not been applied very often to cell membrane analysis (28, 29).
As opposed to 31P, 2H is a very rare isotope that must be incorporated into the sample to be visible by NMR. Its natural background, on the other hand, is almost null. Throughout the 1970s, wide-line 2H NMR was applied to various cell membrane lipids, starting with bacteria (30, 31), and provided structural and dynamic information about lipid acyl chains by detecting changes in phases, order parameters, spectral moments, and membrane fluidity or curvature that complemented information obtained by 31P NMR. This approach was still used in recent studies to probe whole sea urchin sperm cells (32) and the effect of antimicrobial agents on bacterial cell membranes (23, 33). However, in 2015, in-cell 2H MAS NMR was used to probe bacterial cell membrane lipids and was found to provide the same information as static NMR but within a shorter timeframe (20), making it compatible with cell survival.
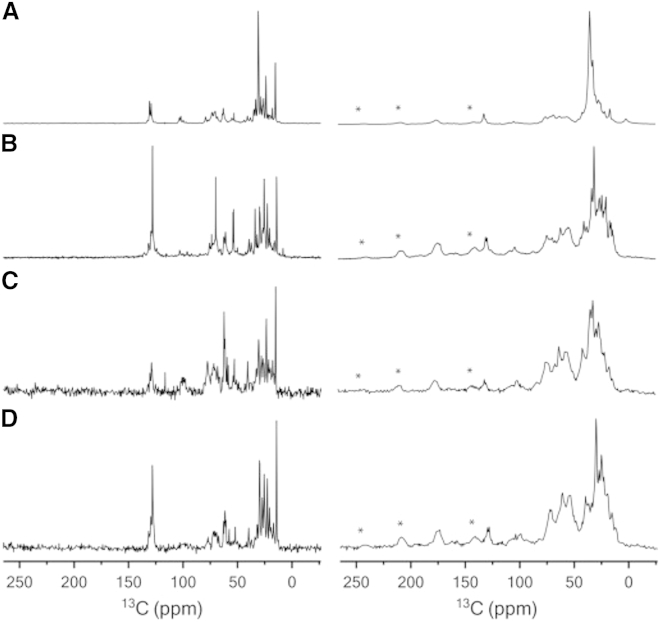
The first attempt to observe whole red blood cells by 1H NMR dates back 60 years ago (1), but 15 years later, the focus shifted toward cell membrane lipids that could be assigned by either proton (2) or carbon (34) NMR. 1H and 13C NMR were used to assign many cell lipids throughout the subsequent decades, and the resolution was greatly improved in the 1990s with the generalization of MAS, allowing solid-state NMR to become a diagnostic tool for the detection of diseases (10, 35). Lipopolysaccharides (LPSs) were also scrutinized by 1H MAS NMR directly on the surface of living bacterial cell membranes (36), which sometimes allowed their acetylation state to be determined in situ (19). More recently, we studied whole microalgae by dynamically filtered 13C MAS NMR (37). Using polarization transfer schemes that are sensitive to molecular dynamics, such as refocused-insensitive nuclei enhanced by polarization transfer (RINEPT), which depends on scalar couplings, and cross-polarization (CP), which depends on dipolar couplings that can be averaged in the presence of motion, we were able to select mobile or rigid molecules, respectively. This strategy allowed us to identify the rigid cell-wall carbohydrates and membrane proteins, as well as the more mobile cell membrane lipids (see Fig. 1).
Figure 1.
(A–D) Dynamically filtered 13C NMR spectra of (A) Nannochloropsis oculata, (B) Pavlova lutheri, (C) wild-type, and (D) cell-wall-depleted Chlamydomonas reinhardtii. Dynamic and rigid molecules are detected by RINEPT (left) and CP (right), respectively. Spinning side bands are indicated by asterisks. Reprinted from (37) with permission from Elsevier.
