Abstract
The increasing awareness of RNA’s central role in biology calls for a new understanding of how RNAs, like proteins, recognize biological partners. Because RNA is inherently flexible, it assumes a variety of conformations. This conformational flexibility can be a critical aspect of how RNA attracts and binds molecular partners. Structurally, RNA consists of rigid basepaired duplexes, separated by flexible non-basepaired regions. Here, using an RNA system consisting of two short helices, connected by a single-stranded (non-basepaired) junction, we explore the role of helix length and junction sequence in determining the range of conformations available to a model RNA. Single-molecule Förster resonance energy transfer reports on the RNA conformation as a function of either mono- or divalent ion concentration. Electrostatic repulsion between helices dominates at low salt concentration, whereas junction sequence effects determine the conformations at high salt concentration. Near physiological salt concentrations, RNA conformation is sensitive to both helix length and junction sequence, suggesting a means for sensitively tuning RNA conformations.
Introduction
Recognition of RNAs expanding biological roles demands an understanding of its interactions with partner molecules. To date, most knowledge of how macromolecules recognize partners is derived from studies of proteins. Two alternative mechanisms have emerged (1). In the induced fit model, the ligand alters the conformation of its macromolecular binding partner. At the other extreme, the conformational capture model requires that the unbound macromolecule fluctuate through various conformations. The ligand binds only when the macromolecule fluctuates through the correct structure. For protein systems the conformational capture mechanism effectively describes several important classes of ligand binding (1, 2). Similar questions are now being asked about mechanisms for RNA-ligand binding. In particular, a number of recent works suggest that the conformational capture mechanism is exploited in the initial recognition of ligands by riboswitches (3, 4, 5, 6, 7). Although a combination of both induced fit and conformational capture is often required to describe full molecular recognition pathways, understanding the conformational capture mechanism, in particular, is crucial for gaining insight into the early stages of RNA-ligand binding.
Because double-stranded RNA is relatively rigid, the flexibility required for conformational fluctuations arises from non-basepaired regions in the structure (8). For example, unpaired junctions lacking stable structures can serve as flexible hinges that bring together two sides of a tertiary contact (9, 10). In other cases, the dynamics of single-stranded chains can drive conformational switching, such as in RNA splicing or riboswitch gene regulation (11). To further explore the factors that support conformational fluctuations in RNA, it is critical to understand the link between sequence, structure, and the equilibrium conformational fluctuations of RNA. However, few traditional experiments can probe these flexible states, and molecular dynamics simulations, which can assist in modeling dynamic regions, are limited in accuracy (12).
A major goal of this work is to begin to understand the connection between RNA structural motifs such as duplexes, the nucleotide sequence of non-basepaired regions, and the range of conformations through which an RNA can fluctuate. To explore these effects we employ a model RNA construct consisting of two RNA helices connected by a single-stranded RNA linker that serves as a junction, representing a model helix-junction-helix (HJH) construct (13).
Before discussing the behavior of the HJH constructs, it is first useful to briefly review the response of each independent motif, helix, or strand, to changing ionic conditions. The negative charge associated with the sugar-phosphate backbone is an important factor for determining any RNA structure. In salt-containing solutions, positively charged counterions are attracted to the RNA and reduce the overall electrostatic potential. Studies of isolated, short duplexes clearly illustrate ion valence and concentration-dependent changes in RNA’s ion atmosphere (14, 15, 16), which, in turn mediate the repulsion between RNA elements (14, 17). These latter interactions depend on helix length because end effects alter the potential around duplexes (18, 19), on length scales on the order of 5 bp (20). Although most past studies focused on longer duplexes, e.g., 25 bp, end effects may also be important because the duplexes found in functional, biologically relevant RNAs tend to be short (only ∼4 bp (21, 22, 23)). Single-stranded RNA (or DNA) is also affected by ions through charge screening. Intrastrand repulsion is reduced as cation concentrations are increased, effectively rendering the molecule more flexible (24, 25, 26). Base stacking also strongly influences the behavior of single-stranded regions. For example, stacking interactions present in poly(A), but not poly(U), make it more rigid at a given salt concentration (26, 27, 28). Often, electrostatic effects and stacking cannot be separated: ions can affect single-strand flexibility by decreasing electrostatic repulsion or by stabilizing base-stacking interactions. Finally, the specific association of ions may alter single-strand conformation, for example Mg2+ ions can bind directly to poly(A) (29, 30), stabilizing its helical conformation. The growing interest in ion-dependent conformations of single-stranded RNA is underscored by numerous studies measuring the effects of ions on homopolymers (26, 28, 29, 30).
Here, we examine the role of both duplex length and junction sequence in determining the overall conformations of the HJH construct. Single-molecule Förster resonance energy transfer (smFRET) studies of double-labeled, freely diffusing RNAs sensitively report ion-dependent conformational changes. By examining the changing conformations of these simple HJH motifs as a function of increasing salt, and comparing these changes with known responses of the elements in isolation, we gain insight into the rich behaviors that arise from their unique combinations. At low salt, we find that RNA conformation is sensitive to neither helix length nor junction sequence, although at high salt it depends only on junction sequence. In solutions containing near physiological salt concentrations, however, we find an unexpected decrease in the efficiency of energy transfer for our RNA constructs, which depends on both junction sequence and helix length. We propose a model consistent with the observed trends. Our results suggest that tradeoffs between helix length and junction sequence provide a way to tune the unfolded state conformations of RNA, critical for any process that relies on conformational fluctuations.
Materials and Methods
Sequence design
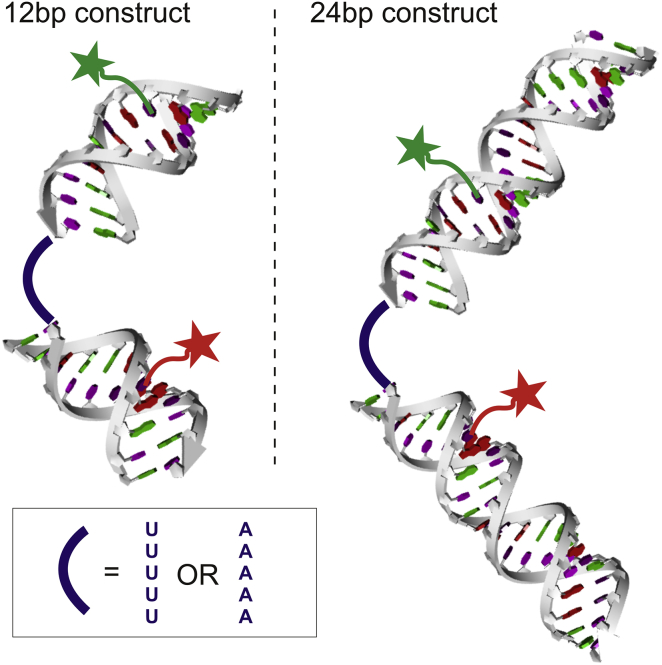
RNA constructs consist of three strands annealed together to form two duplexes connected by a 5-nucleotide single-stranded linker (Fig. 1). The full construct forms a helix-junction-helix type structure (HJH). Varying helix length to be either 12 or 24 bp and junction sequence to be either poly(U)5 or poly(A)5 provides four distinct RNA constructs. The DINAMelt web server (31) was used to ensure that unwanted hairpin and dimer formation was less favorable than the desired duplex assembly. To provide structural information, we measure FRET between two fluorophores incorporated into the RNA. Sites for fluorescent labels were selected to be 8 bp away from the junction region to keep EFRET values in the linear regime for the particular dye pair in use. Although slight differences exist between the simulated accessible volume of the dyes for 12 and 24 bp helix lengths, differences in the measurements do not appear to be significant (discussed in more detail in the Supporting Material and depicted in Fig. S1); therefore, choosing sites to be a fixed distance from the junction region distinguishes effects caused by the label’s environment from effects caused by extending the helices. We chose the Alexa Fluor – Cy5 FRET pair because previous studies using this pair attached internally to RNA duplexes validated the assumption that the dyes are freely rotating (32). Finally, label sites were chosen so that incomplete annealing or the presence of unlabeled strands results in either zero FRET or no observable signal. Refer to the Supporting Material for details about duplex sequence and label sites.
Figure 1.
Model RNA constructs consist of three strands annealed together to form two helices connected by a single-stranded junction. Individual helices are 12 or 24 bp in length and the junction consists of 5 nt of either poly(U) or poly(A). Fluorescent label sites are illustrated by green (donor) and red (acceptor) stars. To see this figure in color, go online.
Sample preparation
RNA molecules were purchased desalted and high-performance liquid chromatography purified from IDT (Coralville, IA). An internal amino-C6-dT nucleobase was included in the RNA sequence at the desired labeling site. Modified RNA strands were reconstituted in 100 mM phosphate buffer (pH 8.3). Fluorescent dyes were dissolved in dimethyl sulfoxide and promptly mixed with the appropriate RNA strand in a 10:1 dye/RNA molar ratio (Alexa Fluor 488 TFP (Life Technologies, Grand Island, NY) was used as donor and Cy5 NHS ester (GE Healthcare) as the acceptor). This mixture was left overnight on a rocker at 4°C. The third unmodified strand was reconstituted in 50 mM potassium 3-(N-morpholino)propanesulfonic acid (K-MOPS) buffer containing 900 mM KCl and 0.1 mM EDTA. RNA strands were mixed together in a 1:1.5:2 ratio of donor/unlabeled/acceptor strand, annealed at 95°C for 2 min and then cooled slowly in a water bath for 40 min. Samples were run on a Micro Bio-Spin 6 chromatography column (BioRAD, Hercules, CA) to separate RNA from unreacted dye. RNA samples were concentrated to 5–10 μM, and then divided into aliquots and stored at −20°C.
On the day of each experiment, one aliquot was thawed and diluted by 150× in buffer containing 100 mM KCl, 50 mM K-MOPS, pH 7, and 20 μM EDTA. For constructs with a poly(A) junction, the sample was annealed at 90°C for 2 min, and then slowly cooled to room temperature over 50 min. For poly(U) junctions, the annealing had no effect on measurement outcomes. This daily stock sample was diluted 1000-fold into the desired buffer for each smFRET measurement. Measurement buffers consisted of 50 mM K-MOPS, pH 7, 20 μM EDTA, and the desired amount of added KCl, or MgCl2. Values of ion concentrations quoted in this work do not include the additional 24 mM K+ ions contributed from the buffer, unless stated otherwise.
Single-molecule FRET
Single-molecule measurements on freely diffusing RNA were performed on a house-built microscope with confocal detection. Fluorescent samples were loaded into a chambered coverglass and illuminated with a 488 nm laser through a 60× 1.2 NA objective. The fluorescence emission was collected through the same objective and split into donor and acceptor channels using a 550 nm longpass dichroic. The donor channel contained an additional 530/30 bandpass emission filter, whereas the acceptor channel contained a 630 longpass emission filter. Two 50 nm optical fibers provided confocal detection and photons were detected by two avalanche photodiodes (SPCM-AQR-14, Perkin Elmer, Santa Clara, CA). Data were acquired using a Flex-2kD correlator card (Correlator.com) in photon counting mode for which photon arrival times were sampled in 25 ns intervals, and then processed further using house-written MATLAB (The MathWorks, Natick, MA) scripts. For each salt concentration, single-molecule data were collected for 30 min. At least two independent measurements were taken to estimate the variance in the measurement. The raw photon counts were converted into the number of photons measured per 1 ms in the donor and acceptor channels, and only events above a threshold on total intensity of 20 counts/ms. The efficiency of energy transfer (EFRET) was calculated as the background-subtracted ratio of the acceptor intensity to the total intensity. A histogram of EFRET values was fitted to three gaussians, the middle of which represents the signal of interest (Figs. S2–S9 show typical histograms collected over 30 min for each salt concentration and RNA construct). The low EFRET peak represents either donor-only constructs or ones with an inactive acceptor. A small population with high EFRET (∼0.8–0.9) was also observed, but the salt dependence was minimal. Therefore, in this work we focus on the mid-FRET peak, which has a strong salt dependence.