Cell wall and extracellular matrix
In addition to membranes, many algae, fungi, and plant cells possess a cell wall composed of polysaccharides and (in bacteria) polysaccharides and peptidoglycan. As early as 1979 (38), the peptidoglycan of Staphylococcus aureus in intact cells was studied by 15N and 13C NMR, which can be used to assign resonances and measure relaxation times, and thus probe chain motion and packing. In the 1980s, the development of MAS, CP-MAS, and high-resolution solid-state NMR shifted the focus from lipids toward more rigid molecules such as polysaccharides and membrane proteins, and investigators sought to determine the atomic structures of these complex molecules. The advent of MAS and CP-MAS allowed a much more precise characterization of the molecular structure and metabolism of bacterial cell walls. The groups of Jake Schaefer and Lynette Cegelski examined lyophilized isolated cell walls, and sometimes intact whole cells, by 15N, 13C, 19F, and 31P MAS NMR, measuring accurate distances within the peptidoglycan and between the peptidoglycan and antibiotic molecules (39, 40, 41, 42, 43, 44). This strategy was recently extended to composition analyses of freeze-dried, isolated extracellular matrix from bacterial biofilms of E. coli (45) and Vibrio cholerae (46) by 15N, 13C, and 31P MAS NMR.
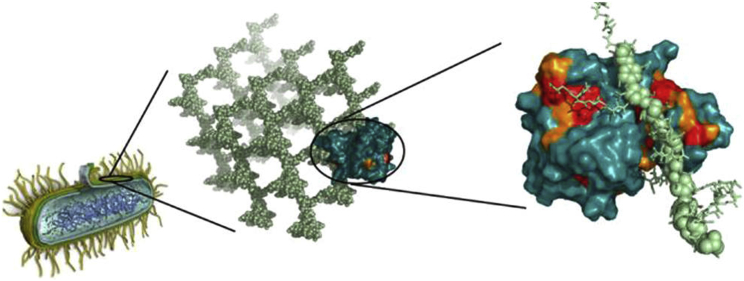
Various isolated (but hydrated) cell walls have also been probed by 13C MAS NMR and DNP. The group of Jean-Pierre Simorre compared the organization of peptidoglycan in various bacteria, including its metal-binding sites and constants, cross-linkage, flexibility, and dynamics (18, 47). More recently, the same group (48) used 15N, 13C, and 31P to determine the high-resolution structure of a complex made up of the Bacillus subtilis peptidoglycan and the L,D-transpeptidase protein (see Fig. 2). At the same time, the group of Mei Hong studied the organization of polysaccharides in the cell wall of the plant Arabidopsis thaliana, including interactions, dynamics, and hydration (49, 50), with the help of solid-state 1H and 13C NMR techniques. With the additional benefit of DNP and an enhancement factor of 30 at 600 MHz, they were able to determine that the functional binding target of the protein expansin in the A. thaliana cell wall is a cellulose that is enriched in hemicellulose xyloglucan and has a different structure compared with bulk cellulose (51).
Figure 2.
Cartoon showing the interactions between the highly cross-linked peptidoglycan of B. subtilis and the L,D-transpeptidase that performs this cross-linking. Reprinted with permission from Schanda et al. (48). Copyright 2014 American Chemical Society. To see this figure in color, go online.
Membrane proteins
The first in-cell NMR study of a membrane protein is a special case, since Halobacterium salinarium is an archaeon in which the sole membrane protein, bacteriorhodopsin (bR), occupies nearly half of the purple membrane surface and is organized as a regular, homogeneous, and rigid 2D hexagonal lattice. In 1983, the group of Bob Griffin began an extensive study of bR by using MAS NMR to determine the bR retinal protonation state (52). The quest for intermediate structural states during the bR photocycle continues today, and 15N and 13C solid-state NMR, as well as DNP, have recently allowed investigators to disentangle several coexisting states, including some that had never been observed previously, thanks to an enhancement factor of up to 90 at 380 MHz (17).
In 2011, Riqiang Fu and collaborators (16) published the first in-cell solid-state structural NMR study of a single membrane protein, the small transmembrane domain of the human lipoprotein receptor 11 (LR11) embedded in the purified Escherichia coli membrane where it was expressed. Several labeling schemes were tested (e.g., uniform labeling versus specific labeling of the alanines) and many parameters were optimized to reduce the intensity of lipid signals versus protein signals. Purified membrane signals were also compared with signals of the same protein reconstituted in detergent micelles.