Results
Effect of helix length
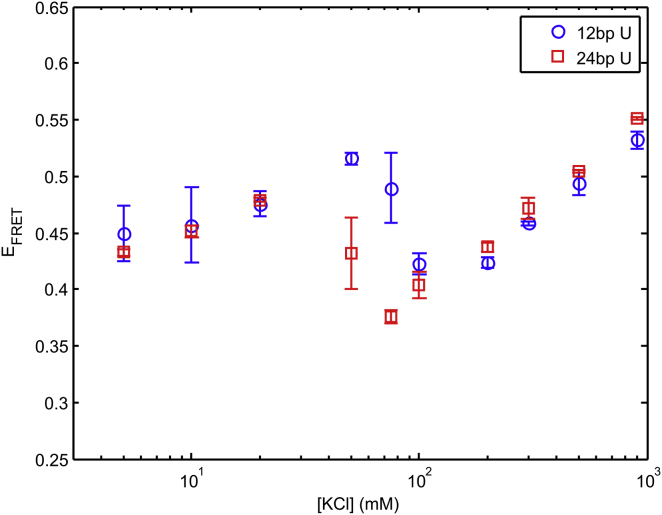
We first investigated the effects of RNA helix length on the conformation of our HJH construct. The short double helices we employ can be considered rigid due to the long persistence length of duplex RNA (∼250 bp (33, 34)); however, as a result of end effects their electrostatic properties may vary with helix length (18). To examine the impact of helix length on RNA conformation, we use smFRET to compare RNA constructs in which a poly(U)5 junction is flanked by either 12 or 24 bp helices. This junction sequence represents the simplest case for these studies because base-stacking interactions are absent in poly(U). For both 12 and 24 bp helix constructs, the donor and acceptor dyes were attached 8 bp away from the junction region, one on each flanking helix (Fig. 1). Fig. 2 shows the EFRET of attached dyes in each of these constructs, measured as a function of KCl in solution. Unexpectedly, the EFRET is nonmonotonic with [KCl], in contrast to the monotonic changes observed for isolated poly(U)40 reported in (26). In addition, the convergence of the two curves at both low and high salt suggests no dependence on helix length in these regimes. The impact of helix length on RNA conformation emerges at intermediate salt concentrations, between 30 and 100 mM monovalent (KCl) concentrations, as the EFRET trends from increasing to decreasing. The reversal in EFRET occurs at lower salt when the construct contains longer 24 bp helices as opposed to 12 bp helices. Although end-fraying may be a concern for such short duplexes, we expect this effect to be more pronounced for the 12 bp construct, where the dye is much closer to the blunt end (fraying at the end closest to the junction is less likely due to the stabilizing effect of the linker (35)). The agreement between the EFRET values for both constructs, at the extremes of salt, suggests that fraying does not contribute substantially to the measured effects.
Figure 2.
Helix length comparison of EFRET for poly(U) junction constructs in KCl. To see this figure in color, go online.
Effect of junction sequence
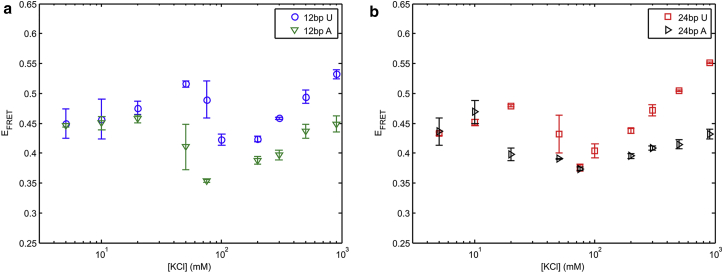
Next, we investigated the sequence dependence of RNA flexibility in solutions containing KCl. The change from a poly(U) junction to poly(A) introduces the possibility of base-stacking interactions (27, 36). To investigate the effects of base stacking on RNA conformation, we measured the KCl dependence of EFRET on RNA constructs with a poly(A) junction and compared results with data shown in Fig. 2 for poly(U). Fig. 3, a and b, respectively show results for the 12 and 24 bp helix lengths. Note that the poly(U) data are repeated from Fig. 2 for ease of comparison. Again, we observe the striking nonmonotonic behavior for the poly(A) junctions that is present in the poly(U) data; however, the new junction sequence alters the EFRET in several ways. At low salt the EFRET values are equivalent for a given helix length, suggesting that junction sequence has little effect when salt concentrations are low (<10–30 mM KCl). At intermediate salt concentrations, the turnover in EFRET of the poly(A) junction construct is shifted to lower salt when compared to the construct containing the poly(U) junction. Finally, at even higher salt (>200 mM KCl), the EFRET once again increases, however the EFRET for the poly(A) construct is always lower than for the poly(U) construct. Except for the values at high salt, the poly(A) constructs exhibit similar trends to those seen with the poly(U) linker: helix length effects manifest at intermediate salt concentrations, in this case between 20 and 100 mM KCl (Fig. S10 shows the poly(A) helix length comparison).
Figure 3.
Junction sequence comparison in KCl for (a) 12 bp and (b) 24 bp helix lengths. To see this figure in color, go online.
Effects of MgCl2
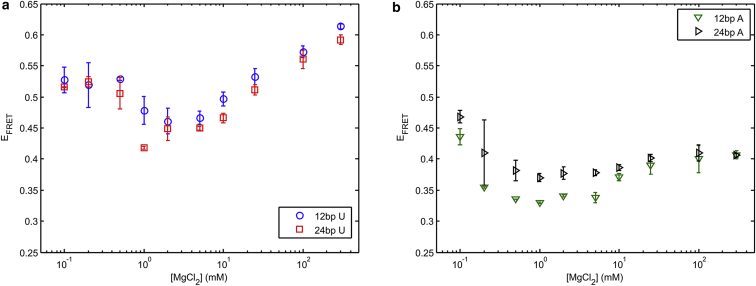
Mg2+ ions stabilize RNA tertiary structure and are essential for RNA functions such as ribozyme catalysis (37). To investigate the effect of Mg2+ ions on RNA structure, we measured HJH conformations as a function of increasing [MgCl2] for all four of our RNA constructs: 12 or 24 bp helices and poly(U) or poly(A) junctions. The only monovalent ions present here are the ∼24 mM K+ present in the buffer. With poly(U) junctions, the effects of MgCl2 echo those in KCl (Fig. 4 a), but are shifted to lower salt concentration: at low salt, the EFRET of both 12 and 24 bp constructs start at the same values, undergo a decrease at 0.5–1 mM MgCl2, and increase again at higher [MgCl2]. At high salt the EFRET values appear independent of helix length. Furthermore, although the differences are less pronounced in MgCl2 than in KCl, the decrease in EFRET occurs at slightly lower [MgCl2] for the longer helix than for the shorter helix. Helix length effects disappear above 2 mM MgCl2.
Figure 4.
Helix length comparison of EFRET in MgCl2 for (a) poly(U) and (b) poly(A) junctions. To see this figure in color, go online.
In the poly(A) constructs, EFRET begins to decrease at the lowest MgCl2 concentrations measured (∼0.2 mM, Fig. 4 b), following the trend observed in KCl that the reversal in EFRET occurs at lower salt for the poly(A) linker than for the poly(U). Helix length effects manifest differently for poly(A) in MgCl2 in that they occur over a larger range (0.1–10 mM MgCl2). Above 10 mM MgCl2, however, these length effects disappear.
Results summary
SmFRET data were acquired for four different RNA constructs in buffered solutions containing either K+ or Mg2+ counterions, over a wide range of ionic concentrations. General trends are most clearly identified in the monovalent ion series. At low [KCl], the EFRET value is independent of both helix length and junction sequence for all constructs studied. As [KCl] increases, EFRET displays nonmonotonic behavior. Finally at higher [KCl], the increasing EFRET is independent of helix length, but does depend on junction sequence. Similar trends are observed as a function of increasing [MgCl2].
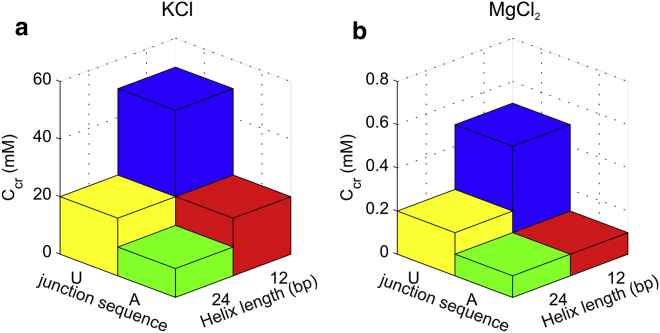
To facilitate comparison between different constructs, we introduced Ccr, the critical salt concentration where the first reversal in EFRET is observed (or the salt concentration where EFRET reaches its local maximum.) We postulate in the following section that this abrupt reversal in EFRET reflects the initiation of a transition where the molecules reorient to adopt a different conformational state, hence Ccr provides a metric for comparison across conditions. Fig. 5 illustrates the effect of both junction sequence and helix length on this conformational rearrangement and motivates the following key observations. First, Ccr can be reduced by either increasing helix length or by introducing base stacking to the junction (changing it from poly(U) to poly(A)). In monovalent salt solutions, these effects appear to be of comparable scale. Second, we observe that helix length effects for poly(U) are similar but less pronounced in MgCl2 than in KCl. Finally, poly(A) constructs in MgCl2 have indistinguishable Ccr, possibly because salt concentrations below 0.1 mM MgCl2 were not measured. With the concept of Ccr, we organize the RNA constructs in terms of their propensity to initiate this transition. In KCl we have 24A<12A∼24U<12U. The order is similar in MgCl2, except that the sequence effects are more pronounced giving 24A∼12A<24U<12U.
Figure 5.
Summary of the critical salt concentration (Ccr) dependence on helix length and junction sequence in (a) KCl and (b) MgCl2. To see this figure in color, go online.
Discussion
Salt concentration-dependent behavior of isolated RNA elements
All of the RNA constructs used in this study contain two structural elements: helices and single strands. Our hypothesis is that each element contributes in a distinct way to overall RNA conformation, tuning the behavior of the system. Before we discuss the implications of our results from the combined system, it is important to review the effects of salt concentration on each individual component. We therefore start by expanding on the introduction and discussing the salt dependence of isolated helix repulsion and on the sequence dependence of RNA homopolymers.
Interactions between isolated short helices
The interaction potential between particles in solution can be described by the second virial coefficient (A2). When A2 = 0, the particles are noninteracting, A2 > 0 represents repulsion, and A2 < 0 denotes an attraction. The salt dependence of A2 was reported previously for isolated RNA helices (14, 17), showing that repulsion between isolated helices in solution decreases with increasing salt concentration. A2 for 25 bp RNA helices drops to zero at ∼150 mM KCl, after which increasing salt concentration leads to attraction in the form of end-to-end stacking (17). End-to-end stacking of two separate RNA molecules is not a concern in the dilute solutions used in this study, and the likelihood of tethered helices stacking within one molecule is low (discussed in the Supporting Material). Nevertheless, the 150 mM threshold represents a useful cutoff and describes where repulsion no longer dominates interhelical interactions for long RNA helices.