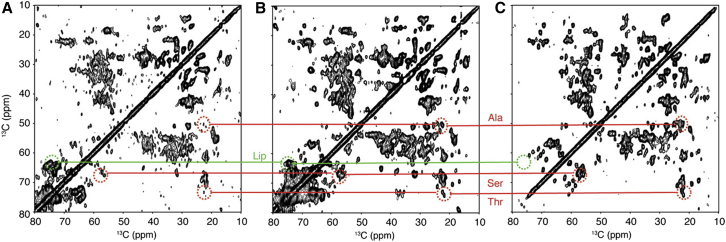
The following year, in-cell 13C and 15N MAS solid-state NMR was applied to observe two small soluble proteins in E. coli, FKBP and Trx, which are invisible by solution-state NMR because interactions with other molecules slow their tumbling prohibitively (9). Using a similar approach, the group of Marc Baldus tackled the structure of the bacterial membrane protein lipid A deacylase of Pseudomonas aeruginosa (PagL) embedded in the E. coli outer membrane where it was expressed (53). Protein expression and labeling schemes were improved, and whole cells were compared with purified membranes and reconstituted proteoliposomes (see Fig. 3), allowing the assignment of PagL signals. Other signals were identified from LPS and peptidoglycan, as well as from another membrane protein, the endogenous Braun lipoprotein. Sample stability was also assessed, and it was found that whole cells were only stable for 36 h, whereas purified membranes were stable for about a week at low temperature. The same sample was subjected to DNP at low temperature, with an enhancement factor of 30 at 400 MHz, allowing faster data acquisition (and hence preservation of membrane integrity) as well as the detection of previously unseen low-abundance molecules such as nucleic acids (54).
Figure 3.
(A–C) 13C-13C NMR spectra of whole cells (A), cell envelopes (B), and (U-13C,15N)-labeled PagL-containing proteoliposomes (C). Characteristic crosspeaks of Ala, Ser, and Thr residues of PagL, and endogenous E. coli lipids (Lip) are indicated. Reprinted with permission from (53). To see this figure in color, go online.
High-field, solid-state DNP spectrometers have only been commercially available since 2009, and the first applications to cellular samples were mostly proofs of concept showing that despite the current resolution limitations at low temperature and the concomitant freezing of dynamics, the benefit of signal/noise ratio enhancement (on the order of 10–100, with an average of 30) allowed signals to be observed from very-low-concentration molecules. For example, in 2011, the group of Hartmut Oschkinat was able to detect by in-cell DNP the protein neurotoxin II bound to its naturally low-concentration receptor, the acetylcholine receptor in the synaptic membrane of the Pacific electric ray (55). In 2012, the same group was able to compare the structure of the membrane-associated protein mistic, determined by DNP directly in isolated native E. coli membranes, with a structure previously determined in detergent micelles (56). In a similar approach, the group of Ayyalusamy Ramamoorthy demonstrated the feasibility of using DNP on the membrane-anchored cytochrome b5, which contains a large soluble domain, in native E. coli cells, and compared the results with solid-state NMR data obtained in other membrane mimetics (57).
So far, in-cell solid-state MAS NMR and DNP have mostly been used to validate structures that were previously determined in model systems. For example, various structures of the M2 proton channel of the influenza A virus had been suggested from experiments performed in liquid crystalline lipid bilayers, detergent micelles, and detergent-based crystals (58). Solid-state NMR performed in native E. coli membranes could validate the tetrameric structure that had been proposed based on the results of solid-state NMR performed in synthetic bilayers (59). The diacylglycerol kinase structure had also been determined in detergent micelles, and Shi et al. (60) compared it with data obtained from the same protein in E. coli membranes, using modified amino acids incorporating 19F labels. In the same line of thought, the structure of Anabaena sensory rhodopsin, which was previously determined in lipid bilayers, was validated against a structure determined in E. coli inner membranes. It was found that Anabaena sensory rhodopsin organized into trimers in both environments, but formed different crystal lattices (61). Baldus et al. (62) recently used in-cell solid-state NMR and DNP data to validate a proposed model structure, without relying on any previously obtained NMR data. Applied to the megadalton bacterial type IV secretion system core complex (T4SScc), their approach showed that T4SScc was well folded in its E. coli membranes.