Interhelical interactions are also affected by helix length via end effects (19). These effects are particularly relevant because nature uses short helices in functional RNAs (21, 22, 23). Modeling DNA as a uniformly charged cylinder, Allison (20) showed that potentials along the cylinder’s surface are reduced for short helices but the differences between helix lengths become less significant as the salt concentration is increased. Simulations of ion distributions around DNA helices show that helix length effects emerge at a length <24 bp (38).
To verify whether similar end effects exist for RNA helices we used the Adaptive Poisson-Boltzmann Solver (APBS) (39) to simulate electrostatic potentials around isolated RNA helices of 12 and 24 bp in length (see the Supporting Material for details, and results in Fig. S11). Although the nonlinear Poisson Boltzmann equation has certain caveats when applied to nucleic acids (i.e., it underestimates the number of ions close to the surface (40)), we simply use it to gain insight into the electrostatics of end effects. Furthermore, we focus on monovalent salt conditions because predictions using the Poisson-Boltzmann equation are believed to be more accurate for monovalent than divalent ions (16). These simulations show similar end effects in RNA as were observed for DNA: the potential of 12 bp RNA helices is smaller in magnitude than that of 24 bp helices, and end effects become less pronounced with increasing salt concentration. In addition, the simulations suggest that these differences arise from a reduction in radial rather than axial potential. The potential along the helical axis is essentially unaffected by helix length (Fig. S12).
Unpaired single strands
Past work on poly(U) homopolymers suggests that it follows random coil behavior, and molecular flexibility increases with salt (26, 28). In contrast, (27, 41, 42) show the propensity of adjacent nucleotides of poly(A) to stack, causing it to form short-range helical structures. The stacking propensity of poly(A) may also be influenced by the salt concentration (43). The stark differences between conformations of poly(A) and poly(U) homopolymers discussed in the Introduction suggest that the junction sequence will profoundly affect the overall conformation of the RNA construct.
Unique behavior emerges as structural elements are combined in monovalent salt
When we combine RNA structural elements using our HJH construct, the unexpected behavior shown in Figs. 2 and 3 emerges. To interpret, we divide the data into three experimentally distinct regimes: low salt (increasing EFRET), mid-salt (decreasing EFRET), and high salt (increasing EFRET). The boundaries between these regimes depend in a nontrivial way on the construct. Because these regions are most distinctly separated in solutions containing KCl, we will focus the first part of the discussion on the monovalent ion dependence of HJH conformations. We propose a model that is consistent with our observations, and suggests how a unique combination of helix length, junction sequence, and ion identity can affect RNA conformation.
Individual structural elements dominate at limiting [KCl]
We begin by examining the limiting cases of low and high [KCl], respectively. At low salt (<10–20 mM KCl) neither junction sequence nor helix length affects the EFRET values (Figs. 2, 3, and S10). In this regime, helix repulsion is significant, and the overall electrostatic potential is minimized in a conformation in which the two helices are pushed as far from each other as possible. Despite any potential end effects, the strength of the helix repulsion is so high that the junction is fully stretched, limiting the available helix conformations to the same subset of states independent of helix length or junction sequence.
Next, we describe the high salt limiting case. Above 150 mM KCl, EFRET is independent of helix length (Figs. 2 and S10). This observation is not surprising because end effects diminish at high salt ((20) and Fig. S12) thus local electrostatic potentials near the junction region should not depend on helix length. Additionally, in light of past measurements of A2, which show that interhelical repulsion is insignificant above 150 mM KCl, albeit for longer helices (17), therefore we expect helix repulsion itself to be negligible at high salt concentrations.
In this regime, the data show the dramatic impact of junction sequence on EFRET (Fig. 3). Most obviously, the poly(A) containing constructs are always more extended (lower EFRET) than those containing poly(U). To compare the salt dependence of EFRET for the poly(U) junction with that previously measured for (isolated) single-stranded poly(U)40 (26), we approximate the data as linear with the logarithm of salt concentration. The comparison confirms that junction conformation dominates in the high salt regime. These two data sets display the same power law dependence of EFRET on [salt] (Fig. S13), confirming that the salt-dependent EFRET changes at high salt, where helix repulsion is negligible, are caused by the ion-dependent changes in the persistence length of poly(U) (26).
We cannot make the same comparison for poly(A) junctions because we do not have corresponding FRET results as we did in the case for poly(U). However, our data show that the fluorescent dyes are always closer together for the poly(U) constructs than for the poly(A) constructs at high salt, consistent with an increase in poly(A) base stacking with salt concentration (43), which drives the flanking helices toward coaxial conformations. In contrast the flexible poly(U) linker allows the flanking helices (and associated fluorophores) to approach one another more closely. Thus, our data are consistent with a picture where the junction sequence determines the RNA conformation at high salt.
Decreasing EFRET at physiological monovalent concentrations explained by flip in relative helix orientation
At low salt, we have shown that helix repulsion dominates RNA conformation; although at high salt, junction sequence dominates. Next, we address the biologically important, mid-salt regime, which includes physiological monovalent concentrations (∼100 mM (44)). Helix repulsion will be reduced relative to the low salt case, and junction dynamics may become visible, allowing helices to explore more conformational space and making end effects more prevalent. Therefore, this regime is characterized by the interplay between all three contributing structural factors: helix repulsion, end effects, and junction dynamics.
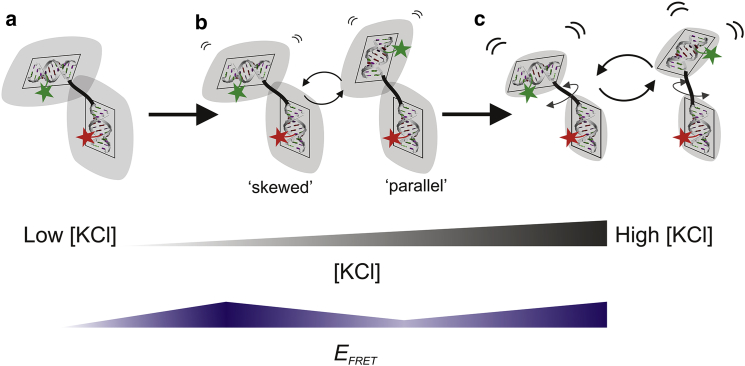
In this regime, we observe a striking decrease in EFRET, which, assuming freely rotating dyes (see Supporting Material for a discussion of this assumption), indicates that the dyes are moving farther apart. We propose a model in which the decrease in EFRET results from two types of interchanging helix conformations, one with a larger EFRET. The population of conformations with higher EFRET are not favored at low salt, and only become accessible as salt is increased above Ccr. Because EFRET histograms show a shifting peak position rather than two separate peaks, these populations must be rapidly interchanging on a timescale faster than the measurement time (45). One potential scenario is that helices can flip from a subset of skewed to parallel conformations at Ccr (Fig. 6). This flip causes the average distance between dyes to increase. A complete picture of how salt may affect the combined HJH RNA is described in the caption for Fig. 6.
Figure 6.
Schematic of proposed model describing mid-salt decrease in EFRET. In this picture, colored stars represent dye labels and shaded gray areas represent the boundary where electrostatic potential of individual helices drops to a finite cutoff. (a) At low [KCl], helix repulsion is high (large overlap between shaded areas). Consequently, the junction is extended to its maximum. Helices are fixed in skewed conformations due to the off-center attachment of the single-stranded RNA linker. (b) As salt is increased to Ccr, repulsion is reduced enough for junction dynamics to let the helices flip between skewed and parallel conformations. The non-right-angled nature of the RNA helix cross section may cause these two distinct subsets of helix orientations. The parallel conformations have dyes farther apart and thus a lower EFRET than the skewed conformations. (c) Increasing salt even further screens helix repulsion completely and junction dynamics alone determine RNA conformation. To see this figure in color, go online.
Our proposed model relies on the assumption that skewed helix orientations are preferred at low salt. Such asymmetric conformations are not unprecedented. In previous work on DNA helices tethered by an uncharged synthetic linker, the off-center attachment points of the linker cause equilibrium distributions of helix orientation to fluctuate around a conformation where the interhelical angle is ∼150° (46). We expect our RNA constructs to exhibit a similar angular bias because the attachment of the RNA single-stranded junction is also offset from the helical axis. Although the DNA helix can be modeled as a right-angle cylinder (its bases are tilted by only −6°), the cross section of the RNA helix is more trapezoidal (its bases are tilted by 16°–19° (47), illustrated in Fig. 6). This tilt of the RNA bases may cause the discrete transition observed at intermediate salt concentrations (Fig. 6 b), rather than the continuous one observed for tethered DNA helices (46). Verification is needed to confirm whether skewed conformations are favored at low salt, and whether a free-energy barrier exists between skewed and parallel states. However, a detailed analysis of the energetics of our RNA constructs is beyond the scope of this work.
Proposed model is consistent with dependence of EFRET on junction sequence, helix length, and MgCl2
The proposed model can explain the observed trends in EFRET as both junction sequence and helix length are varied: Ccr is reduced for poly(A) relative to poly(U) and for 24 vs. 12 bp helices. Sequence dependence is straightforward: stacking of adjacent poly(A) nucleotides drives the junction toward a helical conformation. Because stacking favors parallel helix conformations, less salt is required to flip the helices out of the skewed conformation, explaining the reduced Ccr for poly(A) constructs relative to poly(U).
APBS simulations show that end effects alter the radial electrostatic potential of a helix, whereas the axial potentials are relatively unchanged (Fig. S12). If helices are skewed at low salt, they will interact with one another along their radial directions (i.e., refer to Fig. 6 a). Longer helices have a larger radial potential, so the skewed conformation will be slightly disfavored compared to the shorter helices. Thus, as salt concentration is increased and overall repulsion is reduced, the longer helices will prefer to flip earlier to maximize the distance between the long sides of either helix, causing a reduction in Ccr.
Mg2+ ions affect RNA flexibility in at least two ways. First, at a given salt concentration, they screen RNA’s negatively charged backbone more efficiently than do monovalent ions. Second, they stabilize poly(A) stacking. All the same trends observed for poly(U) in KCl are also detected in MgCl2. Not surprisingly, much less Mg2+ is required than K+ to initiate the transition from skewed to flipped, consistent with many previous observations showing greater effectiveness for charge screening of Mg2+. On the contrary, for poly(A), Ccr is indistinguishable for both helix lengths. This result is consistent with the high sensitivity of poly(A) base stacking to Mg2+ ions. Because stacking is strongly preferred in the presence of Mg2+, helices are driven to the parallel conformation, regardless of end effects. In fact, the measured EFRET of poly(A) constructs agree remarkably well with the simulated EFRET computed from a model where a fully A-form junction connects two 24 bp helices (see Supporting Material).
Helix length and junction sequence can bias conformations of unfolded RNA
In the conformational capture picture of riboswitch ligand binding, the distribution of conformations in the unfolded state determines the function of the RNA molecule. Environmental factors that shift the distribution toward its ligand-bound state will increase binding rates, whereas factors that shift the distribution toward an unbound state will reduce the likelihood of binding. For example, recent work has shown that Mg2+ stabilizes a pseudoknot-like structure in the SAM-II riboswitch, favoring SAM binding (4). Furthermore, the A-rich tail of the class I prequeuosine riboswitch exhibits a dynamic helical structure in solution that the authors propose increases the efficiency of ligand binding with respect to a disordered structure (48). Preorganization of binding partners is not unique to RNA. Evidence from smFRET measurements suggests that NaCl facilitates DNA duplex formation in solution by favoring preformed helical structures of single strands, which can then form a duplex through a docking type interaction (49, 50). In general, this type of preorganization may be critical in tipping the free-energy balance between bound and unbound states of RNA. In this work, we report that helix length and junction sequence affect the available conformations of a model unfolded RNA. We can bias the conformations of RNA (decreasing Ccr) by either decreasing the helix length or by mutating the junction residues from U to A. We propose that these two factors can be exploited to preorganize the structures of unfolded RNA and may be used to influence the interactions between RNA and its binding partners. For example, structural elements of riboswitches may contribute to biasing their conformations toward either ligand bound or unbound states.