Conclusions
Investigators in structural biology are making a significant step forward by tackling structures of molecules in their native environment, and solid-state NMR can contribute to this quest to determine the structures of cell-envelope-associated molecules. Although NMR has been used to study lipids in cellular membranes since 1967, progress in NMR techniques is now allowing researchers to gather much more precise information compatible with medical studies. Technical improvements in solid-state NMR have enabled us to focus on relatively mobile cellular molecules, such as lipids, as well as on much more rigid molecules, such as membrane proteins and cell wall and extracellular matrix components that were previously not accessible by any other structure-determination technique.
It is in the field of membrane protein structure determination that in-cell solid-state NMR has made the most significant leap in the past 5 years. Although most of the membrane proteins studied by in-cell solid-state NMR and DNP have already been studied in model systems, thanks to advances in this field, investigators have been able to optimize sample preparation, protein overexpression, isotopic labeling, NMR acquisition parameters, and general study protocols. In addition, they have provided much appreciated in-cell validations of model structures, and are now starting to tackle in-cell membrane protein complexes that have never been studied by NMR before.
Fast methods that benefit from MAS and DNP, among other technical developments, allow the acquisition of full sets of data within a timeframe that is compatible with cell survival. Therefore, the next step, which seems to be in reach, will be to develop in vivo solid-state NMR for the structural study of membrane-bound biomolecules.
Acknowledgments
This work was supported by the CNRS (UMR 7099), the Université Paris Diderot, the Labex Dynamo (ANR-11-LABX-0011-01), the Natural Science and Engineering Research Council of Canada, and a French Ministère de l’Enseignement Supérieur et de la Recherche scholarship awarded to X.L.W.
Editor: Brian Salzberg.
References
- 1.Odeblad E., Lindström G. Some preliminary observations on the proton magnetic resonance in biologic samples. Acta Radiol. 1955;43:469–476. doi: 10.3109/00016925509172514. [DOI] [PubMed] [Google Scholar]
- 2.Chapman D., Kamat V.B., Penkett S.A. Nuclear magnetic resonance spectroscopic studies of erythrocyte membranes. Nature. 1967;213:74–75. [Google Scholar]
- 3.Eakin R.T., Morgan L.O., Matwiyoff N.A. Carbon-13 nuclear magnetic resonance spectroscopy of living cells and their metabolism of a specifically labeled 13C substrate. FEBS Lett. 1972;28:259–264. doi: 10.1016/0014-5793(72)80726-9. [DOI] [PubMed] [Google Scholar]
- 4.Serber Z., Keatinge-Clay A.T., Dötsch V. High-resolution macromolecular NMR spectroscopy inside living cells. J. Am. Chem. Soc. 2001;123:2446–2447. doi: 10.1021/ja0057528. [DOI] [PubMed] [Google Scholar]
- 5.Bodart J.-F., Wieruszeski J.-M., Lippens G. NMR observation of Tau in Xenopus oocytes. J. Magn. Reson. 2008;192:252–257. doi: 10.1016/j.jmr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Li C., Charlton L.M., Pielak G.J. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J. Am. Chem. Soc. 2008;130:6310–6311. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maldonado A.Y., Burz D.S., Shekhtman A. In-cell NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011;59:197–212. doi: 10.1016/j.pnmrs.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serber Z., Corsini L., Dötsch V. In-cell NMR spectroscopy. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- 9.Reckel S., Lopez J.J., Dötsch V. In-cell solid-state NMR as a tool to study proteins in large complexes. ChemBioChem. 2012;13:534–537. doi: 10.1002/cbic.201100721. [DOI] [PubMed] [Google Scholar]
- 10.Millis K.K., Maas W.E., Singer S. Gradient, high-resolution, magic-angle spinning nuclear magnetic resonance spectroscopy of human adipocyte tissue. Magn. Reson. Med. 1997;38:399–403. doi: 10.1002/mrm.1910380307. [DOI] [PubMed] [Google Scholar]
- 11.Helmus J.J., Surewicz K., Jaroniec C.P. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold A.A., Byette F., Marcotte I. Solid-state NMR structure determination of whole anchoring threads from the blue mussel Mytilus edulis. Biomacromolecules. 2013;14:132–141. doi: 10.1021/bm301493u. [DOI] [PubMed] [Google Scholar]
- 13.Blaise B.J., Giacomotto J., Elena B. Metabolic profiling strategy of Caenorhabditis elegans by whole-organism nuclear magnetic resonance. J. Proteome Res. 2009;8:2542–2550. doi: 10.1021/pr900012d. [DOI] [PubMed] [Google Scholar]
- 14.Sarou-Kanian V., Joudiou N., Beloeil J.-C. Metabolite localization in living drosophila using high resolution magic angle spinning NMR. Sci. Rep. 2015;5:9872. doi: 10.1038/srep09872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warschawski D.E., Arnold A.A., Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Fu R., Wang X., Tian F. In situ structural characterization of a recombinant protein in native Escherichia coli membranes with solid-state magic-angle-spinning NMR. J. Am. Chem. Soc. 2011;133:12370–12373. doi: 10.1021/ja204062v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj V.S., Mak-Jurkauskas M.L., Griffin R.G. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proc. Natl. Acad. Sci. USA. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kern T., Giffard M., Simorre J.-P. Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J. Am. Chem. Soc. 2010;132:10911–10919. doi: 10.1021/ja104533w. [DOI] [PubMed] [Google Scholar]
- 19.Zandomeneghi G., Ilg K., Meier B.H. On-cell MAS NMR: physiological clues from living cells. J. Am. Chem. Soc. 2012;134:17513–17519. doi: 10.1021/ja307467p. [DOI] [PubMed] [Google Scholar]
- 20.Warnet X.L., Laadhari M., Warschawski D.E. A 2H magic-angle spinning solid-state NMR characterisation of lipid membranes in intact bacteria. Biochim. Biophys. Acta. 2016 doi: 10.1016/j.bbamem.2015.10.020. pii:S0005-2736(15)00360-0. [DOI] [PubMed] [Google Scholar]
- 21.Wagner S., Klepsch M.M., de Gier J.W. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. USA. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin M.T., Sperling L.J., Gennis R.B. A rapid and robust method for selective isotope labeling of proteins. Methods. 2011;55:370–378. doi: 10.1016/j.ymeth.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardy-Laporte C., Arnold A.A., Marcotte I. A 2H solid-state NMR study of the effect of antimicrobial agents on intact Escherichia coli without mutating. Biochim. Biophys. Acta. 2013;1828:614–622. doi: 10.1016/j.bbamem.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs R.E., Oldfield E. NMR of membranes. Prog. NMR Spectrosc. 1981;14:113–136. [Google Scholar]
- 25.Davis D.G., Inesi G. Phosphorus and proton nuclear magnetic resonance studies in sarcoplasmic reticulum membranes and lipids. A comparison of phosphate and proton group mobilities in membranes and lipid bilayers. Biochim. Biophys. Acta. 1972;282:180–186. doi: 10.1016/0005-2736(72)90322-7. [DOI] [PubMed] [Google Scholar]
- 26.Ianzini F., Guidoni L., Yatvin M.B. Effects of decreased pH on membrane structural organization of Escherichia coli grown in different fatty acid-supplemented media: a 31P NMR study. Arch. Biochem. Biophys. 1990;278:1–10. doi: 10.1016/0003-9861(90)90223-l. [DOI] [PubMed] [Google Scholar]
- 27.Chia B.C.S., Lam Y.H., Bowie J.H. A 31P NMR study of the interaction of amphibian antimicrobial peptides with the membranes of live bacteria. Lett. Pept. Sci. 2000;7:151–156. [Google Scholar]
- 28.Moreau C., Cavalier A., Le Rumeur E. Sarcolemma phospholipid structure investigated by immunogold electron microscopy and 31P NMR spectroscopy with lanthanide ions. FEBS Lett. 2001;509:417–422. doi: 10.1016/s0014-5793(01)03199-4. [DOI] [PubMed] [Google Scholar]