It is interesting to note that the average duplex length in long RNAs is only ∼4 bp (21, 22, 23), highlighting the importance of short duplexes in biology. We see differences arise between the 12 and 24 bp duplexes at just below physiological monovalent concentrations (20–100 mM KCl). Because decreasing helix length pushes the EFRET transition to higher salt, we might expect that differences between lengths of even shorter helices would be further propelled toward physiologically relevant salt conditions.
Although the biological importance of single-stranded RNA linkers is clearly important (51, 52, 53), double-stranded junctions are pervasive (54, 55, 56). Changing the topology of the junction can dramatically alter the available conformations of the RNA (55, 56, 57, 58). We anticipate that our observation that RNA conformation is influenced by the length of its helices and the sequence of its junctions would be generally true for all junctions, single or double stranded. The studies of single-stranded junctions described here provide a first step in understanding the underlying behavior of junctions in RNA. Future experiments will continue to explore the link between topology and RNA junction structure.
Conclusions
Here, we described the use of smFRET to compare the salt dependence of RNA conformations with two different helix lengths and junction sequences. These measurements show that different structural elements determine the conformations at extreme monovalent salt concentrations: helix repulsion dominates at low salt, whereas junction sequence dominates at high salt. At intermediate, near physiological concentrations, both the junction sequence and the length of the helices determine available RNA conformations. In the presence of Mg2+ ions, the critical salt concentration dividing low and high salt regions is ∼100 times lower than in the presence of K+ ions, and the increased sensitivity of base stacking in poly(A) causes a dramatic sequence dependence. The influence of helix length and junction sequence on RNA conformation at near physiological salt concentrations suggests a biologically relevant mechanism for tuning the available conformations of unfolded RNA. In light of the conformational capture mechanism, helix length and junction sequence may both be important factors in directing interactions between RNA and its binding partners.
Author Contributions
L.P. designed the research, J.L.S performed the research and analyzed the data. Both authors wrote the article.
Acknowledgments
The authors thank the Pollack lab members George Calvey, Yujie Chen, Andrea Katz, Suzette Pabit, Alex Plumridge, and Josh Tokuda for useful discussions.
This work was supported by the National Institutes of Health (R01-GM085062).
Editor: Timothy Lohman.
Footnotes
Supporting Materials and Methods, thirteen figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01115-7.
Supporting Citations
References (59, 60, 61, 62) appear in the Supporting Material.
Supporting Material
References
- 1.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Csermely P., Palotai R., Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoddard C.D., Montange R.K., Batey R.T. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haller A., Rieder U., Micura R. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 5.Heppell B., Blouin S., Lafontaine D.A. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 6.Wilson R.C., Smith A.M., Foster M.P. Tuning riboswitch regulation through conformational selection. J. Mol. Biol. 2011;405:926–938. doi: 10.1016/j.jmb.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serganov A., Patel D.J. Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol. 2012;22:279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowakowski J., Tinoco I., Jr. RNA structure in solution. In: Neidle S., editor. Oxford Handbook of Nucleic Acid Structure. Oxford University Press; New York: 1999. pp. 567–602. [Google Scholar]
- 9.Szewczak A.A., Cech T.R. An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA. 1997;3:838–849. [PMC free article] [PubMed] [Google Scholar]
- 10.Schlatterer J.C., Kwok L.W., Pollack L. Hinge stiffness is a barrier to RNA folding. J. Mol. Biol. 2008;379:859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dethoff E.A., Chugh J., Al-Hashimi H.M. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Šponer J., Banáš P., Otyepka M. Molecular dynamics simulations of nucleic acids. From tetranucleotides to the ribosome. J. Phys. Chem. Lett. 2014;5:1771–1782. doi: 10.1021/jz500557y. [DOI] [PubMed] [Google Scholar]
- 13.Herschlag D., Allred B.E., Gowrishankar S. From static to dynamic: the need for structural ensembles and a predictive model of RNA folding and function. Curr. Opin. Struct. Biol. 2015;30:125–133. doi: 10.1016/j.sbi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabit S.A., Qiu X., Pollack L. Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA. Nucleic Acids Res. 2009;37:3887–3896. doi: 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirmizialtin S., Pabit S.A., Elber R. RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations. Biophys. J. 2012;102:819–828. doi: 10.1016/j.bpj.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirmizialtin S., Silalahi A.R., Fenley M.O. The ionic atmosphere around A-RNA: Poisson-Boltzmann and molecular dynamics simulations. Biophys. J. 2012;102:829–838. doi: 10.1016/j.bpj.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pabit S.A., Sutton J.L., Pollack L. Role of ion valence in the submillisecond collapse and folding of a small RNA domain. Biochemistry. 2013;52:1539–1546. doi: 10.1021/bi3016636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballin J.D., Wilson G.M. Role and applications of electrostatic effects on nucleic acid conformational transitions and binding processes. In: Tadashi M., editor. Application of Thermodynamics to Biological and Materials Science. InTech; West Palm Beach, FL: 2011. pp. 129–174. [Google Scholar]
- 19.Nicolai T., Mandel M. The ionic strength dependence of the second virial coefficient of low molar mass DNA fragments in aqueous solutions. Macromolecules. 1989;22:438–444. [Google Scholar]
- 20.Allison S.A. End effects in electrostatic potentials of cylinders - models for DNA fragments. J. Phys. Chem. 1994;98:12091–12096. [Google Scholar]