- 29.Umegawa Y., Yamaguchi T., Matsuoka S. Centerband-only analysis of rotor-unsynchronized spin echo for measurement of lipid 31P chemical shift anisotropy. Magn. Reson. Chem. 2015;53:514–519. doi: 10.1002/mrc.4247. [DOI] [PubMed] [Google Scholar]
- 30.Oldfield E., Chapman D., Derbyshire W. Lipid mobility in Acholeplasma membranes using deuteron magnetic resonance. Chem. Phys. Lipids. 1972;9:69–81. doi: 10.1016/0009-3084(72)90034-5. [DOI] [PubMed] [Google Scholar]
- 31.Stockton G.W., Johnson K.G., Bloom M. Deuterium NMR study of lipid organization in Acholeplasma laidlawii membranes. Nature. 1977;269:267–268. [Google Scholar]
- 32.Garnier-Lhomme M., Grélard A., Larijani B. Probing the dynamics of intact cells and nuclear envelope precursor membrane vesicles by deuterium solid state NMR spectroscopy. Biochim. Biophys. Acta. 2007;1768:2516–2527. doi: 10.1016/j.bbamem.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Pius J., Morrow M.R., Booth V. 2H solid-state nuclear magnetic resonance investigation of whole Escherichia coli interacting with antimicrobial peptide MSI-78. Biochemistry. 2012;51:118–125. doi: 10.1021/bi201569t. [DOI] [PubMed] [Google Scholar]
- 34.Metcalfe J.C., Birdsall N.J., Partington P. 13C NMR spectra of lecithin vesicles and erythrocyte membranes. Nature. 1971;233:199–201. doi: 10.1038/233199a0. [DOI] [PubMed] [Google Scholar]
- 35.Husted C., Montez B., Oldfield E. Carbon-13 “magic-angle” sample-spinning nuclear magnetic resonance studies of human myelin, and model membrane systems. Magn. Reson. Med. 1993;29:168–178. doi: 10.1002/mrm.1910290204. [DOI] [PubMed] [Google Scholar]
- 36.Jachymek W., Niedziela T., Kenne L. Structures of the O-specific polysaccharides from Yokenella regensburgei (Koserella trabulsii) strains PCM 2476, 2477, 2478, and 2494: high-resolution magic-angle spinning NMR investigation of the O-specific polysaccharides in native lipopolysaccharides and directly on the surface of living bacteria. Biochemistry. 1999;38:11788–11795. doi: 10.1021/bi990673y. [DOI] [PubMed] [Google Scholar]
- 37.Arnold A.A., Genard B., Marcotte I. Identification of lipid and saccharide constituents of whole microalgal cells by 13C solid-state NMR. Biochim. Biophys. Acta. 2015;1848(1 Pt B):369–377. doi: 10.1016/j.bbamem.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Lapidot A., Irving C.S. Nitrogen-15 and carbon-13 dynamic nuclear magnetic resonance study of chain segmental motion of the peptidoglycan pentaglycine chain of 15N-Gly- and 13C2-Gly-labeled Staphylococcus aureus cells and isolated cell walls. Biochemistry. 1979;18:1788–1796. doi: 10.1021/bi00576a024. [DOI] [PubMed] [Google Scholar]
- 39.Jacob G.S., Schaefer J., Wilson G.E., Jr. Direct measurement of peptidoglycan cross-linking in bacteria by 15N nuclear magnetic resonance. J. Biol. Chem. 1983;258:10824–10826. [PubMed] [Google Scholar]
- 40.Garbow J.R., Jacob G.S., Schaefer J. Protein dynamics from chemical shift and dipolar rotational spin-echo 15N NMR. Biochemistry. 1989;28:1362–1367. doi: 10.1021/bi00429a063. [DOI] [PubMed] [Google Scholar]
- 41.Tong G., Pan Y., Schaefer J. Structure and dynamics of pentaglycyl bridges in the cell walls of Staphylococcus aureus by 13C-15N REDOR NMR. Biochemistry. 1997;36:9859–9866. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 42.Singh M., Kim S.J., Schaefer J. REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant. Biochim. Biophys. Acta. 2015;1848(1 Pt B):363–368. doi: 10.1016/j.bbamem.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nygaard R., Romaniuk J.A., Cegelski L. Spectral snapshots of bacterial cell-wall composition and the influence of antibiotics by whole-cell NMR. Biophys. J. 2015;108:1380–1389. doi: 10.1016/j.bpj.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romaniuk J.A., Cegelski L. Bacterial cell wall composition and the influence of antibiotics by cell-wall and whole-cell NMR. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20150024. doi: 10.1098/rstb.2015.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCrate O.A., Zhou X., Cegelski L. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J. Mol. Biol. 2013;425:4286–4294. doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichhardt C., Fong J.C., Cegelski L. Characterization of the Vibrio cholerae extracellular matrix: a top-down solid-state NMR approach. Biochim. Biophys. Acta. 2015;1848(1 Pt B):378–383. doi: 10.1016/j.bbamem.2014.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi H., Ayala I., Hediger S. Solid-state NMR on bacterial cells: selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization. J. Am. Chem. Soc. 2013;135:5105–5110. doi: 10.1021/ja312501d. [DOI] [PubMed] [Google Scholar]
- 48.Schanda P., Triboulet S., Simorre J.-P. Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan. J. Am. Chem. Soc. 2014;136:17852–17860. doi: 10.1021/ja5105987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dick-Pérez M., Zhang Y., Hong M. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011;50:989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
- 50.White P.B., Wang T., Hong M. Water-polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J. Am. Chem. Soc. 2014;136:10399–10409. doi: 10.1021/ja504108h. [DOI] [PubMed] [Google Scholar]
- 51.Wang T., Park Y.B., Hong M. Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls. Proc. Natl. Acad. Sci. USA. 2013;110:16444–16449. doi: 10.1073/pnas.1316290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harbison G.S., Herzfeld J., Griffin R.G. Solid-state nitrogen-15 nuclear magnetic resonance study of the Schiff base in bacteriorhodopsin. Biochemistry. 1983;22:1–5. doi: 10.1021/bi00270a600. [DOI] [PubMed] [Google Scholar]
- 53.Renault M., Tommassen-van Boxtel R., Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Renault M., Pawsey S., Baldus M. Solid-state NMR spectroscopy on cellular preparations enhanced by dynamic nuclear polarization. Angew. Chem. Int. Ed. Engl. 2012;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- 55.Linden A.H., Lange S., Oschkinat H. Neurotoxin II bound to acetylcholine receptors in native membranes studied by dynamic nuclear polarization NMR. J. Am. Chem. Soc. 2011;133:19266–19269. doi: 10.1021/ja206999c. [DOI] [PubMed] [Google Scholar]
- 56.Jacso T., Franks W.T., Reif B. Characterization of membrane proteins in isolated native cellular membranes by dynamic nuclear polarization solid-state NMR spectroscopy without purification and reconstitution. Angew. Chem. Int. Ed. Engl. 2012;51:432–435. doi: 10.1002/anie.201104987. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto K., Caporini M.A., Ramamoorthy A. Cellular solid-state NMR investigation of a membrane protein using dynamic nuclear polarization. Biochim. Biophys. Acta. 2015;1848(1 Pt B):342–349. doi: 10.1016/j.bbamem.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cross T.A., Sharma M., Zhou H.X. Influence of solubilizing environments on membrane protein structures. Trends Biochem. Sci. 2011;36:117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miao Y., Qin H., Cross T.A. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew. Chem. Int. Ed. Engl. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi P., Li D., Tian C. In situ 19F NMR studies of an E. coli membrane protein: in situ NMR of membrane proteins and unnatural amino acid. Protein Sci. 2012;21:596–600. doi: 10.1002/pro.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward M.E., Wang S., Ladizhansky V. In situ structural studies of Anabaena sensory rhodopsin in the E. coli membrane. Biophys. J. 2015;108:1683–1696. doi: 10.1016/j.bpj.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplan M., Cukkemane A., Baldus M. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat. Methods. 2015;12:649–652. doi: 10.1038/nmeth.3406. [DOI] [PubMed] [Google Scholar]