- 21.Fontana W., Konings D.A., Schuster P. Statistics of RNA secondary structures. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- 22.Yoffe A.M., Prinsen P., Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc. Natl. Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang L.T., Yoffe A.M., Ben-Shaul A. A sequential folding model predicts length-independent secondary structure properties of long ssRNA. J. Phys. Chem. B. 2011;115:3193–3199. doi: 10.1021/jp110680e. [DOI] [PubMed] [Google Scholar]
- 24.Ha B.Y., Thirumalai D. Electrostatic persistence length of a polyelectrolyte chain. Macromolecules. 1995;28:577–581. [Google Scholar]
- 25.Fixman M. Electrostatic persistence length. J. Phys. Chem. B. 2010;114:3185–3196. doi: 10.1021/jp1005742. [DOI] [PubMed] [Google Scholar]
- 26.Chen H., Meisburger S.P., Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. USA. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seol Y., Skinner G.M., Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys. Rev. Lett. 2007;98:158103–158106. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 28.Seol Y., Skinner G.M., Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys. Rev. Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 29.Kankia B.I. Binding of Mg2+ to single-stranded polynucleotides: hydration and optical studies. Biophys. Chem. 2003;104:643–654. doi: 10.1016/s0301-4622(03)00125-x. [DOI] [PubMed] [Google Scholar]
- 30.Kankia B.I. Inner-sphere complexes of divalent cations with single-stranded poly(rA) and poly(rU) Biopolymers. 2004;74:232–239. doi: 10.1002/bip.20082. [DOI] [PubMed] [Google Scholar]
- 31.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sindbert S., Kalinin S., Seidel C.A. Accurate distance determination of nucleic acids via Förster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 33.Abels J.A., Moreno-Herrero F., Dekker N.H. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 2005;88:2737–2744. doi: 10.1529/biophysj.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kebbekus P., Draper D.E., Hagerman P. Persistence length of RNA. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- 35.Romaniuk P.J., Hughes D.W., Bell R.A. Stabilizing effect of dangling bases on a short RNA double helix as determined by proton nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 1978;100:3971–3972. [Google Scholar]
- 36.Eisenberg H., Felsenfeld G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J. Mol. Biol. 1967;30:17–37. doi: 10.1016/0022-2836(67)90240-9. [DOI] [PubMed] [Google Scholar]
- 37.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olmsted M.C., Anderson C.F., Record M.T., Jr. Monte Carlo description of oligoelectrolyte properties of DNA oligomers: range of the end effect and the approach of molecular and thermodynamic properties to the polyelectrolyte limits. Proc. Natl. Acad. Sci. USA. 1989;86:7766–7770. doi: 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meisburger S.P., Pabit S.A., Pollack L. Determining the locations of ions and water around DNA from X-ray scattering measurements. Biophys. J. 2015;108:2886–2895. doi: 10.1016/j.bpj.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holcomb D., Tinoco I. Conformation of polyriboadenylic acid: pH and temperature dependence. Biopolymers. 1965;3:121–133. [Google Scholar]
- 42.Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J. Mol. Biol. 1966;15:455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- 43.Dewey T.G., Turner D.H. Laser temperature-jump study of stacking in adenylic acid polymers. Biochemistry. 1979;18:5757–5762. doi: 10.1021/bi00593a002. [DOI] [PubMed] [Google Scholar]
- 44.Lodish H., Berk A., Darnell J. W. H. Freeman; New York: 2000. Molecular Cell Biology. [Google Scholar]
- 45.Gopich I.V., Szabo A. Theory of single-molecule FRET efficiency histograms. In: Komatsuzaki T., Kawakami M., Takahashi S., Yang H., Silbey R.J., editors. Single-Molecule Biophysics. John Wiley & Sons; Hoboken, NY: 2011. pp. 245–297. [Google Scholar]
- 46.Bai Y., Chu V.B., Doniach S. Critical assessment of nucleic acid electrostatics via experimental and computational investigation of an unfolded state ensemble. J. Am. Chem. Soc. 2008;130:12334–12341. doi: 10.1021/ja800854u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saenger W. Springer-Verlag; NY: 1984. Principles of Nucleic Acid Structure. [Google Scholar]
- 48.Eichhorn C.D., Feng J., Al-Hashimi H.M. Unraveling the structural complexity in a single-stranded RNA tail: implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic Acids Res. 2012;40:1345–1355. doi: 10.1093/nar/gkr833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holbrook J.A., Capp M.W., Record M.T., Jr. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 50.Dupuis N.F., Holmstrom E.D., Nesbitt D.J. Single-molecule kinetics reveal cation-promoted DNA duplex formation through ordering of single-stranded helices. Biophys. J. 2013;105:756–766. doi: 10.1016/j.bpj.2013.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal M., Lee M., Breaker R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 52.Lodeiro M.F., Filomatori C.V., Gamarnik A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009;83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen P., Shi X., Qin P.Z. A single-stranded junction modulates nanosecond motional ordering of the substrate recognition duplex of a group I ribozyme. ChemBioChem. 2013;14:1720–1723. doi: 10.1002/cbic.201300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goody T.A., Melcher S.E., Lilley D.M. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mustoe A.M., Bailor M.H., Al-Hashimi H.M. New insights into the fundamental role of topological constraints as a determinant of two-way junction conformation. Nucleic Acids Res. 2012;40:892–904. doi: 10.1093/nar/gkr751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailor M.H., Sun X., Al-Hashimi H.M. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 57.Chu V.B., Lipfert J., Herschlag D. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang R.S., Draper D.E. Bend and helical twist associated with a symmetric internal loop from 5S ribosomal RNA. Biochemistry. 1994;33:10089–10093. doi: 10.1021/bi00199a036. [DOI] [PubMed] [Google Scholar]
- 59.Muschielok A., Andrecka J., Michaelis J. A nano-positioning system for macromolecular structural analysis. Nat. Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 60.Wozniak A.K., Schröder G.F., Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc. Natl. Acad. Sci. USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreon A.C., Gambin Y., Deniz A.A. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macke T.J., Case D.A. Vol. 682. American Chemical Society; Washington, D. C.: 1997. pp. 379–393. (Modeling Unusual Nucleic Acid Structures). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.